SUMMARY
In search of molecules involved in the interaction of intestinal nematodes and mammalian mucosal host cells, we performed mass spectrometry to identify excretory/secretory proteins (ESP) from Strongyloides ratti. In addition to other peptides, we detected in the ESP of parasitic female stage peptides homologous to the Caenorhabditis elegans heat shock protein-17, named Sra-HSP-17.1 (~19 kDa) and Sra-HSP-17.2 (~ 18 kDa) with 49% amino acid identity. The full-length cDNAs (483 bp and 474 bp, respectively) were identified and the genomic organization analyzed. To allow further characterization, the proteins were recombinantly expressed and purified. Profiling of transcription by qRT-PCR and of protein by ELISA in various developmental stages revealed parasitic female-specific expression. The sequence analysis of both DNA and amino acid sequence showed two genes share a conserved alpha-crystallin domain and variable N-terminals. The Sra-HSP-17 proteins showed the highest homology to the deduced small heat-shock protein sequence of the human pathogen S. stercoralis. We observed strong immunogenicity of both proteins, leading to high IgG responses following infection of rats. Flow cytometric analysis indicated the binding of Sra-HSP-17s to the monocytes/macrophage lineage but not to peripheral lymphocytes or neutrophils. A rat intestinal epithelial cell line showed dose dependent binding to Sra-HSP-17.1, but not to Sra-HSP-17.2. Exposed monocytes released IL-10 but not TNF-alpha in response to Sra-HSP-17s, suggesting a possible involvement of secreted female proteins in host immune responses.
Keywords: Heat shock proteins, HSP-17, Strongyloides, excretory/secretory proteins, immune response
INTRODUCTION
Parasitic worm infections are one of the most challenging problems in human and veterinary medicine with an estimated cost of £1 billion per annum [1]. The genus Strongyloides contains a number of parasitic species of medical or veterinary importance infecting a wide range of vertebrates. Characteristically, the genus Strongyloides displays parasitic and free-living stages in their life cycle. The parasitic stage dwelling in the intestine constitutes of females only which produce eggs by mitotic parthenogenesis [2, 3]. The human parasite Strongyloides stercoralis is an enteric nematode that has the capability to escape host immune attack and survive within the human small intestine for decades in the case of autoinfection. Strongyloidiasis is widely distributed throughout the tropics and subtropics and infects around 100 million people [4], but the prevalence is likely underestimated since diagnostic tests are insensitive [5].
The transition from free-living larval stages to the parasitic lifestyle in a mammalian host represents a period in which the parasites encounter stress due to dramatic environmental changes. Adaptation of the nematode taxa to a parasitic lifestyle outlines an evolutionary challenge that is likely to go along with gene duplication and subsequent acquisition of novel gene function among its paralogous members [6]. Strongyloides ratti is a natural parasite of rats, closely related to the human parasite S. stercoralis, and is commonly used as laboratory model for Strongyloides infection and comparable parasitological investigations and genetic mapping [7].
The important difference between a free-living organism and a parasite of vertebrates is that the parasite must survive and reproduce in the face of a sophisticated immune response directed against it [8, 9]. It, therefore, has to generate an array of molecules that interfere with the host’s defense system endeavoring to eliminate the unwanted lodger [10]. The ability of helminths to modulate the immune system underpins their longevity in the mammalian host [11, 12]. This modulation is most likely caused by the release of soluble mediators which ligate, degrade or otherwise interact with host immune cells [13, 14].
During infection, both host and pathogen are confronted with dramatic alterations. S. ratti infection was shown to induce Th2 responses characterized by induction of interleukin-4, -13, IgG1, IgG2a and IgE antibody in rats [15, 16] and generation of interleukin-3, -4, -5, and -13 in mice [17]. Recently, it was reported that the S. ratti infection induces expansion of Foxp3+ regulatory T cells in mice [18]. With these conditions, induction of HSP synthesis is vital for pathogen survival. Although immune responses to HSP, which serve as important antigens in defense against infectious agents, have been observed in various experimental infection models, the exact role of HSPs in immunity to microbial infection is poorly understood [19].
The data presented in this study contribute to the elucidation of small heat shock proteins (sHSP) from Strongyloides. sHSPs are ubiquitous, ATP-independent stress response chaperones. They have the smallest monomeric masses of the heat shock protein classes, ranging from 12 kDa to 42 kDa, yet they usually associate into large polydisperse oligomers [20, 21]. There is a growing awareness of parasite HSPs as candidate molecules involved in the parasite–host interaction, therefore we investigated if Sra-HSP-17s (to be designated Sra-HSP-18 and Sra-HSP-19 according to molecular weights or Sra-HSP-17a and Sra-HSP-17b according to the homology to Cel-HSP-17a and Cel-HSP-17b) from Strongyloides are putative links to host’s mucosal defense system. We recently described the small HSP-10 in S. ratti [22]. In the present study, two novel small heat shock proteins (Sra-HSP-17s), from S. ratti were identified in the excretory/secretory products (ESP) of the parasitic female (PF), isolated, and characterized. These proteins represent novel members of nematode-derived sHSP. We demonstrate the protein products of Sra-hsp-17s are expressed in a stage-specific manner. The present report shows the genomic organization of Sra-hsp-17s, recombinant expression and purification of Sra-HSP-17s, as well as their antigenicity. Experiments on the interaction of Sra-HSP-17s with mucosa-associated epithelial and innate immune cells are also shown.
RESULTS
Presence of HSP-17 peptides in ESP of S. ratti parasitic females
SDS-PAGE analysis of highly concentrated ESP from in vitro-cultured PF revealed multiple protein bands between 10-100 kDa, including those at 18-19 kDa (Fig. S1A). The leakiness was ruled out as source of protein release by culture experiments in the presence of the protein-synthesis inhibitor cycloheximide or azide. This treatment resulted in culture supernatants almost without protein, as shown in SDS gels (Fig. S1B). LC–MS/MS analysis of the respective excised bands from the gel followed by a protein database search revealed the presence of two different peptide sequences assigned to putative HSP17. Peptides, characterized by the expressed sequence tag (EST) cluster numbers SR00984 (Contig 767, EST=17) and SR03349 (Contig 820, EST=17) related to sHSPs were abundant only in the PF ESP. The two sequences were homologues of HSP-17 from Caenorhabditis elegans (E-value 2e-20). In addition, a less abundant cluster SR01014 (Contig 834, EST=65), with 98% identity to SR03349, was also identified (Fig. S2A).
Identification of Sra-hsp-17s full-length cDNAs
The partial sequence of the genes represented by the two clusters SR00984 and SR03349 were used to obtain the full-length of the Sra-hsp-17s. The gene products were named Sra-hsp-17.1 and Sra-hsp-17.2, respectively, according to their nearest C. elegans homologues Cel-hsp-17a (450 bp) and Cel-hsp-17b (447 bp). Whereas the Cel-hsp-17s are nearly identical isoforms which differ by only three nucleotides (Fig. S2B), the Sra-hsp-17s are likely two different homologous genes with only 64% identity (Fig.S2C). Full-length Sra-hsp-17.1 and Sra-hsp-17.2 cDNAs were amplified, resulting in 483 bp and 474 bp products, respectively. The translated protein products have no signal peptides for secretion.
Sequence and phylogenetic analyses
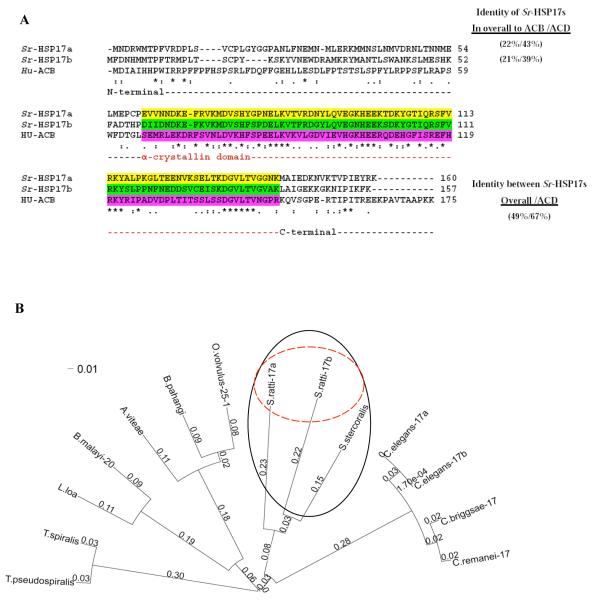
Both Sra-HSP-17s have an HSP20/alpha-crystallin domain (ACD). Structurally, the sHSP family shares a sequence of about 80-100 residues in the C-terminal region that is homologous to ACD from the vertebrate eye lens [23]. The protein sequences of both Sra-HSP-17s were aligned with the human alphaB-crystallin (ABC; Fig. 1A). The aa identity between the Sra-HSP-17s was 49%, being increased in their ACDs to 67% (Fig.1A). The ACDs are covered by residues 61 – 143 and 59 – 141 in the Sra-HSP-17.1 and Sra-HSP-17.2, respectively. The comparison of the deduced complete amino acid sequences of Sra-HSP-17.1 (160 aa, ~18 kDa) and Sra-HSP-17.2 (157 aa, ~ 19 kDa) with members of the sHSP protein family from other nematodes (Fig.S3), showed 39 highly conserved residues, with 19 invariant residues located in the central part (ACD) and the C-terminal regions of the 14 aligned nematodes sHSPs. Clear variation was observed in the N-terminal region of the proteins. The aligned amino acid sequences of all free-living species of the genus Caenorhabditis are highly identical ( ≥ 93%) and differ from the parasitic nematodes. Both Sra-HSP-17s were different from the other nematodes sHSPs, especially in their N-terminal regions, with low overall sequence identities ranging between 21% and 35%. In contrast, the deduced sequence of S. stercoralis sHSP (accession no. N21794; Sst-HSP-17), showed 59% and 61% identity to Sra-HSP-17.1 and Sra-HSP-17.2, respectively, which was even higher than the homology between both Sra-HSP-17s (49%).
Fig. 1. Sequence and phylogenetic analyses.
(A) Alignment of amino acid sequences of Sra-HSP-17.1 and Sra-HSP-17.2 with human alphaB-crystallin (ABC). The percent identities were relative to the complete amino acid sequence and their alpha-crystallin domains (ACD), highlighted in yellow, green and pink, respectively. Symbols: identical (asterisks), no amino acid residue (dashes), conservation of residue size and hydropathy (colons), conservation of size or hydropathy (periods). (B) Phylogenetic tree of the aligned 14 sHSPs orthologs from nematodes including two HSP17-related proteins identified in the S. ratti based on CLUSTAL W. The phylogenetic tree was created using Fast Tree (http://www.microbesonline.org/fasttree/). Fast tree uses the nearest neighbor joining method. The graphic was done using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Bootstrap support values are shown on branches. The compared proteins are: S.stercoralis translated mRNA sequence accession no. N21794, C. elegans HSP17 isoform a accession no. Q20660, C. elegans HSP17 isoform b accession no. Q7JP52, C. briggsae HSP17 accession no. A8XDE7, C. remanei HSP17 accession no. XP_003110614.1, Loa loa sHSP accession no. E1GRM5, Acanthocheilonema viteae Av-25 accession no. Q17102, Brugia malayi HSP20 accession no. A8P0X0, B. pahangi sHSP accession no. CAA61152.1, Trichinella pseudospiralis sHSP accession no. Q000T2, T. spiralis sHSP accession no. Q000T3 and Onchocerca volvulus Ov-25-1 accession no P29778.
Phylogenetic analysis for both DNA and protein sequences produced similar results, of which the latter is displayed in Fig. 1B. Identities and phylogenetic distances of the selected sHSPs showed that the Sra-HSP-17s are more closely related to that of the human pathogen S. stercoralis, than to the HSP-17s of the four Caenorhabditis species, and more distant from the five filariae and the two Trichinella species.
Genomic organization of Sra-hsp-17s
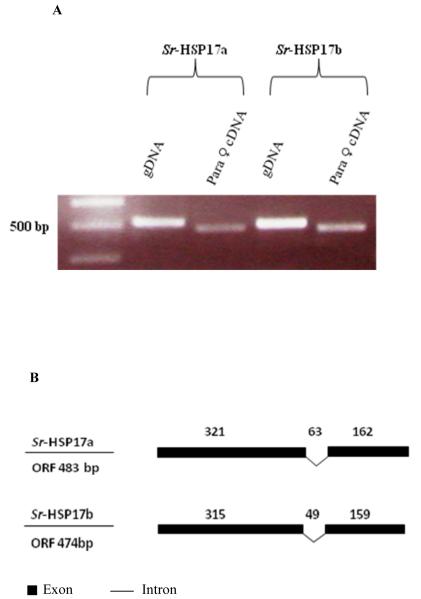
The PCR products (Fig. 2A) showed higher sizes of gDNA products than the PF cDNA products, indicating the presence of at least one short intron in each gene. When cloned, sequenced and aligned, these PCR products confirmed the presence of a single intron (Fig. S4A and S4B) in each shsp located in the second third of the ACDs ORFs with lengths of 63 bp at 321 bases and 49 bp at 315 bases in Sra-hsp-17.1 and Sra-hsp-17.2, respectively (Fig. 2B). Both introns are phase 0 (between two codons); their splice sites sequences followed the GU-AG convention and were found to have similar distributions but differed in position, length and sequence. In addition, the genomic organization of the Sra-hsp-17 genes did not reflect the pseudosymmetrical domain structure of the protein. In our study, we did not identify the gene-link between Sra-hsp-17.1 and Sra-hsp-17.2 using different primers pairs and different PCR conditions, indicating that they are not in tandem or close proximity.
Fig. 2. Genomic organization of Sra-hsp-17s.
(A) Analysis of PCR products on ethidium bromide-stained gel of the amplified products using S. ratti gDNA or PF cDNA. (B) Illustration of the genomic organization of the Sra-hsp-17s from S. ratti. Introns are shown as braked lines and exons are filled boxes. The size of each region (in bases) is indicated above the introns and exons.
Expression and purification of recombinant proteins
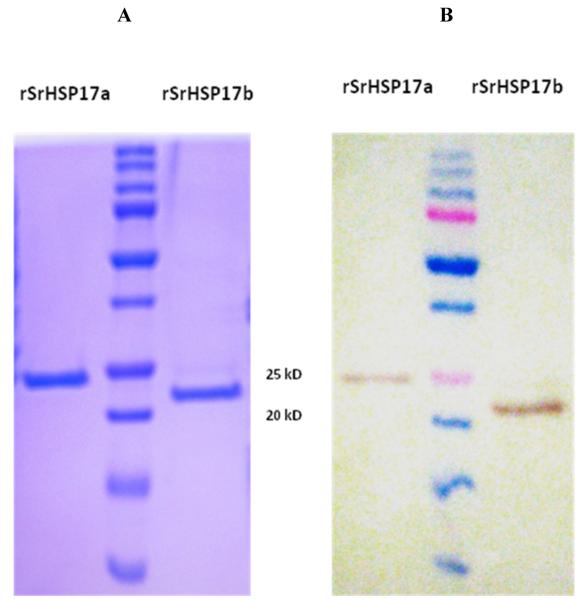
Sra-HSP-17s were recombinantly expressed in E. coli as His-tagged proteins. The bacterial lipopolysaccharide (LPS) was removed to 0.001 - 0.005 EU/μg protein (Table 1), in contrast to unpurified fractions with high LPS levels ( >1 EU/μg protein). The rSra-HSP-17.1 (~23 kDa) and rSra-HSP-17.2 (~22 kDa) were purified to single protein bands shown in Fig. 3A. Western blot analysis with anti-His tag antibodies (Fig. 3B) confirmed the presence of the His-tagged proteins. Since the yield of the purified protein was less than 10% following the removal of the His-tag by enterokinase cleavage, only the His-tagged proteins were used for functional studies.
Table 1. Buffers used in proteins purification procedures and the resulted LPS concentrations that resulted.
All purification procedures were carried out at RT. Dialysis was carried out at 8 °C -10 °C. Buffers were rinsed on ice before use. Highly pure water was used for buffers preparations (Aqua B. Braun, Melsungen AG, Germany).
| Buffer | Constitution | PH | Uses | Time | LPS con. |
|---|---|---|---|---|---|
| B1 | 8 M Urea, 50 mM Tris, 10 mM Imidazole, 500 mM NaCl, 10 % Glycerol, 0.1 % Triton X-100. |
8 | Lysis, binding |
1 h | Not determined |
| B2 | 8 M Urea, 50 mM Tris, 10 mM Imidazole, 500 mM NaCl, 10 % Glycerol, 0.4 % Triton X-114. |
8 | Washing 1 | 3×10 min | Not determined |
| B3 | 8 M Urea, 50 mM Tris, 20 mM Imidazole, 500 mM NaCl, 10 % Glycerol, 0.1 % Triton X-100, 250 μg/ml PMB . |
8 | Washing 2 | 3×10 min | Not determined |
| B4 | 8 M Urea, 50 mM Tris, 250 mM Imidazole, 500 mM NaCl, 10 % Glycerol, 0.1 % Triton X-100. |
8 | Elution | 2×2 min | 0.008-0.02 EU/μg |
| B5 | 2 M urea, 20 mM Tris, 150 mM NaCl, 0.1 % Triton X-100, 30 μg/ml PMB . |
7.8 | Dialysis 3 | 12 h | Not determined |
| B6 | PBS, 30 μg/ml PMB or TBS, 30μg/ml PMB |
7.8 | Dialysis 3 | 2 h | Not determined |
| B7 | PBS or TBS | 7.8 | Final dialysis |
3x2 h | 0.001-0.005 EU/μg |
Fig. 3. Expression and purification of recombinant proteins.
(A) SDS/PAGE and Coomassie staining of rSra-HSP-17.1 and rSra-HSP-17.2, under reducing conditions, purified with a nickel affinity chromatography. (B) Western blot detection of the recombinant His-tag proteins from an identical gel were transferred to a nitrocellulose membrane and treated with anti-His POD.
Chaperone-like activity assay
To determine whether the Sra-HSP-17s possessed the molecular chaperone activity that has been reported for many sHSPs, we studied the ability of both rSra-HSP-17s to alter the thermally-induced aggregation of citrate synthase and malate dehydrogenase substrates, which aggregate irreversibly when incubated at >45°C (Fig. 4A, 4B). In addition, insulin was used as a non-thermal, DTT-aggregated substrate. Addition of different preparations and concentrations of the recombinant parasite proteins did not influence the aggregation of the substrates (Fig. 4C).
Fig. 4. Recombinant Sra-HSP-17s lack the molecular chaperone activity in vitro.
Neither rSra-HSP-17.1 nor rSra-HSP-17.2 were able to reduce the thermally aggregated proteins: (A): citrate synthase (CS) from porcine heart, provided in 2.2 M (NH4)2SO4, 6 mM phosphate, 0.5 mM citrate (Sigma) used in concentration of 75 μg/mL at 45 °C or (B): malate dehydrogenase (MDH) from porcine heart provided in ammonium sulfate (sigma) used at a concentration of 41.5 μg/mL at 48 °C. For both substrates the molar ratios (HSP/substrate) were 1:10 and 1:20. Absorbance was recorded at O.D. 360 nm for insulin and MDH and at O.D 320 nm for CS aggregations each 5 min. for 40 min using ultrospec 2000 spectrophotometer (Pharmacia, Biotech.) for 40 min in 1 mL semi microquartz tubes. (C): Both rSra-HSP-17s showed no inhibition of chemically-induced insulin aggregation at 37 °C in a molar ratio (HSP/insulin) of 1:2.5, 1:5 and 1:10. The bovine pancreas insulin (250 μg/mL), provided in 25mM HEPES pH 8.2 (Sigma), induced for aggregation by 0.02 M DTT.
Production of antibodies and antibody purification
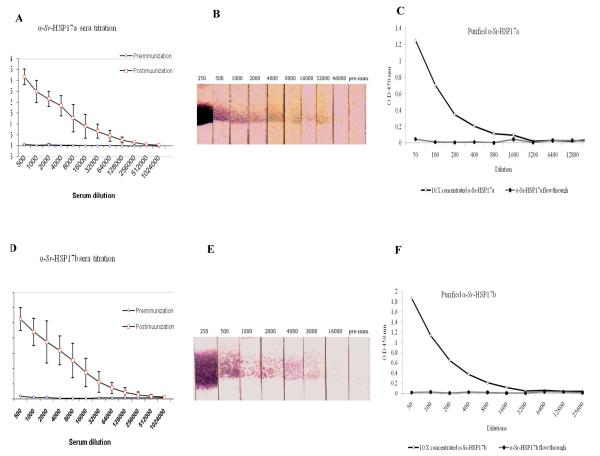
After immunization of seven rats in two independent experiments, with either rSra-HSP-17.1 or rSra-HSP-17.2, post-immunization sera showed levels of IgG antibodies against Sra-HSP-17s still detectable at serum dilutions of >1:64 000 by ELISA (Fig. 5 A, B) and of >1:4000 by Western blots (Fig. 5 C, D). After purification and concentration, the IgG antibodies recognized the rSra-HSP-17s at dilutions >1:800 by ELISA (Fig. 5 E, F). A restricted cross-reactivity of the immune sera against the two Sra-HSP-17s was observed using the immunized sera (at 1:3000) and was found to be much lower after purification of the antibodies at 1:300 (Fig. S5 A, B).
Fig. 5. Recognition of rSra-HSP-17s by IgG in the serum from rSra-HSP-17s-immunized rats.
A, B: The ELISA titration of the immunized sera against the pre-immune sera using rSra-HSP-17s proteins as the antigens. C, D: Western blot titration of the immunized sera using rSra-HSP-17s as the antigens tested with pre-immune serum (1:50) or rSra-HSP-17s-specific rat immune serum taken post immunization (in different dilutions). E, F: ELISA using the recombinant proteins as antigens and titration of the affinity-purified antibodies compared to the concentrated flow through.
Differential transcription of Sra-hsp-17s in S. ratti stages
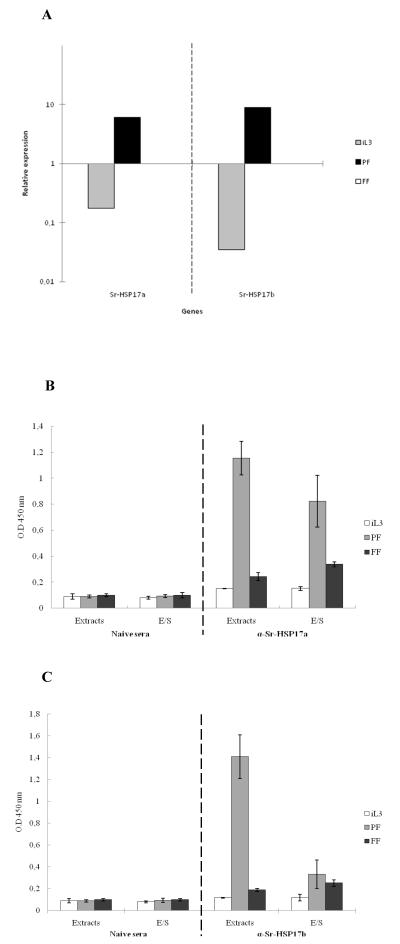
To measure relative levels of gene expression in iL3, PF and FF stages from Strongyloides, all values were expressed as relative quantity of transcripts using the gene-specific transcription levels of the free-living female stage as baseline. In this experiment, gapdh was included as a housekeeping control. The efficiency and linearity of RT-PCR reactions was examined using 10-fold serial dilutions. Following normalization to the expression level of the Sra-gapdh, the transcription levels of the Sra-hsp-17.1 and Sra-hsp-17.2 were elevated more than 6-fold and 9-fold, respectively, in adult PF and highly down-regulated in iL3 stage when compared to expression levels in the FF (Fig. 6A). The differences in the threshold cycle (Ct) values (Table S1) indicated independent transcription of Sra-hsp-17s.
Fig. 6. Stage-associated Sra-HSP-17s.
(A) Relative mRNA expression of Sra-hsp-17s. The data were expressed as the relative quantity of transcripts using the gene-specific transcription levels at the FF stage as baseline. All of the samples were normalized to the expression levels of the constitutatively expressed gene Sra-gapdh. (B) Detection of native Sra-HSP-17.1 using purified antibodies against Sra-HSP-17.1. (C) Detection of native Sra-HSP-17.2 using purified antibodies against Sra-HSP-17.2. Native S. ratti HSP-17s in the ESPs and extracts of the iL3, PF and FF detected by ELISA using Sra-HSP-17s-specific antibodies (1:300). As a negative control, sera from naïve rats before immunization were diluted 1:50. The ESPs and extracts were used as antigens.
Detection of native Sra-HSP-17s in ESP and somatic extracts
Sera from naïve rats did not recognize Sra-HSP-17.1 or Sra-HSP-17.2 in either somatic extracts or in ESP of iL3, PF or FF. By contrast, IgG antibodies from rats immunized with Sra-HSP-17s recognized not only the recombinant proteins, but also the native Sra-HSP-17s from somatic extracts and ESPs of S. ratti stages (Fig. 6 B, C). ELISA tests clearly confirmed the elevated levels of Sra-HSP-17s in PF compared to other stages. Thus, the mean optical density (OD) for Sra-HSP-17.1 was ~ 4.8-fold (extracts) and ~ 2.4-fold (ESP) higher in the PF than in the FF. Similarly, for Sra-HSP-17.2, the OD levels appeared >7.4-fold (extracts) but only > 1.3-fold (ESP) higher in the PF than that in the FF.
Immune recognition of Sra-HSP-17s by sera from Strongyloides-infected rats and exposed humans
Serological analysis was performed to determine if the Sra-HSP-17s are targets for immune recognition during an infection by Strongyloides. ELISA of sera from rats 37 and 112 days after infection revealed time-dependent and significantly increasing IgG reactivity toward both recombinant proteins (P<0.01; Fig. 7A). Furthermore, sera of 50 individuals living in endemic areas for intestinal nematodes, including S. stercoralis, reacted significantly with both Sra-HSP-17s in contrast to 10 non-exposed healthy Europeans, indicating a significant IgG1 recognition (Fig. 7B)
Fig. 7. Immune recognition of Sra-HSP-17s.

(A) Detection of IgG antibody reactivities with rSra-HSP-17.1 and rSra-HSP-17.2 in sera of 5 rats 37 and 112 days after infection with S. ratti as compared with pre-infection sera. (B) IgG1 antibody recognition of rSra-HSP-17s from 50 Africans living in areas endemic for intestinal nematodes including S. stercoralis. Mann-Whitney U test: * P<0.05 and **P<0.01.
Sra-HSP-17s bind to host immune cells
Binding of rSra-HSP-17.1 and rSra-HSP-17.2 to host cells was investigated by flow cytometry (FACS). The host cells were incubated with biotin-labeled Sra-HSP-17.1 or Sra-HSP-17.2. Using freshly isolated human MNC, Sra-HSP-17.2 strongly bound to the monocyte population of the MNC, gated by CD14 staining. In contrast, Sra-HSP-17.1 binds only marginally to the monocytes (Fig. 8A, B). Preincubation with the respective antibodies against the Sra-HSP-17s resulted in an effective reduction (>78% lower fluorescence intensity, FI) of the binding of Sra-HSP-17.2, but only weak reduction of the weakly binding Sra-HSP-17.1. Additionally, significant and dose-dependent reduction of FI with monocytes appeared, due to the competitive effect of unlabeled proteins (Fig. 8C,D). The presence of 1 μg unlabeled Sra-HSP-17.1 with the monocytes beside the biotinylated protein resulted in 55 % reduction of FI and complete inhibition using 2 μg. Similarly, the presence of unlabeled Sra-HSP-17.2 resulted in reduced FI of 45% using 1 μg and 85% using 2 μg unlabeled protein. In contrast, the CD14-negative non-activated lymphocytes showed no binding with both proteins (data not shown). Similarly, CD16-positive neutrophilic granulocytes (PMN) failed to bind to either labeled protein (data not shown). Binding to the monocyte-lineage was confirmed by demonstration of strong binding of CD14-positive macrophage cells (line J774) to both Sra-HSP-17s, pronounced reduction of binding by antibodies against Sra-HSP-17.1 (Fig. 8E, F) and weak reduction by antibodies against Sra-HSP-17.2. Only, Sra-HSP-17.1 showed dose-dependent, moderate binding to the rat intestinal epithelial cell line (IEC-6), which was not inhibitable by preincubation with antibody (Fig. 8G, H). These experiments were repeated three times.
Fig. 8. Binding of Sra-HSP-17.1 and Sra-HSP-17.2 to host immune cells.
(A, B) CD14-positive, gated monocytes. (C, D) Binding competition to monocytes. (E, F) J774 MØ cell line. (G, H) IEC-6 cell line. 2 × 105 cells were incubated at 4 °C for 45 min with biotin-labeled Sra-HSP-17s. Intensity of surface fluorescence (F.I., x-axis) is plotted against cell counts. The noted counts in the figures represent the mean fluorescence index values. Representative results of three independent experiments are shown.
Sra-HSP-17s-induced cytokine release
Sra-HSP-17s were examined for their capacity to induce cytokine secretion in human MNC cultures. Different preparations of rSra-HSP-17s induced a pronounced increase in release of the anti-inflammatory cytokine IL-10 but not of the inflammatory cytokine TNF-alpha (Table 2). Cytokine release was observed in the presence, as well as in the absence, of supplemented PMB, indicating the high purity of the rSra-HSP-17s from LPS (data not shown). In contrast to the cultures exposed to rSra-HSP-17s, the stimulatory effect of the monocyte activator LPS was strongly blocked by PMB.
Table 2. Cytokine responses of MNC exposed to rSra-HSP-17s.
2×105 MNC were cultured for 72 h with rSra-HSP-17.1 or rSra-HSP-17.2 (each at 2 μg/mL) in the presence of polymyxin B (50 μg/mL). As a control, LPS (1 μg/mL) with or without PMB or PHA (1 μg/mL) was used. Cell-free culture supernatants were analysed for released IL-10 and TNF-alpha by commercial ELISAs. Data from two separate experiments are shown (mean ± SD).
| IL-10 (pg/ml) |
TNF-alpha (pg/ml) |
|
|---|---|---|
| unstimulated | 165 ± 86 | 10 ± 14 |
|
| ||
| LPS1) | 1300 ± 66 | 191 ± 88 |
| LPS / polymyxin | 81 ± 78 | 57 ± 66 |
|
| ||
| PHA | 1750 ± 353 | 361 ± 329 |
|
| ||
| Sra-HSP-17.1 | 870 ± 232 | 10 ± 12 |
|
| ||
| Sra-HSP-17.2 | 827 ± 527 | 58 ± 55 |
culture in the absence of polymyxin B
DISCUSSION
In the present study, for the first time, we identified and characterized two novel sHSPs from S.ratti (Sra-HSP-17.1 and Sra-HSP-17.2). The two HSPs were identified by mass spectrometry from culture supernatants of the S. ratti PF stage. The low-molecular weight of 18-19 kDa, further suggested that they belong to the sHSPs family. A number of secreted HSPs have been identified in helminth pathogens such as HSP-18 in ESP from Ostertagia ostertagi [25], the heat shock protein homolog r38 found in the secretions of parasite eggs of Schistosoma mansoni [26], HSP-70 in the secretome of the Fasciola hepatica [27], and HSP-10 and HSP-60 in the secretions of S. ratti recently reported by our group [22]. Interestingly, one of the two hsp-16 genes in C. elegans shows tissue-specific expression in the excretory canal and in few neuronal cells [28]. Since both proteins lack signal peptide, Sra-HSP-17s are possibly secreted by non-classical secretion pathway(s). This has been reported for several HSPs exhibiting no signal peptides – e.g. for HSP-70 secretion by exosome-dependent trafficking [29] or lysosomal endosomes [30, 31].
Secretion of Sra-HSPs by the PF into the intra-host environment of the parasite may be relevant to the parasite–host interactions. The role of small heat shock protein 12.6 from Brugia malayi, which functions as a human IL-10 receptor binding protein [32], in parasite-host interactions has been recently suggested. Further, a role in parasite-host interaction has also been suggested in a recent report on alpha-crystallin [33], which upregulates components of TGF-beta pathway and can enhance the activity of the activating protein-1 (AP-1) by modulating the function of the MAP kinase and thereby retarding the cell migration. It has been suggested that this sHSP negatively regulates pancreatic carcinogenesis.
The full-length cDNAs of the two shsp homologues (Sra-hsp-17s) were identified. The size and sequence properties suggest presence of distinct shsp-17 genes within S. ratti (paralogues), rather than splice variants of the same gene. The primary structures of both Sra- HSP-17s contain the three domains, N-terminal, ACD, and C-terminal domain, confirming the conservation of the ancient ACD domain which is uniform in size (80–100 amino acids) and comprises about one quarter of highly conserved or invariant amino acids.
Phylogenetic comparisons to sHSP originating from other nematodes showed low overall sequence identities (21% - 35%), especially among species at the evolutionary extremes of the phylum Nematoda [34]. However, a higher homology of 59 and 61% identity between Sra-HSP-17.1, Sra-HSP-17.2 and the deduced S. stercoralis sHSP (Sst-HSP-17) partial sequence was observed [35]. Both S. ratti and S. stercoralis are from the same clade (IV). Remarkable homology between the sHSP of the five filarial nematodes (clade III) was found, as well as between the sHSP of the two Trichinella spp. (clade I). The relation (supporting value is 91.4%; Fig. 1B) between the Strongyloides (clade IV) sHSPs and Caenorhabditis (clade V) is closer than to the other parasitic nematodes and might be linked to the presence of FL stages in Strongyloides absent in Brugia and Trichinella that cannot survive outside the host. It has been suggested that Strongyloides belong to the order of Rhabditida, which also contains C. elegans [35], and C. elegans and Strongyloides might be slightly more closely related to each other than any of them is to filaria or Trichinella [36]. However, Strongyloides (Tylenchida ) and Caenorhabditis were classified in different clades in the study of Blaxter et al. [34]. Further, it was stated that the evolutionary status of the family Strongyloididea remained unresolved [37].
Both Sra-hsp-17 genes have a single intron of ancient GU-AG origin interrupting the genes at the ACDs. Interestingly, both genes are located within different large contigs of the S. ratti genome (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/strongyloides), providing additional evidence that the two Sra-hsp-17s are different genes. The HSPs are likely ancient genes that have diverged through evolution as nematodes adapted to parasitism.
Many sHSPs, like Cel-HSP-12.2 and Cel-HSP-12.3 [39], lack chaperone activity due to sequence features of the ACD [40]. Similarly, a recent report indicates HSP-s1 from B. malayi [41] lacks the chaperone activity in vitro, similar to the rSra-HSP-17s that also did not express a chaperone-like activity in vitro. Previous reports suggest that small heat-shock proteins are not necessarily involved in stress responses in parasitic nematodes [42].
The Sra-hsp-17.1 and Sra-hsp-17.2 have stage-specific expression, as they were expressed >6-fold and 9-fold higher in PF in comparison to FF. Both Sra-hsp-17s were conspicuously elevated in PF and down-regulated in iL3 in agreement with previous EST-based transcriptome [43] and microarray [44] studies of S. ratti, which also reported that various orthologs of a C. elegans heat-shock gene hsp-17 were upregulated in PF stage of S. ratti. Further, in microarray analysis, Sra-HSP-17 clusters (SR01014 and SR00984) showed abundant PF-specific expression, but no significant difference in expression under high-immune pressure [45].
The native Sra-HSP-17s in both extracts and ESPs were detected by specific antibodies and were found to be increased in the PF as compared to iL3 and FF. All together, MS/MS analysis, qRT-PCR data, and ELISA analysis of both Sra-HSP-17s confirmed the stage-specificity of Sra-HSP-17s in PF. Stage-related expression of sHSPs has been observed in many nematodes, including Trichinella spiralis [46], B. malayi [41], B. pahangi [47], Nippostrongylus brasiliensis [48], O. ostertagi [25] and Haemonchus contortus [42]. The strong expression of HSPs in the intestine supports the previously framed hypothesis [49] that HSPs are involved in the adaptation of the parasite to its new environment in the intestine. In the present study, the secreted Sra-HSP-17s were recombinantly expressed and purified from contaminating LPS (Table 1). The purified rSra-HSP-17s, were used for immunizing rats, cellular assays, and other functional analyses. Both Sra-HSP-17s, when exposed to the host immune system, were highly immunogenic as demonstrated by strong IgG responses after repeated immunization. In addition, significantly increasing antibody responses against both rSra-HSP-17s were documented after infection of rats with S. ratti, as well as in sera of S. stercoralis-exposed individuals form endemic countries. Interestingly, in an additional experiment, in which rats were infected with S. ratti after prior immunization with rSra-HSP-17s, no further increase of the high antibody responses after infection could be observed (data not shown).
Cellular results obtained in the present study indicated that Sra-HSP-17s interacts and subsequently activates host cells, namely of the monocyte/macrophage lineage and intestinal epithelial cells, however, neither lymphocytes nor granulocytes are affected. Of putative importance, the first-line host cells exposed to Sra-HSP-17s after their release from parasitic females in the intestine, represent the intestinal epithelial cells and these appear to interact with the Sra-HSP-17.1 as indicated by flow cytometry analysis. Directly adjacent to the epithelial cells occur dendritic cells which represent differentiated monocytes. Interestingly, the Sra-HSP-17s can bind differentially to monocytes.
Extensive work has suggested that HSPs may be potent activators of the innate immune system capable of inducing proinflammatory cytokine production by the monocyte-macrophage lineage. Recent evidences suggested that the reported cytokine effects of HSP may be influenced as a result of contaminating LPS and LPS-associated molecules [50, 51]. Our results, however, indicate that both highly purified rSra-HSP-17s, contained none or extremely low endotoxin, were able to induce IL-10 but not TNF-alpha by MNC, importantly, in the presence or absence of the PMB. The effectiveness of PMB was confirmed by neutralization of the LPS-induced cytokines.
In summary, we identified, molecularly characterized, and initiated analysis on the potential biological role of two Strongyloides HSP-17s. The release of the small HSPs from PF, their exposure to the host-local immune system (indicated by binding to host’s immune cells), and generation of pronounced antibody responses illustrate the possible involvement of small HSPs in the local parasite-host interaction.
EXPERIMENTAL PROCEDURES
Parasites
The S. ratti life cycle model was established in our laboratory since ~ 5 years. All animal experiments were approved by and conducted in accordance with guidelines of the appropriate Animal Protection Board of the City of Hamburg (G 21131/591-00.33). Wistar rats (Charles River) were used to maintain the cycle by serial passage as described [52, 53]. For the isolation of the third stage infective larvae (iL3) and the free-living females (FF), fecal pellets were collected on days 6-16 after subcutaneous infection of male Wistar rats with ~ 2500 iL3. Charcoal coprocultures were set up and incubated at 26 °C. The culture dishes were incubated 6 days for the collection of newly generated iL3 or 24 h - 27 h to collect the free-living stages (FLS). The FF were isolated from other FLS by carefully pipetting under the light microscope. For the recovery of iL3 and FLS the standard Baermann isolation technique was used. For the recovery of parasitic females (PF), the rats were infected with ~ 2500 iL3. On day seven post-infection the rats were sacrificed and the PF recovered from the small intestine which was opened longitudinally after cleaning and applied to Baermann funnel for 3 hours.
Somatic extracts and excretory/secretory proteins (ESP) preparation
The freshly harvested iL3, PF and FF were extensively washed in sterile Hanks Balanced Salt Solution (HBSS) supplemented with 100 μg/mL penicillin, 100 units/mL streptomycin (Sigma–Aldrich Chemie, Steinheim, Germany). Somatic extracts were prepared by fast agitation of the worms in the presence of a single steel ball (2.8 mm diameter) using a Precellys Steel Kit (PeqLab Biotech., Erlangen, Germany) in cold phosphate buffer (4 °C) supplemented with 0.1 mM EDTA, 25 mM HEPES for 10 min. The ESP from the same stages: iL3 (4×104/mL), PF (200/mL) and FF (200/mL), which were cultured at 37 °C, were prepared as described before [54]. The incubation times were 24 h for the iL3 and FF and 72 h for PF with changes of media every 24 h. After preparation, extracts and ESP were supplemented with fresh protease inhibitors (Roche Diagnostics, Mannheim, Germany), concentrated (Amicon Ultra 10.000 MWCO filters; Millipore GmbH, Schwalbach, Germany), and the proteins concentrations were determined by standard Bradford assay method. In experiments involving inhibition of the protein synthesis, cycloheximide (Sigma–Aldrich) was added to the culture medium with a final concentration of 70 mM. As metalloproteinase activity was only transiently observed in the culture and no proteinase activity was observed in a substrate gel after exposure of the nematodes to cycloheximide or sodium azide (0.5%), general leakiness of the parasites appeared unlikely (data not shown).
Identification of ESP stage-related proteins by MS/MS
The concentrated ESP were subjected to SDS-PAGE followed by staining with colloidal Coomassie (Invitrogen, Carlsbad, CA, USA). The entire lane was cut into 36 gel blocks of equal size. All gel blocks were then digested with trypsin using published procedures [55]. Peptides derived from in-gel digested proteins were analyzed by online microscale capillary reversed-phase HPLC coupled to a linear ion trap mass spectrometer (LTQ, Thermo Scientific, San Jose, CA, USA). Samples were loaded onto an in-house packed 100 m i.d.×15 cm C18 column (Magic C18, 5 μm, 200 Å, Michrom Bioresource, Auburn, CA, USA) and separated at approximately 500 nl/min with 30 min linear gradients from 5% to 40% acetonitrile in 0.4% formic acid. After each survey spectrum, the six most intense ions per cycle were selected for fragmentation/sequencing. Searches were performed using the ProteinPilot search engine v2.0.1 (Applied Biosystems). The following search parameters were selected: Sample Type: Identification; Cys. Alkylation: Iodoacetamide; Digestion: Trypsin; Instrument: LTQ; Special Factors: Gel-based ID; ID Focus: Amino acid substitutions; Search Effort: Thorough. Proteins were identified based on a minimum of 4.00 ‘unused score’ equivalent to two or more unique peptides of confidence 99.
Data search, sequence and phylogenetic analyses
All MS datasets were searched against combined protein sequence database containing EST sequences from S. ratti and S. stercoralis, as well as the RefSeq protein sequences for C. elegans and C. briggsae. Two EST clusters, from the ESP of the PF only, encodes homologues for Cel-HSP-17s; SR00984 (Contig 767, EST=17), SR03349 (Contig 820, EST=17) were identified and a third less abundant cluster SR01014 (Contig834, EST=65), was identified to has 98% identity to SR03349. The EST database used for this search is available at www.nematode.net [56]. Homology searching on the nucleotide and protein database was carried out using the BLAST program with default settings at NCBI (http://www.ncbi.nlm.nih.gov/). Thereafter, to compare identity and further bioinformatic analysis, sets of software available at the Expert Protein Analysis System (ExPASy) proteomics server (http://expasy.org/tools/), including PROSITE search, alignment, and phylogenetic analysis, of the Swiss Institute of Bioinformatics were used.
RNA isolation, cDNA synthesis and cloning
Total RNA was extracted from PF after 7 days of infection as described [22]. Briefly, after collection, ~ 0.1 g PF was washed extensively in PBS then, homogenized by fast agitation of the worms in the presence of a single steel ball (2.8 mm diameter) using a Precellys Steel Kit (PeqLab Biotech., Erlangen, Germany) using Trizol LS buffer (Invitrogen). Subsequently, RNA was quantified spectrophotometrically, and the quality and integrity of RNA was confirmed via the detection of discrete 18 S and 28 S ribosomal RNA bands on ethidium bromide-stained gel. RNA samples were treated with RNase-free DNase I (Qiagen, Hilden, Germany) and purified with RNeasy MinElute spin column (Qiagen). A total of ~ 5 μg of purified parasite RNA was used to synthesize the first strand of cDNA using SuperScript® III RT (Invitrogen). The manufacturer’s instructions were followed, with the exception of the antisense primer oligo dT-T7I (Table S2) which used in a final concentration of 10 μM. The ESTs clusters were processed as described above, and the longest ESTs were used for the design of forward and reverse primers for the three clusters corresponding to hsp-17. To obtain 3′-cDNA ends specific forward primers af2 and bf2 (Table S2) were used for amplification of Sra-hsp-17.1 and Sra-hsp-17.2 respectively, and the oligodT-T7II was used in both cases as reverse primer. The 5′ ends obtained using the gene specific reverse primers ar2 (Table S2) to amplify Sra-hsp-17.1 and br2 to amplify Sra-hsp-17.2 using the 5′ SL-1 (Table S2), corresponding to the nematode spliced leader sequence [57]. Various Sra-hsp-17 genes PCR products were cloned into pGEM-T Easy vector (Promega Corp.,Madison, USA) and sequenced (LGC Genomics GmbH, Germany). Alignment of the various Sra-hsp-17s cloned fragments resulted in the full length sequences of both genes. Primers including the 3′- and the 5′-ends were designed which included the codon for the initiating methionines and a stop codon. The full length of the Sra-hsp-17.1 and Sra-hsp-17.2 cDNAs were captured by PCR using specific forward primer af1 and reverse primer ar1 and bf1 forward primer and br1(Table S2) as reverse primer respectively. The full length cDNAs were cloned into the pGEM-T Easy vector then the recombinant plasmids were introduced into competent Escherichia coli TOP10 cells (Invitrogen) for amplification. The presence of the insert was confirmed by restriction digestion by NotI (NEB, New England Biolabs GmbH, Germany) observed on ethidium bromide-stained gel and sequencing using the M13 forward-, and genes specific primers.
Genomic organization
For DNA isolation, a pellet of approximately 250,000 S. ratti iL3 was digested for 3-4 h at 56 °C with proteinase K (Qiagen, Hilden, Germany) under constant agitation. RNA free gDNA extracted by standard phenol/chloroform method and treated with RNase A, was then precipitated using NH4-acetate and stored at 4 °C. The specific primer sets af1, ar1 and bf1, br1 (Table S2) were used to amplify the full length genes sequences using 0.1 μg gDNA as a template. The resulting products were cloned into the pGEM-T Easy vector and sequenced. The gDNA and cDNA clones were compared by sequence alignment and analysed by ethidium bromide-stained gel.
Expression of recombinant proteins
A pair of primers was designed to generate a fragment which encodes the full length sequence of the Sra-hsp-17.1. The 5′ sense primer af4 (Table S2) contained a recognition site for HindIII (upper case), an enterokinase cleavage site to remove the His-tag (lower case) and 11 bp (underlined in the primer sequence) of the cDNA which included the codon for the initiating methionine. The 3′ anti-sense primer ar4 contained a recognition site for BamHI and 24 bp of cDNA, which included a stop codon. To express the Sra-hsp-17.2, the cDNA amplified by PCR using the forward primer bf4 contained a recognition site for NdeI (upper case), an enterokinase cleavage site (lower case) and 12 bp (underlined in the primer sequence) of the cDNA for annealing, and the reverse primer br4 contained a BamHI recognition site and 18 bp of cDNA for annealing. The amplified fragments were ligated into PGEM-T Easy vector. Then, the plasmids were isolated and digested with the corresponding restriction enzymes (mentioned above). The restriction fragments purified from the gel (QIAquick; Qiagen) were subcloned into the PJC45 expression vector [59], and the sequences were confirmed again by restriction digestion and sequencing using a PJC45 specific forward primer PJC45f (Table S2). The recombinant plasmids were then transformed into an E. coli Star BL21 DE3. The expression of polyhistidine-recombinant proteins were induced at OD600 ~ 0.25 with 0.2 mM IPTG for 8 h in 30 °C to avoid over expression. The induced cells were suspended and lysed in 5x (v/v) B1 lysis buffer (Table 1) followed by sonication at intervals (Branson Sonifier-250) while kept on ice for four min. The supernatants containing the recombinant Sra-HSP-17s proteins products (rSra-HSP-17.1 and rSra-HSP-17.2) were purified by affinity column chromatography using the profinity TM IMAC Ni–NTA resin (Bio-RAD Laboratories, Germany). The bound proteins were washed, eluted and then dialyzed by serial buffers (Table 1). Subsequently, purified proteins (in 1 X GIBCO DPBS, Invitrogen or in sterilized TBS), were concentrated and stored in 4 °C or −20 °C. The purity of the recombinantly expressed proteins was analyzed by 15 % SDS-PAGE stained with Coomassie blue staining, and the His-tagged protein was detected by Western blotting onto a nitrocellulose membrane applying standard procedures using anti-His POD. Tag-off TM High Activity rEK kit (Novagen, USA) was used to cut the His-tag from the recombinant proteins under the manufacturer’s protocol.
Endotoxin removal
LPS was removed by adding Triton X-114 in the washing procedure of the Ni agarose-bound recombinant protein and the antibiotic PMB, purchased from Sigma–Aldrich (Germany), as part of the purification process (Table 1). Parallel fractions were purified with the same method but without using PMB or Triton X-114. Protein concentrations were determined by the Bradford assay (Bio-Rad) and endotoxin concentrations were quantified by standard Limulus amoebocyte lysate chromogenic endpoint assay (LAL assay) according to the manufacturer’s protocol (Limulus Amebocytes Lysate, QCL-1000, Lonza, Walkersville).
In vitro chaperone-like activity assay
The chaperone-like activity of recombinant Sra-Hsp-17s was studied by measuring the capability to prevent the aggregation of the well known substrates purchased from Sigma, citrate synthase as described [46], malate dehydrogenase as [60] or by thermal aggregation induction and to inhibit DTT-induced insulin aggregation, as previously described [61]. Aggregation of substrates was measured in the presence of different concentrations of rSra-HSP-17.1 or rSra-HSP-17.2.
Generation of antisera, titration and antibody purification
Antibodies against rSra-HSP-17.1 and against rSra-HSP-17.2 were generated by immunization of 10 week-old male Wistar rats with the 20 μg recombinant proteins in alum/PBS s.c. in the neck. The animals were boosted two times in 14 days intervals as described before [22]. Sera collected from immunized rats at day 14 post prime and at day 14 post boost were tested with the corresponding rSra-HSP-17 by ELISAs and Western blots. Antibodies were affinity-purified using a modified method described before [62]. Briefly, after electrophoretic transfer, nitrocellulose membranes were blocked for 1 h using 5% milk/PBS, 5 mm wide strip was longitudinally excised with a new razor blade strip and incubated overnight at 4 °C in 1:5000 anti-His POD diluted in 2.5% milk/PBS/0.05% Tween20. The rest of the membrane was incubated in 1:50 immunized rat sera in 2.5% milk/PBS/0.05% Tween 20 with constant agitation. The clearly visualized band of the excised strip was aligned to the rest of the membrane and the corresponding strip which contained the antibody bound to protein was excised and washed 5 times in PBS/0.05% Tween 20 for 5 min each. Subsequently, the pure antibodies were eluted by incubation in 1 mL of 0.2 M glycine (pH 2.6)/0.05% Tween 20 with constant agitation for 10 min. on ice and then directly concentrated. The buffer was replaced by PBS/ 0.1% BSA (pH 7.5), and the antibodies were stored at – 20 °C. Purified antibodies were analysed by ELISA for (i) detection of native Sra-HSP-17s proteins in the somatic extracts and ESP of the developmental stages and (ii) neutralization experiments in FACS analysis.
Relative mRNA quantification by qRT-PCR
After 7 days of infection S. ratti iL3, PF and FF cDNAs were synthesized from high-quality RNAs as described above. ABI PRISM® 7000 SDS/Relative quantification system (Applied Biosystems, Foster, CA, USA) was used to perform quantitation of gene expression of Sra-hsp-17.1 and Sra-hsp-17.2. Sra-gapdh was included as housekeeping gene. Specific Sra-hsp-17.1 forward af3 (Table S2) and reverse ar3 primers and Sra-hsp-17.2 forward bf3 and reverse br3 primers were used respectively to amplify 150 bp amplicons. The primers used to amplify 154 bp of Sra-gapdh were forward gapdhf and reverse gapdhr. The qualities of primers were analyzed using the Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.htmL). PCR was performed using the qPCR Core kit for SYBR®Green I (Eurogentec S.A.) following the manufacturers recommendations. PCR-efficiency was determined for all PCRs using serial 10-fold dilutions of cDNA [63]. For measurement of gene expression levels, each sample was tested in triplicate, using positive and template-free controls beside negative reverse transcription controls. The specificity and identity of individual amplicons were verified by melting curve analysis and then the PCR products were evaluated by agarose gel electrophoresis. Relative transcriptional differences were calculated from normalized values following the protocol described [64].
Detection of native Sra-HSP-17s
To test the presence of the Sra-HSP-17s in the extracts and ESP of the S. ratti stages, the purified antibodies from the rat sera were used in the ELISA analysis at a 1:300 dilution. 96-well polystyrene microtiter plates (Maxi-Sorb, Nunc) were coated with 200 ng protein either in extracts or ESP per well, blocked with 5% BSA/PBS, and HRP-conjugated goat-anti-rat IgG (Dianova, Hamburg, Germany) was used as a secondary antibody (1:5000). Sera taken from naïve rats served as the negative controls. Optical density was measured at 450 nm.
Immune recognition by ELISAs with rat and human sera
Sera were taken from 5 rats before, 37 days and 112 days after infection with S. ratti iL3, 50 individuals residing in endemic West African areas for nematode infections including strongyloidiasis, and 10 healthy Europeans. Taking of blood samples for research purpose was approved by the Ethics Commission of the Medical Board, Hamburg and by ethic committees and medical authorities in the respective countries. A semi-quantitative analysis of serum IgG antibody levels was performed with modifications using ELISA as previously described [65, 66]. In brief, polystyrene microtiter plates (Maxi-Sorb, Nunc) were coated with Strongyloides recombinant proteins at a concentration of 200 ng/well in carbonate buffer (pH 9.6), sealed with saran wrap and incubated overnight at 4 °C. After removal of unbound protein by washing three times with PBS/0.05% (v/v) Tween 20, the plates were blocked with 5% (w/v) BSA in PBS for one hour at 37 °C. Sera from rats were diluted 1:150, 1:300 and 1:900 and sera from humans were diluted 1:500, 1:1000 and 1:2000 in PBS/0.5% BSA and were incubated at 37 °C for one hour. Unspecifically bound proteins were removed by three washing steps. For detection of bound rat IgG, anti-rat IgG peroxidase–conjugated antibody was applied to a final concentration of 1:5,000. For human sera anti-human IgG1 peroxidase– conjugate antibody was applied to a final concentration of 1:500, and tetramethylbenzidine was used as substrate. Data are expressed as endpoint titers derived from titration curves [66].
Cells preparation
Venous blood samples obtained from healthy volunteers in agreement with institutional guidelines served as a source for peripheral blood mononuclear cells (MNC) and polymorphonuclear cells (PMN). Samples were collected in tubes containing 0.106 mol/L trisodium citrate solution and 1 mL citrate solution (Sarstedt, Germany). Erythrocytes were sedimented from blood samples by addition of equal amounts of 6% hydroxyethyl starch for 40 min at RT. After washing the buffy-coat cells in PBS for 10 min at 400 g, MNC were separated from PMN by Ficoll-Histopaque discontinuous density centrifugation using 3 mL Mono-Poly Resolving Media (MP Biomedicals, Sweden) (density of 1.114 g/mL) overlaid with 3 mL Lymphoflot (Bio-Rad) (density of 1.077 g/mL) centrifuging at 600 g for 30 min. The aspirated MNC and PMN were carefully harvested, washed and resuspended in RPMI 1640 (MNC) and Ca++-containing HBSS (PMN), respectively, at a concentration of 4×106 cells/mL and kept on ice. Murine MØ cell line J774 cells, purchased from the German Collection of Microorganisms and Cell Culture (Braunschweig, Germany), were cultured and passaged in RPMI 1640, 2 mM L-glutamine, 5% inactivated FCS, 25 mM HEPES supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin. The rat small intestinal epithelial cell line (IEC-6) purchased from the European Collection of Cell Cultures (ECACC), UK., was passaged [67] in DMEM, 2 mM glutamine, 5% inactivated fetal calf serum (FCS), 0.1 IU/ml insulin, supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin. The confluent cells were detached by treatment with accutase in Dulbecco’s PBS (PAA Laboratories, Pasching, Austria).
Analysis of cell binding by flow cytometry
To determine the binding properties of the parasite proteins, rSra-HSP-17s were biotinylated using the EZ-Link® Sulfo-NHS-Biotinylation kit (Thermo Scientific, USA) and subsequently purified from the excess of biotin; biotin conjugation was determined according to manufacturer’s instructions. 2-5 μg biotinylated proteins were incubated with 2 × 105 cells/100 μL of human MNC and PMN or macrophage cell line J774 for 45 min at 4 °C to prevent internalization.
Biotinylated rSra-HSP-17s were used directly or after pre-incubation with corresponding purified antibodies at a final dilution 1:20 for 15 min at RT, prior to incubation with the respective cells. For further confirmation of the binding specificity, the biotinylated proteins were incubated with MNC in the presence of 1 or 2 μg of unlabeled corresponding protein. After washing with PBS, samples were incubated with streptavidin-Alexa Fluor 647 (Invitrogen) for 30 min on ice. Samples that were incubated only with streptavidin-Alexa Fluor 647 were used as the negative controls. Anti-CD14-FITC, anti-CD16-PE and ICAM-1were applied to gate the monocytes/macrophages, neutrophils and viable IEC-6, respectively, serving as positive controls. Samples were washed two more times and analyzed by flow cytometry on a FACScalibur cytometer (BD Biosciences), measuring 20,000 events from the gated populations. Flow cytometry data were analyzed with CellQuestPro.
Cytokine ELISAs
Freshly prepared MNC were cultured in RPMI 1640 supplemented with 2 mM L-glutamine, 5% inactivated FCS, 25 mM HEPES, 100 U/mL penicillin, 100 μg/mL streptomycin, and plated in 96-well culture plates (Nunc). 2×105 cells per well were cultured in duplicates with or without addition of 50 μg/mL PMB and exposed to the rSra-HSP-17s (2 μg/mL) or to LPS (1 μg/mL) or phytohaemagglutinin (2 μg/mL) (PHA; Murex Diagnostics Ltd, Dartford, England) at 37 °C/5% CO2 for 72 h. Culture medium was used as a negative control. In the cell-free cultures supernatants IL-10 and TNF-alpha levels were determined by ELISA according the manufacturer’s instructions (eBioscience and R&D Systems)[68]. The absorbance of the final reactant was determined at 450 nm using an ELISA plate reader. Four experiments have been performed at different conditions.
STATISTICAL ANALYSIS
Data were processed and analyzed by Statview software. The log values of the OD data are presented in box plots and whiskers as medians and percentiles (10th, 25th, 50th, 75th, and 90th) [68]. Statistical differences between groups were analyzed by the Mann-Whitney U test. P values <0.05 were taken as moderate, P <0.01 as strong evidence of significance.
Supplementary Material
ACKNOWLEDGEMENTS
The study was in part supported by the Deutsche Forschungsgemeinschaft (DFG projects BR 1020/4-1 and LI 793/5-0). The doctoral student Abuelhassan Elshazly Younis is a member of the Faculty of Science at Aswan, Egypt and supported by the Egyptian Ministry of Higher Education. MP is member of Hamburg School for Structure and Dynamics in Infection financed by the Hamburg Ministry of Science and the Joachim Hertz Stiftung as part of the Hamburg Initiative for Excellence in Science. MM is supported by NIAID grant to MM. We thank Christina M. Taylor for technical assistance. The veterinary team of the Bernhard Nocht Institute for Tropical Medicine is acknowledged.
Abbreviations
- ESP
excretory/secretory proteins
- iL3
infective larvae
- PF
parasitic female
- FLS
free-living stages
- FF
free-living females
- sHSP
small heat shock protein
- ACD
alpha-crystallin domain
- PMB
polymyxin B
- MNC
mononuclear cells
- PMN
polymorphonuclear cells
Footnotes
Database: Nucleotide sequences for Strongyloides ratti heat shock protein-17.1 and 17.2 , have been submitted to the GenBank Database under the accession numbers HQ848950 and HQ848951
SUPPORTING INFORMATION The following supplementary material is available:
This supplementary material can be found in the online version of this article.
REFERENCES
- 1.Newton SE, Munn EA. The development of vaccines against gastrointestinal nematode parasites, particularly Haemonchus contortus. Parasitol Today. 1999;15:116–122. doi: 10.1016/s0169-4758(99)01399-x. doi: S0169-4758(99)01399-X [pii] [DOI] [PubMed] [Google Scholar]
- 2.Viney ME. A genetic analysis of reproduction in Strongyloides ratti. Parasitology. 1994;109(Pt 4):511–515. doi: 10.1017/s0031182000080768. [DOI] [PubMed] [Google Scholar]
- 3.Viney ME. Exploiting the life cycle of Strongyloides ratii. Parasitol Today. 1999;15:231–235. doi: 10.1016/s0169-4758(99)01452-0. doi: S0169475899014520 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Concha R, Harrington W, Jr., Rogers AI. Intestinal strongyloidiasis: recognition, management, and determinants of outcome. J Clin Gastroenterol. 2005;39:203–211. doi: 10.1097/01.mcg.0000152779.68900.33. doi: 00004836-200503000-00004 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Kramme S, Nissen N, Soblik H, Erttmann K, Tannich E, Fleischer B, Panning M, Brattig N. Novel real-time PCR for the universal detection of Strongyloides spp. J Med Microbiol. 2010 doi: 10.1099/jmm.0.025338-0. doi: jmm.0.025338-0 [pii]10.1099/jmm.0.025338-0. [DOI] [PubMed] [Google Scholar]
- 6.Mello LV, O’Meara H, Rigden DJ, Paterson S. Identification of novel aspartic proteases from Strongyloides ratti and characterisation of their evolutionary relationships, stage-specific expression and molecular structure. BMC Genomics. 2009;10:611. doi: 10.1186/1471-2164-10-611. doi: 1471-2164-10-611 [pii]10.1186/1471-2164-10-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nemetschke L, Eberhardt AG, Viney ME, Streit A. A genetic map of the animal-parasitic nematode Strongyloides ratti. Mol Biochem Parasitol. 2010;169:124–127. doi: 10.1016/j.molbiopara.2009.10.008. doi: S0166-6851(09)00248-5 [pii]10.1016/j.molbiopara.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Wakelin D, Walliker D. Genetics of host and parasite--implications for immunity, epidemiology and evolution. Parasitology. 1996;112(Suppl):S1–4. [PubMed] [Google Scholar]
- 9.Wakelin D. Immunology and genetics of zoonotic infections involving parasites. Comp Immunol Microbiol Infect Dis. 1996;19:255–265. doi: 10.1016/0147-9571(96)00017-3. doi: 0147957196000173 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Nagaraj SH, Gasser RB, Ranganathan S. Needles in the EST haystack: large-scale identification and analysis of excretory-secretory (ES) proteins in parasitic nematodes using expressed sequence tags (ESTs) PLoS Negl Trop Dis. 2008;2:e301. doi: 10.1371/journal.pntd.0000301. doi: 10.1371/journal.pntd.0000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behnke JM, Barnard CJ, Wakelin D. Understanding chronic nematode infections: evolutionary considerations, current hypotheses and the way forward. Int J Parasitol. 1992;22:861–907. doi: 10.1016/0020-7519(92)90046-n. doi: 0020-7519(92)90046-N [pii] [DOI] [PubMed] [Google Scholar]
- 12.Maizels RM, Yazdanbakhsh M. Immune regulation by helminth parasites: cellular and molecular mechanisms. Nat Rev Immunol. 2003;3:733–744. doi: 10.1038/nri1183. doi: 10.1038/nri1183 nri1183 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Lightowlers MW, Rickard MD. Excretory-secretory products of helminth parasites: effects on host immune responses. Parasitology. 1988;96(Suppl):S123–166. doi: 10.1017/s0031182000086017. [DOI] [PubMed] [Google Scholar]
- 14.Hewitson JP, Grainger JR, Maizels RM. Helminth immunoregulation: the role of parasite secreted proteins in modulating host immunity. Mol Biochem Parasitol. 2009;167:1–11. doi: 10.1016/j.molbiopara.2009.04.008. doi: S0166-6851(09)00122-4 [pii] 10.1016/j.molbiopara.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkes CP, Bleay C, Paterson S, Viney ME. The immune response during a Strongyloides ratti infection of rats. Parasite Immunol. 2007;29:339–346. doi: 10.1111/j.1365-3024.2007.00945.x. doi: PIM945 [pii] 10.1111/j.1365-3024.2007.00945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paterson S, Wilkes C, Bleay C, Viney ME. Immunological responses elicited by different infection regimes with Strongyloides ratti. PLoS One. 2008;3:e2509. doi: 10.1371/journal.pone.0002509. doi: 10.1371/journal.pone.0002509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eschbach ML, Klemm U, Kolbaum J, Blankenhaus B, Brattig N, Breloer M. Strongyloides ratti infection induces transient nematode-specific Th2 response and reciprocal suppression of IFN-gamma production in mice. Parasite Immunol. 2010;32:370–383. doi: 10.1111/j.1365-3024.2010.01199.x. doi: PIM1199 [pii] 10.1111/j.1365-3024.2010.01199.x. [DOI] [PubMed] [Google Scholar]
- 18.Blankenhaus B, Klemm U, Eschbach ML, Sparwasser T, Huehn J, Kuhl AA, Loddenkemper C, Jacobs T, Breloer M. Strongyloides ratti Infection Induces Expansion of Foxp3+ Regulatory T Cells That Interfere with Immune Response and Parasite Clearance in BALB/c Mice. J Immunol. 2011;186:4295–4305. doi: 10.4049/jimmunol.1001920. doi: jimmunol.1001920 [pii] 10.4049/jimmunol.1001920. [DOI] [PubMed] [Google Scholar]
- 19.Zugel U, Kaufmann SH. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin Microbiol Rev. 1999;12:19–39. doi: 10.1128/cmr.12.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laganowsky A, Benesch JL, Landau M, Ding L, Sawaya MR, Cascio D, Huang Q, Robinson CV, Horwitz J, Eisenberg D. Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function. Protein Sci. 2010;19:1031–1043. doi: 10.1002/pro.380. doi: 10.1002/pro.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jehle S, Rajagopal P, Bardiaux B, Markovic S, Kuhne R, Stout JR, Higman VA, Klevit RE, van Rossum BJ, Oschkinat H. Solid-state NMR and SAXS studies provide a structural basis for the activation of alphaB-crystallin oligomers. Nat Struct Mol Biol. 2010;17:1037–1042. doi: 10.1038/nsmb.1891. doi: nsmb.1891 [pii] 10.1038/nsmb.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tazir Y, Steisslinger V, Soblik H, Younis AE, Beckmann S, Grevelding CG, Steen H, Brattig NW, Erttmann KD. Molecular and functional characterisation of the heat shock protein 10 of Strongyloides ratti. Mol Biochem Parasitol. 2009;168:149–157. doi: 10.1016/j.molbiopara.2009.07.007. doi: S0166-6851(09)00191-1 [pii] 10.1016/j.molbiopara.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Kim KK, Kim R, Kim SH. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 24.Bagneris C, Bateman OA, Naylor CE, Cronin N, Boelens WC, Keep NH, Slingsby C. Crystal structures of alpha-crystallin domain dimers of alphaB-crystallin and Hsp20. J Mol Biol. 2009;392:1242–1252. doi: 10.1016/j.jmb.2009.07.069. doi: S0022-2836(09)00936-X [pii] 10.1016/j.jmb.2009.07.069. [DOI] [PubMed] [Google Scholar]
- 25.Vercauteren I, De Maere V, Vercruysse J, Stevens M, Gevaert K, Claerebout E. A small heat shock protein of Ostertagia ostertagi: stage-specific expression, heat inducibility, and protection trial. J Parasitol. 2006;92:1244–1250. doi: 10.1645/GE-871R.1. [DOI] [PubMed] [Google Scholar]
- 26.Cai Y, Langley JG, Smith DI, Boros DL. A cloned major Schistosoma mansoni egg antigen with homologies to small heat shock proteins elicits Th1 responsiveness. Infect Immun. 1996;64:1750–1755. doi: 10.1128/iai.64.5.1750-1755.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MW, Menon R, Donnelly SM, Dalton JP, Ranganathan S. An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica: proteins associated with invasion and infection of the mammalian host. Mol Cell Proteomics. 2009;8:1891–1907. doi: 10.1074/mcp.M900045-MCP200. doi: M900045-MCP200 [pii] 10.1074/mcp.M900045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shim J, Im SH, Lee J. Tissue-specific expression, heat inducibility, and biological roles of two hsp16 genes in Caenorhabditis elegans. FEBS Lett. 2003;537:139–145. doi: 10.1016/s0014-5793(03)00111-x. doi: S001457930300111X [pii] [DOI] [PubMed] [Google Scholar]
- 29.Lancaster GI, Febbraio MA. Exosome-dependent trafficking of HSP70: a novel secretory pathway for cellular stress proteins. J Biol Chem. 2005;280:23349–23355. doi: 10.1074/jbc.M502017200. doi: M502017200 [pii] 10.1074/jbc.M502017200. [DOI] [PubMed] [Google Scholar]
- 30.Mambula SS, Stevenson MA, Ogawa K, Calderwood SK. Mechanisms for Hsp70 secretion: crossing membranes without a leader. Methods. 2007;43:168–175. doi: 10.1016/j.ymeth.2007.06.009. doi: S1046-2023(07)00126-0 [pii] 10.1016/j.ymeth.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calderwood SK, Mambula SS, Gray PJ., Jr. Extracellular heat shock proteins in cell signaling and immunity. Ann N Y Acad Sci. 2007;1113:28–39. doi: 10.1196/annals.1391.019. doi: annals.1391.019 [pii] 10.1196/annals.1391.019. [DOI] [PubMed] [Google Scholar]
- 32.Gnanasekar M, Anandharaman V, Anand SB, Nutman TB, Ramaswamy K. A novel small heat shock protein 12.6 (HSP12.6) from Brugia malayi functions as a human IL-10 receptor binding protein. Mol Biochem Parasitol. 2008;159:98–103. doi: 10.1016/j.molbiopara.2008.02.010. doi: S0166-6851(08)00061-3 [pii] 10.1016/j.molbiopara.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng M, Chen PC, Xie S, Zhao J, Gong L, Liu J, Zhang L, Sun S, Ma H, Batra SK, et al. The small heat shock protein alphaA-crystallin is expressed in pancreas and acts as a negative regulator of carcinogenesis. Biochim Biophys Acta. 2010;1802:621–631. doi: 10.1016/j.bbadis.2010.04.004. doi: S0925-4439(10)00083-9 [pii] 10.1016/j.bbadis.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- 35.Moore TA, Ramachandran S, Gam AA, Neva FA, Lu W, Saunders L, Williams SA, Nutman TB. Identification of novel sequences and codon usage in Strongyloides stercoralis. Mol Biochem Parasitol. 1996;79:243–248. doi: 10.1016/0166-6851(96)02659-x. doi: 016668519602659X [pii] [DOI] [PubMed] [Google Scholar]
- 36.Anderson RC. Nematode parasites of vertebrates : their development and transmission. C.A.B.International; 1992. [Google Scholar]
- 37.Holterman M, van der Wurff A, van den Elsen S, van Megen H, Bongers T, Holovachov O, Bakker J, Helder J. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown Clades. Mol Biol Evol. 2006;23:1792–1800. doi: 10.1093/molbev/msl044. doi: msl044 [pii] 10.1093/molbev/msl044. [DOI] [PubMed] [Google Scholar]
- 38.Blaxter ML. Nematoda: genes, genomes and the evolution of parasitism. Adv Parasitol. 2003;54:101–195. doi: 10.1016/s0065-308x(03)54003-9. [DOI] [PubMed] [Google Scholar]
- 39.Kokke BP, Leroux MR, Candido EP, Boelens WC, de Jong WW. Caenorhabditis elegans small heat-shock proteins Hsp12.2 and Hsp12.3 form tetramers and have no chaperone-like activity. FEBS Lett. 1998;433:228–232. doi: 10.1016/s0014-5793(98)00917-x. doi: S0014-5793(98)00917-X [pii] [DOI] [PubMed] [Google Scholar]
- 40.Kokke BP, Boelens WC, de Jong WW. The lack of chaperonelike activity of Caenorhabditis elegans Hsp12.2 cannot be restored by domain swapping with human alphaB-crystallin. Cell Stress Chaperones. 2001;6:360–367. doi: 10.1379/1466-1268(2001)006<0360:tlocao>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raghavan N, Ghosh I, Eisinger WS, Pastrana D, Scott AL. Developmentally regulated expression of a unique small heat shock protein in Brugia malayi. Mol Biochem Parasitol. 1999;104:233–246. doi: 10.1016/s0166-6851(99)00150-4. doi: S0166685199001504 [pii] [DOI] [PubMed] [Google Scholar]
- 42.Hartman D, Cottee PA, Savin KW, Bhave M, Presidente PJ, Fulton L, Walkiewicz M, Newton SE. Haemonchus contortus: molecular characterisation of a small heat shock protein. Exp Parasitol. 2003;104:96–103. doi: 10.1016/s0014-4894(03)00138-3. doi: S0014489403001383 [pii] [DOI] [PubMed] [Google Scholar]
- 43.Thompson FJ, Mitreva M, Barker GL, Martin J, Waterston RH, McCarter JP, Viney ME. An expressed sequence tag analysis of the life-cycle of the parasitic nematode Strongyloides ratti. Mol Biochem Parasitol. 2005;142:32–46. doi: 10.1016/j.molbiopara.2005.03.006. doi: S0166-6851(05)00101-5 [pii] 10.1016/j.molbiopara.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Evans H, Mello LV, Fang Y, Wit E, Thompson FJ, Viney ME, Paterson S. Microarray analysis of gender- and parasite-specific gene transcription in Strongyloides ratti. Int J Parasitol. 2008;38:1329–1341. doi: 10.1016/j.ijpara.2008.02.004. doi: S0020-7519(08)00074-X [pii] 10.1016/j.ijpara.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 45.Thompson FJ, Barker GL, Hughes L, Viney ME. Genes important in the parasitic life of the nematode Strongyloides ratti. Mol Biochem Parasitol. 2008;158:112–119. doi: 10.1016/j.molbiopara.2007.11.016. doi: S0166-6851(07)00335-0 [pii] 10.1016/j.molbiopara.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 46.Wu Z, Nagano I, Boonmars T, Takahashi Y. Thermally induced and developmentally regulated expression of a small heat shock protein in Trichinella spiralis. Parasitol Res. 2007;101:201–212. doi: 10.1007/s00436-007-0462-6. doi: 10.1007/s00436-007-0462-6. [DOI] [PubMed] [Google Scholar]
- 47.Devaney E, Egan A, Lewis E, Warbrick EV, Jecock RM. The expression of small heat shock proteins in the microfilaria of Brugia pahangi and their possible role in development. Mol Biochem Parasitol. 1992;56:209–217. doi: 10.1016/0166-6851(92)90170-o. [DOI] [PubMed] [Google Scholar]
- 48.Tweedie S, Grigg ME, Ingram L, Selkirk ME. The expression of a small heat shock protein homologue is developmentally regulated in Nippostrongylus brasiliensis. Mol Biochem Parasitol. 1993;61:149–153. doi: 10.1016/0166-6851(93)90168-w. [DOI] [PubMed] [Google Scholar]
- 49.Jecock RM, Devaney E. Expression of small heat shock proteins by the third-stage larva of Brugia pahangi. Mol Biochem Parasitol. 1992;56:219–226. doi: 10.1016/0166-6851(92)90171-f. [DOI] [PubMed] [Google Scholar]
- 50.Tsan MF, Gao B. Cytokine function of heat shock proteins. Am J Physiol Cell Physiol. 2004;286:C739–744. doi: 10.1152/ajpcell.00364.2003. doi: 10.1152/ajpcell.00364.2003 286/4/C739 [pii] [DOI] [PubMed] [Google Scholar]
- 51.Gao B, Tsan MF. Induction of cytokines by heat shock proteins and endotoxin in murine macrophages. Biochem Biophys Res Commun. 2004;317:1149–1154. doi: 10.1016/j.bbrc.2004.03.160. doi: 10.1016/j.bbrc.2004.03.160 S0006291X04006680 [pii] [DOI] [PubMed] [Google Scholar]
- 52.Viney ME, Lok JB. Strongyloides spp. WormBook. 2007:1–15. doi: 10.1895/wormbook.1.141.1. doi: 10.1895/wormbook.1.141.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keiser J, Thiemann K, Endriss Y, Utzinger J. Strongyloides ratti: in vitro and in vivo activity of tribendimidine. PLoS Negl Trop Dis. 2008;2:e136. doi: 10.1371/journal.pntd.0000136. doi: 10.1371/journal.pntd.0000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maruyama H, El-Malky M, Kumagai T, Ohta N. Secreted adhesion molecules of Strongyloides venezuelensis are produced by oesophageal glands and are components of the wall of tunnels constructed by adult worms in the host intestinal mucosa. Parasitology. 2003;126:165–171. doi: 10.1017/s0031182002002718. [DOI] [PubMed] [Google Scholar]
- 55.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 56.Martin J, Abubucker S, Wylie T, Yin Y, Wang Z, Mitreva M. Nematode.net update 2008: improvements enabling more efficient data mining and comparative nematode genomics. Nucleic Acids Res. 2009;37:D571–578. doi: 10.1093/nar/gkn744. doi: gkn744 [pii] 10.1093/nar/gkn744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hunter T, Bannister WH, Hunter GJ. Cloning, expression, and characterization of two manganese superoxide dismutases from Caenorhabditis elegans. J Biol Chem. 1997;272:28652–28659. doi: 10.1074/jbc.272.45.28652. [DOI] [PubMed] [Google Scholar]
- 58.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. doi: bti770 [pii] 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 59.Clos J, Brandau S. pJC20 and pJC40--two high-copy-number vectors for T7 RNA polymerase-dependent expression of recombinant genes in Escherichia coli. Protein Expr Purif. 1994;5:133–137. doi: 10.1006/prep.1994.1020. doi: S1046-5928(84)71020-5 [pii] 10.1006/prep.1994.1020. [DOI] [PubMed] [Google Scholar]
- 60.Cherepkova OA, Lyutova EM, Eronina TB, Gurvits BY. Chaperone-like activity of macrophage migration inhibitory factor. Int J Biochem Cell Biol. 2006;38:43–55. doi: 10.1016/j.biocel.2005.07.001. doi: S1357-2725(05)00211-6 [pii] 10.1016/j.biocel.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 61.Liao JH, Lee JS, Wu SH, Chiou SH. COOH-terminal truncations and site-directed mutations enhance thermostability and chaperone-like activity of porcine alphaB-crystallin. Mol Vis. 2009;15:1429–1444. [PMC free article] [PubMed] [Google Scholar]
- 62.Muller N, Hemphill A, Imboden M, Duvallet G, Dwinger RH, Seebeck T. Identification and characterization of two repetitive non-variable antigens from African trypanosomes which are recognized early during infection. Parasitology. 1992;104(Pt 1):111–120. doi: 10.1017/s0031182000060856. [DOI] [PubMed] [Google Scholar]
- 63.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. doi: 10.1006/meth.2001.1262 S1046-2023(01)91262-9 [pii] [DOI] [PubMed] [Google Scholar]
- 65.Mpagi JL, Buttner DW, Tischendorf FW, Erttmann KD, Brattig NW. Humoral responses to a secretory Onchocerca volvulus protein: differences in the pattern of antibody isotypes to recombinant Ov20/OvS1 in generalized and hyperreactive onchocerciasis. Parasite Immunol. 2000;22:455–460. doi: 10.1046/j.1365-3024.2000.00325.x. doi: pim325 [pii] [DOI] [PubMed] [Google Scholar]
- 66.Liebau E, Hoppner J, Muhlmeister M, Burmeister C, Luersen K, Perbandt M, Schmetz C, Buttner D, Brattig N. The secretory omega-class glutathione transferase OvGST3 from the human pathogenic parasite Onchocerca volvulus. FEBS J. 2008;275:3438–3453. doi: 10.1111/j.1742-4658.2008.06494.x. doi: EJB6494 [pii] 10.1111/j.1742-4658.2008.06494.x. [DOI] [PubMed] [Google Scholar]
- 67.Quaroni A, Wands J, Trelstad RL, Isselbacher KJ. Epithelioid cell cultures from rat small intestine. Characterization by morphologic and immunologic criteria. J Cell Biol. 1979;80:248–265. doi: 10.1083/jcb.80.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brattig NW, Lepping B, Timmann C, Buttner DW, Marfo Y, Hamelmann C, Horstmann RD. Onchocerca volvulus-exposed persons fail to produce interferon-gamma in response to O. volvulus antigen but mount proliferative responses with interleukin-5 and IL-13 production that decrease with increasing microfilarial density. J Infect Dis. 2002;185:1148–1154. doi: 10.1086/339820. doi: JID010799 [pii] 10.1086/339820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.