Abstract
Purpose
More than 1,300,000 prostate needle biopsies are performed annually in the U.S. with up to 16% incidence of isolated high-grade prostatic intraepithelial neoplasia (HGPIN). HGPIN has low predictive value for identifying prostate cancer (PCA) on subsequent needle biopsies in PSA screened populations. In contemporary series, PCA is detected in about 20% of repeat biopsies following a diagnosis of HGPIN. Further, discrete histological subtypes of HGPIN with clinical implication in management have not been characterized. The TMPRSS2-ERG gene fusion that has recently been described in PCA has also been demonstrated to occur in a subset of HGPIN. This may have significant clinical implications given that TMPRSS2-ERG fusion PCA is associated with a more aggressive clinical course.
Experimental Design
In this study we assessed a series of HGPIN lesions and paired PCA for the presence of TMPRSS2-ERG gene fusion.
Results
Fusion positive HGPIN was observed in 16% of the 143 number of lesions, and in all instances the matching cancer shared the same fusion pattern. 60% of TMPRSS2-ERG fusion PCA had fusion negative HGPIN.
Conclusions
Given the more aggressive nature of TMPRSS2-ERG PCA, the findings of this study raise the possibility that gene fusion positive HGPIN lesions are harbingers of more aggressive disease. To date, pathological, molecular and clinical parameters do not help stratify which men with HGPIN are at increased risk for a cancer diagnosis. Our results suggest that the detection of isolated TMPRSS2-ERG fusion HGPIN would improve the positive predictive value of finding TMPRSS2-ERG fusion PCA in subsequent biopsies.
Introduction
In the United States, approximately 1,300,000 prostate biopsies were performed in 2006 with the detection of 234,460 new cases of prostate cancer [American Cancer Society, Cancer Facts & Figures 2006]. The incidence of isolated high-grade prostatic intraepithelial neoplasia (HGPIN) without carcinoma ranges from <1% to 16%1–5, and the risk of finding carcinoma on subsequent biopsies is 10–39% (median risk of 24%6 depending on the time of repeat biopsy and number of cores7–10. A decline in the predictive value of HGPIN for prostate cancer to about 20% in contemporary needle biopsies is most likely due to extended biopsy techniques that yield higher rates of cancer detection11.
Both HGPIN and prostate adenocarcinoma share molecular anomalies including telomere shortening12, RARβ2 hypermethylation13, allelic imbalances14, and several chromosomal anomalies and c-myc amplification15–17. Overexpression of p1618, reduction of annexin I19 and altered proliferation and apoptosis20 in HGPIN and prostate cancer has also been demonstrated. Table 1 summarizes a selection of molecular alterations identified in HGPIN and prostate cancer.
Table 1.
Molecular evidence of association between HGPIN and prostate cancer (PCA). Numbers of total cases (not foci) of HGPIN per study are in bold.
| Focus and number of HGPIN samples | Technique | Main conclusions | Reference |
|---|---|---|---|
| Telomere shortening as an early somatic DNA alteration in prostate cancer: A total of 6 prostatectomies were evaluated which included 11 HGPIN lesions, and 20 needle biopsies with HGPIN without cancer (n=26) | FISH | Shortening seen in 93% (28/30) of HGPIN lesions is similar to what has been shown in invasive PCA. | Meeker AK et al, Cancer Res 2002 (11). |
| Proliferation and apoptotic markers in normal and premalignant tissue associated with PCA: 13 prostatectomies and 6 cystoprostatectomies were evaluated (n=19) | IHC | Both preneoplastic lesions and normal looking epithelium associated with cancer show altered proliferation and apoptosis | Ananthanarayanan V et al, BMC Cancer 2006 (19). |
| TMPRSS2-ERG in HGPIN: 34 PCA and 19 paired HGPIN were analyzed (n=19). Also 14 BPH and 11 normal as controls. | Real time PCR, sequencing, CGH | 21% of HGPIN lesions harbor the fusion, 50% of PCA, and none of controls | Cerveira N et al, Neoplasia 2006 (21). |
| Quantitative methylation of RARB2: PCA (118 patients), paired HGPIN lesions (n=38), and BPH (30 patients) | Quantitative methylation specific PCR | RARB2 hypermethylation in 97.5% PCA, 94.7% HGPIN, and 23.3% BPH. RARB2 methylation levels correlated with higher pathological stage | Jeronimo C et al, Clin Cancer Res 2004 (12). |
| Annexin I protein expression: PCA (69 prostatectomies), paired HGPIN (n=45), and benign prostate (14 samples) | IHC, real-time PCR | Annexin I was significantly reduced in PCA and HGPIN compared to benign prostate | Kang JS et al, Clin Cancer Res 2002 (18). |
| Overexpression of p16INK4A in HGPIN: 206 patients with clinically localized PCA were screened, a subset with HGPIN (n=154) | IHC | Overexpression of p16INK4A in HGPIN was independent predictor of disease relapse and increased risk of recurrence | Henshall SM et al, Clin Cancer Res 2001 (17). |
| Detection of chromosomal anomalies and c-myc gene amplification in cribriform HGPIN and PCA: A total of 25 prostatectomy specimens were studied, which included 48 foci of HGPIN and 71 foci of PCA (n=25) | FISH | Cribriform HGPIN and cribriform PCA exhibited similar anomalies | Qian J, Jenkins RB and Bostwick DG, Mod Pathol 1997 (16). |
| Detection of c-myc amplification and chromosomal anomalies: HGPIN (48 foci), localized PCA (71 foci), and lymph node metastases (23 foci) in 25 prostatectomies (n=25) | FISH | Gain of chromosome 8 and c-myc amplification are potential markers of PCA progression, HGPIN is likely a precursor | Jenkins RB et al, Cancer Res 1997 (14). |
| Chromosomal anomalies in HGPIN and PCA: 40 radical prostatectomy and pelvic lymphadenectomy specimens studied including 68 foci of HGPIN, 78 foci of PCA, and 8 foci of lymph node metastases (n=40) | FISH | HGPIN and PCA have similar proportions of chromosomal abnormalities, supporting HGPIN as precursor. | Qian J et al, Cancer Res 1995 (15). |
| Assessment of allelic imbalance at 6 polymorphic microsatellite markers: 84 foci of HGPIN 95 foci of PCA from 52 completely embedded, mapped whole mount prostates (n=52) | PCR (majority of cases previously studied by FISH) | Rate of allelic imbalance was similar at 5 of 6 loci studied. Significant genetic heterogeneity seen, suggesting that multiple foci of HGPIN arise independently in prostate | Bostwick DG et al, Cancer 1998 (13). |
Despite the association with prostate cancer, distinct subtypes of HGPIN with clinical relevance (i.e. greater risk of predicting aggressive cancer) have not been characterized. A recent rearrangement involving the androgen-regulated gene TMPRSS2 and members of the ETS transcription factor family has been identified21 and confirmed by multiple other groups22–28. In particular, the TMPRSS2-ERG gene fusion prostate cancer is associated with higher tumor stage and tumor-specific death or metastasis25, 29–31. Two recent studies have demonstrated the presence of TMPRSS2-ERG gene fusion in approximately 20% of HGPIN lesions22, 26.
The purpose of this study was to assess the TMPRSS2-ERG gene fusion status in a large series of HGPIN lesions with paired prostate cancer. Based on the results, we postulate that TMPRSS2-ERG fusion HGPIN is a distinct molecular subtype and its identification indicates the presence of the same genetic aberration in prostate cancer if present. This may impact clinical management of isolated HGPIN in prostate needle biopsies.
Materials and Methods
Case selection
143 HGPIN lesions from equal number of patients were interrogated for the presence of TMPRSS2-ERG gene fusion. This study was conducted under the IRB protocol 2006-P-000715/1 BWH at Brigham and Women’s Hospital. The HGPIN lesions were represented on 22 tissue microarrays (TMA) from prostatectomy specimens (96/143), 34 prostate needle biopsies, and 13 full section prostatectomy samples. Of these, 87% (124/143) had paired prostate cancer. The remaining 19 cases demonstrated isolated HGPIN without evidence of concurrent cancer, and included two cases of HGPIN with adjacent atypical small acinar proliferation10, 32. Clinical and pathologic demographics were available for 93 of the 143 patients. These included 70 of 124 HGPIN lesions with paired prostate cancer as follows: 40 of 96 patients represented in the TMAs, all 34 patients represented in the needle biopsies, and 9 of 13 patients represented in prostatectomy samples. The mean age at presentation was 60 years with a mean pre-operative PSA of 16.5 ng/ml. There were 30% Gleason grade ≤6, 51% Gleason grade 7, and 19% Gleason grade ≥8 prostate cancers.
Pathologic analysis
The morphological diagnosis was confirmed on H&E stained paraffin sections by two pathologists (J-MM and SP) prior to assessment of gene fusion by fluorescent in-situ hybridization (FISH) on a step section, corresponding to one unstained section at identical level obtained at the time of initial tissue sectioning. HGPIN lesions were differentiated into four morphological subtypes: tufting, flat, micropapillary, and cribriform33, 34. In a subset of cases with equivocal diagnosis, immunohistochemistry (IHC) for prostatic basal cells was performed. These were 6 needle biopsy cases with atypical small acinar proliferation (ASAP) for which IHC helped to confirm the diagnosis of prostate cancer. For that purpose, additional unstained slides were deparaffinized in xylene and rehydrated in graded ethanols. The tissue level of the immunohistochemical study was identical to the original H&E. Pressure-cooking was applied as the antigen retrieval method. Primary antibodies against p63 (1:50 dilution of clone 4A4, NeoMarkers, Fremont, CA) and high molecular weight cytokeratin (1:200 dilution of clone 34βE12, DAKO, Carpinteria, CA) for the detection of basal cells were applied with over night incubation at 4°C in a humid chamber. Immunohistochemistry was performed with the avidin-biotin peroxidase technique.
Assessment of TMPRSS2-ERG fusion status using an interphase FISH assay
We have previously described a dual-color interphase break-apart FISH assay to indirectly assess the fusion of TMPRSS2-ERG25, 26, 29. Briefly, two differentially labeled probes were designed to span the telomeric and centromeric neighboring regions of the ERG locus. Using this break-apart probe system a nucleus without ERG rearrangement demonstrates two pairs of juxtaposed red and green signals, forming yellow fusion signals. A nucleus with an ERG break-apart (reflecting a TMPRSS2-ERG fusion) shows split-apart of one juxtaposed red-green signal pair resulting in a single red and green signal for the translocated ERG allele, and a still combined (yellow) signal for the non-translocated ERG allele in each nucleus. The samples were analyzed under a 60× oil immersion objective using an Olympus BX-51 fluorescence microscope equipped with appropriate filters, a CCD (charge-coupled device) camera (Olympus, Center Valley, PA), and the CytoVision FISH imaging and capturing software (Applied Imaging, San Jose, CA). Evaluation of the cases was independently performed by two pathologists (J-MM and SP), both with expertise in analyzing interphase FISH experiments. For each case, we attempted to score at least 50 nuclei. Cases with significant differences between the results of both pathologists were refereed by a third pathologist (MAR).
Results
Of the 143 HGPIN cases, 16% (23/143) demonstrated TMPRSS2-ERG gene fusion. All cases shared the same fusion status with the paired prostate cancer (22/22). There was a single case of TMPRSS2-ERG fusion HGPIN without concurrent adenocarcinoma. The follow-up biopsy of this isolated HGPIN on prostate needle biopsy had not been performed at the time of preparing this manuscript. Of 120 TMPRSS2-ERG fusion negative HGPIN cases, 85% (102/120) had matching adenocarcinoma, and in 32% of these (33/102) the paired prostate cancer demonstrated TMPRSS2-ERG fusion (Figure 1).
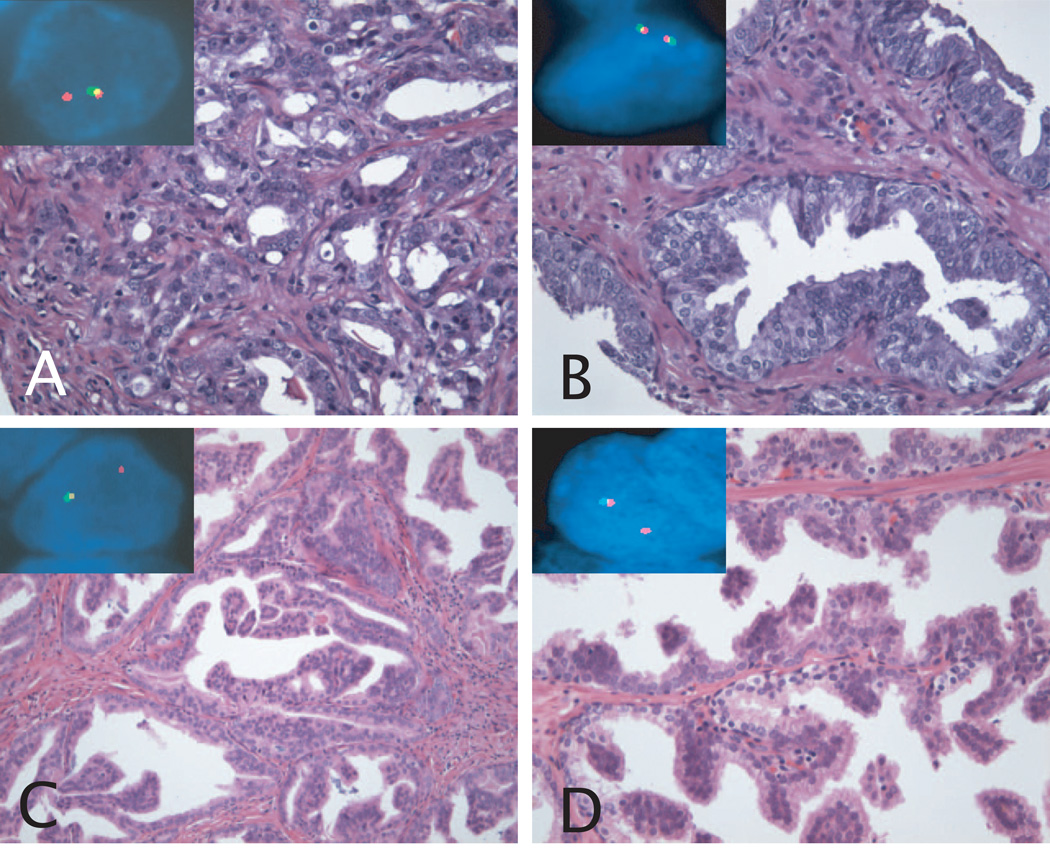
Figure 1. H&E stains and corresponding FISH images of TMPRSS2-ERG fusion assay.
A: TMPRSS2-ERG fusion prostate cancer, Gleason grade 3+4=7. The inset picture shows a nucleus with one yellow and one red signal, demonstrating the presence of TMPRSS2-ERG fusion through deletion. B: Paired HGPIN lesion of prostate cancer in A. The HGPIN features tufting morphology. The inset picture shows a nucleus with two yellow signals, demonstrating absence of genetic aberration. C: TMPRSS2-ERG fusion prostate cancer, Gleason grade 4+4=8. The prostatectomy on this case showed predominant cribriform morphology. The inset picture shows a nucleus with one yellow and one red signal, demonstrating the presence of TMPRSS2-ERG fusion through deletion. D: Paired HGPIN lesion of prostate cancer in C. The HGPIN features tufting and micropapillary morphology. The inset picture shows a nucleus with the same pattern as the matching prostate cancer, demonstrating the presence of TMPRSS2-ERG fusion.
Original magnification of H&E images, 20× objective. Original magnification of FISH images, 60× objective.
Two cases of HGPIN also demonstrated adjacent small atypical glands10, 32. One was fusion positive in both areas (Figure 2A), whereas the other one showed fusion negative HGPIN with adjacent fusion positive atypical glands. Neither case had follow-up re-biopsy at the time of preparing this manuscript. Interestingly, we could identify two cases that showed presence of TMPRSS2-ERG gene fusion HGPIN and adjacent normal epithelium (with no fusion), within the same gland (Figure 2B). Among the morphological subtypes, 31% (44/143) were tufting HGPIN, 4% (6/143) showed flat HGPIN, 2% (3/143) were micropapillary HGPIN, 1% (1/143) cases had cribriform HGPIN morphology, and 62% (89/143) combined more than one of the above subtypes.
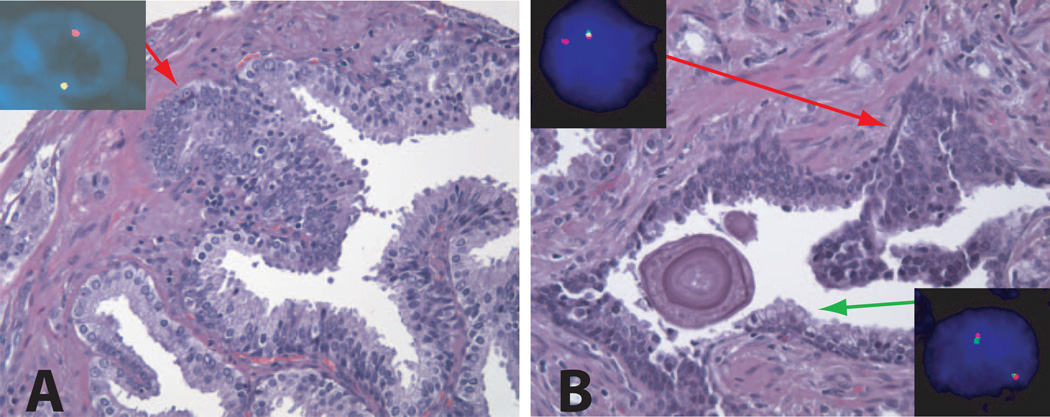
Figure 2. H&E stain and corresponding FISH image of TMPRSS2-ERG fusion assay.
A: HGPIN lesion with adjacent atypical small acinar proliferation. This may represent either outpouching area or tangential section of HGPIN, or true early invasive adenocarcinoma. The red arrow points this area. The inset picture shows a nucleus with one yellow and one red signal, demonstrating the presence of TMPRSS2-ERG fusion through deletion.
Original magnification of H&E images, 20× objective. Original magnification of FISH images, 60× objective.
B: HGPIN and normal prostatic epithelium in the same gland. Red and green arrows point representative areas of HGPIN and normal prostatic epithelium, respectively. The inset pictures show a nucleus of normal epithelium with juxtaposed red-green signal pair (upper left), and a nucleus of HGPIN with one yellow and one red signal, demonstrating TMPRSS2-ERG fusion through deletion (lower right). The surrounding prostatic cancer, mostly Gleason pattern 4, also shared the same gene fusion pattern.
Original magnification of H&E images, 20× objective. Original magnification of FISH images, 60× objective.
Discussion
Several suggested protocols for management of isolated HGPIN in prostate needle biopsies exist. They vary from repeat biopsy at three to six months, at six to twelve months, or at three years35–37. The most aggressive protocol suggests repeat biopsies at three to six-month intervals for two years, thereafter every year for life7. Recent data suggest that the incidence of prostatic adenocarcinoma after the initial diagnosis of isolated HGPIN in needle biopsies is lower than previously reported10, 11, and despite molecular data on HGPIN, biomarkers with direct clinical application have not been used to stratify the risk for subsequent detection of adenocarcinoma. In addition, morphologic features and extent of HGPIN show inconsistent data to predict risk of consecutive prostate cancer. Therefore, the clinical management of patients with isolated HGPIN is problematic and to date, no treatment is indicated after this diagnosis is rendered.
It is valid to speculate that stratification of different subtypes of HGPIN at the molecular level (i.e. TMPRSS2-ERG fusion HGPIN) may be needed for potential prognostic implications, and in view of clinical trials for chemoprevention of prostate cancer where the inclusion criteria is the diagnosis of isolated HGPIN38, 39.
Our results may help in prognostication of a subset of isolated HGPIN lesions, that is, those harboring the TMPRSS2-ERG gene fusion. We have recently postulated that the TMPRSS2-ERG gene fusion is a clonal, early pathogenic event in prostate cancer26, 40. Evidence supporting this hypothesis is that in most instances the gene fusion is homogenously present throughout the cancer within a tumor nodule, is not identified in benign prostatic tissue, and is detected only in a subset of HGPIN lesions. Another group has also confirmed the presence of TMPRSS2-ERG gene fusion in HGPIN using polymerase chain reaction (PCR) technique22. Interestingly, both studies show approximately 20% gene fusion positivity among a small series of HGPIN.
In the current study, the incidence of TMPRSS2-ERG gene fusion HGPIN is 16%, in 143 cases. Given that all TMPRSS2-ERG gene fusion HGPIN lesions share the same fusion pattern with matching cancer, and no fusion positive HGPIN lesions were associated with paired TMPRSS2-ERG fusion negative prostate cancer, we demonstrate that the presence of TMPRSS2-ERG gene fusion HGPIN is always indicative of a prostate cancer bearing the same genetic aberration. Conversely, TMPRSS2-ERG fusion prostate cancer may present with fusion negative HGPIN. Possible scenarios that could explain this finding are that fusion negative HGPIN does either not precede TMPRSS2-ERG fusion prostate cancer, or that TMPRSS2-ERG fusion HGPIN was not sampled if we consider the presence of gene fusion heterogeneity in HGPIN as a possibility. In our previous work26, 41 we had made these observations. However, in the series reported by Cerveira et al22, PCR assessment yielded two cases where the fusion transcript was detected in HGPIN, but not in the concurrent cancer of the same gland. In the present study we have screened a significantly larger number of HGPIN lesions using FISH, the gold standard method to detect these molecular alterations, and we have not observed such combination. This discrepancy could be due to artifact in the PCR assay, or as a consequence of TMPRSS2-ERG heterogeneity in prostate cancer, where the fusion positive area of tumor may have not been sampled. Although TMPRSS2-ERG gene fusion heterogeneity in prostate cancer is out of the scope of the current study, it is pertinent to mention that in our most recent study, 41% of radical prostatectomy high stage cases (at least pT2c) demonstrated interfocal clonal heterogeneity40, also described by Mehra et al42 and Furusato et al43. This fact may have significant clinical implications for follow-up biopsy and treatment strategies, in the context of isolated TMPRSS2-ERG fusion HGPIN.
Taking these results together, we consider that TMPRSS2-ERG fusion HGPIN is a true precursor of a subset of TMPRSS2-ERG prostate cancer, and the presence of the former is always indicative of the latter. Remarkably, we identified two cases where TMPRSS2-ERG fusion HGPIN was showing either early invasion (see Figure 2A) or coexistence with normal epithelium in the same gland (see Figure 2B). This morphologic/gene fusion status correlation further supports our statement, as well as the hypothesis of HGPIN to cancer progression (in this case, of those lesions harboring the TMPRSS2-ERG fusion). These observations are clinically relevant since there is emerging data supporting that TMPRSS2-ERG fusion prostate cancer is associated with worse prognosis, namely, higher tumor stage and tumor-specific death or metastasis24, 25, 29, 31, 44, 45. Hence, the finding of isolated TMPRSS2-ERG fusion HGPIN in needle biopsies may have the highest predictive value for further detection of fusion positive prostate cancer with the significant clinical implication noted above.
Based on the results of our recent work on morphological features associated with TMPRSS2-ERG fusion prostate cancer46, we also considered a potential correlation between the morphology of HGPIN and the TMPRSS2-ERG fusion status. However, 62% of HGPIN cases combined two or more of the morphologic subtypes, hampering a significant association.
Although prospective studies with follow-up of isolated TMPRSS2-ERG gene fusion HGPIN are needed to modify the current approach of management of isolated HGPIN, our results show convincing evidence that fusion positive HGPIN lesions are consistently associated with TMPRSS2-ERG prostate cancer. To further support our findings, studies with follow-up of patients with isolated TMPRSS2-ERG fusion HGPIN or TMPRSS2-ERG fusion HGPIN with adjacent small atypical glands like one or our cases, are underway as part of an Early Detection Research Network (EDRN) protocol. Further, evaluation of the status of TMPRSS2-ERG fusion could also modify inclusion criteria in the aforementioned clinical trials. Moreover, the development of non-invasive (i.e. urine based) diagnostic tests for fusion transcripts could also help in these protocols47.
In summary, we have assessed the largest series of HGPIN lesions for TMPRSS2-ERG fusion status to date and confirmed a prevalence of 16%, similar to previously reported series. In all instances, fusion positive HGPIN is associated with concurrent TMPRSS2-ERG prostate cancer. Given the worse prognosis linked to the latter, detection of isolated TMPRSS2-ERG fusion HGPIN may help us stratify patients into a discrete risk group.
Acknowledgments
Research supported by the NIH Prostate SPORE at the Dana-Farber/Harvard Cancer Center NCI P50 CA090381(M.A.R.), R01AG21404 (M.A.R), German Research Foundation PE1179/1-2) and Department of Defense Grant (PC050965) (S.P.), NIH Grant UO1 CA 113913 for the BID EDRN (Harvard/Michigan Prostate Cancer Clinical Center), and UCSF Prostate Cancer SPORE, NIH Grant P50CA89520 (P.L.P., J.S.).
The authors are grateful to Chungdak Namgyal of the DFHCC TMA core facility, and Laura A. Johnson and Christopher LaFargue for technical support critical to this study.
References
- 1.Bostwick DG, Qian J, Frankel K. The incidence of high grade prostatic intraepithelial neoplasia in needle biopsies. J Urol. 1995;154(5):1791–1794. [PubMed] [Google Scholar]
- 2.Langer JE, Rovner ES, Coleman BG, et al. Strategy for repeat biopsy of patients with prostatic intraepithelial neoplasia detected by prostate needle biopsy. J Urol. 1996;155(1):228–231. [PubMed] [Google Scholar]
- 3.Moore CK, Karikehalli S, Nazeer T, Fisher HA, Kaufman RP, Jr, Mian BM. Prognostic significance of high grade prostatic intraepithelial neoplasia and atypical small acinar proliferation in the contemporary era. J Urol. 2005;173(1):70–72. doi: 10.1097/01.ju.0000148260.69779.c5. [DOI] [PubMed] [Google Scholar]
- 4.Naya Y, Ayala AG, Tamboli P, Babaian RJ. Can the number of cores with high-grade prostate intraepithelial neoplasia predict cancer in men who undergo repeat biopsy? Urology. 2004;63(3):503–508. doi: 10.1016/j.urology.2003.09.066. [DOI] [PubMed] [Google Scholar]
- 5.Vis AN, Van Der Kwast TH. Prostatic intraepithelial neoplasia and putative precursor lesions of prostate cancer: a clinical perspective. BJU Int. 2001;88(2):147–157. doi: 10.1046/j.1464-410x.2001.02295.x. [DOI] [PubMed] [Google Scholar]
- 6.Epstein JI, Herawi M. Prostate needle biopsies containing prostatic intraepithelial neoplasia or atypical foci suspicious for carcinoma: implications for patient care. J Urol. 2006;175(3 Pt 1):820–834. doi: 10.1016/S0022-5347(05)00337-X. [DOI] [PubMed] [Google Scholar]
- 7.Davidson D, Bostwick DG, Qian J, et al. Prostatic intraepithelial neoplasia is a risk factor for adenocarcinoma: predictive accuracy in needle biopsies. J Urol. 1995;154(4):1295–1299. [PubMed] [Google Scholar]
- 8.Herawi M, Kahane H, Cavallo C, Epstein JI. Risk of prostate cancer on first re-biopsy within 1 year following a diagnosis of high grade prostatic intraepithelial neoplasia is related to the number of cores sampled. J Urol. 2006;175(1):121–124. doi: 10.1016/S0022-5347(05)00064-9. [DOI] [PubMed] [Google Scholar]
- 9.Kronz JD, Allan CH, Shaikh AA, Epstein JI. Predicting cancer following a diagnosis of high-grade prostatic intraepithelial neoplasia on needle biopsy: data on men with more than one follow-up biopsy. Am J Surg Pathol. 2001;25(8):1079–1085. doi: 10.1097/00000478-200108000-00014. [DOI] [PubMed] [Google Scholar]
- 10.Netto GJ, Epstein JI. Widespread high-grade prostatic intraepithelial neoplasia on prostatic needle biopsy: a significant likelihood of subsequently diagnosed adenocarcinoma. Am J Surg Pathol. 2006;30(9):1184–1188. doi: 10.1097/01.pas.0000213324.97294.54. [DOI] [PubMed] [Google Scholar]
- 11.Schlesinger C, Bostwick DG, Iczkowski KA. High-grade prostatic intraepithelial neoplasia and atypical small acinar proliferation: predictive value for cancer in current practice. Am J Surg Pathol. 2005;29(9):1201–1207. doi: 10.1097/01.pas.0000168178.48535.0d. [DOI] [PubMed] [Google Scholar]
- 12.Meeker AK, Hicks JL, Platz EA, et al. Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis. Cancer research. 2002;62(22):6405–6409. [PubMed] [Google Scholar]
- 13.Jeronimo C, Henrique R, Hoque MO, et al. Quantitative RARbeta2 hypermethylation: a promising prostate cancer marker. Clin Cancer Res. 2004;10(12 Pt 1):4010–4014. doi: 10.1158/1078-0432.CCR-03-0643. [DOI] [PubMed] [Google Scholar]
- 14.Bostwick DG, Shan A, Qian J, et al. Independent origin of multiple foci of prostatic intraepithelial neoplasia: comparison with matched foci of prostate carcinoma. Cancer. 1998;83(9):1995–2002. doi: 10.1002/(sici)1097-0142(19981101)83:9<1995::aid-cncr16>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins RB, Qian J, Lieber MM, Bostwick DG. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer research. 1997;57(3):524–531. [PubMed] [Google Scholar]
- 16.Qian J, Bostwick DG, Takahashi S, et al. Chromosomal anomalies in prostatic intraepithelial neoplasia and carcinoma detected by fluorescence in situ hybridization. Cancer research. 1995;55(22):5408–5414. [PubMed] [Google Scholar]
- 17.Qian J, Jenkins RB, Bostwick DG. Detection of chromosomal anomalies and c-myc gene amplification in the cribriform pattern of prostatic intraepithelial neoplasia and carcinoma by fluorescence in situ hybridization. Mod Pathol. 1997;10(11):1113–1119. [PubMed] [Google Scholar]
- 18.Henshall SM, Quinn DI, Lee CS, et al. Overexpression of the cell cycle inhibitor p16INK4A in high-grade prostatic intraepithelial neoplasia predicts early relapse in prostate cancer patients. Clin Cancer Res. 2001;7(3):544–550. [PubMed] [Google Scholar]
- 19.Kang JS, Calvo BF, Maygarden SJ, Caskey LS, Mohler JL, Ornstein DK. Dysregulation of annexin I protein expression in high-grade prostatic intraepithelial neoplasia and prostate cancer. Clin Cancer Res. 2002;8(1):117–123. [PubMed] [Google Scholar]
- 20.Ananthanarayanan V, Deaton RJ, Yang XJ, Pins MR, Gann PH. Alteration of proliferation and apoptotic markers in normal and premalignant tissue associated with prostate cancer. BMC Cancer. 2006;6:73. doi: 10.1186/1471-2407-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science (New York, NY. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 22.Cerveira N, Ribeiro FR, Peixoto A, et al. TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006;8(10):826–832. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iljin K, Wolf M, Edgren H, et al. TMPRSS2 Fusions with Oncogenic ETS Factors in Prostate Cancer Involve Unbalanced Genomic Rearrangements and Are Associated with HDAC1 and Epigenetic Reprogramming. Cancer research. 2006;66(21):10242–10246. doi: 10.1158/0008-5472.CAN-06-1986. [DOI] [PubMed] [Google Scholar]
- 24.Mehra R, Tomlins SA, Shen R, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20(5):538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 25.Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG Fusion-Associated Deletions Provide Insight into the Heterogeneity of Prostate Cancer. Cancer research. 2006;66(17):8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 26.Perner S, Mosquera JM, Demichelis F, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31(6):882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 27.Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer. 2006 doi: 10.1002/gcc.20329. [DOI] [PubMed] [Google Scholar]
- 28.Yoshimoto M, Joshua AM, Chilton-Macneill S, et al. Three-Color FISH Analysis of TMPRSS2/ERG Fusions in Prostate Cancer Indicates That Genomic Microdeletion of Chromosome 21 Is Associated with Rearrangement. Neoplasia. 2006;8(6):465–469. doi: 10.1593/neo.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26(31):4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 30.Nam RK, Sugar L, Wang Z, et al. Expression of TMPRSS2 ERG Gene Fusion in Prostate Cancer Cells is an Important Prognostic Factor for Cancer Progression. Cancer Biol Ther. 2007;6(1) doi: 10.4161/cbt.6.1.3489. [DOI] [PubMed] [Google Scholar]
- 31.Rajput AB, Miller MA, De Luca A, et al. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007 doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kronz JD, Shaikh AA, Epstein JI. High-grade prostatic intraepithelial neoplasia with adjacent small atypical glands on prostate biopsy. Hum Pathol. 2001;32(4):389–395. doi: 10.1053/hupa.2001.23522. [DOI] [PubMed] [Google Scholar]
- 33.Bostwick DG, Amin MB, Dundore P, Marsh W, Schultz DS. Architectural patterns of high-grade prostatic intraepithelial neoplasia. Hum Pathol. 1993;24(3):298–310. doi: 10.1016/0046-8177(93)90041-e. [DOI] [PubMed] [Google Scholar]
- 34.Eble JN, Sauter G, Epstein JI, Sesterhann IAE. Pathology and Genetics of Tumours of the Male Urinary System and Male Gential Organs. Lyon: IARC Press; 2004. World Health Organization Classificationn of Tumours. [Google Scholar]
- 35.Aboseif S, Shinohara K, Weidner N, Narayan P, Carroll PR. The significance of prostatic intra-epithelial neoplasia. Br J Urol. 1995;76(3):355–359. doi: 10.1111/j.1464-410x.1995.tb07714.x. [DOI] [PubMed] [Google Scholar]
- 36.Ellis WJ, Brawer MK. Repeat prostate needle biopsy: who needs it? J Urol. 1995;153(5):1496–1498. [PubMed] [Google Scholar]
- 37.Maatman TJ, Papp SR, Carothers GG, Shockley KF. The critical role of patient follow-up after receiving a diagnosis of prostatic intraepithelial neoplasia. Prostate Cancer Prostatic Dis. 2001;4(1):63–66. doi: 10.1038/sj.pcan.4500496. [DOI] [PubMed] [Google Scholar]
- 38.Klein EA. Chemoprevention of prostate cancer. Annu Rev Med. 2006;57:49–63. doi: 10.1146/annurev.med.57.121304.131435. [DOI] [PubMed] [Google Scholar]
- 39.Price D, Stein B, Sieber P, et al. Toremifene for the prevention of prostate cancer in men with high grade prostatic intraepithelial neoplasia: results of a double-blind, placebo controlled, phase IIB clinical trial. J Urol. 2006;176(3):965–970. doi: 10.1016/j.juro.2006.04.011. discussion 70-1. [DOI] [PubMed] [Google Scholar]
- 40.Barry M, Perner S, Demichelis F, Rubin MA. Interfocal heterogeneity for TMPRSS2-ERG fusion in multifocal prostate cancer. Urology. 2007 doi: 10.1016/j.urology.2007.08.032. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomlins SA, Mehra R, Rhodes DR, et al. Integrative molecular concept modeling of prostate cancer progression. Nature genetics. 2007;39(1):41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 42.Mehra R, Han B, Tomlins SA, et al. Heterogeneity of TMPRSS2 Gene Rearrangements in Multifocal Prostate Adenocarcinoma: Molecular Evidence for an Independent Group of Diseases. Cancer research. 2007;67(17):7991–7995. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- 43.Furusato B, Gao CL, Ravindranath L, et al. Mapping of TMPRSS2-ERG fusions in the context of multi-focal prostate cancer. Mod Pathol. 2008;21(2):67–75. doi: 10.1038/modpathol.3800981. [DOI] [PubMed] [Google Scholar]
- 44.Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2007 doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Cai Y, Ren C, Ittmann M. Expression of Variant TMPRSS2/ERG Fusion Messenger RNAs Is Associated with Aggressive Prostate Cancer. Cancer research. 2006;66(17):8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 46.Mosquera JM, Perner S, Demichelis F, et al. Morphological features of TMPRSS2-ERG gene fusion prostate cancer. J Pathol. 2007 doi: 10.1002/path.2154. [DOI] [PubMed] [Google Scholar]
- 47.Laxman B, Tomlins SA, Mehra R, et al. Noninvasive detection of TMPRSS2:ERG fusion transcripts in the urine of men with prostate cancer. Neoplasia. 2006;8(10):885–888. doi: 10.1593/neo.06625. [DOI] [PMC free article] [PubMed] [Google Scholar]