Abstract
The Red Queen hypothesis proposes that coevolution of interacting species (such as hosts and parasites) should drive molecular evolution through continual natural selection for adaptation and counter-adaptation1–3. Although the divergence observed at some host-resistance4–6 and parasite-infectivity7–9 genes is consistent with this, the long time periods typically required to study coevolution have so far prevented any direct empirical test. Here we show, using experimental populations of the bacterium Pseudomonas fluorescens SBW25 and its viral parasite, phage Φ2 (refs 10, 11), that the rate of molecular evolution in the phage was far higher when both bacterium and phage coevolved with each other than when phage evolved against a constant host genotype. Coevolution also resulted in far greater genetic divergence between replicate populations, which was correlated with the range of hosts that coevolved phage were able to infect. Consistent with this, the most rapidly evolving phage genes under coevolution were those involved in host infection. These results demonstrate, at both the genomic and phenotypic level, that antagonistic coevolution is a cause of rapid and divergent evolution, and is likely to be a major driver of evolutionary change within species.
According to the Red Queen hypothesis, biotic interactions are a fundamental driver of molecular evolution2. The Red Queen hypothesis posits that for a given species, its effective environment is likely to be comprised of the other species in the ecosystem, such that an adaptation increasing the fitness of one species necessarily causes a decline in fitness of those species with which it interacts1,3. Such coevolutionary interactions give rise to continual natural selection for adaptation and counter-adaptation by ecologically interacting species1,3, thereby driving molecular evolution2. Nowhere are such evolutionary dynamics thought to be so prevalent as in interactions between hosts and virulent parasites, in which selection is strongly antagonistic yet closely coupled12. Comparative studies have found particularly high rates of molecular evolution in genes associated with infection7–9 or resistance to infection4–6. However, there have been no direct empirical tests of whether antagonistic host–parasite coevolution accelerates molecular evolution in parasite genomes, and whether such evolution is particularly rapid at genes determining infectivity.
Here we use experimental evolution of populations of the bacterium Pseudomonas fluorescens SBW25 and its viral parasite, phage Φ2. We have previously demonstrated that P. fluorescens and Φ2 undergo a persistent coevolutionary ‘arms race’ with reciprocal selection for the evolution of new resistance and infectivity phenotypes through time in bacteria and phage, respectively10,13, but the link between this rapid phenotypic evolution and the underlying pattern of molecular evolution has not been resolved. Crucially, it is possible to separate bacteria and phage when transferring populations to fresh media14, which allows one partner to be held evolutionarily constant while the other partner is allowed to evolve15–17. Initially isogenic, replicate populations of P. fluorescens and Φ2 were propagated by serial transfer under two conditions: (1) evolution, in which the bacterial genotype was held constant and only the phage was allowed to adapt, and (2) coevolution, in which both the bacterium and the phage were allowed to evolve adaptations and counter-adaptations. At the end of the selection experiment we obtained whole-genome sequences of phage populations by high coverage second-generation sequencing to determine the identity and frequency of mutations in each population. Mutations were partitioned into synonymous and non-synonymous changes; very few synonymous mutations were observed and only non-synonymous mutations were used in analyses (see Supplementary Information; note that each indel (that is, insertions or deletions) was counted as one mutation regardless of its length). From these data we calculated the number of sites that had acquired mutations in each population relative to the ancestral reference Φ2 sequence (obtained as part of this study; see Supplementary Information), and from allele frequencies, the genetic distance of each population from the ancestral Φ2 sequence, the genetic divergence among populations and the genetic diversity within each population.
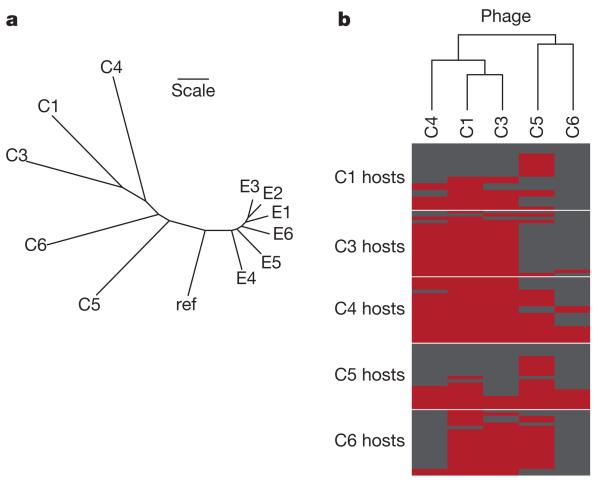
Coevolved phage populations showed twice the genetic distance from the ancestor as that of the evolved populations (average pairwise genetic distances: coevolved, 22.7 ± 1.9 standard error (s.e.); evolved, 11.1 ± 0.4 s.e.; t = 6.64, d.f. = 4.46, P < 0.01), shown also by the increased branch lengths for coevolved populations in the phylogenetic tree in Fig. 1a. Similarly, coevolved populations had a greater number of sites exhibiting mutations than evolved populations (coevolved, mean 52.8, range 46–60; evolved, mean 37.5, range 29–42; likelihood-ratio test (LRT) = 14.3, P < 0.001). Furthermore, far greater genetic divergence was observed among replicate coevolved populations than was observed among replicate evolved populations (ΦST = 0.45 for coevolved populations versus ΦST = 0.06 for evolved populations (Supplementary Table 1), in which ΦST is a measure of the proportion of the total molecular variation attributable to differences among populations18). The tree in Fig. 1a also shows that replicate populations from the same treatment grouped together genotypically. This topology is not due to co-ancestry as all populations were split at the start of the experiment. Instead, the topology reflects parallel evolution: selection acting independently at the same sites among replicate populations. Thus, the tree reflects three key evolutionary patterns. Specifically, the extent to which replicates: (1) followed a similar trajectory away from the ancestral sequence, presumably as they adapted to laboratory conditions; (2) evolved similarly among replicates within a treatment but differently in response to consistent differences between treatments; and (3) showed independent evolution within each replicate, and at a far higher rate in the coevolved than the evolved treatment.
Figure 1. Genetic and phenotypic responses to selection.
a, Phylogenetic tree for evolved (E1–6) and coevolved (C1, C3–6) phage populations and ancestral reference genotype (ref) based on Euclidean distances calculated from the frequency and identity of mutations in each population. Scale bar indicates a Euclidean distance of one. b, The phage-infectivity range based on the ability of each coevolved population to infect 20 bacterial clones from each host population. Infection by phage is shown in red, and resistance by hosts is shown in grey. The dendrogram indicates phenotypic similarity between phage populations.
Increased genetic divergence between parasite populations due to coevolution is likely to be driven primarily by divergent selection on infectivity traits. To address this prediction, we phenotypically characterized the infectivity profile of each phage population. Specifically, we used cross-infection experiments to test whether phage from each population were able to infect hosts from all coevolved populations. We found that coevolved phage populations varied in terms of both the range and identity of host genotypes that they were able to infect (Fig. 1b), but that phage from evolved populations failed to infect any coevolved hosts (data not shown). Indeed, phenotypic divergence of the infectivity profile of coevolved phage populations closely matched the genetic divergence of the phage genomes as demonstrated by the similar topologies of trees constructed using genetic or phenotypic traits (Fig. 1b).
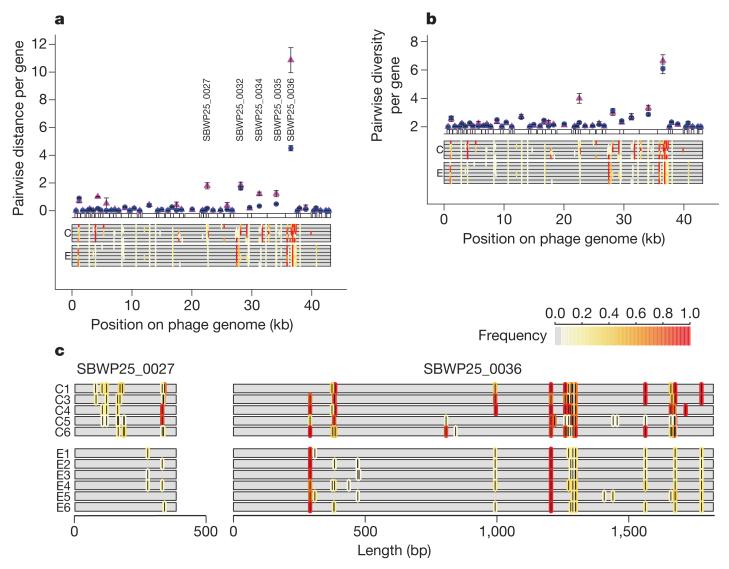
The increased rate of molecular evolution observed in the coevolved populations was not distributed uniformly across the phage genome (Fig. 2a). Four genes showed significantly increased molecular evolution in coevolved versus evolved phage genomes. SBWP25_0036 (EMBL accession FN594518), which encodes a tail-fibre protein (gp49), had a greater number of sites with mutations in the coevolved versus evolved treatment, and, based on the allele frequencies of mutations at these sites, a substantially higher divergence from the ancestral genotype (number of mutational sites: coevolved, mean 17.6, range 15–20; evolved, mean 12.7, range 11–14; LRT = 4.43, P < 0.05; pairwise genetic distance: coevolved, 10.86±0.90 s.e.; evolved, 4.51 ± 0.19 s.e.; t = 7.69, d.f. = 4.38, P < 0.01). SBWP25_0027, which encodes a structural protein (gp40), also had a greater number of sites with mutations and a higher divergence from the ancestor in the coevolved than the evolved populations (number of mutational sites: coevolved, mean 5.8, range 4–7; evolved mean 1.0, range 0–2; LRT = 20.9, P < 0.001; pairwise genetic distance: coevolved, 1.79 ± 0.23 s.e.; evolved, 0.17 ± 0.05 s.e.; t = 7.69, d.f. = 4.38, P < 0.01). SBWP25_0034 and SBWP25_0035, which encode internal virion structural proteins gp47 and gp48, respectively, also showed higher rates of molecular evolution in coevolved populations, although to a lesser extent than SBWP25_0027 and SBWP25_0036 (pairwise genetic distance: SBWP25_0034 coevolved, 1.21 ± 0.13 s.e.; evolved, 0.34 ± 0.06 s.e.; t = 6.79, P < 0.01; SBWP25_0035 coevolved, 1.23 ± 0.22 s.e.; evolved, 0.48± 0.06 s.e.; t = 3.61, P < 0.05). SBWP25_0027 and SBWP25_0036 had a similar density of mutations (Fig. 2c) as each other, but, because of its smaller size, SBWP25_0027 contributed less to the overall divergence of the coevolved genomes from the ancestor than SBWP25_0036. Consistent with the observed evolution of the phage-infectivity range (Fig. 1b), all four of these proteins are predicted to be involved in host attachment19. In tailed bacteriophages, attachment is a two-step process consisting of an initial reversible adsorption by tail fibres, followed by irreversible adhesion by structural proteins20. The average size of deletions in the tail fibre gene (SBWP25_0036) was positively correlated to the number of bacterial genotypes the phage populations could infect (infectivity range) (Supplementary Fig. 1), suggesting that tail fibres are under strong directional selection for reduced protein length during coevolution but that the precise genetic changes varied between populations. Shortened tail fibres also evolved in the evolution treatment, although to a lesser degree than under coevolution and without a concomitant increase in the infectivity range (Supplementary Fig. 1), suggesting that to some extent shorter tail fibres may also be a general adaptation to laboratory conditions, perhaps through increasing adsorption efficiency. SBWP25_0032, encoding a tail tubular protein (gp45), showed divergence from the ancestor in both evolved and coevolved treatments, but at different sites in each treatment. SBWP25_0027 (gp40) and SBWP25_0036 (gp49) accounted for most of the divergence between replicate, coevolved phage populations (Supplementary Fig. 2).
Figure 2. Patterns of molecular evolution in the Φ2 genome.
a, b, Pairwise genetic distance between each phage population and the ancestral genotype (a), and genetic diversity within each phage population (b). Symbols denote means ± s.e.m. of replicate populations within the coevolved (magenta; n = 5) and the evolved (blue; n = 6) treatments. The locations of mutations within each population are shown as bars underneath each coding sequence, with the colour of each bar indicating the frequency of each mutation within each population (white, rare; red, common). c, Magnified view of identity and frequency of mutations in each population for SBWP25_0027 (gp40) and SBWP25_0036 (gp49). bp, base pairs.
Coevolved populations also showed higher genetic diversity within populations than did evolved populations (Supplementary Table 1). This was predominantly due to variation in SBWP25_0027 (gp40), as SBWP25_0036 (gp49) displayed within-population diversity in both treatments (Fig. 2b). This high genetic diversity at SBWP25_0036 is surprising given the apparent directional selection for reduced tail-fibre protein length during coevolution. This suggests that SBWP25_0036 polymorphisms may be transient and the result of recurrent continuing selective sweeps, and/or clonal interference. Alternatively, both SBWP25_0036 and SBWP25_0027 may be subject to diversifying or fluctuating selection within populations. Together these genes (SBWP25_0036 and SBWP25_0027) are believed to control host adhesion19; thus, it is possible that diversity at these genes may determine fine-scale host-specificity differences between phage genotypes. Such phenotypic differences between individual phage clones from the same population have been observed in a previous study in this system16.
Overall, our results are consistent with accelerated evolution in the coevolution treatment that is driven by selective effects, rather than purely demographic differences between treatments. Demographic effects, such as reduced generation time or population size, or reduced fidelity of DNA replication, would have led to a genomewide increase in divergence and diversity, which was not observed. By contrast, genetic divergence and diversity for most phage genes were roughly similar in the two treatments (Fig. 2), indicating selection under coevolution on specific infectivity genes/traits, such as that for tail-fibre protein length or infectivity range (Supplementary Fig. 1). Furthermore, whereas greater genetic divergence among coevolved populations (Fig. 1a) could be explicable simply if coevolved populations are also smaller, and hence more susceptible to genetic drift, this explanation is incompatible with the higher genetic diversity observed in SBWP25_0027 in coevolved populations (Fig. 2b). In line with this, there was no significant difference in phage population size between treatments over the course of the experiment (log10 (plaque-forming units (p.f.u.) ml−1) averaged through time: coevolved, 7.39 ± 0.14 s.e.; evolved, 7.51 ± 0.08 s.e.; t = 0.71, d.f. = 9, P = 0.5).
Our results highlight coevolution as a fundamental driver of molecular evolution, and emphasize the utility of genome re-sequencing for quantifying evolutionary dynamics in experimental evolution21. We directly demonstrate that antagonistic coevolution accelerates molecular evolution and can generate genetic divergence both between and within populations. By contrast, populations adapting to a fixed host genotype showed a remarkable degree of parallel evolution, indicating genetic constraints on the evolutionary trajectories taken by replicate populations22,23. Coevolutionary interactions between species are likely therefore to be responsible for rapid evolutionary change within species, potentially causing sufficient between-population genetic divergence to drive speciation itself24.
METHODS
Selection experiment
A single bacterial colony of P. fluorescens SBW25 was isolated from laboratory stocks by plating on KB agar, and a single plaque of Φ2 was isolated from an agar overlay of P. fluorescens infected with a sample of Φ2 from laboratory stocks. Each was grown up overnight and used to found 12 populations with 107 SBW25 cells and 104 phage particles. Cultures were propagated in 30-ml glass universals containing 6 ml KB media by serial transfer for 24 days in a static incubator at 28 °C. Transfers for the coevolved treatment involved transferring 60 μl (1%) of culture to a fresh KB microcosm every 48 h. The evolved treatment involved isolating phage populations using 0.1 vol. chloroform and centrifuging at 14,000g for 2 min, and then inoculating fresh microcosms with 60 μl (1%) of the phage population plus 107 ancestral SBW25 cells every 48 h. Every two transfers, phage densities were estimated by plating dilutions of each population onto KB agar plates with a semi-solid overlay bacterial lawn.
Molecular methods
The complete sequence of the ancestral Φ2 clone was obtained through a whole-genome shotgun library approach. Φ2 DNA was isolated from an infected SBW25 culture after lysis with 0.1 vol. chloroform, centrifugation at 14,000g for 2 min, and extraction from the supernatant as previously described25. The purified phage DNA was used to produce 2–4 kb and 4–6 kb size-fractionated subclone libraries in pCR4-TOPO (Invitrogen). Sequencing was performed using ABI BigDye Terminator cycle sequencing kits with sample analysis performed on ABI 3730×l instruments. The finished sequence was generated from a total of 1,104 good quality reads (93% pass rate; 867,704 bp; theoretical coverage of ×20) including 363 finishing reads used to close gaps and span low coverage regions. The genome was assembled using Phrap and was tested by BLASTN against the P. fluorescens SBW25 genome sequence to ensure that no chromosomal sequences had been incorporated into the phage alignment. The sequence was annotated in Artemis26 (Wellcome Trust Sanger Institute) where putative coding sequences (CDS) were investigated by BLASTP against the non-redundant (nr) NCBI protein database. Highest homologies were found to LKA1 bacteriophage, and CDS named accordingly. The genome has been submitted to EMBL under accession number FN594518.
Phage DNA was extracted from selection experiment populations as earlier (sufficient quality and quantity of DNA for subsequent pyrosequencing was obtained for five of six coevolved populations and for all evolved populations). Shotgun single-stranded template DNA (sstDNA) libraries were prepared for five coevolved populations (C1, C3–6) and six evolved populations (E1–6) by nebulising DNA and ligating multiplex identifier tags according to manufacturer’s instructions. Libraries were then sequenced on a Roche 454 Titanium pyrosequencer. The average sequence coverage for each library ranged from ×74–354. Reads were mapped to the Φ2 reference sequence and mutations were identified and their frequencies calculated using the Roche Newbler mapping tool. Although some mutations were fixed in individual populations, no single mutation was fixed across all populations, indicating that the reference sequence used was that of the ancestral genotype.
Infectivity profiles
Twenty independent bacterial colonies were isolated from each of the five coevolved populations by plating on KB agar. Samples from each coevolved and evolved phage population were isolated using 0.1 vol. chloroform and centrifugation at 14,000g for 2 min to produce phage supernatants. An infectivity profile for a phage population was determined by drying lines of phage on KB agar plates, and perpendicularly streaking all isolated bacterial colonies across it. A bacterial colony was deemed susceptible if it showed any inhibition of growth after encountering the line of phage10.
Population genetic methods
The average number of pairwise genetic differences between each population and the ancestral genotype was calculated from the frequency of each mutation identified by the Newbler mapping tool. To minimise potential bias due to variation in sequence coverage between libraries, all non-synonyomous mutations identified were used, including potential sequencing errors present at low frequency; although the results were essentially identical if analyses were limited to only high-confidence mutations or to non-synonymous and synonymous mutations combined. (Because the frequency of sequencing errors and synonymous mutations were low they contributed little to genetic distance measures.) Indels, regardless of size, were counted as a single mutation. The number of sites with mutations was calculated based on sites with a mutation frequency of >5%. This threshold was used to avoid including sequencing errors, which were likely to have occurred, on average, at comparable magnitudes in both treatments (for further discussion of handling sequencing errors in heterogeneous population samples see ref. 27). The significance of different genetic distances between treatments was determined using Welch two sample t-tests, and for numbers of sites containing mutations using generalized linear models with a Poisson error distribution. A phylogenetic tree was constructed using Euclidean genetic distance (the square root of pairwise differences), which is suggested as an appropriate metric for molecular variation data18. We note that it is possible that a small number of the mutations that we observed could have arisen in the original outgrowth from which all the populations were initiated; thus, some allelic variation could have been shared by all the populations at the beginning of the selection experiment. However, any such effect would not be sufficient to explain our results, as replicate populations from the same treatment would only group together within the tree (as observed in Fig. 1a) where selection increases the frequency of the same allele independently in replicate populations in one treatment but not in the other treatment. Analysis of molecular variation (AMOVA) was conducted to apportion molecular variation into components due to variation within populations, among populations and between treatments18. To achieve this, AMOVA was conducted on populations in each treatment separately and on all populations combined. All analyses were conducted in R v2.9 and Perl v5.8.
Acknowledgements
We are grateful to M. Begon and G. Hurst for comments on previous drafts of this manuscript. We acknowledge funding from the Natural Environment Research Council, the Wellcome Trust, the Leverhulme Trust and the European Research Council. We are grateful for technical assistance from staff at the Centre for Genomic Research, University of Liverpool.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information The ancestral genome sequence of phage Φ2 has been submitted to the EMBL Nucleotide Sequence Database under accession number FN594518.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Van Valen L. A new evolutionary law. Evol. Theory. 1973;1:1–30. [Google Scholar]
- 2.Van Valen L. Molecular evolution as predicted by natural selection. J. Mol. Evol. 1974;3:89–101. doi: 10.1007/BF01796554. [DOI] [PubMed] [Google Scholar]
- 3.Stenseth NC, Smith JM. Coevolution in ecosystems: Red Queen evolution or stasis? Evolution. 1984;38:870–880. doi: 10.1111/j.1558-5646.1984.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 4.Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 5.Hedrick PW. Evolutionary genetics of the major histocompatibility complex. Am. Nat. 1994;143:945–964. [Google Scholar]
- 6.Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 7.Blanc G, et al. Molecular evolution of Rickettsia surface antigens: evidence of positive selection. Mol. Biol. Evol. 2005;22:2073–2083. doi: 10.1093/molbev/msi199. [DOI] [PubMed] [Google Scholar]
- 8.Mu J, et al. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nature Genet. 2006;39:126–130. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- 9.Barrett LG, et al. Diversity and evolution of effector loci in natural populations of the plant pathogen Melampsora lini. Mol. Biol. Evol. 2009;26:2499–2513. doi: 10.1093/molbev/msp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckling A, Rainey PB. Antagonistic coevolution between a bacterium and a bacteriophage. Proc. R. Soc. Lond. B. 2002;269:931–936. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brockhurst MA, Morgan AD, Fenton A, Buckling A. Experimental coevolution with bacteria and phage: the Pseudomonas fluorescens–Φ2 model system. Infect. Genet. Evol. 2007;7:547–552. doi: 10.1016/j.meegid.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nature Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- 13.Brockhurst MA, Morgan AD, Rainey PB, Buckling A. Population mixing accelerates coevolution. Ecol. Lett. 2003;6:975–979. [Google Scholar]
- 14.Morgan A, Gandon S, Buckling A. The effect of migration on local adaptation in a coevolving host–parasite system. Nature. 2005;437:253–256. doi: 10.1038/nature03913. [DOI] [PubMed] [Google Scholar]
- 15.Morgan A, Buckling A. Relative number of generations of hosts and parasites does not influence parasite local adaptation in coevolving populations of bacteria and phages. J. Evol. Biol. 2006;19:1956–1963. doi: 10.1111/j.1420-9101.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- 16.Poullain V, Gandon S, Brockhurst MA, Buckling A, Hochberg ME. The evolution of specificity in evolving and coevolving antagonistic interactions between a bacteria and its phage. Evolution. 2008;62:1–11. doi: 10.1111/j.1558-5646.2007.00260.x. [DOI] [PubMed] [Google Scholar]
- 17.Benmayor R, Hodgson DJ, Perron GG, Buckling A. Host mixing and disease emergence. Curr. Biol. 2009;19:764–767. doi: 10.1016/j.cub.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitocondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ceyssens PJ, et al. Genomic analysis of Pseudomonas aeruginosa phages LKD16 and LKA1: establishment of the ΦKMV subgroup within the T7 supergroup. J. Bacteriol. 2006;188:6924–6931. doi: 10.1128/JB.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calendar RL. The Bacteriophages. Oxford Univ. Press; 2005. [Google Scholar]
- 21.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 22.Bull JJ, et al. Exceptional convergent evolution in a virus. Genetics. 1997;147:1497–1507. doi: 10.1093/genetics/147.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wichman HA, Badgett MR, Scott LA, Boulianne CM, Bull JJ. Different trajectories of parallel evolution during viral adaptation. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- 24.Thompson JN. The Geographic Mosaic of Coevolution. Univ. of Chicago Press; 2005. [Google Scholar]
- 25.Santos MA. An improved method for the small scale preparation of bacteriophage DNA based on phage precipitation by zinc chloride. Nucleic Acids Res. 1991;19:5442. doi: 10.1093/nar/19.19.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 27.Barrick JE, Lenski RE. Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb. Symp. Quant. Biol. 2009 Sep 23; doi: 10.1101/sqb.2009.74.018. doi:10.1101/sqb.2009.74.018. [DOI] [PMC free article] [PubMed] [Google Scholar]