Abstract
Decades of research in rodent models has shown that early postnatal overnutrition induces excess adiposity and other components of metabolic syndrome that persist into adulthood. The specific biologic mechanisms explaining the persistence of these effects, however, remain unknown. On postnatal day 1 (P1), mice were fostered in control (C) or small litters (SL). SL mice had increased body weight and adiposity at weaning (P21), which persisted to adulthood (P180). Detailed metabolic studies indicated that female adult SL mice have decreased physical activity and energy expenditure but not increased food intake. Genome-scale DNA methylation profiling identified extensive changes in hypothalamic DNA methylation during the suckling period, suggesting that it is a critical period for developmental epigenetics in the mouse hypothalamus. Indeed, SL mice exhibited subtle and sex-specific changes in hypothalamic DNA methylation that persisted from early life to adulthood, providing a potential mechanistic basis for the sustained physiological effects. Expression profiling in adult hypothalamus likewise provided evidence of widespread sex-specific alterations in gene expression. Together, our data indicate that early postnatal overnutrition leads to a reduction in spontaneous physical activity and energy expenditure in females and suggest that early postnatal life is a critical period during which nutrition can affect hypothalamic developmental epigenetics.
Environmental influences on the development of body weight regulatory mechanisms may be an important factor in the worldwide obesity epidemic (1,2). Evidence in humans indicates that overnutrition during early postnatal life can permanently alter body weight regulation, increasing susceptibility to obesity throughout life (3,4). Accordingly, various animal models have been developed to explore the effects of infant overnutrition on lifelong obesity risk. Artificial feeding of rodent pups by intragastric cannula provides clear evidence for sustained effects of early postnatal overnutrition (5) but requires raising newborn rodents in isolation, which itself has long-term consequences. Overfeeding dams during lactation, with a high-fat diet, for example, could indirectly overnourish pups. Indeed, two recent rodent studies (6,7) report that the obesogenic effect of maternal high-fat diet occurs specifically during the suckling period. Pups from high-fat-fed dams are not consistently heavier at weaning, however (8,9), indicating that maternal overnutrition does not reliably induce early postnatal overnutrition.
In the rodent small litter model of early postnatal overnutrition (10), offspring from several litters born on the same day are randomized and fostered to either normal size (control [C]) or small litters (SL). Suckling in SL is naturalistic, easy to implement, and consistently induces early postnatal overnutrition, providing an apt model in which to study potential long-term effects of infantile overnutrition by excessive formula feeding (11). The early postnatal exposure induces elevated body weight and adiposity that persists to adulthood (10–12), with concomitant increases in plasma insulin (11,13) and leptin concentrations (13) and impaired glucose tolerance (11,13,14). It remains unresolved, however, whether the sustained increase in adiposity of adult SL rodents results from increased energy intake or decreased energy expenditure (13,15). Moreover, the fundamental mechanisms by which the metabolic effects of SL exposure persist to adulthood are unknown.
Environmental influences on developmental epigenetics (16,17) provide a likely mechanism. Epigenetic mechanisms regulate mitotically heritable alterations in gene expression potential that are not caused by changes in DNA sequence (18) and are known to play key roles in brain development (19). DNA methylation, the most stable epigenetic modification (20), is a likely mechanism to explain effects that persist for a lifetime (2). Given its central role in regulating food intake and energy expenditure (21), the hypothalamus is an obvious tissue in which to explore a potential epigenetic basis for induced alterations in body weight regulation. We therefore set out to determine 1) whether the persistently elevated adiposity of SL mice is caused by increased food intake or decreased energy expenditure, and 2) whether early postnatal overnutrition causes persistent changes in hypothalamic epigenetic regulation that may perpetuate altered body weight regulation.
RESEARCH DESIGN AND METHODS
For the litter size studies, virgin FVB/NJ females (The Jackson Laboratory) were mated with FVB/NJ males at age 8 weeks. In each batch, 14–15 mating pairs were set up on the same day; four independent batches of mice were studied over the course of 2 years. On postnatal day 1 (P1), pups from all litters born on the same day (P0) were weighed, sexed, and pooled randomly. Only pups from a birth litter size of 6–12 were included. Foster dams received either four (SL) or nine (C) pups. There were two females and two males in each SL and four to five females and males in each C litter. Litter assignment was performed systematically to balance body weight at P1. At P21, offspring from both groups were weaned onto a fixed-formula, soy protein–free diet (2020X; Harlan Teklad); females were housed two to five per cage, and males were housed individually. Body composition, food intake, energy expenditure, and physical activity were measured at P21–P25 and approximately P180. The P21 vs. P0 methylation-specific amplification and microarray hybridization (MSAM) comparisons used female C57BL/6J mice, and the pyrosequencing validation studies were performed in C57BL/6J and FVB/NJ mice of both sexes (The Jackson Laboratory). Sex was confirmed by PCR amplification of Sry. All applicable institutional and governmental regulations concerning the ethical use of animals were followed during this research. The protocol was approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine. All mice were housed in a temperature-controlled facility (22°C), provided free access to food and water, and maintained on a 12-h light cycle.
Body composition.
Body composition was determined by quantitative magnetic resonance (EchoMRI-100; Echo Medical Systems) according to the manufacturer’s instructions.
Food intake, energy expenditure, and physical activity.
Prior to metabolic study, mice were acclimatized to single housing in Comprehensive Laboratory Animal Monitoring System (CLAMS) cages (Columbus Instruments, Columbus, OH) for 3 days. Mice were subsequently transferred to calorimetry cages for 4 days, during which food intake, energy expenditure (by indirect calorimetry), and physical activity were monitored in real time (22). Only data from the last 3 days (6:00 a.m. to 6:00 a.m.) were used. Upon weaning (P21), mice were housed in a normal cage for 1 day before being introduced to the CLAMS cages. Hence, the metabolic measurements did not commence until P25.
Genome-scale DNA methylation profiling.
MSAM was performed as previously described (23,24), using a starting quantity of 0.5 µg genomic DNA. In both MSAM experiments, two independent cohybridizations (biological replicates) were performed on a custom 2×105k array (Agilent Technologies). The array includes 90,694 probes covering 86% (23,742) of the 27,675 potentially informative SmaI/XmaI intervals between 200 bp and 2 kb in the mouse genome (average 3.8 probes per interval) (25). Fold change and P value were calculated as mean and median, respectively, of all probes in each interval; “hits” were called for fold change >2 or <0.5 and P < 0.0001 in both cohybridizations. Gene Ontology analysis was performed using the Gene Ontology enRIchment anaLysis and visuaLizAtion tool (GOrilla) (26), in each case using the potentially informative genes on the array as the background set. SmaI/XmaI cut sites were annotated by RefGene according to NCBI36/mm8 mouse genome (Feb 2006 release), according to the schema in Supplementary Fig. 2. Relevant details of the MSAM experiments and the hybridization data are available in the GEO database: P21 vs. P0, www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32475; and SL vs. C, P180, www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32477.
Quantitative analysis of DNA methylation.
Site-specific analysis of CpG methylation was performed by bisulfite pyrosequencing (23,24). For validation of MSAM hits, pyrosequencing assays were designed to cover both informative SmaI/XmaI sites when possible. A hit was considered validated if either assay showed a substantial methylation difference in the same direction as in MSAM. Sensitivity and linearity of each pyrosequencing assay were confirmed by running methylation standards (27). Pyrosequencing primers and annotations of the CpG sites examined are listed in Supplementary Tables 3 and 4, respectively.
Gene expression profiling and analysis.
Total hypothalamic RNA was isolated by the RNeasy kit (Qiagen). RNA quality control, cRNA preparation, labeling, and microarray hybridization were conducted as previously described (28). cRNA was hybridized onto Illumina MouseWG6 v2 Expression BeadChips (Illumina) following the manufacturer’s protocol. Signal intensities extracted from Illumina Genome Studio software were preprocessed using LUMI (29) and the R statistical package (http://www.r-project.org/), including probes with a detection P value ≤0.05 in at least half of the samples. These 20,791 probe signals were then quantile normalized. To test for effects of group, sex, and group × sex, we performed two-way ANOVA using Genomics Suite Software, version 6.6 β (Partek). Contrasts were applied to all groups to identify differentially expressed transcripts, using an α level of 0.05 after Benjamini-Hochberg false discovery rate adjustment. Network analyses were performed using IPA (Ingenuity Systems). Relevant details of the expression microarray experiment and the hybridization data are available in the GEO database: www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40616.
Quantitative analysis of gene expression.
Expression levels of specific genes were determined by quantitative PCR. Total hypothalamic RNA was isolated by the RNeasy kit (Qiagen), and cDNA was synthesized using M-MLV Reverse Transcriptase (Promega) with random primers (Life Technologies). Gene expression levels were determined with either TaqMan (Life Technologies) or Sybr Green (Life Technologies), using the 2−ΔΔCt method (assay details provided in Supplementary Table 6). In all cases, ActB was used as an endogenous control.
General statistical methods.
Enrichment of SmaI/XmaI cut sites relative to genomic regions was analyzed by χ2 test. Group differences in body weight at P1 were analyzed by two-tailed t test. Body composition data at P25 and P180 were analyzed by ANOVA (SAS Proc Mixed). Body weight data at P21, P60, P120, and P180 were analyzed by repeated-measures ANOVA (SAS Proc Mixed, compound symmetry covariance structure) with age in the repeated effect. For the CLAMS studies, hourly measurements of food intake, energy expenditure, and activity counts for three consecutive days were averaged into one 24-h record for each mouse. Repeated-measures ANOVA used the full power of these time-series data while recognizing the nonindependence of the 24 multiple measures within each mouse. Analysis of food intake, energy expenditure, and physical activity were performed both with and without lean mass and fat mass included as independent variables to adjust for group differences in body size and composition (30). Group differences in DNA methylation by pyrosequencing were analyzed by repeated-measures ANOVA, with CpG site as the repeated effect (Supplementary Table 2). Loci that showed significant group effects on methylation but no significant group × age interaction were considered persistently altered by SL suckling. Requiring the same group difference, in the same direction, at both ages (in independent sets of mice) affords substantial protection against type 1 error; these analyses were therefore not otherwise adjusted for multiple testing. Akaike information criterion (AIC) model selection by adjusted R2 (SAS Proc Reg) was performed based on individual average dark-period energy expenditure and physical activity data.
RESULTS
Early postnatal overnutrition reduces adult energy expenditure in females.
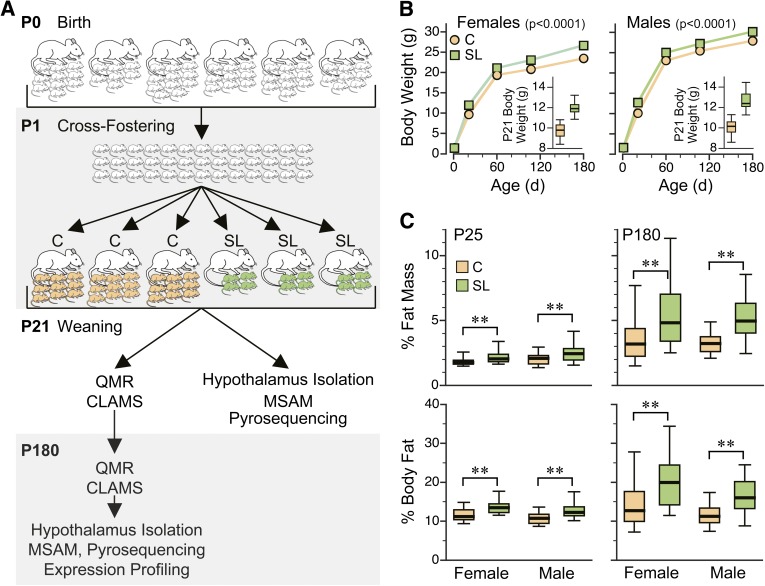
We used the SL mouse model (Fig. 1A) to study persistent effects of overnutrition during the suckling period. We studied four independent batches (groups of litters cross-fostered at one time) over 2 years, including offspring from 24 C and 26 SL litters total. Consistent with previous studies, SL mice were heavier at P21 and remained so into adulthood (P < 0.0001 in both females and males) (Fig. 1B). Although the increase in adult body weight was modest, effects on body composition were substantial. Both male and female SL adults had 50% higher fat mass and percent body fat compared with C mice (P < 0.005 in all comparisons) (Fig. 1C). There were no group differences in lean mass. Clearly, suckling in a small litter induces persistent changes in regulatory mechanisms that affect adult body composition.
FIG. 1.
SL mice are persistently heavier and fatter than C mice. A: Overview of the litter size experiment. FVB mice were cross-fostered at P1 and randomly assigned to SL (green) or C (orange) litters. Quantitative magnetic resonance (QMR) and CLAMS measurements were performed after weaning (P21–P25) and at P180. Hypothalami were isolated at approximately P25 and P180. Four independent batches of SL and C mice were studied over the course of 2 years. B: Body weight of SL and C mice did not differ at P1 (P > 0.7). SL mice showed significantly higher body weight at P21 (insets), which was maintained to adulthood (P180) (P < 0.0001 in both females and males). Data are presented as means ± SEM of 20–94 mice in each group, sex, and age. (Error bars are smaller than symbols.) Box plots (insets) represent median (mid-line), 25th–75th percentiles (box), and 5th–95th percentiles (whiskers). C: Both male and female SL mice have higher adiposity at P25 (left panel) (P < 0.01). By P180 (right panel), group differences in body composition are much greater both in absolute terms (top panel) (P < 0.002) and as percent of body weight (bottom panel) (P < 0.004). Plots represent 20–30 mice of each group, sex, and age (**P < 0.01). d, days.
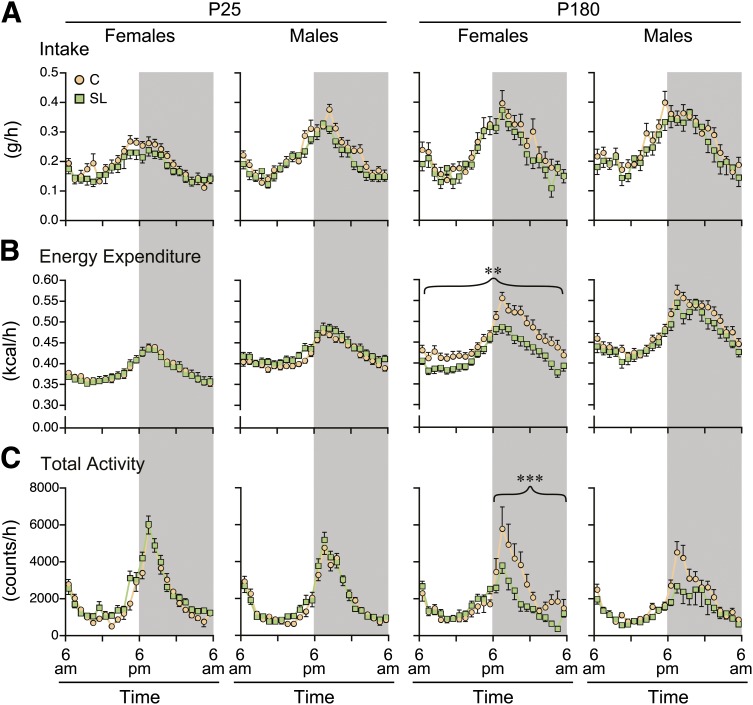
To determine whether these changes involve alterations in food intake and/or energy expenditure, we used metabolic cages to simultaneously monitor food intake, energy expenditure, and voluntary physical activity. In an attempt to identify persistent metabolic differences that might explain the sustained group differences in adiposity, we performed the metabolic measurements shortly after weaning (P25) and in adulthood (P180). (Again, these data represent four batches of mice studied over the course of 2 years.) After appropriate least squares normalization for lean mass and fat mass (30), food intake of SL mice tended, surprisingly, to be slightly lower than that of C mice at both P25 and P180 (Fig. 2A), but these differences were not statistically significant. Energy expenditure (normalized for lean mass and fat mass [30]) was nearly identical between SL and C mice at P25 (Fig. 2B). At P180, however, energy expenditure of SL females was significantly lower than that of C females (P = 0.002); this group difference was significant during both the light and dark periods. Resting metabolic rate was estimated as the lowest average energy expenditure within 1 h for each mouse. After least squares normalization for lean mass and fat mass, female mice showed no group differences in resting metabolic rate at either age. Resting metabolic rate of SL males, however, was higher at P25 (P = 0.02) and lower at P180 (P = 0.03) relative to C males. There were no group differences in respiratory exchange ratio. Group differences in voluntary physical activity, again normalized for lean mass and fat mass, were consistent with those in energy expenditure: none were found at P25, but SL females were significantly less active than C females at P180, specifically during the dark period (group × light interaction P = 0.0009) (Fig. 2C). It is noteworthy that adult SL females were less physically active even after including body weight and body composition in the model; hence, their lower activity was not caused by their excess adiposity. (For comparison, unnormalized data on food intake, energy expenditure, and physical activity are shown in Supplementary Fig. 1.) Including physical activity in the model for energy expenditure of P180 females drastically reduced the significance of the SL effect (from P = 0.002 to P = 0.01), suggesting that physical activity explains much of the group difference in energy expenditure. Together, these data indicate that the persistent alterations in energy balance of female SL mice are due not to excess food intake but, rather, to reduced energy expenditure. The sex specificity of this effect may be related to the male-specific decline of physical activity with age (Supplementary Fig. 1C).
FIG. 2.
Adult SL females have reduced energy expenditure and physical activity. A: Hourly data on food intake are presented as least squares means, adjusting for lean mass and fat mass. Light/dark cycle is indicated by shading. Food intake of female and male SL mice did not differ from that of C mice at any age. A, B, and C: mean ± SEM of 20–30 mice in each group, sex, and age. B: Hourly data on energy expenditure are presented as least squares means, adjusting for lean mass and fat mass. No group differences were found at P25. At P180, SL females had significantly lower energy expenditure (P = 0.002), and this group difference was seen during both the light and dark cycles. SL males likewise tended to have lower energy expenditure at P180, but this difference was not statistically significant. C: Hourly data on physical activity are presented as least squares means, adjusting for lean mass and fat mass. No group differences were found at P25. At P180, SL females had significantly lower activity, specifically during the dark cycle (P = 0.0009). SL males likewise tended to have lower activity at P180, but this difference was not statistically significant (**P < 0.01; ***P < 0.001).
Extensive epigenetic development occurs in the early postnatal hypothalamus.
Ontogenic periods when epigenetic mechanisms are being established or undergoing maturation constitute critical periods during which environmental influences can cause persistent changes in epigenetic regulation (31,32). To determine whether the suckling period might be a critical period for developmental epigenetics in the hypothalamus, we tested for changes in hypothalamic DNA methylation. We used MSAM, which is based on sequential digestion of genomic DNA with the methylation-sensitive and -insensitive isoschizomers SmaI and XmaI (23,33). Two independent P21 vs. P0 MSAM cohybridizations (incorporating a dye swap) were performed.
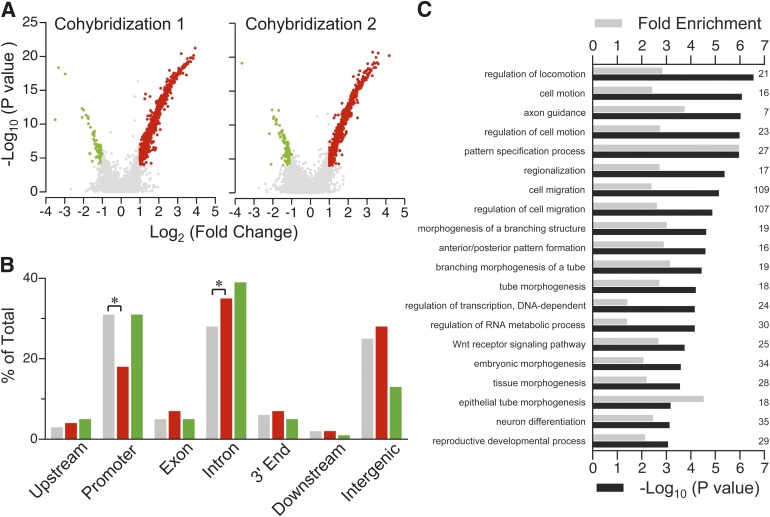
Using stringent criteria validated in our previous studies (24,33), 868 SmaI/XmaI intervals changed methylation from P0 to P21. Only 31 intervals lost methylation (Fig. 3A), and the genomic distribution of associated SmaI/XmaI cut sites was not different from that on the array (Fig. 3B and Supplementary Fig. 2). Methylation increased at 837 intervals (Fig. 3A); associated cut sites were significantly underrepresented at promoters (P < 0.0001) and overrepresented in introns (P < 0.0001) (Fig. 3B). In a larger number of P0 and P21 mice, we used bisulfite pyrosequencing (23,33) to measure P0–P21 changes in hypothalamic CpG methylation at 10 intervals identified by MSAM; all 10 validated (100%) (7 are shown in Fig. 4). Additionally, since the P21 vs. P0 MSAM studies were performed in C57BL/6J mice, we confirmed (at a subset of loci) that these methylation changes also occur in both sexes of FVB/NJ mice (the strain used for the litter size studies) (Supplementary Fig. 3).
FIG. 3.
Extensive DNA methylation changes in the early postnatal hypothalamus. A: Volcano plots of the two independent P21 vs. P0 MSAM cohybridizations. Red dots indicate probes showing increased DNA methylation, green dots indicate probes showing decreased DNA methylation, and gray dots indicate no change. DNA methylation increases predominated; few genomic intervals showed decreased DNA methylation. B: Relative to all genomic intervals on the array (gray), those showing increased DNA methylation (red) were significantly enriched in introns and depleted in promoter regions (*P < 0.0001). The genomic distribution of intervals that lost methylation from P0 to P21 (green) did not differ from that of all intervals on array. C: Enriched gene ontology process categories (P < 10−3) of genes associated with methylation increases are almost all related to development. (Numbers on the right side indicate how many genes in the target set are associated with each ontology.)
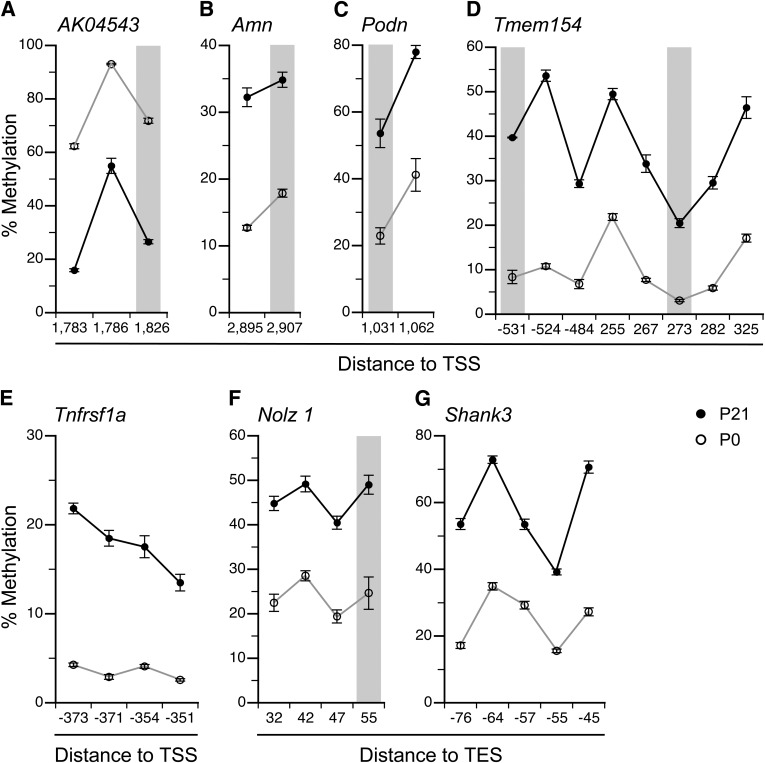
FIG. 4.
Validation of P21 vs. P0 MSAM by bisulfite pyrosequencing. A: At AK04543, the P0-P21 methylation decrease identified by MSAM was confirmed. At all other regions analyzed, methylation increases identified by MSAM were confirmed: Amn (B), Podn (C), Tmem154 (D), Tnfrsf1a (E), Nolz1 (F), and Shank3 (G). Gray columns indicate the SmaI/XmaI cut sites. Data are represented as means ± SEM of 5–10 mice per age. CpG site locations are provided relative to transcription start site (TSS) or transcription end site (TES).
Functional significance of the developmental changes in DNA methylation was evaluated by gene ontology analysis. Relative to all potentially informative genes on the array, no enriched ontologies were found for the few genes associated with intervals that lost methylation. Genes that gained methylation, however, were significantly enriched for 20 biologic process categories (Fig. 3C); of these, 17 are explicitly related to development, including neurodevelopmental processes such as axon guidance and neuron differentiation. Postnatal development of the mouse hypothalamus clearly involves functionally important epigenetic changes. This may be a critical period during which environment can influence these processes, with long-term consequences.
Early postnatal overnutrition causes persistent and sex-specific alterations in hypothalamic DNA methylation and gene expression.
We therefore examined DNA methylation differences among SL and C hypothalami. Intrigued by the large group differences in energy expenditure and physical activity in P180 females, we used MSAM to compare hypothalamic DNA methylation of SL and C females at P180. Two independent SL vs. C cohybridizations were performed, with each hypothalamic DNA sample pooled from five females drawn from different foster litters. The results, however, provided no evidence of persistent group differences in hypothalamic DNA methylation.
Reasoning that DNA methylation changes in SL mice might be too subtle to detect by MSAM, we used bisulfite pyrosequencing to examine a panel of candidate genes. Since genomic regions undergoing methylation change from P0 to P21 are most likely to show persistent effects of overnutrition during this period (2), most of the genomic regions that we selected were those identified in our P21 vs. P0 MSAM experiments. In addition to 10 of those already validated (Fig. 4), we examined six hits near genes previously reported to change expression in hypothalamus from P0 to P21 (34) and two showing interindividual variation in DNA methylation. Additionally, promoters of a few genes critical to hypothalamic function and development (Agrp, Fto, and Pomc) were included. In total, 24 loci (named according to the nearest gene) were selected (Supplementary Table 1).
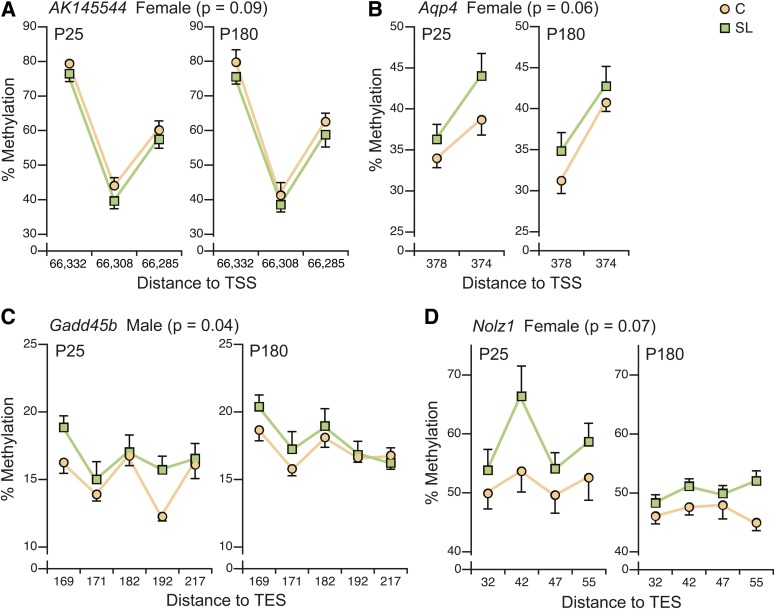
Of these, 15 showed no DNA methylation differences between SL and C hypothalamus at P25 (Supplementary Fig. 4 and Supplementary Table 2) and were not examined at P180. The remaining nine loci were examined at both P25 and P180 (Supplementary Fig. 5 and Supplementary Table 2). Considering this an exploratory data analysis, we set an α-level of 0.1 for main effects and 0.2 for interactions. The initial model including both sexes showed significant main effects for age but not group (SL vs. C); interestingly, however, four loci showed significant group × sex interactions (Supplementary Table 2). We therefore performed sex-specific analyses. In females, AK145544, Aqp4, and Nolz1 and in males Gadd45b showed main effects of group that did not differ by age (no group × age interaction). Plots of average site-specific methylation at these loci (Fig. 5) illustrate subtle but persistent differences in DNA methylation. Since most of these changes were found in females, we used quantitative real-time RT-PCR to measure gene expression of AK145544, Aqp4, and Nolz1 in hypothalamus of P180 females but found no significant group differences. To test whether differences in methylation and expression at these loci could explain individual variation in adult physical activity or energy expenditure, we applied the AIC (35) to identify the best model for each. In addition to methylation and expression of the three genes, SL group membership, lean mass, and fat mass were included as potential explanatory variables. Physical activity was not significantly predicted by any model. Remarkably, however, the best model for energy expenditure (Supplementary Fig. 6) included expression of all three genes and methylation at Nolz1 and Aqp4 and predicted 55% of the variation in P180 energy expenditure in females (P = 0.025). Hence, group differences in DNA methylation and gene expression at these loci, though subtle, may explain some of the observed alterations in energy balance.
FIG. 5.
Early postnatal overnutrition causes persistent and sex-specific changes in hypothalamic DNA methylation. A: In SL females, a persistent reduction in DNA methylation was found at AK145544 (P = 0.09). At all other loci, DNA methylation was higher in SL mice: Aqp4 in females (P = 0.06) (B), Gadd45b in males (P = 0.04) (C), and Nolz1 in females (P = 0.07) (D). n = 12 per group per sex at P25, n = 6 per group per sex at P180. CpG site locations are provided relative to transcription start site (TSS) or transcription end site (TES).
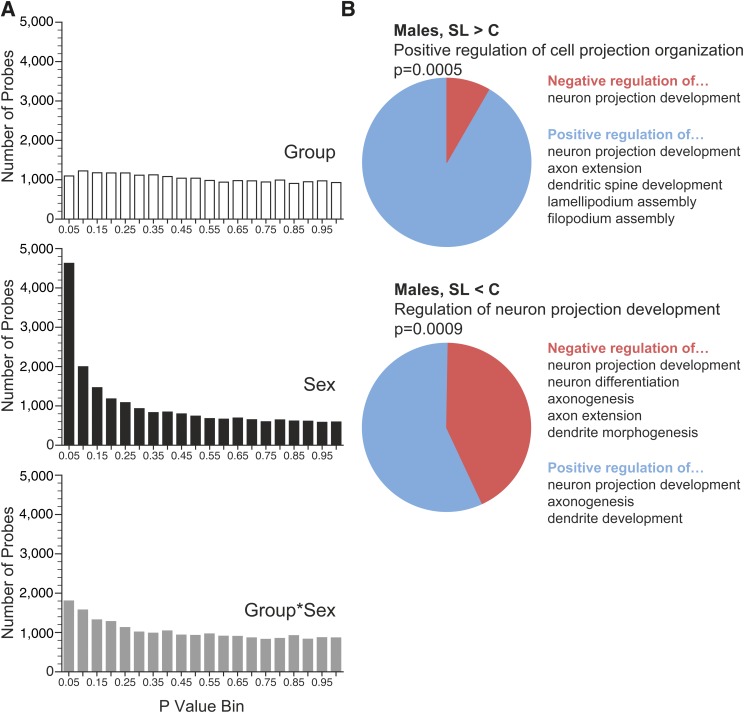
As a complementary approach to identify genes with persistent alterations in epigenetic regulation, we profiled gene expression in P180 hypothalamus of SL and C mice. Three males and three females were studied in each group (12 arrays total). The results showed strong effects of sex (Fig. 6A), with 342 transcripts showing significant differences (false discovery rate <0.05). Although none of the group or group × sex effects survived multiple testing correction, there was an enrichment of low P value probes in the group × sex analysis (Fig. 6A), suggesting subtle sex-specific effects. We therefore performed gene ontology analysis on the 37 and 732 transcripts with at least a 20% group difference (P < 0.05) in females and males, respectively. (The reference list comprised the 14,628 transcripts significantly expressed in hypothalamus.) No enriched gene ontology terms were found in females. In males, however, for both the 381 genes upregulated and the 351 downregulated in SL hypothalamus, the foremost biological process related to formation of neuronal projections. Examination of the gene ontology terms associated with the genes comprising these enrichments (Fig. 6B and Supplementary Table 5) suggests a subtle shift in expression profile that may favor neuronal remodeling in the hypothalamus of adult SL males. Additionally, in an analysis of gene networks associated with the expression changes in male hypothalamus (Supplementary Fig. 7), two of the top three networks were related to cellular development and nervous system development. These networks are centered on Atn1 and dynein, respectively (both regulators of neurodegeneration), again supporting potential alterations of neuronal remodeling in SL males.
FIG. 6.
Evidence of sex-specific gene expression alterations in P180 SL hypothalamus. A: P value distribution of array probes by group, sex, and group × sex shows a strong effect of sex. Also, the group × sex interaction shows greater enrichment of low P values than group alone. B: Results of gene ontology analysis in males. Among both the 381 genes upregulated in SL males (SL > C) and the 351 downregulated in SL males (SL < C), the strongest gene ontology association related to development of neuronal projections. (See also Supplementary Table 5.)
DISCUSSION
Here we showed that early postnatal overnutrition, known to permanently increase body weight and adiposity, also reduces voluntary physical activity and energy expenditure in adult females. These physiological changes were associated with persistent alterations in hypothalamic DNA methylation at specific loci. Overall, these findings provide support for the hypothesis that early postnatal overnutrition causes subtle but widespread changes in hypothalamic epigenetic regulation that persist to influence adult energy balance.
Our study addresses a key outstanding question: whether the persistently altered energy balance of SL rodents is due to increased food intake or decreased physical activity. Previous studies reported increased food intake (13,36) and energy expenditure (37) in adult SL rodents. Those conclusions, however, were based on nonnormalized data, disregarding the altered weight and body composition of SL rodents. Here, we used least squares means to appropriately adjust expenditure and intake data for body weight and composition (30). Compared with C mice of the same weight and body composition, adult SL mice were not hyperphagic (Fig. 2A). Their energy expenditure, however, again compared with C mice of the same weight and body composition, was lower (Fig. 2B), significantly so in females. Hence, our data provide strong evidence that reduced energy expenditure, not increased food intake, explains the increased adiposity of female adult SL mice.
In addition to food intake and energy expenditure, however, there are other determinants of energy balance, such as nutrient absorption, which were not measured in this study. Also, it is possible that group differences in central regulation of food intake may have been unmasked if mice were provided a highly palatable diet (38). Other than physical activity, we did not measure additional determinants of energy expenditure, such as brown adipose tissue activity. These shortcomings may explain why the excess adiposity of male SL mice occurred without measurable differences in physical activity or energy expenditure. (Notably, a recent study found age-associated alterations in thermogenic capacity of brown adipose tissue in male SL rats [39], consistent with our finding that resting metabolic rate is increased at P21 but decreased at P180 in male SL mice.) In females, the reduced energy expenditure was largely explained by reduced physical activity (Fig. 2C). In an earlier report in rats, prenatal undernutrition likewise caused persistent reductions in locomotor activity, most prominently in females (40). Given the worldwide trends of decreasing physical activity (41), it is crucial to determine whether, in humans as in rodents, nutrition during early life modulates voluntary physical activity for a lifetime.
Despite its importance in central regulation of food intake and energy expenditure (21), our understanding of the molecular mechanisms driving functional development of the hypothalamus remains limited. Mouse hypothalamic development continues into early postnatal life, a critical period for formation of leptin-sensitive neuroanatomic projections that function in energy homeostasis (42) and major alterations of hypothalamic gene expression (34). Here, we have shown for the first time that during this same period widespread changes in DNA methylation—mostly increases—are underway. The association of these methylation increases with genes involved in neural development (Fig. 3C) suggests a process of postnatal epigenetic maturation. Because projections from the arcuate nucleus of the hypothalamus to other brain regions form prenatally in primates but postnatally in rodents (43), it is often proposed that hypothalamic development during the suckling period in the mouse is comparable with that in a third-trimester human. It is currently unknown, however, whether and when the epigenetic maturation we have documented in the postnatal mouse occurs in humans. Moreover, our findings that postnatal overnutrition leads to a decrease in physical activity in female mice raises the question of whether the mouse is a good model for physical activity in humans. Although we currently have only a rudimentary understanding of the neurobiological regulation of spontaneous physical activity (44), the hypothalamus and other brain regions are known to be involved, as are several highly conserved neuropeptides including cholecystokinin, corticotrophin-releasing hormone, leptin, and orexins.
It was recently reported that SL rats have alterations in hypothalamic DNA methylation (45,46). Those studies, however, assessed DNA methylation only at P21. Our data therefore provide the first evidence that early postnatal overnutrition induces persistent epigenetic changes in the hypothalamus. Additionally, unlike previous studies on related models (45–48), rather than focus on single CpG sites we performed integrated analysis of all CpG sites represented in each assay because 1) regional changes in DNA methylation are more likely to affect gene expression and 2) concordant changes at multiple adjacent sites are less likely to occur by chance. Notably, contrary to the previous report of increased DNA methylation (at 2 of 20 CpG sites measured) at the Pomc promoter in the hypothalamus of P21 SL rats (45), our methylation assay spanning five nearby CpG sites found no SL vs. C differences at P25 (Supplementary Fig. 4).
We developed the strategy of examining genomic regions undergoing DNA methylation changes from P0 to P21, based on the conjecture that these changes may be susceptible to environmental influences. Indeed, 4 of 21 regions undergoing P0–P21 DNA methylation change showed evidence of persistent methylation differences between SL and C mice, supporting the utility of this approach. Hence, the ∼900 loci that we report that undergo postnatal methylation changes may provide useful candidate regions for future studies of environmental influences on hypothalamic developmental epigenetics.
With the potential exception of AK145544, all four genes with persistent changes in hypothalamic DNA methylation in SL hypothalamus play important roles in neural development or function (49–51). At each of these genes, the methylation change in SL mice was modest (Fig. 5); the cumulative effect of such changes at hundreds or thousands of genes, however, could be considerable. This interpretation is supported by the AIC model selection (Supplementary Fig. 6), which included as significant predictors of adult energy expenditure expression of three and methylation at two of the genes that we identified, in most cases with F values comparable with that of lean body mass.
The results of transcriptional profiling in P180 hypothalamus mirrored our DNA methylation analyses in detecting subtle, widespread, and sex-specific alterations in gene expression. Analyzing the corpus of genes with potentially altered expression in male SL hypothalamus identified highly significant gene ontology enrichments pertaining to regulation of neuronal projections (Fig. 6). The adult rodent hypothalamus maintains significant synaptic plasticity (52); our data suggest that early postnatal overnutrition in males may persistently augment this capability. Adult mice that become obese owing to a high-fat diet, conversely, appear to have reduced hypothalamic neurogenesis (53).
All the potential explanatory effects we found—changes in energy expenditure, physical activity, DNA methylation, and gene expression—were sex specific. The long-term consequences of early life exposures have long been recognized to differ by sex (16). Our findings of sexual dimorphism in the epigenetic responses to early postnatal environment suggest that nutrition may interact with the epigenetic mechanisms regulating hormone-dependent sexualization of the neonatal hypothalamus (54). In fact, the sex differences found here might provide an answer as to why the lower physical activity in SL females arose only in adulthood. In male mice, physical activity declined with age in both groups, but in females this decline was seen only in SL mice (Supplementary Fig. 1C). Our results may suggest, therefore, that postnatal overnutrition is leading to masculinization of the CNS pathways that regulate age-related changes in physical activity.
Encouraged by earlier studies that gained insights into hypothalamic developmental epigenetics (55), we too studied DNA methylation in whole hypothalamus. The interpretation of our data is therefore complicated by the heterogeneity of the hypothalamus at both the regional and the cellular level. The hypothalamus is comprised of distinct regions, or “nuclei,” with specialized functions, gene expression patterns (21), and epigenetic regulation (56). Additionally, the nervous system includes diverse cell types; the simplest classification distinguishes neurons and glia, which are epigenetically distinct (57,58). To improve our understanding of how early postnatal overnutrition causes persistent changes in regulation of body weight and body composition, it will be advantageous to characterize epigenetic effects within specific hypothalamic nuclei and specific cell types. For example, based on our current data we cannot exclude the possibility that the persistent alterations in DNA methylation that we identified represent a shift in the proportion of hypothalamic cell types rather than induced alterations in epigenetic regulation within specific cell types. Moreover, since early postnatal life is a critical period for not only epigenetic but also neuroanatomic development (42), studying these processes in an integrated fashion will likely be necessary to gain a clear understanding of how early postnatal nutrition affects the establishment of hypothalamic body weight regulation.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (1R01DK081557) and the U.S. Department of Agriculture (USDA) (CRIS 6250-51000-055) (to R.A.W.). Body composition and CLAMS studied were performed in the Mouse Metabolic Research Unit at the USDA/Agricultural Research Service (ARS) Children’s Nutrition Research Center, which is supported by funds from the USDA ARS.
No potential conflicts of interest relevant to this article were reported.
G.L. and J.J.K. performed experiments and wrote the manuscript. W.Z. and E.L. performed experiments. G.K.-R. performed bioinformatic analyses. M.S.B. performed experiments. M.L.F. provided critical guidance on experimental procedures and edited the manuscript. R.A.W. designed the study and wrote the manuscript. R.A.W. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank Adam Gillum (USDA/ARS Children’s Nutritional Research Center [CNRC]) for assistance with the figures and Firoz Vohra (USDA/ARS CNRC) for assistance with the CLAMS studies.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db12-1306/-/DC1.
REFERENCES
- 1.McAllister EJ, Dhurandhar NV, Keith SW, et al. Ten putative contributors to the obesity epidemic. Crit Rev Food Sci Nutr 2009;49:868–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr 2007;27:363–388 [DOI] [PubMed] [Google Scholar]
- 3.Koletzko B, von Kries R, Closa R, et al. European Childhood Obesity Trial Study Group Lower protein in infant formula is associated with lower weight up to age 2 y: a randomized clinical trial. Am J Clin Nutr 2009;89:1836–1845 [DOI] [PubMed] [Google Scholar]
- 4.Stettler N. Nature and strength of epidemiological evidence for origins of childhood and adulthood obesity in the first year of life. Int J Obes (Lond) 2007;31:1035–1043 [DOI] [PubMed] [Google Scholar]
- 5.West DB, Diaz J, Woods SC. Infant gastrostomy and chronic formula infusion as a technique to overfeed and accelerate weight gain of neonatal rats. J Nutr 1982;112:1339–1343 [DOI] [PubMed] [Google Scholar]
- 6.Oben JA, Mouralidarane A, Samuelsson AM, et al. Maternal obesity during pregnancy and lactation programs the development of offspring non-alcoholic fatty liver disease in mice. J Hepatol 2010;52:913–920 [DOI] [PubMed] [Google Scholar]
- 7.Sun B, Purcell RH, Terrillion CE, Yan J, Moran TH, Tamashiro KL. Maternal high-fat diet during gestation or suckling differentially affects offspring leptin sensitivity and obesity. Diabetes 2012;61:2833–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregersen S, Dyrskog SE, Storlien LH, Hermansen K. Comparison of a high saturated fat diet with a high carbohydrate diet during pregnancy and lactation: effects on insulin sensitivity in offspring of rats. Metabolism 2005;54:1316–1322 [DOI] [PubMed] [Google Scholar]
- 9.Tamashiro KL, Terrillion CE, Hyun J, Koenig JI, Moran TH. Prenatal stress or high-fat diet increases susceptibility to diet-induced obesity in rat offspring. Diabetes 2009;58:1116–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennedy GC. The development with age of hypothalamic restraint upon the appetite of the rat. J Endocrinol 1957;16:9–17 [DOI] [PubMed] [Google Scholar]
- 11.Aubert R, Suquet JP, Lemonnier D. Long-term morphological and metabolic effects of early under- and over-nutrition in mice. J Nutr 1980;110:649–661 [DOI] [PubMed] [Google Scholar]
- 12.Fiorotto ML, Burrin DG, Perez M, Reeds PJ. Intake and use of milk nutrients by rat pups suckled in small, medium, or large litters. Am J Physiol 1991;260:R1104–R1113 [DOI] [PubMed] [Google Scholar]
- 13.Rodrigues AL, de Moura EG, Passos MC, et al. Postnatal early overfeeding induces hypothalamic higher SOCS3 expression and lower STAT3 activity in adult rats. J Nutr Biochem 2011;22:109–117 [DOI] [PubMed] [Google Scholar]
- 14.Waterland RA, Garza C. Early postnatal nutrition determines adult pancreatic glucose-responsive insulin secretion and islet gene expression in rats. J Nutr 2002;132:357–364 [DOI] [PubMed] [Google Scholar]
- 15.Martins MR, Vieira AK, de Souza EP, Moura AS. Early overnutrition impairs insulin signaling in the heart of adult Swiss mice. J Endocrinol 2008;198:591–598 [DOI] [PubMed] [Google Scholar]
- 16.Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol 2009;5:401–408 [DOI] [PubMed] [Google Scholar]
- 17.Waterland RA, Garza C. Potential mechanisms of metabolic imprinting that lead to chronic disease. Am J Clin Nutr 1999;69:179–197 [DOI] [PubMed] [Google Scholar]
- 18.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003;33(Suppl):245–254 [DOI] [PubMed] [Google Scholar]
- 19.Fagiolini M, Jensen CL, Champagne FA. Epigenetic influences on brain development and plasticity. Curr Opin Neurobiol 2009;19:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 2009;10:295–304 [DOI] [PubMed] [Google Scholar]
- 21.Flier JS. Obesity wars: molecular progress confronts an expanding epidemic. Cell 2004;116:337–350 [DOI] [PubMed] [Google Scholar]
- 22.Choi CS, Savage DB, Abu-Elheiga L, et al. Continuous fat oxidation in acetyl-CoA carboxylase 2 knockout mice increases total energy expenditure, reduces fat mass, and improves insulin sensitivity. Proc Natl Acad Sci USA 2007;104:16480–16485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen L, Kondo Y, Guo Y, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet 2007;3:2023–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waterland RA, Kellermayer R, Laritsky E, et al. Season of conception in rural gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet 2010;6:e1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kellermayer R, Balasa A, Zhang W, et al. Epigenetic maturation in colonic mucosa continues beyond infancy in mice. Hum Mol Genet 2010;19:2168–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 2009;10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen L, Guo Y, Chen X, Ahmed S, Issa JP. Optimizing annealing temperature overcomes bias in bisulfite PCR methylation analysis. Biotechniques 2007;42:48–58, 50, 52 passim [DOI] [PubMed] [Google Scholar]
- 28.Bucasas KL, Franco LM, Shaw CA, et al. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J Infect Dis 2011;203:921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics 2008;24:1547–1548 [DOI] [PubMed] [Google Scholar]
- 30.Kaiyala KJ, Schwartz MW. Toward a more complete (and less controversial) understanding of energy expenditure and its role in obesity pathogenesis. Diabetes 2011;60:17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 2003;23:5293–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci 2004;7:847–854 [DOI] [PubMed] [Google Scholar]
- 33.Waterland RA, Kellermayer R, Rached MT, et al. Epigenomic profiling indicates a role for DNA methylation in early postnatal liver development. Hum Mol Genet 2009;18:3026–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimogori T, Lee DA, Miranda-Angulo A, et al. A genomic atlas of mouse hypothalamic development. Nat Neurosci 2010;13:767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akaike H. A new look at statistical model identification. IEEE Trans Automat Contr 1974;19:716–723 [Google Scholar]
- 36.Plagemann A, Harder T, Rake A, et al. Perinatal elevation of hypothalamic insulin, acquired malformation of hypothalamic galaninergic neurons, and syndrome x-like alterations in adulthood of neonatally overfed rats. Brain Res 1999;836:146–155 [DOI] [PubMed] [Google Scholar]
- 37.Wiedmer P, Klaus S, Ortmann S. Energy metabolism of young rats after early postnatal overnutrition. Br J Nutr 2002;88:301–306 [DOI] [PubMed] [Google Scholar]
- 38.Levin BE, Govek E. Gestational obesity accentuates obesity in obesity-prone progeny. Am J Physiol 1998;275:R1374–R1379 [DOI] [PubMed] [Google Scholar]
- 39.Xiao XQ, Williams SM, Grayson BE, et al. Excess weight gain during the early postnatal period is associated with permanent reprogramming of brown adipose tissue adaptive thermogenesis. Endocrinology 2007;148:4150–4159 [DOI] [PubMed] [Google Scholar]
- 40.Vickers MH, Breier BH, McCarthy D, Gluckman PD. Sedentary behavior during postnatal life is determined by the prenatal environment and exacerbated by postnatal hypercaloric nutrition. Am J Physiol Regul Integr Comp Physiol 2003;285:R271–R273 [DOI] [PubMed] [Google Scholar]
- 41.Hills AP, Okely AD, Baur LA. Addressing childhood obesity through increased physical activity. Nat Rev Endocrinol 2010;6:543–549 [DOI] [PubMed] [Google Scholar]
- 42.Bouret SG, Draper SJ, Simerly RB. Trophic action of leptin on hypothalamic neurons that regulate feeding. Science 2004;304:108–110 [DOI] [PubMed] [Google Scholar]
- 43.Grove KL, Grayson BE, Glavas MM, Xiao XQ, Smith MS. Development of metabolic systems. Physiol Behav 2005;86:646–660 [DOI] [PubMed] [Google Scholar]
- 44.Garland T, Jr, Schutz H, Chappell MA, et al. The biological control of voluntary exercise, spontaneous physical activity and daily energy expenditure in relation to obesity: human and rodent perspectives. J Exp Biol 2011;214:206–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plagemann A, Harder T, Brunn M, et al. Hypothalamic proopiomelanocortin promoter methylation becomes altered by early overfeeding: an epigenetic model of obesity and the metabolic syndrome. J Physiol 2009;587:4963–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plagemann A, Roepke K, Harder T, et al. Epigenetic malprogramming of the insulin receptor promoter due to developmental overfeeding. J Perinat Med 2010;38:393–400 [DOI] [PubMed] [Google Scholar]
- 47.Coupé B, Amarger V, Grit I, Benani A, Parnet P. Nutritional programming affects hypothalamic organization and early response to leptin. Endocrinology 2010;151:702–713 [DOI] [PubMed] [Google Scholar]
- 48.Vucetic Z, Kimmel J, Totoki K, Hollenbeck E, Reyes TM. Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology 2010;151:4756–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma DK, Jang MH, Guo JU, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 2009;323:1074–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manley GT, Binder DK, Papadopoulos MC, Verkman AS. New insights into water transport and edema in the central nervous system from phenotype analysis of aquaporin-4 null mice. Neuroscience 2004;129:983–991 [DOI] [PubMed] [Google Scholar]
- 51.Urbán N, Martín-Ibáñez R, Herranz C, et al. Nolz1 promotes striatal neurogenesis through the regulation of retinoic acid signaling. Neural Dev 2010;5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinto S, Roseberry AG, Liu H, et al. Rapid rewiring of arcuate nucleus feeding circuits by leptin. Science 2004;304:110–115 [DOI] [PubMed] [Google Scholar]
- 53.McNay DE, Briançon N, Kokoeva MV, Maratos-Flier E, Flier JS. Remodeling of the arcuate nucleus energy-balance circuit is inhibited in obese mice. J Clin Invest 2012;122:142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lenz KM, McCarthy MM. Organized for sex-steroid hormones and the developing hypothalamus. Eur J Neurosci 2010;32:2096–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chahrour M, Jung SY, Shaw C, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 2008;320:1224–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoivik EA, Bjanesoy TE, Mai O, et al. DNA methylation of intronic enhancers directs tissue-specific expression of steroidogenic factor 1/adrenal 4 binding protein (SF-1/Ad4BP). Endocrinology 2011;152:2100–2112 [DOI] [PubMed] [Google Scholar]
- 57.Iwamoto K, Bundo M, Ueda J, et al. Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome Res 2011;21:688–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skene PJ, Illingworth RS, Webb S, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell 2010;37:457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]