Abstract
The DNA damage response (DDR) arrests cell-cycle progression until damage is removed. DNA damage-induced cellular senescence is associated with persistent DDR. The molecular bases that distinguish transient from persistent DDR are unknown. Here we show that a large fraction of exogenously-induced persistent DDR markers are associated with telomeric DNA in cultured cells and mammalian tissues. In yeast, a chromosomal DNA double-strand break (DSB) next to telomeric sequences resists repair and impairs DNA ligase 4 recruitment. In mammalian cells, ectopic localization of telomeric factor TRF2 next to a DSB induces persistent DNA damage and DDR. Linear telomeric DNA, but not circular or scrambled DNA, induces a prolonged checkpoint in normal cells. In terminally-differentiated tissues of old primates, DDR markers accumulate at telomeres which are not critically short. We propose that linear genomes are not uniformly reparable and telomeric DNA tracts, if damaged, are irreparable and trigger persistent DDR and cellular senescence.
Keywords: Telomeres, DNA damage response, TRF2, DNA repair, cellular senescence, ageing
Cellular senescence is a stable condition in which cells are unable to further divide1, 2. In addition to suppressing cancer development3–6, cellular senescence also has been demonstrated to contribute to organismal ageing and healthspan7. Its establishment has been observed following telomere shortening below a crucial length8–10, upon oncogene activation11, 12 or exposure to exogenous DNA damaging agents13. These events are associated with the activation of DNA damage response (DDR) pathways. Indeed, ageing is associated with increased frequency of DDR-positive cells in a variety of different organs, tissues and cell types14–17.
The DDR apparatus coordinates cellular DNA repair activities promptly upon DNA damage detection and transiently arrests cell-cycle progression in proliferating cells (checkpoint function) until DNA damage has been removed in full18. The molecular mechanisms underlying the observed prolonged, seemingly permanent, DDR activation in senescent cells are unclear. DDR persistency, usually detected in the form of distinct nuclear DDR foci which label genomic DNA damage sites18, 19, may be consequent to persistent DNA lesions. However, the efficacy, rather than the efficiency, of normal mammalian cells to repair DNA damage has not been thoroughly investigated.
RESULTS
Normal human fibrobasts cannot repair exogenous DNA damage in full and retain persistent DDR foci
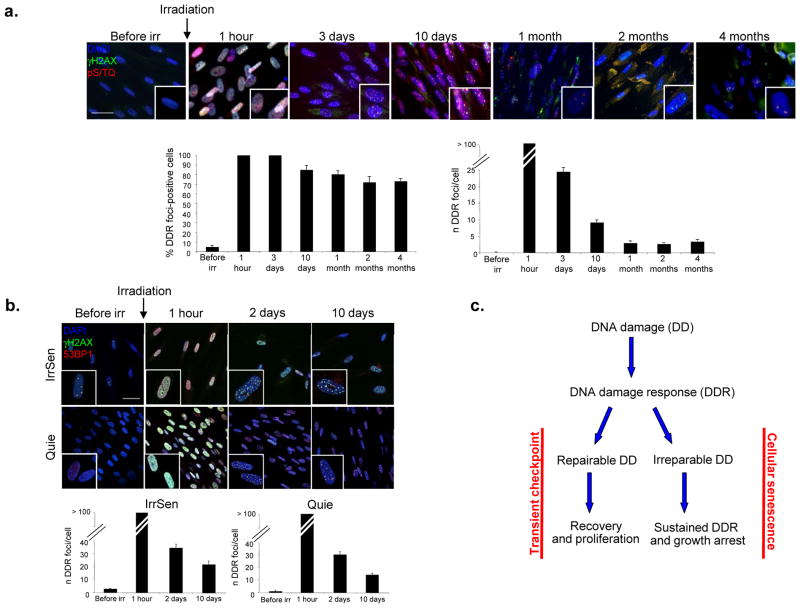
We exposed non-proliferating (quiescent) early passage normal human diploid fibroblasts (HDFs) to ionizing radiations (IR, 20 Gy). As quiescent cells do not proliferate, no telomere shortening, a potential trigger of DDR activation and cellular senescence, can occur. Cells were then stained before and at various time points after IR for evidence of DDR activation in the form of distinct nuclear foci containing the phosphorylated histone H2AX (γH2AX) and proteins phosphorylated by the activated form of ataxia-telangiectasia mutated (ATM) or the ATM-and Rad3-related protein (ATR) (pS/TQ). We observed that, despite an efficient wave of repair leading to a progressive reduction in time of IR-induced DDR foci, measured both as percentage of DDR-positive cells and as number of DDR foci per cell, a small but significant number of focal DDR signals persists in the majority of irradiated cells, even four months post-IR, the last time point studied (Fig. 1a). These few but persisting DDR signalling events cause the establishment of cellular senescence in these cells (hereafter named IrrSen), as demonstrated by high senescence-associated β-galactosidase (SA-β-gal) activity and inability to incorporate BrdU, despite replating and release from contact inhibition (Suppl. Fig. S1a). Transient inhibition of ATM kinase activity by a small molecule compound leads to rapid DDR inactivation, escape from senescence, and increased levels of KI-67, a marker of cell proliferation (Suppl. Fig. S1b and data not shown). These results indicate that sustained DDR is constantly and actively maintained and that this signalling is necessary for IR-induced senescence maintenance.
Figure 1. IR induces persistent DDR activation and cellular senescence.
a. IR generates persistent DDR. Top, images show DDR foci induction and resolution in early passage quiescent (contact-inhibited) BJ human fibroblasts following exposure to 20 Gy IR. Persistent DDR, in the form of γH2AX and pS/TQ foci, is still detectable even 4 months after IR. Bottom, bar graphs show the fraction of γH2AX foci-positive cells (± s.e.m.) (on the left) and the average number of γH2AX foci (± s.e.m.) per cell (on the right), at the indicated time points. More than 100 discrete foci cannot be counted accurately due to their proximity (1 hour time point). (For the quantification shown, around 100 cells per time point were analysed; scale bar = 20 μm) b. IrrSen cells are able to resolve additional IR-induced (10 Gy) DNA damage to an extent similar to Quie cells, as shown by the comparable kinetics of resolution of 53BP1 and γH2AX foci per cell over time after IR. (For the quantification shown, around 100 cells per time point were analysed; scale bar = 50 μm) c. Model, two opposite outcomes are possible upon DNA damage generation.
Persistent DDR activation can also be observed upon exposure to 20 Gy fractionated in 10 days (2 Gy each day) or, to a comparatively lower extent, upon a single 2 Gy dose (Suppl. Fig. S1c), in 20 Gy-irradiated proliferating cells (Suppl. Fig. S1d, e), in an independent HDF strain (MRC5; Suppl. Fig. S2a), in cells expressing telomerase (BJ hTERT; Suppl. Fig. S2b), in cells maintained at low (3%) oxygen tension (Suppl. Fig. S2c), and in proliferating cells treated with bleomycin, a DNA damaging agent also used in cancer therapy20 (Suppl. Fig. S3a). Thus, upon exposure to DNA damaging agents under different conditions, while the majority of DDR foci are transient and thus inconsequential for cell proliferation, the few DDR foci that persist are sufficient to maintain cellular senescence and impair the ability of cells to recover and proliferate.
We also observed, consistent with a recent report21, that IrrSen cells display focal accumulation of the activated form of CHK2 (CHK2 pT68), which colocalizes with persistent γH2AX foci, while freshly-irradiated cells show a more diffuse nuclear staining (Suppl. Fig. S3b). This suggests that DNA damage that is not promptly resolved causes downstream DDR factors (such as CHK2) to be retained longer at lesion sites.
As persistent DDR foci are those that ultimately determine the fate of a cell, it is important to understand their nature. We first tested whether the observed persistence is the result of compromised DNA repair capacity of senescent cells. IrrSen and Quie HDFs were exposed in parallel to the same dose of IR and DNA damage repair kinetics were monitored by DDR foci detection and quantification. We discovered that γH2AX and p53-binding protein 1 (53BP1) foci induced in IrrSen HDFs progressively disappear with a kinetic similar to that observed in Quie HDFs (Fig. 1b). Therefore, senescent cells are DNA repair proficient and, since they can form both repairable transient DDR foci and persistent ones, the observed DDR persistency cannot be a feature of cells unable to repair the damage, rather it is the consequence of the irreparability of individual DNA breaks. Overall, these results indicate that following DNA damage generation, the choice between senescence or proliferation is dictated by the presence, or absence, of even few irreparable DNA lesions (Fig. 1c).
Persistent DDR foci are localized at telomeres
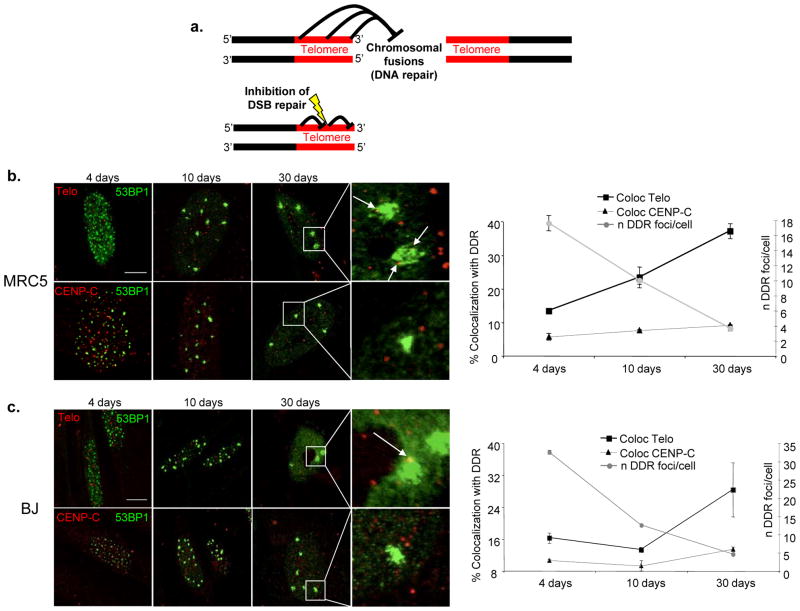
The molecular bases that distinguish repairable/transient DDR foci from irreparable/persistent DDR foci are unknown. We hypothesised that persistent DDR foci may result from DNA damage occurring at genomic loci that resist cellular DNA repair activities. We also reasoned that, if such loci were not negatively selected during evolution, they should provide a selective advantage to the cell. Telomeres are genomic loci made of repetitive DNA sequences, all with the same orientation, coated by specific proteins that function to inhibit DNA repair at chromosome termini in order to prevent chromosomal fusions and consequent genome instability22. We reasoned that telomeric repeats and factors, while acting cumulatively at the chromosome distal end, may also prevent DNA repair throughout their length, thus inhibiting repair of DNA damage generated within repeats (see model in Fig. 2a). In support of this model, in in vitro DNA repair assays using human cell extracts, telomeric DNA repeats resist Non-Homologous End-Joining (NHEJ) at their 3′ end23, 24.
Figure 2. Persistent DDR is preferentially associated with telomeric DNA.
a. A model of inhibition of DNA repair at telomeres. Top, Telomeric repeats prevent DNA end joining (DNA repair by NHEJ) at their distal end in order to prevent chromosomal fusions. Bottom, in the same manner, stretches of telomeric repeats may prevent DNA end joining (DNA repair by NHEJ) of DNA damage generated within repeats across the telomere length. b–c. Persistent DDR foci colocalize with telomeres in IrrSen human diploid fibroblasts (HDFs). Left, representative pictures of colocalizations between DDR, detected as 53BP1 foci, and telomeres, detected using a telomeric PNA probe (Telo), or centromeres, detected by antibodies raised against a centromeric protein (CENP-C), at the indicated time points following IR in MRC5 (b.) (scale bar = 10 μm) and in BJ (c.) (scale bar = 20 μm) cells. 30 days after IR, IrrSen HDFs show the highest degree of colocalizations between 53BP1 and the telomeric PNA probe. Arrows indicate telomeric signals colocalizing with 53BP1 foci. Right, graphs show the percentage of colocalizations (± s.e.m.) between 53BP1 foci and telomeric (Telo) or centromeric (CENP-C) regions, and the average number of 53BP1 foci per cell (± s.e.m.) at the indicated time points after IR. (For the quantifications shown, around 50–200 cells per time point were analysed).
IR is expected to generate DNA damage and DDR foci randomly. While most DNA damage is repaired and the total number of DDR foci progressively declines, if telomeres resist repair, the fraction of persisting DDR foci at telomeres should progressively increase over time. We therefore performed immunofluorescence stainings against the DDR factor 53BP1, in conjunction with fluorescence in situ hybridization (FISH) using a telomeric Cy3-conjugated peptide-nucleic acid (PNA) probe (immunoFISH) in two independent HDF strains (BJ and MRC5), at different time points following exposure to IR. As a control, cells were stained for CENP-C, a marker of centromeres, genomic regions also made of repetitive sequences, like telomeres. By using an unbiased method of colocalization analysis based on imaging software, we discovered that, while the number of DDR foci per cell progressively declines, the fraction of those that colocalize with a telomeric signal gradually increases. Thirty days after IR, up to 40% of 53BP1 foci that have not been repaired are detected at telomeres, while much fewer are at centromeres in both cell strains (Figs. 2b, c). Such DDR accumulation is highly significant as telomeres represent a very small fraction of the genome (around 0.02 %). Similar results were obtained using γH2AX as an independent DDR marker (Suppl. Fig. S4a). These conclusions are not restricted to IR, since also bleomycin induces persistent DDR foci that colocalize with telomeric signals to a similar extent (Suppl. Fig. S4b). In addition, a fractionated dose of 20 Gy (2 Gy each day for 10 days) generates a similar number of persistent DDR foci at telomeres (Suppl. Fig. S4c). This suggests that the observations made with a single 20 Gy dose are unlikely to be due to acute generation of excessive DNA damage and potential squelching of DNA repair factors. Furthermore, when cells were analysed 30 days after a single dose of 2 Gy, although persistent DDR foci were, as expected, lower in number (Suppl. Fig. S1c), their fraction at the telomeres was similar to that observed after a 20 Gy dose (Suppl. Fig. S4c).
Part of these results could be reproduced using very similar experimental settings25.
Persistent DDR foci are physically associated with chromosomal telomeres
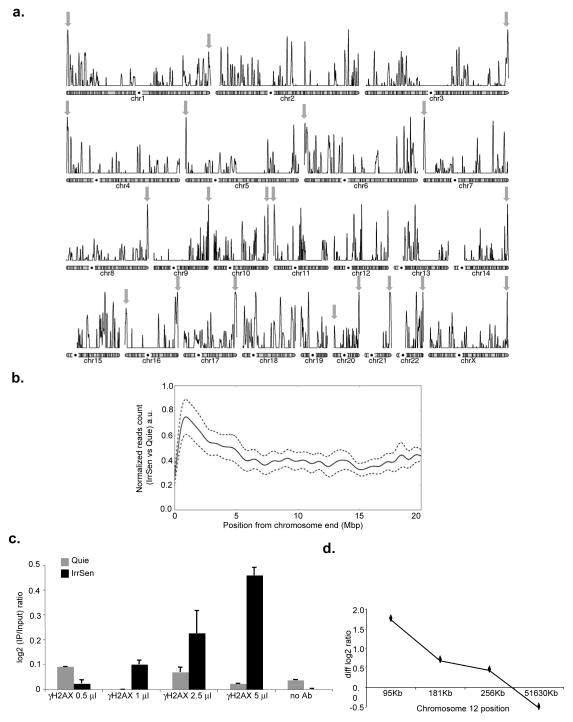
To independently interrogate the preferential localization of persistent DDR marks in the genome, we mapped γH2AX genome-wide in IrrSen and control Quie cells by chromatin immunoprecipitation experiments and next generation sequencing (ChIPseq). Analysis of individual chromosomes showed that the highest peak of each chromosome arm was within 5 Mbp from the chromosome end at 20 out of 41 mapped chromosome ends (Fig. 3a) and, overall, among the 10 highest peaks, 8 were within 5 Mbp of a chromosome end. Furthermore, by compiling the signals of IrrSen over Quie cells from all chromosome arms according to their distance from the chromosome end, we observed a statistically significant enrichment for γH2AX in the most distal 5 Mbp of mapped DNA (Fig. 3b).
Figure 3. Persistent DDR is physically associated with telomeric DNA.
a. Chromosomal view of the enrichment of γH2AX in IrrSen vs Quie cells. Individual chromosomes representation of the enrichment of γH2AX in IrrSen BJ hTERT cells. Grey arrows indicate the highest peak of each chromosome arm within 5 Mbp from a chromosome end. b. Enrichment of γH2AX in IrrSen vs Quie cells at chromosome ends. Solid line represents the accumulation pattern of γH2AX over 20 Mbp from the chromosomes end. The profile shows an enrichment peak at the most terminal region (0–5 Mbp). The enrichment is significantly different when compared to the accumulation in a more internal region (15–20 Mbp), (Mann-Whitney-U p-value = 0, n=2.3e6). 95% credibility intervals are shown as dashed lines; a.u. means arbitrary units. c. Increasing amounts of antibodies against γH2AX immunoprecipitate increasing amounts of subtelomeric DNA from IrrSen BJ cells compared to non-irradiated Quie cells, as assayed on telomere 12p by ChIP and qPCR using a primer pair distant 95 Kb from chromosome end. Triplicate qPCR reactions were carried out and the averaged results are plotted as the log2 ratio between IP and input (n = 2). d. γH2AX and subtelomeric DNA are physically associated in IrrSen BJ cells. Enrichment of γH2AX was assayed on telomere 12p by ChIP and qPCR with previously independently-validated primer pairs19 at indicated increasing distances from the chromosome end. Graph shows subtracted log2 ratios between IP and input of IrrSen minus Quie cells (diff log2 ratio). Triplicate qPCR reactions were carried out and the averaged results are plotted (n = 2).
In addition, we performed ChIP assays followed by quantitative real-time PCR (qPCR) using an independent set of PCR primers, previously validated in replicative senescent HDFs19. We observed that increasing amounts of antibodies against γH2AX immunoprecipitate increasing amounts of subtelomeric DNA in IrrSen HDFs but not in non-irradiated Quie HDFs (Fig. 3c) and that such enrichment decreases from the chromosome terminal region towards the centromere (Fig. 3d), in a manner similar to that observed in cells undergoing telomere uncapping following removal of the telomeric repeat binding factor 2 (TRF2) (Suppl. Fig. S4d). Overall, these results confirm the preferential localization of persistent DDR at telomeres and demonstrate that DDR association results from physical association, not mere cytological proximity, with the telomeres.
Exogenously-induced persistent DDR foci colocalize with telomeres also in vivo
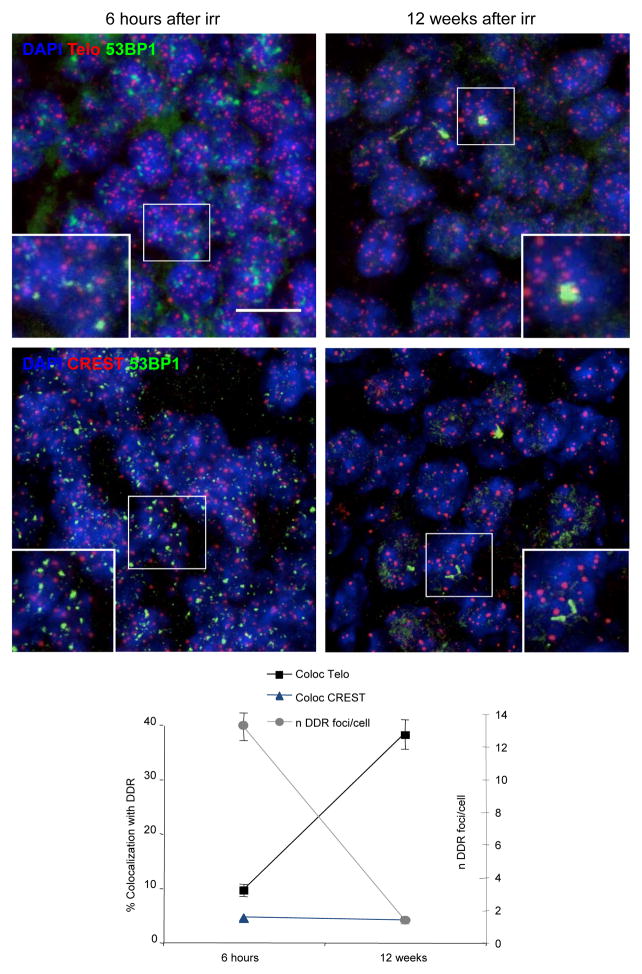
We next extended our analyses to determine whether also in vivo DNA damage can persist and preferentially localise at telomeres. We thus exposed mice to whole-body IR and monitored DDR foci appearance and progressive resolution in terminally differentiated neurons in the brain hippocampus. We observed that a DDR in the form of distinct 53BP1 foci is robustly detected in hippocampal neurons immediately after IR. Twelve weeks after treatment, while most DDR foci had disappeared, remaining 53BP1 foci were still detectable (Fig. 4), consistent with26. We then tested by immunoFISH whether these persistent DDR foci colocalize with telomeric DNA. We discovered that nearly 40% of 53BP1 foci colocalize with a telomeric signal after twelve weeks post irradiation, while CREST antibodies labelling centromeres rarely do (Fig. 4). Thus, also in vivo in terminally differentiated non-proliferating cells, a fraction of DNA breaks keeps signaling and a significant portion of them localizes at telomeres.
Figure 4. IR generates persistent DDR at telomeres in vivo.
DDR, in the form of 53BP1 foci, telomeres, as detected by a telomeric PNA probe (Telo), or centromeres, visualized with antibodies against centromeric proteins (CREST), were analysed in hippocampal neurons of irradiated adult mice at the indicated time points post IR. Top, representative pictures show activation of DDR immediately after IR, persistence of individual foci and their colocalizations with telomeres at 12 weeks (last time point analyzed) after IR. Bottom, graph shows the average number of 53BP1 foci per cell in DDR-positive cells and the percentage of colocalizations (± s.e.m.) between 53BP1 foci and telomeric DNA (Telo) or centromeres (CREST). (For the quantification shown, around 400 cells per time point were analysed: n = 3: scale bar = 10 μm).
DDR foci persistence at telomeres is neither due to TRF2 loss nor telomeres heterochromatin structure
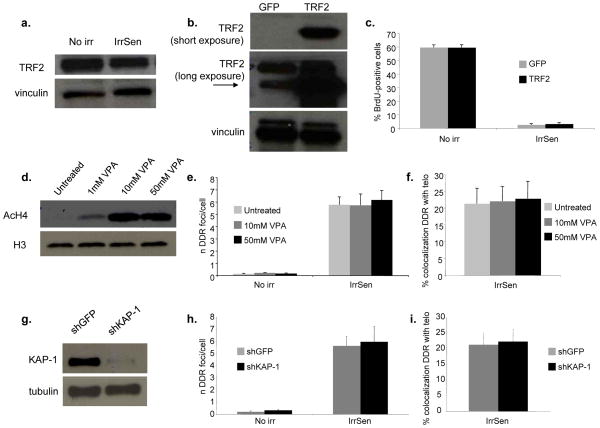
TRF2 is a mammalian protein that directly binds to telomeric DNA and its loss triggers DDR activation at telomeres8, 27, 28. Furthermore, it has been shown to be downregulated during replicative senescence29. We therefore tested whether persistent DDR foci at telomeres in IrrSen cells were associated with TRF2 loss. First, we observed that the abundance of TRF2 in the cell is not significantly affected upon irradiation (Fig. 5a). In addition, persistent DDR foci (pS1981 ATM) in IrrSen cells colocalize with TRF2 focal signals or telomeric DNA to the same extent (Suppl. Fig. S5a and Figs. 2b, c). Thus, DDR activation in IrrSen cells is not associated with detectable TRF2 loss or mislocalization. Nevertheless, to functionally test this possibility, we overexpressed TRF2 in HDFs and exposed them to IR (Fig. 5b). We observed that TRF2 overexpression does not prevent the appearance of persistent DDR foci at telomeres (Suppl. Figs. S5b–d) or the establishment of cellular senescence, as determined by BrdU incorporation rates (Fig. 5c) and SA-β-gal activity (Suppl. Fig. S5e).
Figure 5. TRF2 overexpression does not prevent senescence establishment and heterochromatin disruption does not prevent the persistence of DDR at telomeres.
a. TRF2 expression is not altered in IrrSen cells. Immunoblot shows TRF2 protein levels in IrrSen BJ hTERT, compared to non-irradiated cells (No irr). Vinculin was used as a loading control. b. Immunoblot showing TRF2 expression in BJ hTERT cells infected with either TRF2- or GFP-expressing lentiviruses. Vinculin was used as a loading control. c. TRF2 overexpression does not prevent senescence establishment. TRF2 and GFP overexpressing BJ hTERT cells were irradiated (20 Gy) and analyzed 30 days later. Bar graphs show the percentage of BrdU-positive cells (± s.e.m.) (For the quantification shown, around 400 cells per sample were analysed). d. Heterochromatin disruption by VPA treatment does not significantly affect the number of persistent DDR foci and their colocalization with telomeres. Immunoblot shows the increased levels of acetylated histone H4 (AcH4) in BJ hTERT cells treated with the indicated concentration of VPA, compared to untreated control. H3 was used as a loading control. e. VPA-treated cells were irradiated with 20 Gy and analyzed 30 days later. Bar graphs show the number of 53BP1 foci per cell and (f.) the percentage of 53BP1 foci colocalizing with a telomeric PNA probe (± s.e.m.), in cells treated with the indicated doses compared to untreated control. (For the quantification shown, around 30–100 cells per sample were analyzed). g. KAP-1 knock down does not significantly affect the number of persistent DDR foci and their colocalization with telomeres. Immunoblot shows the expression of KAP-1 in shKAP-1 and shGFP BJ hTERT. Tubulin was used as a loading control. h. Cells were irradiated with 20 Gy and analyzed 30 days later. Bar graphs show the number of 53BP1 foci per cell and (i.) the percentage of 53BP1 foci colocalizing with a telomeric PNA probe (± s.e.m.). (For the quantification shown, around 30–100 cells per sample were analyzed).
We next tested if persistent DDR at telomeres was related to the heterochromatic structure of chromosome ends30. We therefore perturbed heterochromatin with a histone deacetyalse (HDAC) inhibitor, valproic acid (VPA)31 (Fig. 5d), or by knocking down KAP-1 (Fig. 5g), an important mediator of the activity of ATM in heterochromatin32, and we exposed these and control cells to IR. We observed that neither treatments make a significant impact on the numbers of persistent DDR foci per cell (Figs. 5e, h) or the fraction of DDR foci at telomeres (Figs. 5f, i). Thus, persistent DDR is not the consequence of the heterochromatic nature of the telomeres.
Telomeric DNA next to a chromosomal DSB prevents its repair
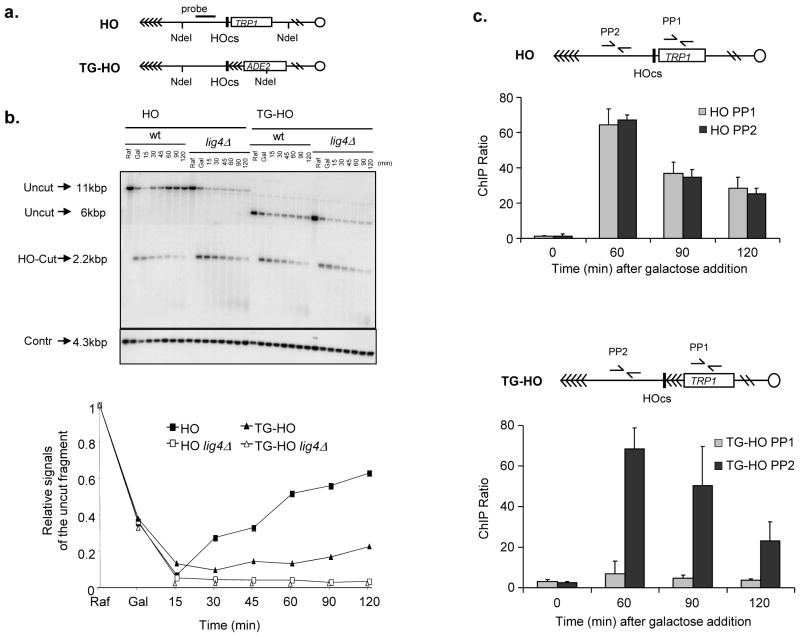
We next aimed to directly test the hypothesis that telomeric repeats resist DNA-double strand break (DSB) repair. For this purpose, we employed one of the most robust and best characterized systems commonly used to assess DSB repair in a chromosomal context in a living cell: the inducible expression of the HO endonuclease in Saccharomyces cerevisiae cells. We used two isogenic strains: one carrying the HO cleavage site next to a stretch of telomeric repeats, and one carrying the HO site but lacking adjacent telomeric repeats33, 34 (Fig. 6a). As expected, the HO-induced DSB generated in G1 in a non-telomeric region is efficiently repaired by NHEJ. Strikingly, however, the HO break next to telomeric repeats is not, and repair is suppressed to the same extent observed in DNA ligase 4-deleted (lig4Δ ) strains, which we used as a control (Fig. 6b). Thus, a chromosomal DSB next to telomeric repeats resists DNA repair. To investigate which is the specific step that is impaired in the DNA repair process, we set up conditions for ChIP assays followed by qPCR to detect ligase 4 (the enzyme responsible for DNA ligation in NHEJ) at the HO cut site flanked, or not, by telomeric repeats. We observed that, while ligase 4 is efficiently recruited to the DSB site, such recruitment is dramatically reduced at the chromosome end flanked by telomeric repeats (Fig. 6c).
Figure 6. Lack of repair of a chromosomal DSB adjacent to telomeric DNA repeats and impaired ligase 4 recruitment.
a. Schematic of the HO (RMY169 and RMY169 lig4Δ )34 or the TG-HO (UCC5913 and UCC5913 lig4Δ )33 system on yeast chromosome VII. HOcs represents the cutting site for the HO endonuclease, which is flanked by 81 bp TG sequence (represented by arrows) in the TG-HO strain. b. HO-cut formation and repair in strains carrying the HO or the TG-HO system at chromosome VII. G1-arrested cell cultures in YEP+raffinose (Raf) were supplemented with galactose (Gal) to induce HO expression. After 1 hour of induction, cells were washed and transferred to YEPD in the presence of α-factor to maintain the G1 arrest. Genomic DNA prepared at different time points after galactose removal were subjected to southern blot analysis with the probe indicated in (a). Top, the probe reveals an uncut fragment in the absence of HO-cut or after the break had been repaired by NHEJ (Uncut), whereas the HO-induced DSB results in the formation of an HO-cut DNA fragment (HO-Cut). (Contr) represents the loading control. Bottom, quantification of a representative experiment. Three independent experiments were performed with similar results. c. Ligase 4 is efficiently recruited to the DSB site only when it is not flanked by telomeric repeats. HO expression was induced at time zero by galactose addition to G1-arrested cells carrying the HO (top) or TG-HO (bottom) system. Lig4 recruitment was analyzed by ChIP and qPCR. Data are expressed as relative fold enrichment (± s.d.) of PP1 or PP2 over CON signal after normalization to input signals for each primer set (n = 4).
Ectopic localization of TRF2 next to a DSB impedes repair and fuel prolonged DDR activation
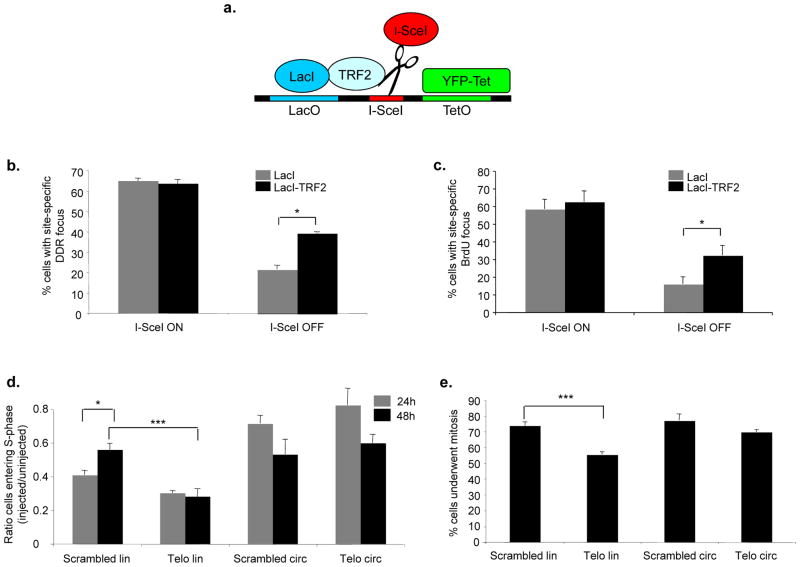
Irreparability may be a feature of telomeric DNA per se or of the proteins that bind to it. Mammalian TRF2 was previously shown to prevent chromosomal fusions in vivo27, 35 and to inhibit NHEJ in vitro23, 24. We therefore generated a gene fusion product between the lactose inhibitor (LacI) and a truncated form of TRF2 (LacI-TRF2), lacking its DNA binding domain — a similar fusion protein was previously proven to be functional in telomere studies36. We expressed this fusion protein in a mouse fibroblast cell line (NIH 2/4) carrying a single integrated cut site for the inducible endonuclease I-SceI, flanked by lactose operator repeats on one side and by tetracycline operator repeats on the other37 (Fig. 7a). The expression of the tetracycline repressor fused to the yellow fluorescent protein (YFP-Tet) allows the visualization of this genomic locus in the nucleus. Since the expression of LacI-TRF2 allows the accumulation of TRF2 next to an exposed non-telomeric DNA end, this structure mimics, to an extent, a telomere that bears telomeric proteins yet lacks telomeric DNA. Upon induction of the fusion product between I-SceI endonuclease and the red fluorescent protein and glucocorticoid receptor ligand binding domain (RFP-I-SceI-GR) (see Figure legend 7a for details)37, a local DDR is triggered, as shown by γH2AX focus formation colocalizing with YFP-Tet punctuated signal (Suppl. Fig. S6a). Upon RFP-I-SceI-GR inactivation, in cells expressing LacI alone, cellular DNA repair activities reduce the percentage of DDR-positive cells at the locus studied (from 65% to 22%). However, in LacI-TRF2 expressing cells, DDR focus persists in a significantly larger fraction of cells compared to the LacI control (40% vs 22%, respectively) (Fig. 7b). This is despite similar initial cutting rates (63% vs 65%). The generation of a physical DNA damage and its subsequently repair mirrors DDR focus formation, as shown by the presence of exposed single-stranded DNA detected by BrdU staining under non-denaturing conditions (Fig. 7c). As an additional control, the expression of LacI fused to the cyan fluorescent protein or LacI alone allows DNA damage foci resolution to a similar extent (Suppl. Fig. S6b), suggesting that the inhibition of DNA repair mediated by LacI-TRF2 is specific and not due to steric hindrance. Importantly, this activity acts locally in cis only, as irradiated cells expressing LacI or LacI-TRF2 show comparable DDR foci resolution rates in the nucleus (Suppl. Fig. S6c). Therefore, TRF2, a crucial component of telomeres, is sufficient to control DNA repair and DDR focus persistence when ectopically expressed next to a DNA break.
Figure 7. Ectopic TRF2 modulates DNA repair and DDR focus persistence, and exposed telomeric DNA ends cause a prolonged checkpoint.
a. Schematic of the integrated locus studied in NIH 2/4 cells37. Upon transfection, LacI or LacI-TRF2 binds to the lactose operator (LacO) repeats, YFP-Tet binds to the tetracycline operator (TetO) repeats, and RFP-I-SceI-GR cuts the specific site between the two sets of repeats. b. Quantification of cells positive for γH2AX at the I-SceI-locus (± s.e.m.) expressing LacI or LacI-TRF2, as detected by immunofluorescence confocal microscopy. I-SceI ON corresponds to 3 hours after RFP-I-SceI-GR induction, I-SceI OFF corresponds to 24 additional hours after removal of inducing agent. I-SceI site was detected as a distinct focus double-positive for YFP-Tet and anti-LacI antibody signals (* p value < 0.05; for the quantification shown, around 100 cells per sample were analyzed; n = 2). c. Quantification of cells positive for a BrdU signal (± s.e.m.) at the I-SceI-locus expressing LacI or LacI-TRF2, as detected by BrdU immunostaining under non-denaturing conditions and confocal microscopy. Values were normalized on the fraction of cells that had incorporated BrdU. (* p value < 0.05; for the quantification shown, around 100 cells per sample were analyzed; n = 2). d. Linearized telomeric DNA triggers a prolonged cell cycle arrest. Bar graph shows the ratio (± s.e.m.) between the percentages of injected cells that underwent DNA synthesis (assayed by BrdU incorporation) and the percentages of uninjected cells in the same experiment, at 24 or 48 hours after microinjection (* p-value < 0.05; *** p-value < 0.001; for the quantification shown, around 200–400 cells per time point, per DNA type were analyzed; n = 3). e. Linearized telomeric DNA impedes cell proliferation. Bar graph shows the percentages (± s.e.m.) of cells that underwent mitosis at 48 hours post-microinjection (*** p-value < 0.001; for the quantification shown, around 200 cells per DNA type were analyzed; n = 3). Passage through mitosis was monitored by detection in the cytoplasm of nucleus-injected IgG 48 hours after microinjection, as detected by immunofluorescence.
Exposed telomeric DNA ends in a cell cause prolonged cell-cycle arrest
According to our model, telomeric DNA ends exposed by DNA damage resist repair and they are thus expected to cause a more protracted DNA damage-induced checkpoint compared to non-telomeric ones. To functionally test this, we microinjected plasmids carrying a stretch of 24 telomeric (TTAGGG) or scrambled (TGAGTG) tandem-repeats in HDFs nuclei. The plasmids were injected either linearized at the 3′ end of the repeats or in their circular form, as a control (Suppl. Fig. S7a) – these linear plasmids were previously tested in vitro and the telomeric one was demonstrated to resist repair by NHEJ23. Importantly, when introduced in mammalian cells, these exogenous telomeric, but not scrambled, DNA repeats are recognized by endogenous telomeric proteins, as demonstrated by ChIP (Suppl. Fig. S7b). We then tested the impact of the exposure in the nucleus of HDFs of these two types of DNA ends on progression through S-phase and mitosis. Consistent with published evidence38, linear DNA plasmids inhibited DNA synthesis, indicating the activation of a DNA damage-induced checkpoint within 24 hours of microinjection, while circular plasmids made only a minor impact (Fig. 7d). However, after additional 24 hours, while cells microinjected with linear scrambled DNA initiated cell-cycle reentry into S-phase, as demonstrated by BrdU incorporation rates similar to circular DNA-injected cells, the cells carrying the linear telomeric DNA persisted in their growth arrest (Fig. 7d). Re-entry in the cell cycle is not due to plasmid degradation, as assessed by qPCR amplification of the plasmids at 48 hours after microinjection (Suppl. Fig. S7c). Consistent with an inhibition of proliferation, also passage through mitosis, as independently monitored by the detection in the cytoplasm of nucleus-injected IgG, was impaired in linear telomeric DNA-injected cells, but was unaffected in cells injected with scrambled DNA ends (Fig. 7e). Consistent with checkpoint enforcement and resolution, p53 phosphorylation on Serine 15 (an ATM- and ATR-dependent phosphorylation event) is initially triggered to a similar extent upon both telomeric or scrambled linear DNA injection, while at a later time point only cells injected with a telomeric linear DNA show a sustained DDR signal (Suppl. Fig. S7d). Therefore, exposed telomeric DNA ends trigger a protracted DNA damage-induced checkpoint that prevents cell-cycle progression.
Persistent DDR markers, in in vitro irradiated HDFs and in in vivo irradiated mouse neurons, accumulate at telomeres which are not preferentially short
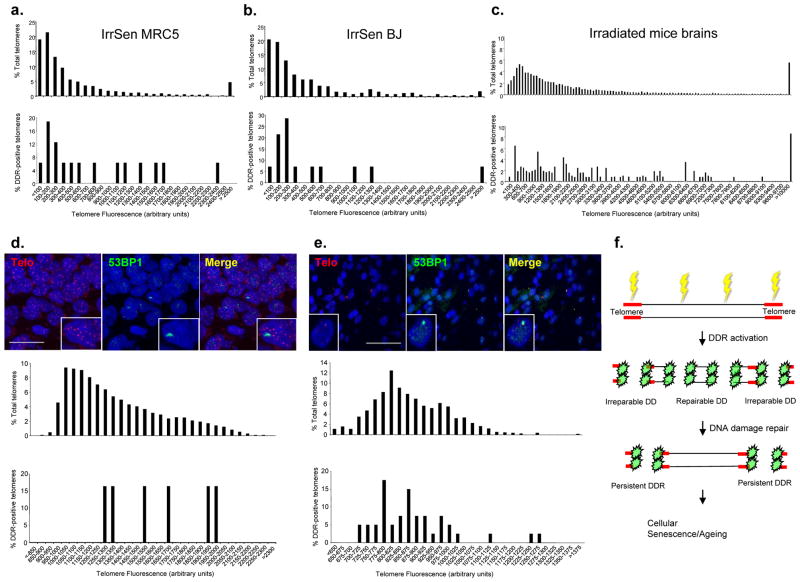
As telomere shortening triggers DDR8, 9, we tested whether DDR foci at telomeres in IrrSen cells were preferentially associated with short telomeres. By quantifying the telomeric probe signal intensities and distributing them in a plot according to their length, we observed no preferential accumulation of DDR markers at short telomeres in in vitro irradiated HDFs (IrrSen MRC5 and BJ) (Figs. 8a, b) and in in vivo irradiated neurons (12 weeks after IR) (Fig. 8c). Therefore, persistent DDR caused by exogenous DNA damaging agents is not triggered by critically short telomeres.
Figure 8. Persistent DDR accumulates at telomeres independently of their lengths, also in ageing primates.
a.–c. Association of persistent DDR at the telomeres is not triggered by telomere shortening. Relative distribution of total telomeres lengths (upper histograms) and of 53BP1 focus-positive telomeres lengths (lower histograms) according to telomeric probe signal intensity (Telomere Fluorescence Arbitrary Units), in IrrSen HDFs (MRC5, a., and BJ, b.; approximately 1000 telomeres per sample were analyzed) and (c.) in in vivo mouse hippocampal neurons 12 weeks after IR (telomeres of approximately 200 cells per sample were analyzed; n = 3). DDR in hippocampal neurons (d.) and in liver hepatocytes (e.) of ageing baboons is not preferentially associated with the shortest telomeres. Top, representative images of telomeric PNA probe (Telo) and 53BP1 foci in hippocampal neurons (scale bar = 20 μm) and liver hepatocytes (scale bar = 50 μm) of aged baboons. Relative distribution of total telomeres lengths (upper histograms) and of 53BP1-focus positive telomeres lengths (lower histograms) according to telomeric probe signal intensity (Telomere Fluorescence Arbitrary Units) in cells from aged baboons. Telomeres from 4 individual baboons were analyzed for hippocampus and from 6 individual baboons for liver. f. Model: DNA damage generated by exogenous sources, such as IR, or in association with ageing, triggers DDR activation throughout the genome. DNA breaks generated along the telomeres are not repaired and fuel a persistent DDR that initiates and maintains cellular senescence.
DDR markers during physiological ageing in primates accumulate at telomeres which are not preferentially short
The observation that DNA damage generated at telomeres resists repair may be of relevance for the study of ageing. Ageing is associated with DNA damage accumulation14–17, and DDR foci colocalizing with telomeres have been observed in vivo in ageing primates14, 17. It is presently unclear whether such DDR is triggered solely by telomere shortening. We therefore performed immunoFISH for 53BP1 foci and telomeric DNA sequences in hippocampal neurons and liver hepatocytes from young and old baboons. Neurons are terminally differentiated cells and are not expected to proliferate and undergo progressive telomere attrition; similarly, liver hepatocytes are expected to undergo no or limited turnover. We discovered that persistent DDR foci accumulate with ageing also in these non-dividing cells (Suppl. Figs. S8a, b). Importantly, when DDR localizes at telomeres, these are not preferentially short (Figs. 8d, e). Therefore, in ageing animals DDR foci accumulate, also in non-proliferating tissues, and this may occur at telomeres that are not critically short.
In summary, we propose that when cells are exposed to exogenous or endogenous sources of DNA damage, DSBs generated throughout the genome will trigger DDR foci formation. The vast majority of lesions will be repaired, leading to DDR foci resolution. However, those few DSBs that happen to occur at telomeres will not be repaired, resulting in persistent DDR foci formation that will fuel the permanent activated DNA damage-induced checkpoint state known as cellular senescence (Fig. 8f).
DISCUSSION
Our results unravel that genomes are not uniformly repairable and that some genomic loci, like telomeric tracts, resist DNA damage repair despite a global cellular competence for DNA repair. Impaired repair of other types of DNA lesions has also been reported at telomeres39, 40. Noteworthy, irreparability of telomeric tracts may be the direct and unavoidable consequence of their functions in preventing chromosomal fusions – a DNA repair event between chromosomes. It follows that the intrinsic vulnerability of chromosome ends, despite its obvious drawbacks, is an evolutionary-selected trait conserved to maintain the linear structure of chromosomes. As our observations hold true in primates, rodents and yeast, the mechanisms uncovered seem to be ancestral and evolutionary conserved.
Ageing has been shown to correlate with, and possibly to be caused by, the accumulation of DNA damage both in the soma and in the stem cells compartments14–17. Our discovery that telomeres resist DNA repair provides a novel interpretation to the observed accumulation of DDR at telomeres in ageing animals, by which DDR may arise independently of telomere attrition. Indeed, DDR foci, previously reported in skin fibroblasts of old baboons14, can also be associated with long telomeres (U. Herbig, unpublished observations). Importantly, our results suggest a model for cellular ageing based on DNA damage accumulation and DDR activation also in non-proliferating cells, such as terminally differentiated cells like neurons, and quiescent stem cells.
It is interesting to mention that a recent report highlighted an inverse correlation between lifespan and telomere length across several mammalian species41: this is consistent with our model in which long telomeres may offer a more abundant target for the accumulation of irreparable DNA damage.
Finally, the notion that DDR activation at critically short telomeres is the trigger of replicative cellular senescence8, 9 together with our novel results implying persistent telomeric DNA damage in cells undergoing cellular senescence caused by DNA damaging agents and during ageing (this manuscript), and the observation that also oncogenes induce persistent DNA damage at telomeres (Suram, A. et al, submitted), allow us to propose a unifying paradigm in which cellular senescence establishment is the result of irreparable telomeric DNA damage generation and consequent persistent DDR signaling.
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturecellbiology/
Methods
Cell Culture
Early passage foreskin fibroblast BJ cells (The American Type Culture Collection, ATCC), BJ hTERT (obtained by retroviral expression of BJ cells with hTERT), lung fibroblast MRC5 cells (ATCC) and HeLa (ATCC) cells were grown under standard tissue culture conditions (or at 3% oxygen tension where indicated). Cells expressing a dominant-negative form of TRF2 (TRF2ΔBΔM) were used as in8. NIH2/4 cells were grown and used as in37 with few modifications available upon request. Bleomycin (Sigma-Aldrich), or its solvent PBS, was used for 3 days at 10 μg/ml in cell culture medium and, after treatment, cell growth was continued in regular culture medium, as in42. Senescence-associated β-galactosidase (SA-β-gal) assay was performed as in43. ATM kinase inhibitor (KU55933, Tocris bioscience), or DMSO as negative control, was used at 10 μM concentration for 72 hours. Valproic acid (Sigma-Aldrich), or its solvent PBS, was used for 16 hours at 1, 10 or 50 mM concentration in cell culture medium.
BrdU staining
Cells were labeled with 10 μg/ml bromodeoxyuridine (BrdU, Sigma) for 16–24 hours and incorporation was evaluated by immunofluorescence after DNA denaturation. For BrdU staining under non-denaturing conditions, cells were stained as in44.
IR
Ionizing radiation was induced by a high-voltage X-rays generator tube (Faxitron X-Ray Corporation). Cultured cells were irradiated with the indicated dose; mice were irradiated with 8 Gy (total body IR) at the age of 2 months using GammaCell 200 and cobalt60 as a source.
Plasmids
pDEA-7Z–derived plasmids containing (TTAGGG)24 (pNB146 plasmid) or (TGAGTG)24 (pNB158 plasmid) as in23 were digested with EcoRI to generate linearized plasmids with telomeric or scrambled repeats at the 3′ end. Cherry-LacI, YFP-Tet and RFP-I-SceI-GR expressing vectors were as in37, 45. LacI-TRF2 vector was generated by cloning the TRF2 coding sequence (amino acids 29–446) at the N-terminus of the LacI vector. TRF2 and GFP expressing lentiviruses were as in44. shKAP-1 and shGFP retroviruses were as in46.
Immunofluorescence and immunoFISH
Cells were fixed and probed as in43. ImmunoFISH with DAKO kit was performed in accordance with the manufacturers’ instructions. Images were acquired using a wide field Olympus Biosystems Microscope BX71 and the MetaMorph software. Confocal sections were obtained with a Leica TCS SP2 AOBS confocal laser microscope by sequential scanning or, in case of immunoFISH analysis, by acquisition of optical z-sections at different levels along the optical axis. Colocalization between DDR and telomeres was assessed by ImageJ software with colocalization ImageJ plug-in on confocal 3D stacks. Two points are considered as colocalized if their respective intensities are higher than the threshold of their channels, and if their ratio (of intensity) is higher than the ratio setting value. Comparative immunofluorescence analyses were performed in parallel with identical acquisition parameters. Telomere length was analyzed by quantification of telomeric signal fluorescence intensities by ImageJ software. For immunofluorescence stainings, brain tissue from four old baboons (325–353 months old) and two young baboons (58 and 88 months old) and liver tissue from ten old baboons (309–356 months old) and twelve young baboons (2–87 months old) were analyzed. For mouse tissues experiments, all in vivo manipulations were approved by the Comité Institutionnel des Bonnes Pratiques Animales en Recherche (CIBPAR) of CHU-Ste-Justine. For baboons tissues experiments, all procedures were approved by the Institutional Animal Care and Use Committee (IACUC). The SBRF animal program has been accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care, International (AAALAC) since 1973.
Antibodies
Anti-γH2AX (immunofluorescence: mouse, Millipore, 05–636, 1:200; ChIP: rabbit, Abcam, ab2893, 5μl/IP); anti-ATM pS1981 (mouse, Rockland, 200-301-400, 1:400; rabbit, Abcam, ab2888, 1:300); anti-pS/TQ (Cell Signalling, 2851, 1:200); anti-53BP1 (mouse, a gift from Thanos Halazonetis, University of Geneva, Geneva, Switzerland, 1:20; rabbit, Novus, NB100-304, 1:200); anti-BrdU (denaturing conditions: Becton Dickinson, 347580, 1:20; non-denaturing conditions: Abcam, ab6326, 1:200); anti-CENP-C (a gift from A. Musacchio, IFOM-IEO-Campus, Milan, Italy, 1:1000); anti-CREST (Antibodies Incorporated, 15–234, 1:100); anti-CHK2 pT68 (Cell Signalling, 2661, Lot 7, 1:100); anti-p53 pSer15 (Cell Signalling, 9284, 1:100); anti-TRF2 (ChIP: Santa Cruz, sc8528, 5μl/IP; immunofluorescence and immunoblot: Upstate, 05–521, 1:500); anti-LacI (Abnova, PAB10255, 1:400); anti-FLAG (Sigma, F3165, 5μl/IP); anti-KAP-1 (Abcam, ab10484, 1:1000), anti-AcH4 (a gift from S. Minucci, IEO, Milan, Italy, 1:1000), anti-H3 (Abcam, ab10799, 1:1000).
Immunoblotting
As in43.
ChIP
In vivo cross-linking, chromatin purification and immunoprecipitations were performed as previously described47.
Illumina Sequencing
As in48, with minor modifications. After PCR amplification, fragments were column purified. Cluster generation was performed and loaded into individual lanes of a flow cell (8 picomoles/sample).
ChIPseq data analysis
Read tags passing standard Illumina quality filter were aligned using BWA 0.5.9 using default parameters49. Two sets of analyses were performed: per-sample processing and senescent vs quiescent enrichment. We named the samples as follows: γS: ChIP of γH2AX for IrrSen cells; IS: Input for IrrSen cells; γQ: ChIP of γH2AX for Quie cells; IQ: Input for Quie cells. Per-sample analysis was performed in order to evaluate γH2AX enrichments at telomeres by “pileup” analysis (see below) and collect data for effective statistics. Each alignment was preprocessed using DSPCHIP 0.8.5b4 (http://code.google.com/p/dspchip) with the following parameters: remove duplicates, quality filter = 15, expected window = 2 Mb, FIR = blackman, pipeline = N. IP vs Input enrichment was produced as intermediate step to study the IrrSen vs Quie enrichment. Processed alignments were processed further with DSPCHIP: γS – IS and γQ – IQ values were calculated and negative values were discarded. Resulting profiles were used to calculate IrrSen vs Quie enrichment, again by subtraction; data were thresholded using Otsu’s method50. Final profiles were plotted on all chromosomes using a modified version of LODPLOT library51 available in R (http://r-cran.org)52. In order to generate the compilative “pileup” analysis of signals from all chromosome ends (Fig. 3b), data from 46 processed chromosome ends were collected up to 20 Mbp from the chromosome end toward the centromere. We sampled preprocessed alignments every100 bp in a 20 Mb window from the telomere. Enrichment for each telomere has been calculated as (γS – IS) – (γQ – IQ). The mean of the compiled signals of 46 chromosomes was calculated along the 20 Mbp span, together with 95% bayesian credibility intervals.
In order to assess the significance of the enrichment at the subtelomeric regions (0–5 Mbp), we considered a distant region as control (15–20 Mbp) and performed a Mann-Whitney-U test (U statistic: 720954291135.5, p-value: 0.0, N=2.3e6).
Quantitative Real-time PCR (qPCR)
qPCR were performed as in53. In experiments in Figs. 3c-d and S4d, TaqMan chemistry PCR system (Applied Biosystems) was used. qPCR was carried out in duplicate and the averaged results are plotted as the difference of the log2 ratio of senescent minus control cells (diff log2 ratio). qPCR amplicons in Figs. 3c-d and S4d were as in19. In experiment in Fig. S1b, TaqMan chemistry PCR system (Applied Biosystems) was used and qPCR was carried out in triplicate. Beta-2-microglobulin (B2M) was used as a control gene for normalization. The following assays were used from Applied Biosystems: Hs00606991_m1 (KI-67), Hs99999907_m1 (B2M). In experiment in Fig. S7b, qPCR was performed on a Roche LightCycler 480 Sequence Detection System. qPCR was carried out in triplicate and the averages were normalized against input. In experiment in Fig. S7c, qPCR was performed on a Roche LightCycler 480 Sequence Detection System and was carried out in triplicate. qPCR primers for Figs. S7b, c:
Proximal Forward: GGCCTCTTCGCTATTACGC
Proximal Reverse: CTCACTGGCCGTCGTTTTAC
Distal Forward: TAAAAGTGCTCATCATTGGAAAAC
Distal Reverse: TGGTGAAAGTAAAAGATGCTGAAG
Microinjection
Microinjections were done using an AIS2 computer assisted micromanipulation system (Luigs and Neumann, Germany), mounted on an inverted Zeiss microscope with a motorized stage. Plasmid DNA was injected in cell nuclei at a concentration of 5 ng/μl, together with 4 mg/ml of IgG rabbit. Cells were serum-starved for 48 hours before injection. After injection, cells were placed in normal medium containing 10% FBS and BrdU, 6 hours or 24 hours later, for 24 hours. 200 cells were microinjected per type; independent experiments were repeated at least three times.
Yeast strains
The strain RMY169 (MATa-inc ade2-101 lys2-801 ura3-52 trp1-Δ 63 his3-Δ 200 ochre leu2-Δ 1::GAL-HO::LEU2 VII-L::TRP1-HOcs-LYS2) was provided by T. Weinert (University of Arizona, Tucson, USA)34. The strain UCC5913 (MATa-inc ade2-101 lys2-801 ura3-52 trp1-Δ 63 his3-Δ 200 leu2-Δ 1::GAL-HO::LEU2 VII-L::ADE2-TG(1-3)-HOcs-LYS2) was provided by D. E. Gottschling (Fred Hutchinson Cancer Research Center, Seattle, USA)33. Cells were grown in YEP medium (1 % yeast extract, 2 % bactopeptone, 50 mg/liter adenine) supplemented with 2 % glucose (YEPD), 2 % raffinose (YEP+raf) or 2 % galactose (YEP+gal).
Analysis of DSB formation and repair in yeast
DSB formation and repair were analyzed in G1-phase arrested RMY169, RMY169 lig4Δ, UCC5913 and UCC5913 lig4Δ strains by Southern blot analysis. The probe used to detect DSB formation and repair corresponds to a 346bp fragment ~500bp centromere-distal to the HO-cut site. As loading control, a 4.3kb fragment about 30kb distal from the left telomere of chromosome VII was used. Quantitative analysis of DSB formation and repair was performed by calculating the ratio of band intensities for the uncut fragment and the loading control.
ChIP - qPCR in yeast
qPCR were performed as in54. After exposure to formaldehyde, chromatin samples were immunoprecipitated with anti-Myc antibodies. Quantification of immunoprecipitated DNA was achieved by qPCR on a Biorad MiniOpticon using primer pairs located ~480-bp centromere-proximal (PP1) or 550-bp centromere-distal (PP2) to the HO cutting site on chromosome VII and at the ARO1 locus of chromosome IV (CON) and normalized to input signal for each primer set; data are expressed as the fold enrichment of PP1 or PP2 over the amount of CON in the immunoprecipitates.
Statistical analysis
Results are shown as means or percentages plus minus standard error of the mean (s.e.m.) or standard deviation (s.d.) as indicated; p-value was calculated by Student’s two-tailed t-test or chi-squared test, respectively. In figure legends, n indicates the number of independent experiments.
ChIPseq Accession number
ChIPseq data are submitted to NCBI-SRA (accession number: SRA049677).
Supplementary Material
Acknowledgments
We thank V. Dall’Olio and L. Tizzoni from IFOM RT-PCR Unit, A. Oldani and all IFOM Imaging Unit, L. Rotta from IFOM Microarray and NGS Unit, IFOM Cell Biology Unit for support; V. Boccardi for discussions; P. Baumann, T. F. Halazonetis, T. Weinert, D. E. Gottschling, E. Soutoglou, E. Gilson, P. Jeggo, A. Musacchio and S. Minucci for sharing reagents; O. Le for mouse brain tissues sectioning; all F.d’A.d.F. lab members for helpful discussions. F.d’A.d.F. laboratory is supported by FIRC (Fondazione Italiana per la Ricerca sul Cancro), AIRC (Associazione Italiana per la Ricerca sul Cancro), European Union (GENINCA, contract number 202230), HFSP (Human Frontier Science Program), AICR (Association for International Cancer Research), EMBO Young Investigator Program and Telethon. M.P.L laboratory is supported by AIRC (grant number 11407), Cofinanziamento 2008 MIUR/Università di Milano-Bicocca and European Union. C.M.B. is supported by a grant from the Canadian Institute of Health Research #IAO-79317. U.H. is supported by a New Scholar Award from the Ellison Medical Foundation (AG-NS-0387-07) and by a grant (R01CA136533) from the National Cancer Institute.
Footnotes
Author Contributions
F.R. generated and assembled data in Figs. 5a–c, 5e–f, 5h–i, 7a–c, 8d, S1b–c, S3b, S4c, S5, S6, S7b–c, S8a; M.C. and M.P.L. generated data in Fig. 6; S.B. performed the microinjection experiments; D.C. performed the analysis of sequencing data and generated data in Figs. 3a–b; J. M. K. generated data in Figs. 8e and S8b; G.B. contributed to the pre-processing and analysis of sequencing data in Figs. 3a–b; M.D. provided technical assistance; V.M. generated data in Figs. 5d and 5g and provided technical assistance; C.M.B. provided irradiated mice brain sections; U.H. provided baboons sections and edited the ms; M.F. generated and assembled data of all remaining figures, performed ChIP assays in mammalian cells, contributed to experimental design and ms writing; F.d’A.d.F. planned and supervised the project and wrote the ms.
The authors declare no conflict of interests.
References
- 1.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 2.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Braig M, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collado M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 6.Michaloglou C, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 7.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.d’Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 9.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 10.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 11.Evan GI, d’Adda di Fagagna F. Cellular senescence: hot or what? Curr Opin Genet Dev. 2009;19:25–31. doi: 10.1016/j.gde.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt CA. Senescence, apoptosis and therapy--cutting the lifelines of cancer. Nat Rev Cancer. 2003;3:286–295. doi: 10.1038/nrc1044. [DOI] [PubMed] [Google Scholar]
- 14.Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 15.Rossi DJ, et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 16.Nijnik A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 17.Jeyapalan JC, Ferreira M, Sedivy JM, Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mech Ageing Dev. 2007;128:36–44. doi: 10.1016/j.mad.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meier A, et al. Spreading of mammalian DNA-damage response factors studied by ChIP-chip at damaged telomeres. EMBO J. 2007;26:2707–2718. doi: 10.1038/sj.emboj.7601719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou BB, Bartek J. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat Rev Cancer. 2004;4:216–225. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]
- 21.Rodier F, et al. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J Cell Sci. 2011;124:68–81. doi: 10.1242/jcs.071340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae NS, Baumann P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 24.Bombarde O, et al. TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends. EMBO J. 2010;29:1573–1584. doi: 10.1038/emboj.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hewitt G. Only telomeres trigger a persistent DNA damage response in stress-induced senescence and during ageing. Nature Communications. 2012 doi: 10.1038/ncomms1708. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le ON, et al. Ionizing radiation-induced long-term expression of senescence markers in mice is independent of p53 and immune status. Aging Cell. 2010;9:398–409. doi: 10.1111/j.1474-9726.2010.00567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 28.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 29.Fujita K, et al. Positive feedback between p53 and TRF2 during telomere-damage signalling and cellular senescence. Nat Cell Biol. 2010;12:1205–1212. doi: 10.1038/ncb2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalo S, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 31.Marchion DC, Bicaku E, Daud AI, Sullivan DM, Munster PN. Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Cancer Res. 2005;65:3815–3822. doi: 10.1158/0008-5472.CAN-04-2478. [DOI] [PubMed] [Google Scholar]
- 32.Goodarzi AA, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 33.Diede SJ, Gottschling DE. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- 34.Michelson RJ, Rosenstein S, Weinert T. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev. 2005;19:2546–2559. doi: 10.1101/gad.1293805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 36.Ancelin K, et al. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol Cell Biol. 2002;22:3474–3487. doi: 10.1128/MCB.22.10.3474-3487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soutoglou E, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang LC, Clarkin KC, Wahl GM. Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G1 arrest. Proc Natl Acad Sci U S A. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen S, Saretzki G, von Zglinicki T. Preferential accumulation of single-stranded regions in telomeres of human fibroblasts. Exp Cell Res. 1998;239:152–160. doi: 10.1006/excr.1997.3893. [DOI] [PubMed] [Google Scholar]
- 40.Rochette PJ, Brash DE. Human telomeres are hypersensitive to UV-induced DNA Damage and refractory to repair. PLoS Genet. 2010;6:e1000926. doi: 10.1371/journal.pgen.1000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomes NM, et al. Comparative biology of mammalian telomeres: hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging Cell. 2011;10:761–768. doi: 10.1111/j.1474-9726.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marusyk A, Wheeler LJ, Mathews CK, DeGregori J. p53 mediates senescence-like arrest induced by chronic replicational stress. Mol Cell Biol. 2007;27:5336–5351. doi: 10.1128/MCB.01316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 44.Ye J, et al. TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell. 2010;142:230–242. doi: 10.1016/j.cell.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 45.Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziv Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 47.Francia S, Weiss RS, d’Adda di Fagagna F. Need telomere maintenance? Call 911. Cell Div. 2007;2:3. doi: 10.1186/1747-1028-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghisletti S, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nobuyuki O. A threshold selection method from gray-level histograms. IEEE Trans Sys Man Cyber. 1979;9(1) [Google Scholar]
- 51.Duffy DL. R package version 1.1. 2007. Lodplot: Plot a genome scan. [Google Scholar]
- 52.Team, R.D.C. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2011. [Google Scholar]
- 53.Di Micco R, et al. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol. 2011;13:292–302. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Viscardi V, Bonetti D, Cartagena-Lirola H, Lucchini G, Longhese MP. MRX-dependent DNA damage response to short telomeres. Mol Biol Cell. 2007;18:3047–3058. doi: 10.1091/mbc.E07-03-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.