i. Summary
This chapter provides protocols necessary for quantifying human, mouse, and non-human primate signal joint T cell receptor excision circles (sjTRECs) produced during TCRA gene rearrangement. These nonreplicated episomal circles of DNA are generated by the recombination process used to produce antigen-specific T cell receptors. The number of sjTRECs per mg of thymus tissue or per 100,000 lysed cells has been shown to be a molecular marker of thymopoiesis and naïve T cells. This technology is beneficial to investigators interested in quantitating the level of naïve T cell production occurring in a variety of systems, and complements traditional phenotypic analyses of thymopoiesis. This chapter specifically describes procedures required for rapid detection and quantitation of sjTRECs in thymus tissue or isolated cells using real-time quantitative PCR. The sjTREC assay system comprises species-specific forward and reverse primers for amplification of a unique site on the T cell receptor δ (TCRD) sjTREC, a fluorescently labeled (FAM/ZEN/IABkFQ) species-specific real-time probe, and a species-specific sjTREC DNA plasmid standard for quantitation.
Keywords: Human, Mouse, Non-Human Primate, Thymopoiesis, T cell receptor, Real-time PCR
1. Introduction
The peripheral T cell pool is established early in fetal development by education of bone marrow-derived T cell progenitors on the thymic stroma, with subsequent emigration of mature naïve T cells to peripheral sites (e.g., lymph node and spleen) (1). This process of thymopoiesis is essential for establishing the peripheral T cell pool early in life, and has recently been shown to continue across the lifespan, with thymic output occurring well into the fourth and fifth decades of life (2–5).
A breakthrough for the study of thymopoiesis and peripheral T cell homeostasis in humans was the development of the T cell receptor excision circle (TREC) assay for the study of thymic function in vivo (6, 7). Kong et al. (1998) (7) showed that excised T cell receptor DNA circles were present in recently produced T cells and that these extrachromosomal DNA circles are the byproduct of TCR gene rearrangement, are not replicated, and are diluted by T cell proliferation. Douek et al. (6, 8) developed a real-time PCR assay for the quantitation of signal joint T cell receptor excision circles (sjTRECs) in humansa and reported that they localize in naïve-phenotype T cells and that their frequency falls in peripheral blood CD4 and CD8 T cells with increasing age. The number of sjTRECs per mg of thymus tissue or per 100,000 lysed cells is used as a molecular marker of thymopoiesis and naïve T cells. Thus, measurement of sjTRECs has provided an invaluable assay for rapid assessment of thymic function and the status of T cell immune reconstitution in humans (6, 9–12).
The widespread use of mouse models in the study of T cell biology and immune reconstitution led to the development of a mouse sjTREC PCR assay by our group, based on the human assay (13). The TCRA gene rearrangement event monitored by the human sjTREC assay is the generation of a unique signal joint between δRec and Jα on extrachromosomal circles of DNA in T cells (8, 14). These pseudogenes are two genetic elements of V and Jα respectively, and are conserved between humans and mice (15). The BALB/c mouse homolog of Jα is 3.1 kb upstream of the most upstream Jα (16). However, there exist three reported murine δRec homologs that can be utilized to generate murine TCRD excision circles with the Jα. δRec1 and δRec2 have been identified in the region upstream of Dδ2 (17), and a putative δRec3 has been described 1.6 kb 3’ of Dδ1 (18). The assay described here detects the unique signal joint formed between the murine Jα and δRec1 because these elements were determined to rearrange at a high frequency in mouse thymus (14). In addition, the mouse primers and probes were designed using the BALB/c mouse TCRA/D gene sequence (Gen-Bank AE008686). Although the assay can be used for other strains, it is optimal for BALB/c mice.
The mouse sjTREC PCR assay has been used in a variety of experimental settings. It has been used to characterize murine thymic function during aging, as well as to study the effect of IL-7 on thymopoiesis and peripheral T cell expansion in young and aged mice (13). Mouse sjTREC PCR analysis has also been used to monitor thymopoiesis in neonates (19), investigate murine models of stem cell transplantation (20), and to measure recovery of thymopoiesis following stress-induced thymic atrophy (21–24). Factors that can influence peripheral sjTREC levels are thymic output, T cell proliferation (either homeostatic or antigen-driven), and trafficking in and out of lymphoid tissues.
As was done with mice, Sodora et al. adapted the human sjTREC assay to monitor thymopoiesis in non-human primates (25). Measurements of sjTREC were specifically used to show that, as in humans, thymopoiesis decreases across the lifespan of non-human primates (rhesus macaques and sooty mangabeys) (25). The non-human primate sjTREC assay has been used to assess thymopoiesis in these species and in cynomolgus monkeys following simian immunodeficiency virus infection and/or interleukin-7 treatment (26–29). Primer and probe sequences are provided in this chapter for sjTREC measurement in rhesus, mangabey, and cynomolgus systems.
This chapter describes a generalized real-time PCR assay for the rapid detection and quantitation of human, mouse, or non-human primate sjTRECs using two readily available real-time thermal cyclers: Bio-Rad iCycler iQ and Bio-Rad CFX 96 Real-Time System. The sjTREC assay system comprises species-specific forward and reverse PCR primers for amplification of a unique site on the T cell receptor δ (TCRD) sjTREC, a fluorescently labeled (FAM/ZEN/IABkFQ) species-specific real-time PCR probe for TCRD sjTREC, and a species-specific sjTREC DNA plasmid standard for quantitation (i.e. human, mouse, non-human primate). The DNA standard (calibrated in number of molecules) and samples are amplified for 45 PCR cycles and quantitated using a real-time thermal cycler. As Taq DNA polymerase amplifies the unique TCRD sjTREC sequence, annealed quiescent probe is digested by the nuclease activity of Taq and FAM fluorescence is liberated. Fluorescence is detected at each cycle and used to calculate molecules of sjTREC using a standard curve generated by the system software.
This chapter provides the procedures required for preparation of samples to be assayed, either genomic DNA extracted from thymus tissue or proteinase K-digested lysates of isolated cells, and preparation of working stocks of the sjTREC DNA plasmid standards. For laboratories setting up the sjTREC PCR assay for the first time, it is important that three to five days be devoted to growing up the sjTREC DNA standard plasmid(s) and freezing down a large supply of pre-diluted aliquots of the standards. This initial investment will save time in the future and generate sufficient aliquots of standards for two to three years worth of assays.
2. Materials
The single most critical parameter when performing quantitative real-time PCR is to avoid contamination of the work area, equipment, reagents, and samples. All reagents should be prepared as described, observing stringent molecular-biology technique. Purchase molecular biology-grade stock reagents and use aerosol-resistant pipet tips for all procedures. Wear gloves for all reagent and buffer preparation and change gloves regularly.
2.1 Miscellaneous Supplies
Pre-weighed thymus tissue biopsy or pre-counted and pelleted cell preparations (i.e. thymocytes or peripheral T cell subsets) (see Note 1)
Sterile forceps
Trizol Reagent (Invitrogen), room temperature
Soft tissue homogenizing CK14 tubes (1.4 mm ceramic beads in 2 mL tubes; Bertin Technologies)
Molecular-biology grade Chloroform
100% and 75% molecular-biology grade ethanol
Sodium citrate at 0.1 M in 10% ethanol
DNase/RNase-free molecular-biology grade water
1.5 mL polypropylene screw-cap tubes (Sarstedt brand suggested)
15 mL polypropylene conical tubes
10 mM Tris-Cl, pH 7.8 in DNase/RNase-free H2O
19.2 mg/mL proteinase K (PCR Grade, Roche)
Yeast tRNA (PCR Grade)
2.2. Species-specific sjTREC plasmid standard
Grow up a stock of human, mouse, or rhesus sjTREC plasmid (Gregory D. Sempowski, Duke University; gregory.sempowski@duke.edu) using standard molecular biology techniques. Determine the concentration in µg/mL of the purified plasmid (see Note 2)
Using the species-specific plasmid molecular weight, determine the number of molecules of sjTREC plasmid per µL in the plasmid standard preparation (see Note 3).
Prepare a stock of 2×1010 sjTREC molecules/µL in DNase/RNase-free water.
Make serial dilutions of 100 µL standard into 900 µL DNase/RNase-free water for 1010, 109, and 108 molecules per 5 µL aliquot. Vortex well to mix and microcentrifuge briefly at maximum speed before opening tubes.
For standard dilutions of 107, 106, 105, 104, 103, and 102 molecules/5 µL, dilute 500 µL of the 108 dilution up to 5 mL in a 15 mL polypropylene conical tube with water containing 30 ng/mL yeast tRNA (see Note 4). Vortex well to mix and centrifuge briefly at maximum speed in a tabletop centrifuge before opening tubes.
Validate prepared standard dilutions by running 5 µL of each dilution (107 through 102) in the sjTREC PCR assay (see Note 5).
Aliquot each standard at 1 mL/tube, but aliquot the last milliliter of each standard at 200 µL/tube. Each 200 µL tube is aliquoted as 15 µL/tube working aliquots as needed. One 15 µL aliquot of each standard will be needed per sjTREC PCR assay run.
Freeze all standard stocks and dilutions immediately at −80°C. Store standards in a separate box away from all other PCR reagents and experimental samples. Label all tubes well and avoid freeze/thaw of the working aliquots of standards.
2.3. PCR Reaction
Platinum Taq DNA polymerase, 50 mM MgCl2, and reaction buffer (Invitrogen or equivalent).
10 mM dNTP mix: To prepare dNTP mix, combine 1 M stock solutions of dATP, dCTP, dGTP, and dUTP at a ratio of 1:100 in DNase/RNase-free water such that each deoxynucleotide is present in the mixture at a final concentration of 10 mM. Mix solution well by vortexing and store up to 2 years at −80°C in 500 µL aliquots.
96-well PCR plates and PCR plate strip caps (see Note 6)
2.4. Species-specific Primers
12.5 µM Stocks of species-specific sjTREC primers (see Notes 7, 8, 9)
Reconstitute lyophilized primers with water to a stock concentration of 125 µM. Store 50 µl aliquots at −80°C.
Prepare a working stock by diluting 1:10 with water to a final concentration of 12.5 µM, aliquot at 200 µL/tube, and store at −80°C.
2.5. Species-specific Probes
5 µM Stock of species-specific sjTREC probe (see Notes 7, 8, 9)
Real-time PCR probes (Integrated DNA Technologies) contain a 5’ reporter fluorochrome (FAM), an internal quencher (ZEN), and a 3’ dark quencher (IABkFQ). This double-quenching design allows for lower background and higher signal than single-quench probes.
Reconstitute lyophilized probe with water to a stock concentration of 50 µM. Store 50 µL aliquots at −80°C.
Prepare a working stock by diluting 1:10 with water to a final concentration of 5 µM, aliquot at 200 µL/tube, and store at −80°C.
2.6. Required Equipment
55°C water bath
Tabletop centrifuge with swinging 96-well plate holders and 15 mL tube holders
Microcentrifuge
Thermomixer (Eppendorf)
Precellys-24 Homogenizer (Bertin Technologies; or similar)
PCR setup hood with UV lamp (optional)
Bio-Rad iCycler iQ Thermal Cycler or Bio Rad CFX96 Real-Time System (or similar) with optical system and filter sets for detection of FAM
3. Methods
To avoid contamination of the work area, equipment, reagents, and samples all reagents should be prepared as described, observing stringent molecular-biology technique. Use aerosol-resistant pipet tips for all procedures, wear gloves for all procedures and change gloves when transitioning between setting up the PCR reaction mix, the addition of the samples, and the addition of the standards.
3.1 Preparation of thymus tissue genomic DNA
Using sterile forceps, transfer frozen tissue biopsy to ceramic bead tube containing 1 mL Trizol reagent.
Load tubes into Precellys-24 homogenizer and process for 20 seconds at 5,000 rpm.
Using a micropipettor with a 200 µL tip, transfer tissue homogenate into a pre-labeled 1.5 mL microcentrifuge tube.
Add 200 µL of chloroform to each microcentrifuge tube containing the 1 mL homogenized tissue/Trizol. Cap tubes well and vortex for 15 sec. Incubate at 15° to 30°C for 15 min.
Microcentrifuge samples 15 min at 12,000 rpm, 2° to 8°C. After centrifugation, the aqueous phase will contain RNA, the interphase will contain cellular proteins and some DNA, and the organic (pink) phase will contain genomic DNA.
Carefully remove aqueous phase using a micropipettor with a 200 µL tip. Aqueous phase can be stored in a cryovial at −80°C if future extraction of RNA is desired.
Add 300 µL of 100% ethanol to the interphase/organic phase remaining in the microcentrifuge tube. Vortex gently. Incubate samples 2 to 3 min at 15° to 30°C.
Microcentrifuge 5 min at 12,000 rpm, room temperature.
Remove Trizol/ethanol supernatant to an appropriate waste container.
Add 1 mL 0.1 M sodium citrate/10% ethanol to each sample tube. Incubate samples 30 min at 15° to 30°C with periodic mixing.
Microcentrifuge 5 min at 12,000 rpm, room temperature. Repeat 0.1 M sodium citrate wash as in step 10.
Resuspend DNA in 1 mL 75% ethanol. Incubate 10 to 20 min at 15° to 30°C with periodic mixing.
Microcentrifuge 5 min at 12,000 rpm, room temperature. Remove supernatant using a micropipettor with a 200 µL tip.
Briefly dry pellet (no more than 5 min) under vacuum in a Speedvac evaporator or air-dry in a fume hood. Dissolve pellet in 200–500 µL water. Record the precise volume used to dissolve the pellet. If DNA does not dissolve, place samples at 55°C for 10 min to increase solubility. Cool on ice, vortex, microcentrifuge briefly at maximum speed to collect solution at bottom of tube, and transfer solubilized DNA to a fresh 1.5 mL microcentrifuge tube.
Quantitate DNA using a UV spectrophotometer (read absorbance at 260 nm). Multiply A260 × dilution × 50 to determine DNA concentration in µg/mL. Multiply concentration by the total volume of DNA to determine total DNA yield. Divide the total µg of DNA by the initial weight of tissue used to determine µg of DNA per milligram of thymus tissue.
Freeze DNA samples at −80°C or proceed to Step 3.3.
3.2 Proteinase K-lysis of isolated lymphocytes
Calculate volume of proteinase K needed per sample by multiplying the total cell count by 0.0001; the result is the amount of proteinase K working solution in µL to add to the pellet for a final concentration of 10,000 cells/µL proteinase K.
Immediately before use, dilute the 19.2 mg/mL stock solution of proteinase K 1:200 with 10 mM Tris-Cl, pH 7.8. Layer this solution (using the volume calculated in Step 1) on top of each pellet without letting the pipet tip touch the pellet. Vortex tube and flick down.
Place tubes in a Thermomixer at 56°C and shake for 1 hour at 1200 rpm.
Turn Thermomixer up to 95°C for 10 min to inactivate the proteinase K. Vortex tubes and microcentrifuge 1 min at 12,000 rpm.
Store lysates at −80°C or proceed to Step 3.3.
3.3 Quantitation of TCR delta excision circles by real-time PCR using the Bio-Rad iCycler IQ or CFX96 real-time thermal cycler machines
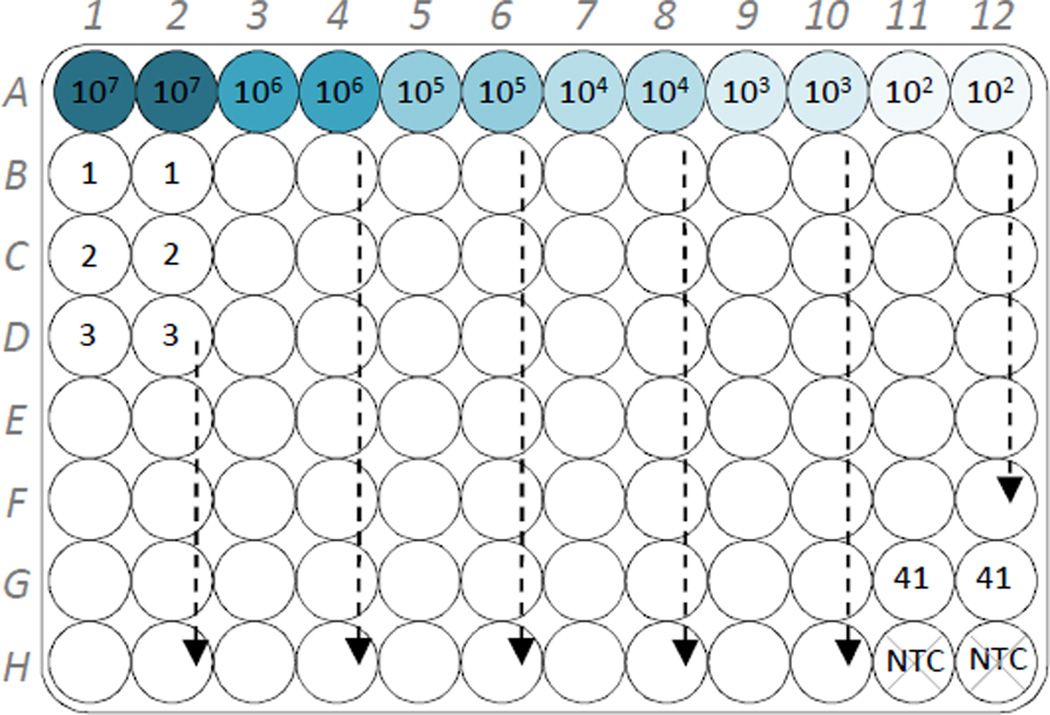
Design the plate layout for the sjTREC PCR assay (Fig. 1). Determine number of samples, standards, and no-template controls (NTC) to be run in duplicate. Add two extra wells to the total number of wells needed to allow for pipetting error.
- Prepare PCR reaction mix (see Note 10). For each of the wells to be included in the assay (as determined in step 1), add the following to a 1.5 mL tube to prepare a PCR master mix: 12.125 µL H2O
- 2.5 µL Platinum Taq buffer
- 1.75 µL 50 mM MgCl2
- 0.5 µL 10 mM dNTP mix
- 1.0 µL forward primer working solution
- 1.0 µL reverse primer working solution
- 1.0 µL FAM probe
- 0.125 µL Platinum Taq enzyme
Vortex gently to mix well, then microcentrifuge briefly at maximum speed to collect the solution at the bottom of the tube.
Add 20 µL of PCR reaction mix to each well according to the plate layout (see step 1 and Fig. 1).
Add 5 µL of DNase /RNase-free water to each no-template control (NTC) well, according to the plate layout, and cap the wells.
- Add 5 µL of each experimental sample (either 1 µg of DNA in 5 µL water or the volume equivalent (5 µL) of 50,000 cells of a proteinase K cell lysate) into duplicate wells according to the plate layout. Cap wells after each row.
-
6.1Prepare DNA samples by diluting 3 µg of each sample to a final volume of 15 µL with water. Use 5 µL (1 µg DNA) of diluted DNA per well in the sjTREC PCR assay.
-
6.1
In a separate clean area, add 5 µL of each pre-diluted standard (102 to 107 molecules/5 µL) to duplicate wells according to the plate layout. Cap wells (see Note 11).
Gently vortex plate and centrifuge 5 min at 1500 rpm in a tabletop centrifuge with 96-well plate holders.
- Program the BioRad iCycler or CFX96 system for a 25 µL sample as follows:
- Cycle 1: (1 time) Step 1, 95°C for 10 min
- Cycle 2: (45 times) Step 1, 95°C for 15 sec; Step 2, 60°C for 1 min
- Cycle 3: (1 time) Step 1, 4°C HOLD
Enable real-time data collection of FAM signal during Cycle 2 and use the “heated lid” option. Place plate in the thermal cycler, lock lid, and start run (see Note 12).
When the run is complete, remove the 96-well plate from the thermal cycler and discard.
Set the threshold at the midpoint of the linear amplification range of the standards (see system software manual). Analyze the r2 and y intercept of the standard curve (see Note 13).
Fig. 1.
Suggested layout of duplicate sjTREC standards (107 to 102 molecules), no-template controls (NTC), and experimental samples (1 to 41) on a 96-well PCR plate.
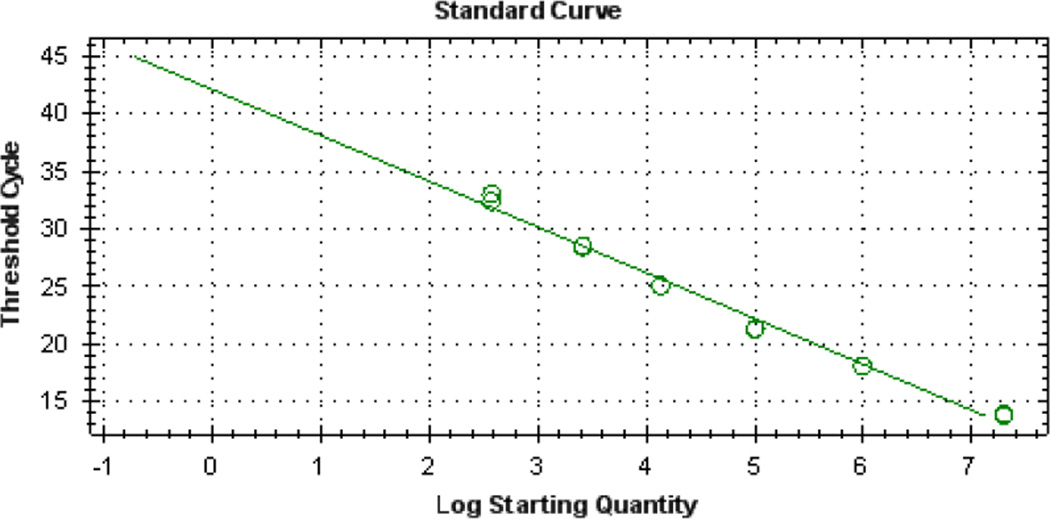
Fig. 2.
Representative mouse sjTREC standard curve generated using the CFX96 Real-Time System.
Acknowledgements
This work was supported by NIH grants AG-025150 and AI-067798 and was performed in the Regional Biocontainment Laboratory at Duke which received partial support for construction from NIH/NIAID (UC6-AI-058607).
Footnotes
Fresh thymus tissue samples (~100 mg) should be snap-frozen in a dry ice/ethanol bath and stored at −80°C or in liquid nitrogen until ready for TREC assay. Isolated cell samples (250,000 minimum) should be thawed at 37°C, washed with 10 mL PBS, and then pelleted by centrifugation in a tabletop centrifuge at 1,500 rpm for 5 min at 4°C.
The biggest source of contamination is the plasmid that contains the mouse sjTREC DNA standard. Avoid working with the plasmid or aliquots of diluted standard when containers of other assay reagents or experimental samples are open.
The weight of one molecule of the Duke University sjTREC plasmid is as follows: mouse = 2.25 × 10−18 g; human = 4.19×10−18 g; non-human primate (rheseus) = 3.37 × 10−18g.
Yeast tRNA is required to stabilize the diluted plasmid.
The Ct values for the standards should be evenly spaced over the 5-log curve. Ten-fold dilutions read out with a 3 cycle difference in Ct. The y intercept of the curve should be 45 ± 3, and have an r2 > 0.995. Quantitated and prediluted aliquots of sjTREC plasmid DNA are available for calibration purposes (Gregory D. Sempowski, Duke University).
It is important to use plates and caps designed for the specific thermal cycler being used, as these machines are calibrated for the specific material, density, color, and refractive angle of the plasticware.
Mouse forward sjTREC primer: 5’- CAT TGC CTT TGA ACC AAG CTG -3’. Mouse reverse sjTREC primer: 5’- TTA TGC ACA GGG TGC AGG TG -3’. Mouse probe: 5'- /56-FAM/CA GGG CAG G/ZEN/T TTT TGT AAA GGT GCT CAC TT/3IABkFQ/ -3'.
Human forward sjTREC primer: 5’- CAC ATC CCT TTC AAC CAT GCT -3’. Human reverse sjTREC primer: 5’- GCC AGC TGC AGG GTT TAG G -3’. Human probe: 5’- /56-FAM/AC ACC TCT G/ZEN/G TTT TTG TAA AGG TGC CCA CT/3IABkFQ/ -3'.
Non-human primate (Rhesus, NHP) forward sjTREC primer: 5’- CAC ATC CCT TTC AAC CAT GCT -3’. NHP reverse sjTREC primer: 5’- GCC AGC TGC AGG GTT TAG G -3’. NHP Probe: 5'- /56-FAM/AC GCC TCT G/ZEN/G TTT TTG TAA AGG TGC TCA CT/3IABkFQ/ -3'.
Prepare PCR reaction mix and add to plate in a PCR hood or other ultraclean PCR preparation area. Do not, under any circumstances, use sjTREC standards in this setup area.
It is very important to have a separate work area for the sjTREC standards. The plasmid is highly concentrated, and contamination of experimental samples or PCR master mix reagents is likely unless separate work areas are established.
Refer to the Bio-Rad iCycler iQ Real-Time PCR Detection System or CFX Manager software user’s manual for specifics on programming and operation of the iCycler or CFX96 real-time thermal cyclers.
Raw fluorescence data will be presented as Ct values, i.e., the PCR cycle at which the FAM reporter signal crosses the threshold setting. System software calculates Ct values of experimental samples and then converts them to number of molecules of sjTREC by comparing sample Ct to the standard curve. No template control wells should have a Ct value of 45 (negative). Sample values will be reported as number of sjTREC molecules per µg of DNA, or number of sjTREC molecules per 50,000 cells. Results (sjTREC/µg DNA or sjTREC/50,000 cells) will vary depending on the experimental samples. Typical sjTREC levels in normal BALB/c thymus tissue and isolated CD4 and CD8 splenocytes throughout aging are detailed in the initial publication of the mouse sjTREC PCR assay (13). The iCycler iQ and the CFX96 systems will give similar results. The linear range of detection of human, mouse and NHP sjTREC in 1 µg DNA or 50,000 cells is 10,000,000 to 100 molecules. A representative standard curve from the CFX96 Real-Time System is shown in Fig. 2. Typical runs on the BioRad systems use auto-calculated baseline determination, and the threshold value is generally placed between 17 and 28. PCR efficiency should be >80%. If these criteria are not met, then the run should be repeated with fresh aliquots of all reagents.
References
- 1.Haynes BF, Denning SM, Le PT, Singer KH. Human intrathymic T cell differentiation. Semin Immunol. 1990;2:67–77. [PubMed] [Google Scholar]
- 2.Flores KG, Li J, Sempowski GD, Haynes BF, Hale LP. Analysis of the human thymic perivascular space during aging. J Clin Invest. 1999;104:1031–1039. doi: 10.1172/JCI7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jamieson BD, Douek DC, Killian S, Hultin LE, Scripture-Adams DD, Giorgi JV, Marelli D, Koup RA, Zack JA. Generation of functional thymocytes in the human adult. Immunity. 1999;10:569–575. doi: 10.1016/s1074-7613(00)80056-4. [DOI] [PubMed] [Google Scholar]
- 4.Sempowski GD, Hale LP, Sundy JS, Massey JM, Koup RA, Douek DC, Patel DD, Haynes BF. Leukemia inhibitory factor, oncostatin M, IL-6, and stem cell factor mRNA expression in human thymus increases with age and is associated with thymic atrophy. J Immunol. 2000;164:2180–2187. doi: 10.4049/jimmunol.164.4.2180. [DOI] [PubMed] [Google Scholar]
- 5.Lynch HE, Goldberg GL, Chidgey A, Van den Brink MR, Boyd R, Sempowski GD. Thymic involution and immune reconstitution. Trends Immunol. 2009;30:366–373. doi: 10.1016/j.it.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douek DC, McFarland RD, Keiser PH, Gage EA, Massey JM, Haynes BF, Polis MA, Haase AT, Feinberg MB, Sullivan JL, Jamieson BD, Zack JA, Picker LJ, Koup RA. Changes in thymic function with age and during the treatment of HIV infection. Nature. 1998;396:690–695. doi: 10.1038/25374. [DOI] [PubMed] [Google Scholar]
- 7.Kong F, Chen CH, Cooper MD. Thymic function can be accurately monitored by the level of recent T cell emigrants in the circulation. Immunity. 1998;8:97–104. doi: 10.1016/s1074-7613(00)80462-8. [DOI] [PubMed] [Google Scholar]
- 8.Douek DC, Vescio RA, Betts MR, Brenchley JM, Hill BJ, Zhang L, Berenson JR, Collins RH, Koup RA. Assessment of thymic output in adults after haematopoietic stem-cell transplantation and prediction of T-cell reconstitution. Lancet. 2000;355:1875–1881. doi: 10.1016/S0140-6736(00)02293-5. [DOI] [PubMed] [Google Scholar]
- 9.Davis CC, Marti LC, Sempowski GD, Jeyaraj DA, Szabolcs P. Interleukin-7 permits Th1/Tc1 maturation and promotes ex vivo expansion of cord blood T cells: a critical step toward adoptive immunotherapy after cord blood transplantation. Cancer research. 2010;70:5249–5258. doi: 10.1158/0008-5472.CAN-09-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Page KM, Mendizabal AM, Prasad VK, Martin PL, Parikh S, Wood S, Sempowski GD, Szabolcs P, Kurtzberg J. Posttransplant autoimmune hemolytic anemia and other autoimmune cytopenias are increased in very young infants undergoing unrelated donor umbilical cord blood transplantation. Biol Blood Marrow Transplant. 2008;14:1108–1117. doi: 10.1016/j.bbmt.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C, Chen BJ, Deoliveira D, Sempowski GD, Chao NJ, Storms RW. Progenitor cell dose determines the pace and completeness of engraftment in a xenograft model for cord blood transplantation. Blood. 2010;116:5518–5527. doi: 10.1182/blood-2009-12-260810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarzotti-Kelsoe M, Win CM, Parrott RE, Cooney M, Moser BK, Roberts JL, Sempowski GD, Buckley RH. Thymic output, T-cell diversity, and T-cell function in long-term human SCID chimeras. Blood. 2009;114:1445–1453. doi: 10.1182/blood-2009-01-199323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sempowski GD, Gooding ME, Liao HX, Le PT, Haynes BF. T cell receptor excision circle assessment of thymopoiesis in aging mice. Mol Immunol. 2002;38:841–848. doi: 10.1016/s0161-5890(01)00122-5. [DOI] [PubMed] [Google Scholar]
- 14.Hockett RD, Jr, Nunez G, Korsmeyer SJ. Evolutionary comparison of murine and human delta T-cell receptor deleting elements. The New biologist. 1989;1:266–274. [PubMed] [Google Scholar]
- 15.de Villartay JP, Hockett RD, Coran D, Korsmeyer SJ, Cohen DI. Deletion of the human T-cell receptor delta-gene by a site-specific recombination. Nature. 1988;335:170–174. doi: 10.1038/335170a0. [DOI] [PubMed] [Google Scholar]
- 16.Toda M, Fujimoto S, Iwasato T, Takeshita S, Tezuka K, Ohbayashi T, Yamagishi H. Structure of extrachromosomal circular DNAs excised from T-cell antigen receptor alpha and delta-chain loci. Journal of molecular biology. 1988;202:219–231. doi: 10.1016/0022-2836(88)90453-6. [DOI] [PubMed] [Google Scholar]
- 17.Takeshita S, Toda M, Yamagishi H. Excision products of the T cell receptor gene support a progressive rearrangement model of the alpha/delta locus. Embo J. 1989;8:3261–3270. doi: 10.1002/j.1460-2075.1989.tb08486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott JF, Rock EP, Patten PA, Davis MM, Chien YH. The adult T-cell receptor delta-chain is diverse and distinct from that of fetal thymocytes. Nature. 1988;331:627–631. doi: 10.1038/331627a0. [DOI] [PubMed] [Google Scholar]
- 19.Min B, McHugh R, Sempowski GD, Mackall C, Foucras G, Paul WE. Neonates support lymphopenia-induced proliferation. Immunity. 2003;18:131–140. doi: 10.1016/s1074-7613(02)00508-3. [DOI] [PubMed] [Google Scholar]
- 20.Chen BJ, Cui X, Sempowski GD, Gooding ME, Liu C, Haynes BF, Chao NJ. A comparison of murine T-cell-depleted adult bone marrow and full-term fetal blood cells in hematopoietic engraftment and immune reconstitution. Blood. 2002;99:364–371. doi: 10.1182/blood.v99.1.364. [DOI] [PubMed] [Google Scholar]
- 21.Billard MJ, Gruver AL, Sempowski GD. Acute endotoxin-induced thymic atrophy is characterized by intrathymic inflammatory and wound healing responses. PloS one. 2011;6:e17940. doi: 10.1371/journal.pone.0017940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi X, Zhang P, Sempowski GD, Shellito JE. Thymopoietic and bone marrow response to murine Pneumocystis pneumonia. Infect Immun. 2011;79:2031–2042. doi: 10.1128/IAI.01213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen BJ, Deoliveira D, Spasojevic I, Sempowski GD, Jiang C, Owzar K, Wang X, Gesty-Palmer D, Cline JM, Bourland JD, Dugan G, Meadows SK, Daher P, Muramoto G, Chute JP, Chao NJ. Growth hormone mitigates against lethal irradiation and enhances hematologic and immune recovery in mice and nonhuman primates. PloS one. 2010;5:e11056. doi: 10.1371/journal.pone.0011056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruver AL, Ventevogel MS, Sempowski GD. Leptin receptor is expressed in thymus medulla and leptin protects against thymic remodeling during endotoxemia-induced thymus involution. J Endocrinol. 2009;203:75–85. doi: 10.1677/JOE-09-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sodora DL, Douek DC, Silvestri G, Montgomery L, Rosenzweig M, Igarashi T, Bernacky B, Johnson RP, Feinberg MB, Martin MA, Koup RA. Quantification of thymic function by measuring T cell receptor excision circles within peripheral blood and lymphoid tissues in monkeys. Eur J Immunol. 2000;30:1145–1153. doi: 10.1002/(SICI)1521-4141(200004)30:4<1145::AID-IMMU1145>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 26.Bona R, Macchia I, Baroncelli S, Negri DR, Leone P, Pavone-Cossut MR, Catone S, Buffa V, Ciccozzi M, Heeney J, Fagrouch Z, Titti F, Cara A. T cell receptor excision circles (TRECs) analysis during acute intrarectal infection of cynomolgus monkeys with pathogenic chimeric simian human immunodeficiency virus. Virus research. 2007;126:86–95. doi: 10.1016/j.virusres.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 27.Aspinall R, Pido-Lopez J, Imami N, Henson SM, Ngom PT, Morre M, Niphuis H, Remarque E, Rosenwirth B, Heeney JL. Old rhesus macaques treated with interleukin-7 show increased TREC levels and respond well to influenza vaccination. Rejuvenation research. 2007;10:5–17. doi: 10.1089/rej.2006.9098. [DOI] [PubMed] [Google Scholar]
- 28.Muthukumar A, Zhou D, Paiardini M, Barry AP, Cole KS, McClure HM, Staprans SI, Silvestri G, Sodora DL. Timely triggering of homeostatic mechanisms involved in the regulation of T-cell levels in SIVsm-infected sooty mangabeys. Blood. 2005;106:3839–3845. doi: 10.1182/blood-2005-01-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sodora DL, Milush JM, Ware F, Wozniakowski A, Montgomery L, McClure HM, Lackner AA, Marthas M, Hirsch V, Johnson RP, Douek DC, Koup RA. Decreased levels of recent thymic emigrants in peripheral blood of simian immunodeficiency virus-infected macaques correlate with alterations within the thymus. J Virol. 2002;76:9981–9990. doi: 10.1128/JVI.76.19.9981-9990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]