Abstract
Human killer cell immunoglobulin-like receptors (KIR) recognize A3/11, Bw4, C1 and C2 epitopes carried by mutually exclusive subsets of HLA-A, B, and C allotypes. Chimpanzee and orangutan have counterparts to HLA-A, B, and C, and KIR that recognize the A3/11, Bw4, C1 and C2 epitopes, either individually or in combination. Because rhesus macaque has counterparts of HLA-A and B, but not HLA-C, we expected that rhesus KIR would better recognize HLA-A and B, than HLA-C. Comparison of the interactions of nine rhesus KIR3D with 95 HLA isoforms, showed the KIR have broad specificity for HLA-A, B, and C, but vary in avidity. Considering both the strength and breadth of reaction, HLA-C was the major target for rhesus KIR, followed by HLA-B, then HLA-A. Strong reactions with HLA-A were restricted to the minority of allotypes carrying the Bw4 epitope, whereas strong reactions with HLA-B partitioned between allotypes having and lacking Bw4. Contrasting to HLA-A and B, every HLA-C allotype bound to the nine rhesus KIR. Sequence comparison of high- and low-binding HLA allotypes revealed the importance of polymorphism in the helix of the α1 domain and the peptide-binding pockets. At peptide position 9, nonpolar residues favor binding to rhesus KIR, whereas charged residues do not. Contrary to expectation, rhesus KIR bind more effectively to HLA-C, than to HLA-A and B. This property is consistent with MHC-C having evolved in hominids to be a generally superior ligand for KIR than MHC-A and MHC-B.
Keywords: KIR receptors, MHC, rhesus macaque, NK cells
Introduction
Human killer cell immunoglobulin-like receptors (KIR) form a diverse, polymorphic family of receptors for HLA class I that diversify the repertoire and response of NK cells (Purdy and Campbell, 2009). Genetic studies have found that counterparts to the variable human KIR gene family are present only in species of simian primate (Parham et al., 2010), while functional studies have demonstrated that chimpanzee (Moesta et al., 2009), orangutan (Older Aguilar et al., 2010), and rhesus macaque KIR (Rosner et al., 2011) are MHC class I receptors like their human counterparts. The KIR gene family comprises several phylogenetically distinct lineages that have co-evolved with different epitopes of MHC class I. Human lineage II KIR recognize the Bw4 and A3/11 epitopes carried by HLA-A and B. In contrast, the lineage III KIR recognize the C1 epitope, carried by HLA-C and two HLA-B allotypes, and the C2 epitope carried only by HLA-C (Rajalingam et al., 2004).
Old World monkeys have counterparts to HLA-A and B, but not to HLA-C (Adams and Parham, 2001). Consistent with this distribution, Old World monkeys have diverse lineage II KIR genes, but little lineage III KIR diversity (Blokhuis et al., 2010; Hershberger et al., 2005; Kruse et al., 2010; LaBonte et al., 2001; Palacios et al., 2010; Sambrook et al., 2005). Only hominoid species with a counterpart to HLA-C, chimpanzee, bonobo, gorilla and orangutan, have diverse lineage III KIR genes (Abi-Rached et al., 2010b; Guethlein et al., 2007). These correlations indicate that lineage II KIR evolved to recognize MHC-A and MHC-B, whereas lineage III KIR evolved to recognize HLA-C. In this context, we expected that lineage II rhesus KIR should recognize MHC-A and MHC-B better than MHC-C. A recent analysis of the interaction of nine rhesus lineage II KIR-Fc fusion proteins with 15 rhesus MHC class I variants (6 Mamu-A, 7 Mamu-B, and 2 Mamu-I) expressed by transfected class I-deficient K562 cells, showed that four KIR-Fc (3DLW03, 3DL05, 3DL11, and 3DS05) recognize Mamu-A, but no reactions with Mamu-B or Mamu-I were detected (Rosner et al., 2011). Here we investigated the recognition of human HLA-A, -B, and -C by the same set of rhesus lineage II KIR-Fc.
Materials and Methods
KIR-Fc fusion proteins
KIR-Fc fusion proteins were made as described previously (Rosner et al., 2011). Briefly, the Ig domains and stem were amplified from rhesus KIR cDNA clones. Products were cloned into pGEM-T Easy vector (Promega) and sequenced to check for errors. Inserts were cut with EcoRI and ligated into pFUSE-hIgG1-Fc2 vector (Invitrogen). These constructs were stably transfected in 293 (human embryonic kidney, DSMZ) cells. After a 3 day incubation in ultraCHO serum-free medium (Lonza), supernatant was harvested and KIR-Fc fusion protein was purified using MAbTrap Kit (GE Healthcare). Eluted KIR-Fc fusion protein was concentrated with Amicon Ultra-30 columns (Millipore).
HLA class I binding assays
Binding of KIR-Fc fusion proteins to 29 HLA-A, 50 HLA-B, and 16 HLA-C allotypes was assessed using LABScreen single-antigen bead sets (One Lambda). Measurements were made with Luminex100 as described by (Moesta et al., 2008) and KIR-Fc binding was normalized to that of W6/32, a mouse monoclonal antibody that recognizes all HLA class I isoforms with similar avidity (Barnstable et al., 1978; Brodsky and Parham, 1982a, b).
Determination of HLA class I positions of significance
Based on their interaction with 3DLW03, all tested HLA class I were ranked and divided into groups of high binding (top one third), intermediate binding (middle one third), and low binding (bottom one third) allotypes. Significant differences between the high and low binding groups were tested for at each position of sequence variation, comparing all HLA class I, as well as HLA-A and HLA-B separately. Separate comparison was not possible with HLA-C, as it is not represented in the low binding allotypes. Significance was measured using the chi squared test, accepting two-tailed P values of less than 0.0001 as significant. An additional requirement for a position to be significant was that the most common residue at this position be different for the two groups.
Results
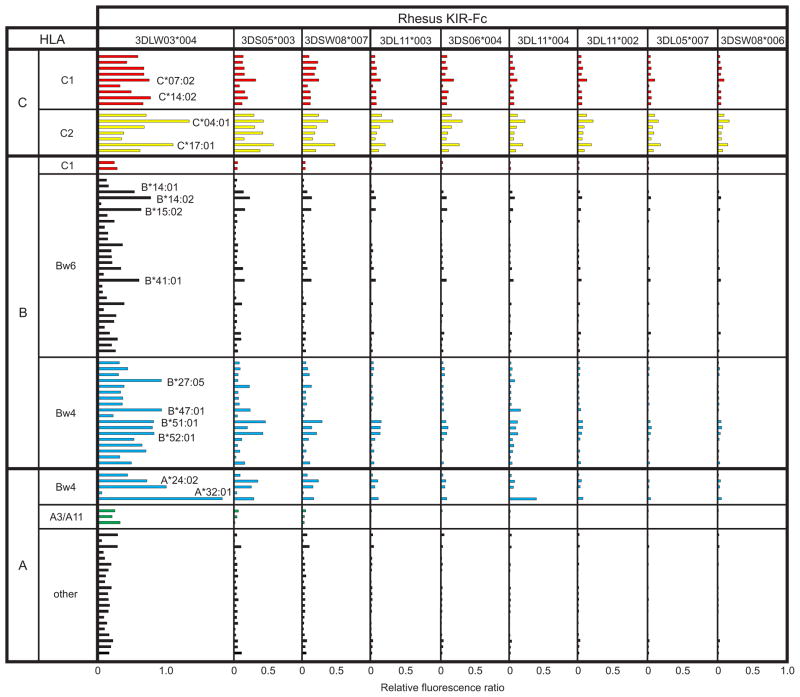
Rhesus lineage II KIR-Fc fusion proteins were tested for binding to a panel of 29 HLA-A, 50 HLA-B and 16 HLA-C allotypes class I using the sensitive, robust, cell-free binding assay we previously described (Moesta et al., 2009; Older Aguilar et al., 2010). All nine rhesus KIR-Fc had demonstrable interactions with HLA class I, but they varied in avidity to form three groups: 3DLW03 stood out for its strong reactions, 3DS05 and 3DSW08 had intermediate reactions, and the remaining six KIR-Fc had weak reactions (Figure 1). Of the four rhesus KIR shown previously to bind Mamu-A ligands, one had high avidity for HLA class I (3DLW03), one had intermediate avidity (3DS05) and two had low avidity (3DL11 and 3DL05). Overall, the binding profiles of the nine rhesus KIR-Fc are similar, and the differences between them can be explained either in terms of avidity or specificity, as previously described for human KIR2DL2 and KIR2DL3 (Moesta et al., 2008). From examining the nine KIR-Fc proteins as a group, it is both striking and unexpected that HLA-C is the dominant target, both in the breadth of the reactions, which embrace all HLA-C allotypes, and their strength. This is particularly evident for the low and intermediate avidity receptors (Figure 1).
Figure 1.
Binding of rhesus macaque KIR to HLA-A, B and C. Within each locus the allotypes are grouped first by epitope and second by numerical order. To account for differences in the amount of HLA class I on each bead, values of KIR-Fc binding were normalized to the binding of W6/32, a monoclonal antibody recognizing a shared (monomorphic) HLA class I epitope. Bw6 is the alternative epitope to Bw4 for HLA-B allotypes. The binding assay was performed as described (Older Aguilar et al., 2010).
Reactions with HLA-A and HLA-B were most clearly revealed by high avidity 3DLW03, but are also partly mirrored in the profiles of the intermediate and weaker KIR (Figure 1). To some extent, many of the HLA-B allotypes bound 3DLW03, but on average the reactions were stronger for Bw4+ HLA-B than Bw4- (Bw6) HLA-B, a discrimination that became more pronounced at the HLA-A locus, where the only strong reactions were with Bw4+ HLA-A allotypes (Figure 2). The binding of 3DLW03 to Mamu-A1*001, which has a Bw4 sequence motif, was shown to be reduced by ~80% when the Bw4 motif was mutated to the Bw6 sequence motif (Rosner et al., 2011). That the mutant retained 20% of the binding shows that the distinction between Bw4 and Bw6 is less clear-cut in rhesus than in human, as is also apparent from the mixed Bw4/Bw6 sequence motifs that occur naturally in some rhesus MHC class I (Rosner et al., 2011). In comparison to the human, chimpanzee, and orangutan KIR that we have studied with this binding assay (Graef et al., 2009; Moesta et al., 2009; Moesta et al., 2008; Older Aguilar et al., 2010), 3DLW03 has the broadest reactivity for HLA-A, B and C (Figure 2).
Figure 2.

Rhesus KIR have broader reactivity with HLA class I than human or orangutan KIR. The rhesus KIR have been grouped according to their avidity: high (3DLW03), intermediate (3DS05, 3DSW08*007) and low (3DL11, 3DS06, 3DL05, 3DSW08*006). Mean binding values are shown for groups of HLA-A, B and C allotypes defined by the presence or absence of epitopes recognized by KIR.
Although surprising, the observed specificity of rhesus KIR3D for HLA class I is very unlikely to be a consequence of experimental artifact. Variation in the representation of the various HLA class I allotypes on the surface of their respective beads is controlled by normalizing KIR-Fc binding to that of the W6/32 monoclonal antibody, which binds a conserved epitope of HLA class I (Brodsky and Parham, 1982a, b). The low background in the binding assay is demonstrated by the many HLA class I allotypes that do not bind to rhesus KIR-Fc (Figures 1 and 2). The strong signal and sensitivity of the assay over two orders of magnitude have been demonstrated in our previous publications, as have correlations of structural polymorphism and patterns of cellular cytotoxicity with the observed expected and unexpected binding reactions (Graef et al., 2009; Moesta et al., 2008; Older Aguilar et al., 2010).
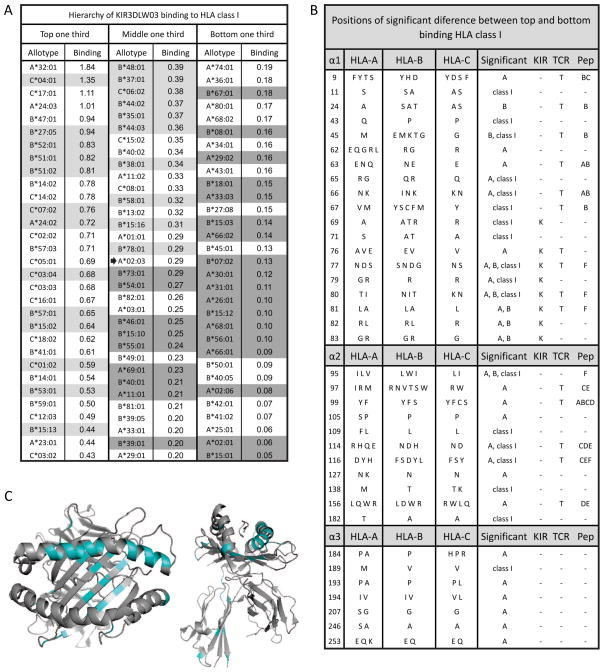
To identify HLA class I polymorphisms that influence binding to 3DLW03, the 95 allotypes were divided into high (top one third), intermediate (middle one third) and low (bottom one third) binding groups (Figure 3A). The high-binding group comprises 4 HLA-A, 14 HLA-B and 13 HLA-C allotypes, whereas the low-binding group comprises 17 HLA-A, 14 HLA-B and no HLA-C allotype. Thus HLA-C allotypes tend towards high binding, HLA-A allotypes tend towards low binding, and HLA-B allotypes evenly distribute between the two categories. The residue frequencies at each position of substitution were compared for the high- (top one third) and low- (bottom one third) binding HLA class I allotypes. At 37 of the 274 positions examined, certain residues were found to be significantly over-represented in either the high-binding or low-binding group of allotypes. These positions comprise 19 in the α1 domain, 11 in the α2 domain and 7 in the α3 domain (Figure 3B). Fourteen of the sites are in the α1 helix, a major contact site with KIR, including residues 76 and 80 that determine the binding of lineage III human KIR2D to C1 and C2 (Mandelboim et al., 1997), and residues 77, 79–83 that determine the binding of lineage II human KIR3D to Bw4 (Sanjanwala et al., 2008) and 3DLW03 to Mamu-A (Rosner et al., 2011). For example, arginine 83, the only Bw4 motif residue necessary for KIR3DL1 interaction (Sanjanwala et al., 2008), is almost universal in the high-binding group, whereas glycine 83 is more common in the low-binding group. In contrast to the concentration of over-represented sites in the α1 helix, position 156 is the only such site in the helix of the α2 domain.
Figure 3.
HLA class I reactivity with rhesus KIR correlates with substitutions in all three extracellular domains. (A) On the basis of their interaction with KIR3DLW03: HLA-A, B and C allotypes were divided into groups of high binding (top one third), intermediate binding (middle one third) and low binding (bottom one third) allotypes. The median value is marked with an arrow and allotypes in grey are those for which peptide information is compared in Figure 4A and 4B. (B) Substitution at 37 positions of polymorphism was significantly different between the high and low binding HLA class I allotypes. The residues present at these sites in the 95 HLA-A, B, and C allotypes are shown. Comparisons were made with all HLA class I and with HLA-A and HLA-B separately, as given under Significant. Separate comparison was not possible with HLA-C, because it is not represented in the low-binding allotypes. Positions that contact KIR (KIR), contact the T-cell receptor (TCR), or form peptide-binding pockets (Pep) are indicated. Significance was measured using the chi squared test, accepting two-tailed P values of less than 0.0001 as significant. Additionally required was that the most common residue in the two groups be different. (C) The 37 significant positions have been colored blue on ribbon diagrams of the α1 and α2 domains (left) and all extracellular domains (right) of the HLA-C*04:01 structure (Fan et al., 2001).
As fifteen of the overrepresented sites, nine in the α1 domain and six in the α2 domain, are residues that form peptide-binding pockets (Figure 3B), we looked for differences in peptide preference that might correlate with binding to 3DLW03. For 50 of the 95 HLA class I allotypes tested there is information on the peptide preference in the SYFPEITHI database (Rammensee et al., 1999) (Figure 4). Of these, 23 were above the median binding level with 3DLW03 (Figure 4A), and 27 were below (Figure 4B). The most obvious difference between the two groups is at P9, the carboxy-terminal anchor residue of the peptide, which is situated within the KIR binding footprint (Boyington and Sun, 2002). None of the high-binding allotypes favor peptides with charged P9 residues, whereas six of the low-binding allotypes do (Figure 4C). Included in their number is HLA-A*11:01, a highly peptide-dependent ligand for human KIR3DL2 (Hansasuta et al., 2004) that binds weakly to the nine rhesus KIR-Fc. These results point to a strong influence of the sequence of the bound peptide on the avidity of the interaction of HLA class I with rhesus lineage II KIR.
Figure 4.
Shown is a comparison of peptide preferences from the SYFPEITHI database (Rammensee et al., 1999) for HLA class I with high (A) and low (B) binding to rhesus 3DLW03 as shown in Figure 3. HLA are listed in order from top to bottom of decreasing interaction with 3DLW03. Anchor residues are boldened and auxiliary anchors are underlined. In part C, the nonpolar residues are replaced with “n”, uncharged polar with “u”, positive with “+”, and negative with “−” at P9. The bound HLA favor nonpolar P9 residues.
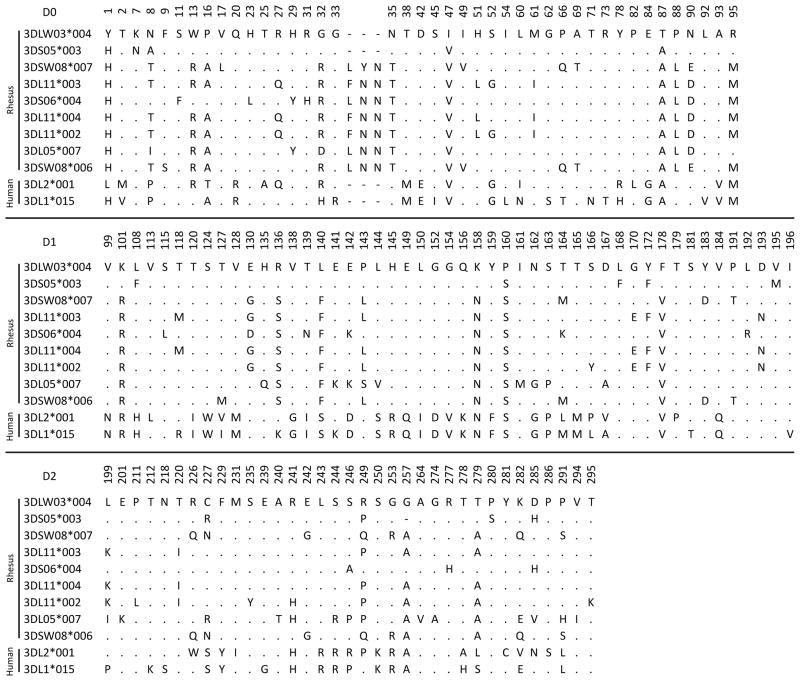
This study demonstrates that rhesus macaque lineage II KIR recognize a broad variety of human HLA-A, B and C allotypes that is not defined by a single epitope or sequence motif, as was observed, but to lesser extent, for certain orangutan (Popy2DLA and 2DLB) and human (KIR2DL2 and KIR2DS4) lineage III KIR (Moesta et al., 2008; Older Aguilar et al., 2010). Such cross-reactivity is consistent with the MHC class I epitopes being structural variations on a common theme. All nine rhesus KIR-Fc tested showed related patterns of reactivity, but 3DLW03 stood out for its high avidity and became the main focus for our analysis. Distinguishing 3DLW03 from seven of the rhesus KIR analyzed here is the absence of three amino acids in the D0 domain following residue 34 (Kruse et al., 2010), and is a property 3DLW03 shares with human lineage II KIR (Figure 5). The loop comprising residues 30–34 of human lineage II KIR is predicted to contact bound HLA class I (Sharma et al., 2009), suggesting the insertion in rhesus macaque KIR could increase the size of the loop and alter interaction with MHC class I. Thus the absence of this insertion could contribute to the high avidity of 3DLW03 for HLA class I. This cannot be the only factor, because 3DS05 lacks the insertion, but has intermediate avidity comparable to 3DSW08 that has the insertion. Other candidates for contributing to the avidity difference are the 35 amino-acid substitutions that distinguish the extracellular domains of 3DLW03 and 3DSW08.
Figure 5.
Positions of amino-acid sequence difference between the nine rhesus macaque lineage II KIR tested for binding to HLA class I and human KIR3DL1*015 and KIR3DL2*001. Rhesus KIR3DLW03*004 is used as the consensus and identity with it is marked with a dot. Gaps are marked with dashes. Numbering is based on 3DL1*015 (Sharma et al., 2009).
Discussion
The striking result of this study is that rhesus lineage II KIR bind more strongly and broadly to HLA-C, than to HLA-A and HLA-B. This result was not anticipated, first because rhesus macaque has counterparts to HLA-A and -B but no counterpart to HLA-C, and second because human lineage II KIR bind to HLA-A and -B epitopes but not to HLA-C epitopes. So how can this superior recognition of an apparently non-physiological ligand be explained?
All human HLA-C allotypes carry either the C1 or C2 epitopes recognized by human lineage III KIR. Of the two, C2 is more recently evolved and present only in human, chimpanzee and gorilla. The more ancient C1 is carried by all orangutan MHC-C and by varying numbers of human, chimpanzee and orangutan MHC-B allotypes. The modern MHC-B and MHC-C genes originated with duplication of a MHC-B like ancestor, following which one daughter locus remained B-like while the other evolved under natural selection in the hominids to become C-like (Fukami-Kobayashi et al., 2005). This history makes it possible that a C1-like epitope was present in the MHC-B like ancestor and passed on to the progenitors of both modern MHC-B and -C. In that case an HLA-C1-like epitope could have been carried by MHC-B in the last common ancestor of Old World monkeys and hominoids and passed on to MHC-B in the modern rhesus population. Thus the increased binding of rhesus macaque lineage III KIR to HLA-C than to HLA-A and -B, could reflect physiological interactions of these KIR with rhesus MHC-B carrying an HLA-C1-like epitope. Indeed, the combination of valine 76 with asparagine 80 that characterizes the human C1 epitope is present in several groups of Mamu-B allotypes, as well as some Mamu-A allotypes (Robinson et al., 2011). In contrast the combination of valine 76 with lysine 80 that characterizes the human C2 epitope is present in neither Mamu-A nor -B. That rhesus KIR react similarly with both C1 and C2 does not preclude a C1-like epitope being the only physiological ligand, because orangutans, which lack C2, have KIR that recognize both physiological C1 and non-physiological C2 (Older Aguilar et al., 2010).
That some orangutan KIR react equally well with C1 and C2, shows that KIR can bind strongly to non-physiological MHC class I. To explain the strong reactions of rhesus KIR with HLA-C, it is therefore not necessary to postulate that rhesus macaque has a physiological MHC-C-like epitope carried by MHC-A or -B. Various lines of evidence suggest that MHC-C differentiated to became a more specialized and better source of KIR ligands than MHC-A and MHC-B (Abi-Rached et al., 2010a; Moesta et al., 2009; Older Aguilar et al., 2010). In this view the interactions of C1 and C2 epitopes carried by MHC-C with lineage III KIR would represent an improvement on the interactions of the lineage II KIR with the Bw4 epitope carried by MHC-A and MHC-B. Although this was naturally a process of co-evolution between cognate KIR and MHC class I, the changes made to the peptide-binding specificity and α1 helix of HLA-C could also have imbued HLA-C with properties that make it an inherently stronger ligand for KIR than HLA-A and HLA-B. That we observe this for KIR derived from the rhesus macaque, a species for which none of the 95 HLA class I alloypes is a physiological ligand, provides further support for the model that MHC-C evolved away from MHC-B under selection for improving its capacity to provide better ligands for KIR.
Acknowledgments
This work was supported by NIH grants AI31168 and AI24258 to P.P.
References
- Abi-Rached L, Kuhl H, Roos C, ten Hallers B, Zhu B, Carbone L, de Jong PJ, Mootnick AR, Knaust F, Reinhardt R, et al. A small, variable, and irregular killer cell Ig-like receptor locus accompanies the absence of MHC-C and MHC-G in gibbons. J Immunol. 2010a;184:1379–1391. doi: 10.4049/jimmunol.0903016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abi-Rached L, Moesta AK, Rajalingam R, Guethlein LA, Parham P. Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells. PLoS Genet. 2010b;6:e1001192. doi: 10.1371/journal.pgen.1001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams EJ, Parham P. Species-specific evolution of MHC class I genes in the higher primates. Immunol Rev. 2001;183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x. [DOI] [PubMed] [Google Scholar]
- Barnstable CJ, Jones EA, Crumpton MJ. Isolation, structure and genetics of HLA-A, -B, -C and -DRw (Ia) antigens. Br Med Bull. 1978;34:241–246. doi: 10.1093/oxfordjournals.bmb.a071504. [DOI] [PubMed] [Google Scholar]
- Blokhuis JH, van der Wiel MK, Doxiadis GG, Bontrop RE. The mosaic of KIR haplotypes in rhesus macaques. Immunogenetics. 2010;62:295–306. doi: 10.1007/s00251-010-0434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol. 2002;38:1007–1021. doi: 10.1016/s0161-5890(02)00030-5. [DOI] [PubMed] [Google Scholar]
- Brodsky FM, Parham P. Evolution of HLA antigenic determinants: species cross-reactions of monoclonal antibodies. Immunogenetics. 1982a;15:151–166. doi: 10.1007/BF00621948. [DOI] [PubMed] [Google Scholar]
- Brodsky FM, Parham P. Monomorphic anti-HLA-A,B,C monoclonal antibodies detecting molecular subunits and combinatorial determinants. J Immunol. 1982b;128:129–135. [PubMed] [Google Scholar]
- Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory eceptor KIR2DL1-HLA-Cw4 complex. Nat Immunol. 2001;2:452–460. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- Fukami-Kobayashi K, Shiina T, Anzai T, Sano K, Yamazaki M, Inoko H, Tateno Y. Genomic evolution of MHC class I region in primates. Proc Natl Acad Sci U S A. 2005;102:9230–9234. doi: 10.1073/pnas.0500770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, Gleimer M, Hammond JA, Guethlein LA, Bushnell DA, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206:2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guethlein LA, Older Aguilar AM, Abi-Rached L, Parham P. Evolution of killer cell Ig-like receptor (KIR) genes: definition of an orangutan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC-C. J Immunol. 2007;179:491–504. doi: 10.4049/jimmunol.179.1.491. [DOI] [PubMed] [Google Scholar]
- Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, Rowland-Jones S, Braud VM. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- Hershberger KL, Kurian J, Korber BT, Letvin NL. Killer cell immunoglobulin-like receptors (KIR) of the African-origin sabaeus monkey: evidence for recombination events in the evolution of KIR. Eur J Immunol. 2005;35:922–935. doi: 10.1002/eji.200425408. [DOI] [PubMed] [Google Scholar]
- Kruse PH, Rosner C, Walter L. Characterization of rhesus macaque KIR genotypes and haplotypes. Immunogenetics. 2010;62:281–293. doi: 10.1007/s00251-010-0433-4. [DOI] [PubMed] [Google Scholar]
- LaBonte ML, Hershberger KL, Korber B, Letvin NL. The KIR and CD94/NKG2 families of molecules in the rhesus monkey. Immunol Rev. 2001;183:25–40. doi: 10.1034/j.1600-065x.2001.1830103.x. [DOI] [PubMed] [Google Scholar]
- Mandelboim O, Reyburn HT, Sheu EG, Vales-Gomez M, Davis DM, Pazmany L, Strominger JL. The binding site of NK receptors on HLA-C molecules. Immunity. 1997;6:341–350. doi: 10.1016/s1074-7613(00)80336-2. [DOI] [PubMed] [Google Scholar]
- Moesta AK, Abi-Rached L, Norman PJ, Parham P. Chimpanzees use more varied receptors and ligands than humans for inhibitory killer cell Ig-like receptor recognition of the MHC-C1 and MHC-C2 epitopes. J Immunol. 2009;182:3628–3637. doi: 10.4049/jimmunol.0803401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- Older Aguilar AM, Guethlein LA, Adams EJ, Abi-Rached L, Moesta AK, Parham P. Coevolution of killer cell Ig-like receptors with HLA-C to become the major variable regulators of human NK cells. J Immunol. 2010;185:4238–4251. doi: 10.4049/jimmunol.1001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios C, Cuervo LC, Cadavid LF. Evolutionary patterns of killer cell Ig-like receptor genes in Old World monkeys. Gene. 2010 doi: 10.1016/j.gene.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Parham P, Abi-Rached L, Matevosyan L, Moesta AK, Norman PJ, Older Aguilar AM, Guethlein LA. Primate-specific regulation of natural killer cells. J Med Primatol. 2010;39:194–212. doi: 10.1111/j.1600-0684.2010.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy AK, Campbell KS. Natural killer cells and cancer: regulation by the killer cell Ig-like receptors (KIR) Cancer Biol Ther. 2009;8:2211–2220. doi: 10.4161/cbt.8.23.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. 2004;172:356–369. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2011;39:D1171–1176. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner C, Kruse PH, Hermes M, Otto N, Walter L. Rhesus Macaque Inhibitory and Activating KIR3D Interact with Mamu-A-Encoded Ligands. J Immunol. 2011;186:2156–2163. doi: 10.4049/jimmunol.1002634. [DOI] [PubMed] [Google Scholar]
- Sambrook JG, Bashirova A, Palmer S, Sims S, Trowsdale J, Abi-Rached L, Parham P, Carrington M, Beck S. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjanwala B, Draghi M, Norman PJ, Guethlein LA, Parham P. Polymorphic sites away from the Bw4 epitope that affect interaction of Bw4+ HLA-B with KIR3DL1. J Immunol. 2008;181:6293–6300. doi: 10.4049/jimmunol.181.9.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Bastard K, Guethlein LA, Norman PJ, Yawata N, Yawata M, Pando M, Thananchai H, Dong T, Rowland-Jones S, et al. Dimorphic motifs in D0 and D1+D2 domains of killer cell Ig-like receptor 3DL1 combine to form receptors with high, moderate, and no avidity for the complex of a peptide derived from HIV and HLA-A*2402. J Immunol. 2009;183:4569–4582. doi: 10.4049/jimmunol.0901734. [DOI] [PMC free article] [PubMed] [Google Scholar]