Abstract
Myopia, or nearsightedness, is the most common human eye disorder in the world, and is a significant global public health concern. Along with cataract, macular degeneration, infectious disease, and vitamin A deficiency, myopia is one of the most important causes of visual impairment worldwide. Severe or high-grade myopia is a leading cause of blindness because of its associated ocular morbidities of retinal detachment, macular choroidal degeneration, premature cataract, and glaucoma. Ample evidence documents the heritability of the non-syndromic forms of this condition, especially for high-grade myopia, commonly referred to as myopic spherical refractive power of 5–6 diopters or higher. Multiple high-grade myopia genetic loci have been identified, and confirmatory studies identifying high-grade and moderate myopia loci have also occurred. In general, myopia susceptibility genes are unknown with few association studies performed, and without confirmation in other research laboratories or testing of separate patient cohorts.
Keywords: myopia, International Myopia Conference Sek-jin Chew lecture, human molecualr genetics, mapping studies, association studies
Prevalence Rates and Public Health Concern
Myopia is the most common human eye disorder in the world, and its public health and economic impact are considerable.(1) The prevalence of myopia varies because of varied definitions, but in the U.S. adult population the estimated prevalence of at least 25% is supported by multiple studies.(2–8) Females are reported to have an earlier onset and a slightly higher prevalence than males.(3,6,7) Whites have a significantly higher prevalence than African Americans. (9–11) Chinese and Japanese populations have high myopia prevalence rates of > 50–70%.(10,12,13) Ashkenazi Jews, especially Orthodox Jewish males, have shown a higher prevalence than other white U.S. and European populations.(11,14)
Myopia is a significant public health problem, as it is associated with increased risk for visual loss.(1,7,15–18) Myopic chorioretinal degeneration is the 4th most frequent cause of blindness leading to visual services and disability registration, and accounts for 8.8% of all causes of blindness. (18) An estimated 5.6% of blindness among schoolchildren in the U.S. is attributable to myopia.(18)
Substantial resources are required for optical correction of myopia with spectacles, contact lenses, and, more recently, surgical procedures such as LASIK. (19,20) The market for optical aids in the US was estimated to exceed $8 billion in annual sales in 1990; most dollars were spent for the correction of myopia. (19,20) The development of methods for preventing or slowing the onset of myopia, or for limiting its progression is of high significance. (1)
Determinants of Refractive Status
The human eye refractive state depends on the coordinated contributions of the refractive powers of the cornea and lens, axial length (AL), refractive indices of the aqueous and vitreous, and the person’s age.(21) The aqueous and vitreous humor effects are constant - both have a refractive index of 1.336. (21) Thus, the major refractive components are the cornea, lens, and AL. The size, shape, and power of all are determined largely by inheritance. (22) Conformational factors such as intrauterine environment, bony orbits, and eyelids may influence eye shape and growth. (21)
Refractive error in the adult population follows a leptokurtic distribution with the peak around emmetropia (plano spherical refraction). (21) Individual refractive components such as dioptric lenticular and corneal powers, anterior chamber depth (in mm), and AL (in mm) follow more bell-shaped distributions. The average refractive error at birth is approximately +1.00 to 2.00 diopters (D) of hyperopia (far-sightedness), and the AL measures approximately 17 mm. By adulthood, the AL grows to about 24 mm.(23) The corneal diameter of the infant is 10mm compared to the adult size of 12mm. Due to steep curvature, corneal power (keratometry) averages +51.00D at birth and flattens to ~ +44.00 D by 6 weeks of age.(24) Lenticular power averages +34.00 D at birth and decreases to +28.00 D by 6 months of age, and to +21.00 D by adulthood.(24) The process of emmetropization, where the refractive components change in a complementary and coordinated fashion as the eye grows, results in minimal changes in refractive error, as the radius of curvature of the cornea decreases, the refractive powers of the lens and cornea decrease, and the AL increases.(15,25) The postnatal human eye normally maintains an AL of within 2% of its optimal focal point.(26) Usually emmetropia- where a clear image is focused on the retina- is reached at by 12 to 18 months, with no substantial refractive change in normal eyes after 16 years.(24)
Types of Myopia
“Juvenile-onset” myopia most often develops and progresses between the ages of 10 and 16 years, whereas “pathologic” or high–grade myopia usually begins to develop in the perinatal period, and is associated with rapid refractive error myopic shifts before 10–12 years of age. (16,27–29)
“Pathologic” or high-grade myopia (refractive spherical dioptric power of −5.00 D or higher) is a major cause of legal blindness in many developed countries.(6,11,14,18,30–32) It affects 27% to 33% of all myopic eyes, corresponding to a prevalence of 1.7% to 2% in the general population of the U.S.(8) High myopia is especially common in Asia.(18,31,33) In Japan, pathologic or high myopia reportedly affects 1–2% of the general population.(10) Comparative prevalence rates from different countries show variability, but confirm that myopia affects a significant proportion of the population in many countries.(8,16,18,31–33)
Ocular Morbidity
Many investigations have reported on the association of high myopia with cataract, glaucoma, retinal detachment (RD), and posterior staphyloma with retinal degenerative changes. (15,26,34–47) High myopia is associated with progressive and excessive elongation of the globe, which may be accompanied by degenerative changes in the sclera, choroid, Bruch’s membrane, retinal pigment epithelium, and neural retina. Various funduscopic changes within the posterior staphyloma develop in highly myopic eyes. These changes include geographic areas of atrophy of the retinal pigment epithelium (RPE) and choroid, lacquer cracks in Bruch’s membrane, subretinal hemorrhage, and choroidal neovascularization (CNV). Among these fundus lesions, macular CNV is the most common vision-threatening complication of high myopia.(37–41,43). Clinical and histopathologic studies have documented CNV in 4–11% of highly myopic eyes. Relative to emmetropic eyes, an approximate 2-fold increased risk of CNV was estimated for eyes with −1.00 to −2.00 D of myopia, a 4-fold increase with −3.00 to – 4.00 D, and a 9-fold increase with −5.00 to −6.00 D.(14,36,43,48) Poor visual outcome following myopic CNV is not uncommon, often affecting relatively young patients.
RD risk is 3–7 times greater for persons with myopia greater than 5.00 D, relative to myopia of less than −5.00 D. (14,26) Myopia of −5.00 to −10.00 D is associated with a higher likelihood of RD.(14,25) The lifetime risk for RD is estimated to be 1.6% for patients with less than −3.00 D of myopia and 9.3% for those with myopia greater than −5.00 D.(46) A sub-group with lattice degeneration and greater than −5.00 D of myopia have a lifetime risk of 35.9%.(46) The prevalence of lattice degeneration increases with higher myopia using AL as a biometric parameter reflective of refractive error.(14,25,49,50)
Glaucoma was observed in 3% of patients with myopia who had ALs of less than 26.5 mm, in 11% with ALs between 26.5 and 33.5 mm, and in 28% of those with longer lengths.(47)
Role of Environment in Myopic Development
The prevalence of myopia in some populations has increased dramatically from one generation to the next in increasingly industrialized settings, or with increased level of educational achievement.(14,51–54) Assessing the impact of inheritance on myopic development may be confounded by children adopting parental behavioral traits associated with myopia, such as higher-than-average near-work activities (i.e. reading).(55) Observational studies of this risk factor do not fully explain the excessive familial clustering of myopia, however. A detailed assessment of confounding effects and interactions between hereditary and environmental influences in juvenile-onset myopia has shown that near work describes very little of the variance in refractive error compared to parental myopia. (56) In addition, near work exerted no confounding influence on the association between parent and child myopia, indicating that children do not become myopic by adopting parental reading habits. More importantly, there was no significant interaction between parental myopia and near work; reading was weakly and equally associated with myopia regardless of the number of myopic parents. This suggests that children inherit myopia as a trait from parents.
Role of Genetics in Myopic Development
Multiple familial aggregation studies report a positive correlation between parental myopia and myopia in their children, indicating heritable myopia susceptibility.(57–61) Children with a family history of myopia had on average less hyperopia, deeper anterior chambers, and longer vitreous chambers even before be-coming myopic. Yap et al. noted a prevalence of myopia in 7-year-old children of 7.3% when neither parent was myopic, 26.2% when one parent was myopic, and 45% when both parents were myopic. (61)
Multiple familial studies support a high genetic effect for myopia.(62–65) Naiglin et al. performed segregation analysis on 32 French multiplex families with high myopia, and determined an autosomal dominant (AD) mode of inheritance.(66) The λs for myopia (the increase in risk to siblings of a person with a disease compared to the population prevalence) has been estimated to be 4.9–19.8 for sibs for high myopia (−6.00 spherical D or greater), and 1.5–3 for low or common myopia (approximately –1.00 to –3.00 spherical D), suggesting a definite genetic basis for high myopia, and a strong genetic basis for low myopia.(67–68) A high degree of familial aggregation of refraction, particularly myopia, was reported in the Beaver Dam Eye Study population after accounting for the effects of age, sex, and education.(69) Segregation analysis suggested the involvement of multiple genes, rather than a single major gene effect.
Twin studies provide the most compelling evidence that myopia is inherited.(70–74) Multiple studies note an increased concordance of refractive error as well as refractive components (AL, corneal curvature, lens power) in monozygotic twins compared to dizygotic twins. (70–73) Sorsby et al. noted a correlation coefficient for myopia of 0 for control pairs, 0.5 for dizygotic twins, and almost 1.0 for monozygotic twins in a study of 78 monozygotic twin pairs and 40 dizygotic twin pairs. (72) Twin studies estimate a high heritability value for myopia, (the proportion of the total phenotypic variance that is attributed to genetic variance) of between 0.5–0.96.(70–74)
Ocular Refractive Component Genetics
Refraction is determined by coordinated contributions of ocular biometric components such as AL, anterior chamber depth (ACD), corneal curvature (keratometry readings in diopters), and lens thickness. Multiple reports have examined familial aggregation and heritability of ocular components. (63,73–75) These may be intimately related to the phenotype of myopia, and the genetic effects of these traits should be examined.
Several studies have reported an inverse relationship of AL to refraction (the longer the eye, the more myopic the refractive error). (76–80) AL is the largest contributor to the determination of refractive error. (81,82) Estimates of heritability for AL range from 40–94%. (63,73,74,84) A study of 3 large Sardinian families found modest evidence for linkage on chromosome 2p24 with a LOD score of 2.64. (84) A chromosome 5q locus for AL was noted in a recent whole genome twin study. (83) AL includes ACD, and studies have shown that increased ACD has an inverse relationship as well to refractive error.(80) The heritability reports for ACD range from 70–94% (63,73,74,80,84), and the same Sardinian study found modest linkage evidence to chromosome 1p32.2 with a LOD score of 2.32.(84)
Corneal curvature steepness is more likely to result in myopia, as hyperopic eyes are more likely to have flatter corneal curvatures. (76,84) Heritability estimates for corneal curvature range from 60 to 92%. (63,73,74,81) The same Sardinian study noted modest linkage of corneal curvature to chromosomes 2p25, 3p26, and 7q22 with LOD scores ranging from 2.34 to 2.50. (81) Increased lens thickness correlates with increased myopia. (80) A di- and monozygotic twin study showed 90–93% heritability for lens thickness.(73)
Molecular Genetic Studies of Human Myopia
Much of the current information on human myopia molecular genetics can be drawn from our studies of familial high myopia, usually defined as myopic spherical refractive error of at least - 5.00 D. An X-linked recessive form of myopia, named the Bornholm (Denmark) eye disease (BED), was designated the first myopia locus (MYP1-OMIM 310460) on chromosome Xq28. (86) Collaborating with BED researchers, we made comparative molecular genetic haplotype and sequence analyses of a large Minnesota family of Danish descent that showed significant linkage of myopia to chromosome Xq27.3-q28. The phenotype of both families appears to be due to a novel cone dysfunction, and not simple myopia. (87) The genetic etiology of each family appears to be distinct, as the haplotypes were different. (87) A report by Michaelides et al. confirms the different X-linked cone dysfunction syndrome with an associated high myopia phenotype as distinct from the BED in 4 families. (88) We identified the first AD locus for non-syndromic high myopia within a 7.6 cm region on chromosome 18p11.31 (MYP2, OMIM 160700) in 7 U.S. families. (89) This locus was confirmed in Chinese Hong Kong and Italian Sardinian cohorts. (90,91) Using the Hong Kong cohort, investigators identified TGIF-beta as the implicated gene for MYP1 using limited SNP association studies and exonic sequencing. (90) Our group fully sequenced TGIF-beta in our cohort of original MYP1 families, and found no associations with high myopia affection status. (92) An Australian lab also investigated the association of TGIF with refraction and ocular biometric measurements in a Caucasian case-control population, and found no association. (93) No association of TGIF to high myopia case control studies was found in Japanese and Chinese cohorts. (94,95) A second locus for AD high myopia mapped to a 30.1 cM region on chromosome 12q21–23 (MYP3, OMIM 603221) in an American family of German-Italian descent also in our laboratory. (96) This locus was confirmed in a high myopia Caucasian British cohort, and in a large German family. (97,98) A statistically suggestive third locus for AD high myopia was reported on chromosome 7q36 in a Caucasian French cohort (MYP4, OMIM 608367).(99) A fourth AD locus on chromosome 17q21–23 (MYP5, OMIM 608474) was determined in a large multigenerational English-Canadian family in our laboratory. (100) We identified a locus for AD high-grade myopia on chromosome 2q37 (MYP12, OMIM 609994) in a large, multigenerational U.S. Caucasian family. (101) This locus was replicated in an Australian study of three multigenerational AD moderate myopia families. (102) We recently published a new locus (MYP17) on chromosome 10q21.2 in a Hutterite (Austrian /German) colony. (103) Loci on chromosomes Xq23–25(104) (MYP13, OMIM 300613) and 4q22–27(105) (MYP11, OMIM 609994) have also recently been identified by Zhang et al. in ethnic Chinese families. (104,105) Loci identified to date for isolated non-syndromic high-grade myopia are primarily AD and highly penetrant.
At least two studies have shown nominal or no linkage of low to moderate myopia to many of the known high myopia loci. Mutti et al. genotyped 53 common myopia families (at least one child with more myopia than −0.75D in each meridian) using the highest intra-interval LOD score microsatellite markers for the chromosome 18p and 12q loci and did not establish linkage. (106) Ibay et al. found no strong evidence of linkage to the chromosomes 18p, 12q, 17q, and 7q in a cohort of 38 Ashkenazi Jewish families with mild/moderate myopia (−1.00D or more). (107) These studies suggest that different genes account for mild/moderate myopia susceptibility or development, or that the genes effects are too small to be detected with the relatively small sample sizes. However, a recent study showed replication of our chromosome 2q37 locus in a cohort of Caucasian Australians with moderate to low myopia. (102)
Three whole-genome mapping studies have identified several candidate gene intervals for common, juvenile-onset myopia using spherical refractive error data. The results of these studies demonstrate the potential for determining molecular genetic factors implicated in myopia at all levels of severity. These studies however, used microsatellite genotyping instead of SNP technology, a limited cohort sample size, and two used homogenous isolated populations. One study was a genome screen of 44 families of Ashkenazi Jewish descent. (108) Individuals with at least –1.00D of myopic spherical refractive error were classified as affected. Their strongest signal localized to chromosome 22q12 (MYP6, OMIM 608908) - (HLOD = 3.56; NPL = 4.62). Eight additional regions (14q, 4q22-q28, 8q22.2, 10q22, 11q23, 13q22, 14q32, and 17qter) had nominal linkage evidence. Hammond et al. evaluated 221 dizygotic twin pairs with moderate myopia and found significant linkage to 4 loci, with a maximum LOD score of 6.1 on chromosome 11p13-MYP7, OMIM 609256).(74) Other identified loci mapped to chromosomes 3q26-MYP8, OMIM 609257 (LOD 3.7), 4q12-MYP9, OMIM 609258 (LOD 3.3), and 8p23-MYP10, OMIM 609259 (LOD 4.1). This group found that the PAX6 gene at the chromosome 11p13 locus showed linkage with 5 SNPs, but no association. They suggested that PAX6 (a major eye development gene) may play a role in myopia development, possibly due to genetic variation in an upstream promoter or regulator. This group later reported that neither the master control genes PAX6 or SOX2 were implicated in common myopia in a large population study cohort. (109) A report replicated the chromosome 8p23 myopia locus (MYP10) in an isolated Pennsylvania Amish population of 34 families. (110) These identified loci are prioritized in our association studies.
Gene Expression and Functional Studies Related to Myopia and Eye Growth
One impediment to correlating genotypic data with actual tissue histopathology in human myopia is that the tissue of interest (i.e. retina/ sclera) can not be directly sampled. Animal models of myopia have been developed to be used as surrogates, although it is unclear how correlative induced myopia in animals may be to physiologic myopia in humans. Animal studies over the past 30 years in juvenile and newborn monkey, tree shrew, and chick models have revealed an active emmetropization mechanism that normally achieves and maintains a match of the eye AL to the eye’s optical power so that the photoreceptors are in focus for distant objects. (111–115) This control mechanism begins in the retina, where neurons (perhaps a subset of amacrine cells detect focused versus defocused images. (116) Constant hyperopic defocus uniformly (across species) produces retinal signals that pass, in a signaling cascade that is not well understood, through the retinal pigment epithelium (RPE) and choroid (116) to remodel the scleral extracellular matrix and cause axial elongation. (117,118) This, in turn, reduces the hyperopic defocus, so that this feedback system is self-limiting, resulting in a match of AL to optical power. Genes expressed in retina, RPE, choroid, and/or sclera that are involved in the normal emmetropization process could be involved in myopia development with irregular expression so that the emmetropization mechanism is disrupted, causing the eye to elongate and become myopic. (117,120–122) The retina is suspected of having the highest likelihood of harboring candidate genes that influence scleral growth based on multiple studies, including optic nerve sectioning in animal models. (123) Genes within significant intervals will be ranked using information from human retinal gene expression databases and publications such as Ret Net (http://www.sph.uth.tmc.edu/RetNet/), (124–126) and retinal SAGE libraries. (127)
Mouse Studies of Myopia
Factors that regulate rate and duration of eye growth in mice have revealed two loci (Eye1 and Eye2) that may be responsible for larger eye size. (128–130) Human syntenic homologous regions are chromosomes 6p, 16q13.3, and 19q13 for Eye2, and chromosome 7q for Eye1.
The first knock-out mouse model for relative myopia (131) was based on form-deprivation experiments in chickens, mice, and rhesus macaque monkeys. The immediate early gene transcription factor ZENK (also known as Egr-1) is implicated in the feedback mechanisms for visual control of axial eye growth and myopia development. (132–134) ZENK is up-regulated in retinal amacrine cells when axial eye growth is inhibited by positive lens wear, and is down-regulated when axial growth is enhanced by negative lenses, suggesting that ZENK is linked to an axial eye growth inhibitory signal. ZENK knockout mice had longer eyes and a relative myopic shift relative to heterozygous and wild-type mice with identical genetic background. (131) ZENK maps to a significantly linked loci at chromosome 5q31.1 from our international consortium dataset. We recently selected ZENK SNPs and performed Taqman genotyping association studies in a Duke myopia dataset, and found no association. We did not find sequence variants after mutation screening. (Data not shown.)
Association Studies for Non-syndromic Myopia
The list of hypothesized candidate genes for myopia, based largely on the current understanding of the pathophysiology of syndromic myopia, is relatively small. The vast majority of the work examining the relationship between candidate genes and individual polymorphisms in candidate genes has been performed on single candidates at a time, to the exclusion of other independent or interacting genes. Two myopia susceptibility genes were identified in Chinese cohorts using the family-based transmission disequilibrium test (TDT) approach (135) - the hepatocyte growth factor (which maps to the homologous human mouse Eye 2 locus at chromosome 7q21.1, and myocillin-a gene which has been implicated in juvenile-onset glaucoma, and can be associated with early-onset myopia. (136,137) Our lab has performed candidate gene SNP genotyping of these two genes using TaqMan on our collection of 120 Duke families (primarily Caucasian) to perform TDT family-based association testing, and we found no association with the same SNPs used in the primary studies. (Data not shown) The collagen type 1 alpha 1 gene (COL1A1) is an extracellular matrix gene expressed in the scleral wall, and maps within the MYP5 locus for high myopia on chromosome 17q22-q23.3. A case-control study of a Japanese cohort showed association with high myopia for this gene with 2 SNPs. (138) Another case control study of mixed ethnicities showed a 2 SNP association to the collagen 2 alpha 1 gene (COL2A1), which maps to chromosome 12q13.11, a locus not associated with myopia to date. (139) We have also performed TaqMan SNP genotyping association studies in our family cohort for these two scleral genes using the same SNPs as the original studies, and have found inconclusive evidence for association to either gene. No functional SNP effects were implicated for any of these candidate genes, and it is unclear how ethnic differences play a role in the degree of associative significance.
Scleral Gene Expression Studies
We published human and mouse ocular tissue expression studies to report candidate genes that may be relevant to myopic ocular shape change as it relates to scleral wall expansion. This was to develop a catalog of genes with an extracellular matrix associated function in sclera, given that many syndromic forms of myopia have connective tissue-associated gene mutations. Genes that are important for constituent organization and maintenance of connective tissue function may be physiologically important in heritable myopic eye development. To expand the candidate gene catalog for myopia susceptibility genes, our laboratory performed the first comprehensive gene expression studies in human sclera, describing genes expressed in a cDNA library, and using microarray technology and reverse-transcription polymerase chain reaction (RT-PCR) of mRNA extracted from human sclera. (140,141) We also described the first comprehensive effort to determine scleral eye growth genes differentially expressed in developmentally staged mice using microarray gene chip technology.(142) Ocular developmental candidate genes (MFRP, SOX2, CHX10) were screened in extreme states of refractive error/ eye growth.
International High Myopia Consortium
Our laboratory established an international consortium for studying the genetics of high-grade myopia. This was accomplished in collaboration with Drs. Thomas Rosenberg of the Gordon Norrie Disease Centre for Genetic Eye Diseases in Hellerup Denmark, Jeremy Guggenheim at the Cardiff University in Wales UK, David Mackey of the University of Melbourne, Australia, and Patrick Calvas and Francois Malacaze of the Toulouse University in France. We submitted an application to the NIH Center for Inherited Disease Research (CIDR) in July 2005 to request WGG of our combined 250+ multiplex families with heritable high-grade myopia. CIDR approval was granted, (CIDR contract # N01-HG-65403). This study was approved by all Institutional Review Boards at each study site.
Overall Dataset
Although several linkage scans for high-grade myopia have been reported, all were based on microsatellite markers and a small number of families. We recently completed whole genome genotyping of our international high myopia dataset with a release of 5,928 out of 6008 SNPs attempted using the Illumina Linkage Panel IVb, which has an average SNP inter-marker distance coverage of 0.65cM, or 521kb. Our dataset consisted of 261 multiplex families (at least two affected individuals per family). A total of 1568 samples were released. For experimental samples, a total of 1400 out of 1457 samples were attempted, 81 out of 81 blind duplicates were attempted, and 87 Centre d'tudes du Polymorphisme Humain, France (CEPH) DNA controls (http://ccr.coriell.org/Sections/Collections/NIGMS/CEPHDNAPools.aspx) were used. Total genotypes released was 9,295,104. There was a missing data rate of 0.20% missing genotypes/ total genotypes. The missing data rate varied by sample tissue-type of 0.28% for buccal, 0.25% for saliva, 0.16% for CEPH, and 0.18% for blood. The Mendelian consistency rate was 99.98%, and the blind duplicate reproducibility rate was 99.99%. To our knowledge, this is the largest dataset used for a linkage study for high myopia. Objective measurements for spherical equivalent, sphere, cylinder, axis, and axial lengths were documented. (Some individuals were plano in each eye and therefore had no measurable refractive error.) Keratometry data was obtained by all sites except Cardiff. Axial length was not obtained at the Cardiff and Hellerup sites, and not consistently measured on subjects at the Duke and Toulouse sites. The diagnosis of myopia was determined by the spherical refractive error. Since our target phenotype is high myopia, we defined affected individuals as having a minus spherical refractive error less than or equal to −5.00 diopters for the less affected eye. Unaffected individuals had a plus spherical refractive error greater than or equal to 0 D. The mean number of subjects per pedigree is 5, the range of spherical refractive error is −50.00D to +7.00D. Table 1 summarizes the phenotypic characteristics of the dataset.
Table 1.
Mean, standard deviation, and sample size (in parentheses) for each ocular measurement from right (OD) and left (OS) eye by ascertainment site. Duke= DUK, Cardiff=CARD, Toulouse= TOU, Hellerup=HEL, Melbourne=MEL.
| Center | DUK | CARD | TOU | HEL | MEL | |
|---|---|---|---|---|---|---|
| Sample size | 656 | 252 | 315 | 163 | 15 | |
|
Sphere (Diopters) |
OD | −4.95± 6.0 (627) |
−6.48±4.62 (234) |
−5.12±5.89 (181) |
−4.73± 5.43 (144) |
−4.03± 2.54 (10) |
| OS | −4.96± 5.93 (627) |
−6.39±4.66 (235) |
−4.83±5.66 (178) |
−5.17± 5.22 (144) |
−3.53± 4.39 (9) |
|
|
Cylinder (Diopters) |
OD | 0.8± 0.94 (614) |
1.09± 1.05 (221) |
0.84±0.97 (153) |
1.12± 0.35 (121) |
2.53± 2.28 (10) |
| OS | 0.77± 0.9 (610) |
1.05± 0.91 (220) |
0.92±1.03 (147) |
1.12± 0.99 (124) |
2.08± 2.14 (9) |
|
|
Axial Length (millimeters) |
OD | 25.48± 2.41 (197) |
-- | 25.10± 2.58 (152) |
-- | 24.76± 0.25 (3) |
| OS | 25.76± 2.45 (193) |
-- | 25.11± 2.68 (151) |
-- | 24.32± 2.17 (3) |
|
|
Keratometry* (Diopters) |
OD | 43.71± 3.57 (244) |
-- | 43.30±2.22 (114) |
43.18± 1.73 (130) |
43.67± 2.23 (5) |
| OS | 43.63± 3.74 (249) |
-- | 43.30± 2.22 (113) |
43.26± 1.63 (132) |
43.11± 1.02 (5) |
|
The mean and standard deviation of the keratometry measurements was based on the average of the keratometry 1 (K1) and keratometry 2 (K2) readings of each participant
Table 2 provides the descriptive statistical summary for each phenotype obtained from all clinical sites.
Table 2.
Summary of mean, standard deviation and heritability for each quantitative phenotype.
| Phenotype | No. of Samples | Mean | Std Dev | Residual heritability (h2r) | P-value of h2r |
|---|---|---|---|---|---|
| Spherical Equivalent (right eye) |
1,258 | −4.57 | 5.54 | 0.24 | 3.09×10−9 |
| Spherical Equivalent (left eye) |
1,258 | −4.6 | 5.51 | 0.23 | 2.76×10−8 |
| Sphere (right eye) | 1,224 | −5.08 | 5.65 | 0.25 | 3.03×10−9 |
| Sphere (left eye) | 1,221 | −5.09 | 5.62 | 0.25 | 1.67×10−8 |
| Cylinder (right eye) | 827 | 1.11 | 1.33 | 0.09 | 0.175 |
| Cylinder (left eye) | 822 | 1.05 | 0.99 | 0.19 | 0.002 |
| Axis (right eye) | 826 | 85.46 | 58.36 | 0.07 | 0.118 |
| Axis (left eye) | 823 | 86.94 | 59.15 | 0.19 | 0.003 |
Both eyes show similar mean and standard deviation (SD) for each phenotypic variable. We also performed polygenic analysis using the SOLAR program to estimate residual heritability for each phenotypic variable. As shown in Table 2, only the right eye cylinder and axis have non-significant residual heritability. Based on our experience with other linkage studies of quantitative traits, the likelihood of identifying linkage regions is higher if the residual heritability of a trait is greater than 20%. From these preliminary data, we believe that spherical equivalence and sphere are the best quantitative traits to analyze. Furthermore, these data prove that these quantitative traits are genetic.
A series of quality control programs were implemented before linkage analyses, which included examining family relationships using the RELPAIR and PREST programs, Mendelian inconsistency testing using PedCheck and PREST software, and Hardy-Weinberg equilibrium testing using the Genetic Data Analysis (GDA) program. (143–146)
The final dataset used for linkage analysis was based on 249 families (137 DUK, 46 CARD, 40 TOU, 25 HEL and 1 MEL) and 5796 markers. The dataset contained 10 Asian, 12 African-American, and 227 Caucasian families.
We performed two point parametric linkage analysis using the FASTLINK and HOMOG programs (http://linkage.rockefeller.edu). (147) Since the mode of inheritance for high myopia is unknown, parametric analyses were performed using both autosomal dominant (AD) and autosomal recessive (AR) models with disease allele frequencies of 0.01 for both models. The MERLIN program was used to perform both parametric and non-parametric multipoint linkage analysis, and the same AD and AR models were assumed in the MERLIN parametric analysis. (148) It is known that linkage disequilibrium (LD) may inflate the type I error of multipoint linkage analysis. (149) The current version of MERLIN has an option to take into account marker-marker LD to reduce its impact on the LOD score. The threshold of correlation due to LD (r2) between markers was set at 0.16 in the multipoint linkage analysis. Overall, and multiple stratified center and race-specific datasets were analyzed, (DUK, CARD, TOU, MEL, HEL and Asian, African-American, Caucasian).
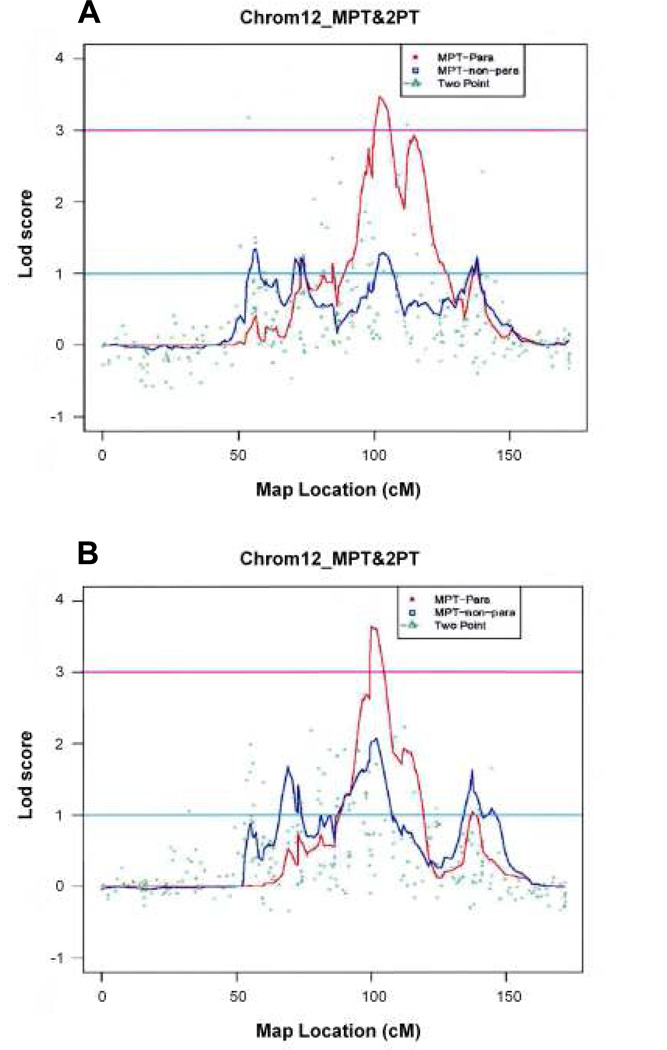
Overall, 61 markers across 19 chromosomes had two point HLOD scores ≥2, and 18 linkage regions from 12 chromosomes had peak multipoint HLOD or NPL scores ≥ 1.5. SNP rs2063239 on chromosome 12 had the highest two point HLOD score (HLOD=4.80). The multipoint analysis corresponded to the two point results for chromosome 12 delineating a 36.59cM linkage region with the SNP peak marker rs337663 (HLOD=3.48). This region overlaps with regions identified by multipoint parametric linkage analyses for DUK (peak HLOD=3.62, rs1489895), Asian (peak HLOD =1.55, rs930248), and Caucasian (peak HLOD=2.34, rs337663) subsets as well as nonparametric linkage analysis for the DUK subset (peak NPL=2.07, rs337663). The chromosome 12q22 linkage region was contracted to a 9.94cM region from rs163016 to rs1520724 where it is shared by four datasets (overall, DUK, Asian, and Caucasian) (see Figure 1). Other regions of interest emanating from the CIDR linkage study were located on chromosomes 2, 5, 9, and X. Two regions were found in chromosomes 2, 5, and X. Several linkage regions were notable for high lod scores in single datasets, including: chromosomes 5 (1st region) and X (2nd region) from DUK; chromosomes 2 (2nd region), 4, and X (1st region) from CARD; chromosome 14 from TOU; chromosome 1 from HEL; and chromosomes 1 and 3 from the Asian dataset. Replicated MYP loci from this study include: MYP1 on chromosome Xq27.3–28; MYP3 on chromosome 12q21–23; MYP5 on chromosome 17q21–23; MYP9 on chromosome 4q12; MYP12 on chromosome 2q37; MYP14 on chromosome 1q36 and MYP15 on chromosome 1q41. The results of this international consortium genotyping are in press. (150)
Figure 1.
The Center for Inherited Research (CIDR) International Family High Myopia linkage study analyses show strong confirmation for linkage of one or more high-grade myopia susceptibility genes to the MYP3 locus at chromosome 12q21–23. The major contributors to this locus with two peaks are from the Duke and Hellerup sites. The Melbourne, Toulouse and Cardiff sites did not have significant linkage to this locus. The strongest consensus linkage region is near 101.97cM, which replicates previous MYP3 locus analyses.
Chromosome 12 Linkage
As mentioned earlier, my lab established linkage for the MYP3 locus to chromosome 12q21–23 in 1998 in a single large, multigenerational Caucasian family with autosomal dominant high-grade myopia. (96) This locus was replicated in 3 independent labs. (97) (Eric Yap-Singapore, and Paul Baird-Australia-personal communication) It was recently replicated in a Hong Kong Chinese cohort (Pang CP, et al. Poster 1397–2007 American Society of Human Genetics Meeting, 2007). Our SNP genotyping of the international myopia cohort showed the strongest linkage region at the MYP3 locus.
These analyses show strong confirmation for linkage of one or more high-grade myopia susceptibility genes to the MYP3 locus. The major contributors to this locus with two peaks are from the Duke and Hellerup sites. The Melbourne, Toulouse and Cardiff sites did not have significant linkage to this locus. Interestingly, a subset of the Cardiff cohort previously showed suggestive microsatellite linkage to this chromosomal interval. For race-specific analysis, Asian and African American categories did not have significant linkage. The strongest consensus linkage region is near 101.97cM, which replicates previous MYP3 locus analyses. This interval was contracted to a 23.5cM region from the original 31 cM microsatellite-derived MYP3 locus. The Duke site provides the highest contribution. Future studies will focus on SNP fine-mapping association analysis of the 23.5 cM interval using Illumina Golden Gate custom chip arrays.
MYP1 Locus on Chromosome Xq27.3–28
We have more fully characterized and refined the interval (Xq27.3–28) for the first high myopia locus MYP1, which is an X-linked high-grade myopia associated with a novel mild cone dysfunction and color vision deficits (protanopia for the Bornholm Eye Disease pedigree, and deuteranopia in my large Minnesota pedigree). (86–88) We have sequence-screened multiple genes (SPRY3, THMLE, TKTL1, RENBP, SYBL1, MECP, CXorf2) for mutations within the MYP1 locus interval. More than 40 of approximately 300 X-linked diseases map to this region. The red and green opsin (visual cone pigment) genes are located in a head-to-tail tandem array on chromosome Xq28. (151) Our most promising candidate gene TEX28 (or CXorf2), is a nested gene within this cone pigment gene array. This gene was more highly expressed in SAGE libraries of the human macular retina relative to peripheral retina (SAGE experiments courtesy of Dr. Cathy Bowes-Rickman-Duke University Eye Center). CXorf2 is comprised of five exons that almost span the entire distance between the protein-coding regions of the opsin genes and a transketolase-related gene, and encodes a polypeptide of 410 amino acid residues. (Figure 2.)
Figure 2.
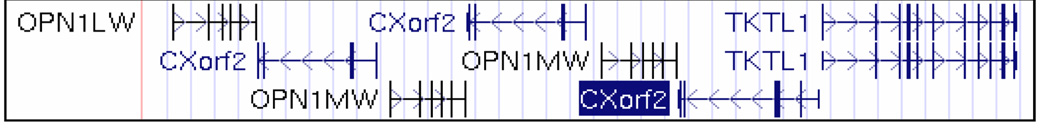
Schematic representation of the arrangement of Opsin gene array and CXorf2 genes on chromosome Xq28 locus source - UCSC genome browser. OPN1LW= opsin 1 low wavelength –red cone pigment gene, OPN1MW =opsin 1 medium wavelength –green cone pigment gene, TKTL1= transketolase-related gene. Vertical bars are exons, horizontal bars are introns, and arrow heads indicate the direction of transcription. A color version of this figure is available online at www.optvissci.com.
We investigated whether the X-linked myopia phenotype with possible cone dysfunction is a consequence of disruption of the CXorf2 gene. More specifically, we screened CXorf2 for any mutations using automated sequencing, and for any variations in copy number at the genomic DNA level using real-time PCR and the commercial NimbleGen comparative genomic hybridization (CGH) SNP panel for the X chromosome. (NimbleGen Systems, Madison WI).
Primer pairs were designed to amplify the identified exons (including at least 100 bp of each intron-exon boundary) of CXorf2. Polymerase chain reactions were performed on genomic DNA and amplified products were visualized using agarose gel electrophoresis. Amplicons were purified with Quickstep™ 2 SOPE™ Resin (Edge BioSystems, Gaithersburg, MD) and sequenced using BigDye™ Terminator on ABI3730 or 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). Sequences were trimmed for quality and aligned using Sequencher™ (Gene Codes, Ann Arbor, MI). Normal and affected individual DNA sequences were compared alongside the known reference sequence (University of California Santa Cruz genome browser- http://genome.ucsc.edu/) for CXorf2.
After preliminary screening, we noted a single nucleotide polymorphism (SNP) in the 5’ untranslated region (UTR) of exon 2 of the CXorf2 gene that segregates with the affection status in the X-linked myopia phenotype with protanopia. This was revealed in all affected males (4) and a carrier female screened in this pedigree, while the unaffected males (3) and marry-in controls (2) did not show this polymorphism. In addition, further screening of 107 external controls did not reveal this polymorphism. A second X-linked myopia family was provided to us for gene screening by the M. Michaelides and Anthony Moore and colleagues at Moorfields Eye Institute in London England with the same phenotype which also showed this UTR polymorphism in the affected males only. 5’UTRs are deemed to be crucial determinants of mRNA stability and translation efficiency. (152,153) Furthermore, polymorphisms in 5’UTRs have been proven to be associated with disease. (154,155) Hence, we suspect that the polymorphism we observe in CXorf2 could be regulatory in nature, and these results indicate that CXorf2 is a potential candidate gene for this phenotype.
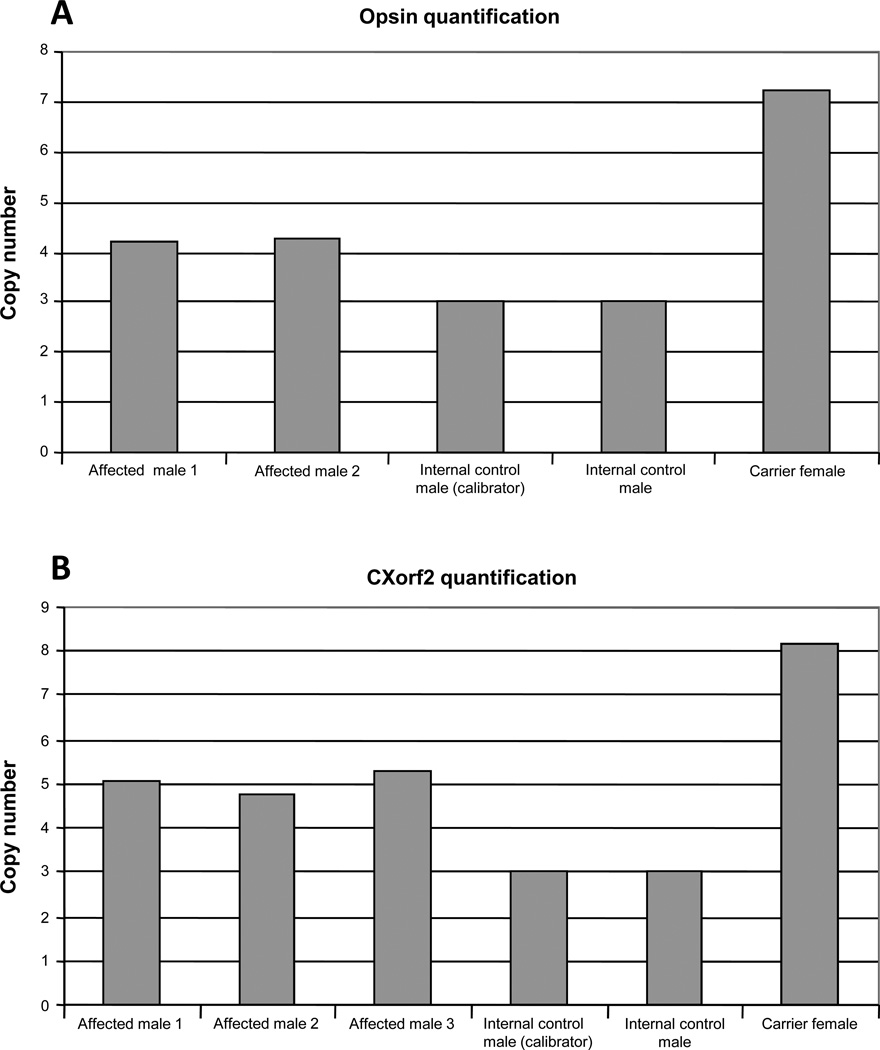
To examine the copy number variation of CXorf2 gene, gene expression assays (Assays-by-Design™) consisting of a mix of unlabeled PCR primers and TaqMan® MGB probe (FAM™ dye-labeled) were designed on CXorf2 sequence to quantitate the pattern of expression at the genomic DNA level. A similar assay targeted on the opsin genes was also designed to validate the sensitivity of the experiment as the number of copies of opsins has been previously determined by single-strand conformational polymorphism testing in the current pedigree. (87) An assay on GAPDH (housekeeping gene) served as an endogenous reference. All real-time PCR reactions were performed with Taqman® Universal PCR Master Mix on ABI7900HT Fast Real-time PCR System (Applied Biosystems, Foster City, CA). Data were analysed using the ‘Comparative CT method’ where the amount of target normalized to an endogenous reference and relative to a calibrator (known individual value) is calculated as 2−(deltadeltaCT) (User bulletin #2, ABI Sequence Detection Systems) [deltadeltaCT is the difference between the normalized CT (cycle threshold) of an unknown sample and the normalized CT of the calibrator]. Results were plotted as the number of copies present in each individual with respect to the assigned number of copies in the calibrator. The assay targeted on the opsin genes (Figure 2) revealed that employing real-time custom gene expression assays is a reliable way to study copy number variation. The number of copies expected to be present for the opsin genes were observed in affected males (4 copies), control males (3 copies) and a carrier female (7 copies). (87) There are minimal studies on CXorf2 in the literature, and only 3 copies of CXorf2 have been reported within the opsin gene array.(156,157) Figure 3a shows quantification of opsin gene expression by real-time PCR. Expression pattern follows the number of copies previously reported by Young et al. (84) Figure 3b shows quantification of CXorf2 gene expression by real-time PCR. All affected males show an expression of up to 5 copies of CXorf2. The calibrator was assigned a value of 3 copies based on and relative to the findings of the number of known copies of opsin gene array. Our results with the assay targeted on the CXorf2 gene indicate that the affected male individuals in this pedigree express up to 5 copies of CXorf2 (Figure 3). This finding was also found in the second pedigree from Moorfields, but was not found in the Bornholm Eye Disease pedigree. The copy number variations were confirmed using the NimbleGen array CGH product as well, which has an average SNP probe spacing for chromosome X of 340bp. (Figure 4). Copy number variations have been proposed to play a role in disease inheritance and susceptibility to disease due to their ability to affect genic dosage. (158) Hence, we believe that these CXorf2 gene alterations may be responsible for this phenotype. (159)
Figure 3.
(A) Quantification of opsin gene expression by real-time polymerase chain reaction. Expression pattern follows the number of copies previously reported by Young et al 2004. 87 (B) Quantification of CXorf2 gene expression by real-time polymerase chain reaction. All affected males show an expression of up to 5 copies of CXorf2. The calibrator was assigned a value of 3 copies based on and relative to the findings of the number of known copies of opsin gene array.
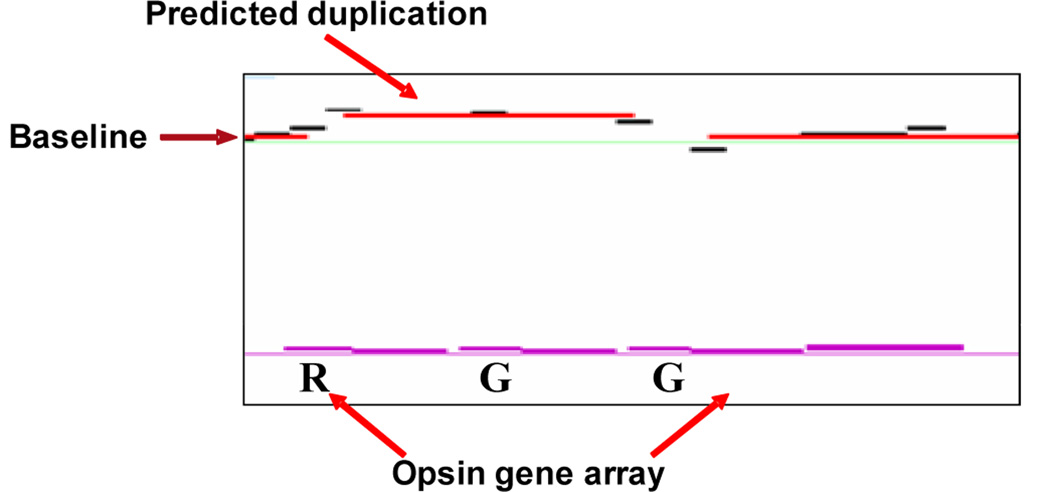
Figure 4.
Array comparative genomic hybridization (CGH) y-axis represents log2 ratio calculated by subtracting the log value of the reference sample intensity from the log of the test sample intensity. R= red opsin gene, G= green opsin gene. A color version of this figure is available online at www.optvissci.com.
MYP2 Locus on Chromosome 18p11.31
We have screened almost all of the known candidate genes within the expanded 7.6cM interval of the MYP2 chromosome 18p11.31, and have found no mutations or associated polymorphisms of significance that travel with the high myopic affection status. None of the sequence variants found co-segregated with the affected phenotype. Specifically, there were no heterozygous or homozygous polymorphisms observed only in affected individuals in any MYP2 pedigree. An interim summary of the screening progress was published in 2004.(160) Since moving to Duke, the CLUL1, MRCL3, MRLC2, YES1, C18ORF2, METTL4, KNTC2, ENOSF1, ADCYAP1, TYMS, CR627458, COLEC12, THOC1, and CETN1 genes were screened in unrelated controls and in representative DNA samples of high myopia affected and unaffected family members from the original 7 MYP2 pedigrees. All candidate genes were screened by direct sequencing. Consistent segregation of a mutation with the disease was not demonstrated in any of the 7 families. Novel SNPs were found and submitted to the dbSNP database.
Despite replication from 2 independent studies for this locus, the results from the international high myopia dataset using the Illumina IVb SNP linkage panel have not supported the presence of a linked interval at chromosome 18p11.31. Rationale for this ranges from limited SNP spacing of the commercial Illumina panel with inadequate interval coverage, the strong likelihood of a monogenic basis for the limited number of families (or single family) used for this initial microsatellite-identified locus, and the possibility of locus race-specific associations (the initial study showed significant linkage in one large Chinese family alone, and one of the replication studies was a Hong Kong Chinese cohort). The literature is replete with examples of initial microsatellite results not confirmed in another sample set or analysis. (161,162)
MYP5 locus on chromosome 17q21–23
Two genes have been screened-DHX40, and SEPT4. No sequence variants were found to segregate or to be associated with the disease phenotype.
As stated before, significant association (p <0.05) under a model assuming dominance to 2 COL1A1 SNPs (rs2075555 – G allele in intron 11 with p = 0.0071 and OR =1.36, and rs2269336 - G allele in the 5” upstream region with p = 0.014 and OR =1.31) was found in a case/ control Japanese high myopia cohort. (139) We genotyped the same 8 SNPs using the TaqMan protocol on 141 high myopia Duke families with no pedigree or Mendelian error inconsistencies previously screened by the CIDR genotyping described above. The SNPs selected are shown in Figure 5, and genotyping results are shown in Table 3.
Figure 5.
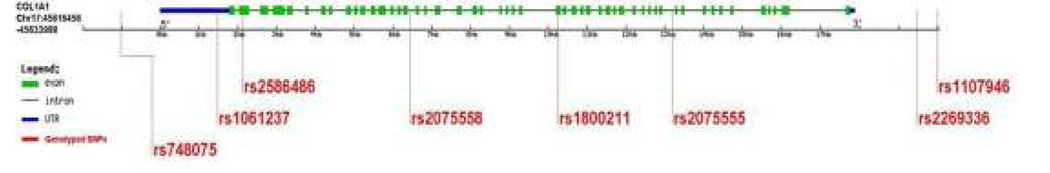
Human COL1A1 gene structure. The size of each exon and intron is not to scale. Vertical lines show 8 single nucleotide polymorphisms (SNPs) with their respective public reference SNP rs number from the dbSNP database.
Table 3.
Association analysis of 8 single nucleotide polymorphisms (SNPs) of COL1A1 gene with high myopia for a dataset of 120 Duke families. Those SNPs in bold had significant association in a Japanese cohort. (Inamori 2007) 138
| SNP rs | Public Position (bp) | Function | Amino Acid Change | Allele | PDT | P-value GenoPDT |
APL |
|---|---|---|---|---|---|---|---|
| rs748075 | 45615428 | 3'Downstream | C/G | 0.32 | 0.62 | 0.99 | |
| rs1061237 | 45617774 | 3' UTR | A/G | 0.35 | 0.44 | 0.87 | |
| rs2586486 | 45618215 | Exon | Lys [K]-->Gln [Q] | G/T | 1 | 1 | 0.29 |
| rs2075558 | 45622584 | Intron | A/C | 0.59 | 0.45 | 0.21 | |
| rs1800211 | 45626379 | Exon | His [H]-->Arg [R] | C/T | 1 | 1 | 0.02 |
| rs2075555 | 45629290 | Intron | G/T | 0.30 | 0.15 | 0.65 | |
| rs2269336 | 45635355 | 5' Upstream | C/G | 0.28 | 0.38 | 0.32 | |
| rs1107946 | 45635989 | 5' Upstream | A/C | 0.41 | 0.44 | 0.9 |
The rs1800211 SNP showed moderate significant association in our cohort, but we did not replicate significance of the 2 SNPs more highly associated in the Japanese cohort. This warrants further investigation, as the rs1800211 SNP is exonic, and exhibits potential functional implications for the protein.
SUMMARY
No results have implicated more than just a single gene, or have expanded into an analysis of a specific pathway. This may implicate additional genes in a pathway also shown to be risk factors for myopia in validation studies and point to potential haplotype-specific treatments. No candidate genes have been shown to account for even a modest fraction of the familial risk of myopia, and for most the data are conflicting about whether a true association exists. The genes underlying all high-grade myopia peaks except possibly for chromosome Xq28 remain unidentified, suggesting substantial genetic heterogeneity. Consideration is needed to assess the role of environmental factors to genetic influences, such as interactions of early-age near-work and genotype. (10,163) Consideration also needs to be given to the identification of phenotypes indicating etiologically homogeneous subgroups, e.g. early age-of-onset, with/without retinal degenerative changes, or classification by individual response to treatments that reduce accommodation to near objects, such as progressive addition lens use. (164) Candidate gene studies underscore that myopia is very complex, in fact, so complex that single candidate gene studies are unlikely to demonstrate the type of relationships needed to account for the majority of susceptibility genes. There is a need for a genome-wide approach, incorporating candidate genes but not restricted to the study of candidate genes, to explore the relative contributions and interactions between known candidate genes and possibly novel genes in increased myopia susceptibility.
ACKNOWLEDGEMENTS
I extend many thanks to the members of the myopia families for their cooperation in the multiple projects. This study was supported by grants from National Institute of Health (NIH) (RO1 EY014685), NIH CIDR contract # N01-HG-65403, and the Research to Prevent Blindness, Inc.
REFERENCES
- 1.Pararajasegaram R. VISION 2020-the right to sight: from strategies to action. Am J Ophthalmol. 1999;128:359–360. doi: 10.1016/s0002-9394(99)00251-2. [DOI] [PubMed] [Google Scholar]
- 2.Vitale S, Ellwein L, Cotch MF, Ferris FL, 3rd, Sperduto R. Prevalence of refractive error in the United States, 1999–2004. Arch Ophthalmol. 2008;126:1111–1119. doi: 10.1001/archopht.126.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Q, Klein BE, Klein R, Moss SE. Refractive status in the Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 1994;35:4344–4347. [PubMed] [Google Scholar]
- 4.Sperduto RD, Seigel D, Roberts J, Rowland M. Prevalence of myopia in the United States. Arch Ophthalmol. 1983;101:405–407. doi: 10.1001/archopht.1983.01040010405011. [DOI] [PubMed] [Google Scholar]
- 5.Angle J, Wissmann DA. The epidemiology of myopia. Am J Epidemiol. 1980;111:220–228. doi: 10.1093/oxfordjournals.aje.a112889. [DOI] [PubMed] [Google Scholar]
- 6.Katz J, Tielsch JM, Sommer A. Prevalence and risk factors for refractive errors in an adult inner city population. Invest Ophthalmol Vis Sci. 1997;38:334–340. [PubMed] [Google Scholar]
- 7.Goss DA, Winkler RL. Progression of myopia in youth: age of cessation. Am J Optom Physiol Opt. 1983;60:651–658. doi: 10.1097/00006324-198308000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Kempen JH, Mitchell P, Lee KE, Tielsch JM, Broman AT, Taylor HR, Ikram MK, Congdon NG, O'Colmain BJ. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol. 2004;122:495–505. doi: 10.1001/archopht.122.4.495. [DOI] [PubMed] [Google Scholar]
- 9.Saw SM, Katz J, Schein OD, Chew SJ, Chan TK. Epidemiology of myopia. Epidemiol Rev. 1996;18:175–187. doi: 10.1093/oxfordjournals.epirev.a017924. [DOI] [PubMed] [Google Scholar]
- 10.Baldwin WR. A review of statistical studies of relations between myopia and ethnic, behavioral, and physiological characteristics. Am J Optom Physiol Opt. 1981;58:516–527. doi: 10.1097/00006324-198107000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Zylbermann R, Landau D, Berson D. The influence of study habits on myopia in Jewish teenagers. J Pediatr Ophthalmol Strabismus. 1993;30:319–322. doi: 10.3928/0191-3913-19930901-12. [DOI] [PubMed] [Google Scholar]
- 12.Saw SM. A synopsis of the prevalence rates and environmental risk factors for myopia. Clin Exp Optom. 2003;86:289–294. doi: 10.1111/j.1444-0938.2003.tb03124.x. [DOI] [PubMed] [Google Scholar]
- 13.Wong TY, Foster PJ, Johnson GJ, Seah SK. Education, socioeconomic status, and ocular dimensions in Chinese adults: the Tanjong Pagar Survey. Br J Ophthalmol. 2002;86:963–968. doi: 10.1136/bjo.86.9.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burton TC. The influence of refractive error and lattice degeneration on the incidence of retinal detachment. Trans Am Ophthalmol Soc. 1989;87:143–155. [PMC free article] [PubMed] [Google Scholar]
- 15.Curtin BJ. Posterior staphyloma development in pathologic myopia. Ann Ophthalmol. 1982;14:655–658. [PubMed] [Google Scholar]
- 16.Curtin B. Myopia: a review of its etiology, pathogenesis and treatment. Surv Ophthalmol. 1970;15:1–17. [Google Scholar]
- 17.Ghafour IM, Allan D, Foulds WS. Common causes of blindness and visual handicap in the west of Scotland. Br J Ophthalmol. 1983;67:209–213. doi: 10.1136/bjo.67.4.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokoro T, Sato A. Results of Investigation of Pathologic Myopia in Japan: Report of Myopic Chorioretinal Atrophy. Tokyo: Ministry of Health and Welfare; 1982. pp. 32–35. [Google Scholar]
- 19.National Advisory Eye Council, Strabismus, Amblyopia and Visual Processing Panel. Vision Research: A National Plan. 1999–2003. NIH Publication 98–4120. Washington, DC: National Institutes of Health; 2004. [Google Scholar]
- 20.Javitt JC, Chiang YP. The socioeconomic aspects of laser refractive surgery. Arch Ophthalmol. 1994;112:1526–1530. doi: 10.1001/archopht.1994.01090240032022. [DOI] [PubMed] [Google Scholar]
- 21.Sorsby A, Leary GA, Richards MJ. Correlation ametropia and component ametropia. Vision Res. 1962;2:309–313. [Google Scholar]
- 22.Tron EJ. The optical elements of the refractive power of the eye. In: Ridley F, Sorsby A, editors. Modern Trends in Ophthalmology. New York: Paul B. Hoeber; 1940. p. 245. [Google Scholar]
- 23.Jansson F. Measurements of intraocular distances by ultrasound. Acta Ophthalmol Suppl. 1963;(suppl 74):1–51. [PubMed] [Google Scholar]
- 24.Gordon RA, Donzis PB. Refractive development of the human eye. Arch Ophthalmol. 1985;103:785–789. doi: 10.1001/archopht.1985.01050060045020. [DOI] [PubMed] [Google Scholar]
- 25.Mutti DO, Mitchell GL, Jones LA, Friedman NE, Frane SL, Lin WK, Moeschberger ML, Zadnik K. Axial growth and changes in lenticular and corneal power during emmetropization in infants. Invest Ophthalmol Vis Sci. 2005;46:3074–3080. doi: 10.1167/iovs.04-1040. [DOI] [PubMed] [Google Scholar]
- 26.Curtin BJ. The Myopias: Basic Science and Clinical Management. Harper & Row; 1985. [Google Scholar]
- 27.Grosvenor T. A review and a suggested classification system for myopia on the basis of age-related prevalence and age of onset. Am J Optom Physiol Opt. 1987;64:545–554. doi: 10.1097/00006324-198707000-00012. [DOI] [PubMed] [Google Scholar]
- 28.Mantyjarvi MI. Changes of refraction in schoolchildren. Arch Ophthalmol. 1985;103:790–792. doi: 10.1001/archopht.1985.01050060050022. [DOI] [PubMed] [Google Scholar]
- 29.Gwiazda J, Thorn F, Bauer J, Held R. Emmetropization and the progression of manifest refraction in children followed from infancy to puberty. Clin Vis Sci. 1993;8:337–344. [Google Scholar]
- 30.Leibowitz HM, Krueger DE, Maunder LR, Milton RC, Kini MM, Kahn HA, Nickerson RJ, Pool J, Colton TL, Ganley JP, Loewenstein JI, Dawber TR. The Framingham Eye Study monograph: An ophthalmological and epidemiological study of cataract, glaucoma, diabetic retinopathy, macular degeneration, and visual acuity in a general population of 2631 adults, 1973–1975. Surv Ophthalmol. 1980;24:335–610. [PubMed] [Google Scholar]
- 31.Lin LL, Chen CJ, Hung PT, Ko LS. Nation-wide survey of myopia among schoolchildren in Taiwan, 1986. Acta Ophthalmol Suppl. 1988;185:29–33. doi: 10.1111/j.1755-3768.1988.tb02657.x. [DOI] [PubMed] [Google Scholar]
- 32.Fledelius HC. Myopia prevalence in Scandinavia. A survey, with emphasis on factors of relevance for epidemiological refraction studies in general. Acta Ophthalmol Suppl. 1988;185:44–50. doi: 10.1111/j.1755-3768.1988.tb02661.x. [DOI] [PubMed] [Google Scholar]
- 33.Wilson A, Woo G. A review of the prevalence and causes of myopia. Singapore Med J. 1989;30:479–484. [PubMed] [Google Scholar]
- 34.Grossniklaus HE, Green WR. Pathologic findings in pathologic myopia. Retina. 1992;12:127–133. doi: 10.1097/00006982-199212020-00009. [DOI] [PubMed] [Google Scholar]
- 35.Gass JDM. Steroscopic Atlas of Macular Diseases: Diagnosis and Treatment. 4th ed. Vol.1. Mosby: St. Louis; 1997. [Google Scholar]
- 36.Steidl SM, Pruett RC. Macular complications associated with posterior staphyloma. Am J Ophthalmol. 1997;123:181–187. doi: 10.1016/s0002-9394(14)71034-7. [DOI] [PubMed] [Google Scholar]
- 37.Rabb MF, Garoon I, LaFranco FP. Myopic macular degeneration. Int Ophthalmol Clin. 1981;21:51–69. doi: 10.1097/00004397-198102130-00006. [DOI] [PubMed] [Google Scholar]
- 38.Noble KG, Carr RE. Pathologic myopia. Ophthalmology. 1982;89:1099–1100. doi: 10.1016/s0161-6420(82)34677-1. [DOI] [PubMed] [Google Scholar]
- 39.Jalkh AE, Weiter JJ, Trempe CL, Pruett RC, Schepens CL. Choroidal neovascularization in degenerative myopia: role of laser photocoagulation. Ophthalmic Surg. 1987;18:721–725. [PubMed] [Google Scholar]
- 40.Hayasaka S, Uchida M, Setogawa T. Subretinal hemorrhages with or without choroidal neovascularization in the maculas of patients with pathologic myopia. Graefes Arch Clin Exp Ophthalmol. 1990;228:277–280. doi: 10.1007/BF00920048. [DOI] [PubMed] [Google Scholar]
- 41.Hotchkiss ML, Fine SL. Pathologic myopia and choroidal neovascularization. Am J Ophthalmol. 1981;91:177–183. doi: 10.1016/0002-9394(81)90170-7. [DOI] [PubMed] [Google Scholar]
- 42.Levy JH, Pollock HM, Curtin BJ. The Fuchs' spot: an ophthalmoscopic and fluorescein angiographic study. Ann Ophthalmol. 1977;9:1433–1443. [PubMed] [Google Scholar]
- 43.Avila MP, Weiter JJ, Jalkh AE, Trempe CL, Pruett RC, Schepens CL. Natural history of choroidal neovascularization in degenerative myopia. Ophthalmology. 1984;91:1573–1581. doi: 10.1016/s0161-6420(84)34116-1. [DOI] [PubMed] [Google Scholar]
- 44.Fried M, Siebert A, Meyer-Schwickerath G. A natural history of Fuchs’ spot: a long-term follow-up study. Doc Ophthalmol Proc Ser. 1981;28:215–221. [Google Scholar]
- 45.Hampton GR, Kohen D, Bird AC. Visual prognosis of disciform degeneration in myopia. Ophthalmology. 1983;90:923–926. doi: 10.1016/s0161-6420(83)80018-9. [DOI] [PubMed] [Google Scholar]
- 46.Perkins ES. Morbidity from myopia. Sight Sav Rev. 1979;49:11–19. [PubMed] [Google Scholar]
- 47.Perkins ES. Glaucoma in the younger age groups. Arch Ophthalmol. 1960;64:882–891. doi: 10.1001/archopht.1960.01840010884008. [DOI] [PubMed] [Google Scholar]
- 48.Vongphanit J, Mitchell P, Wang JJ. Prevalence and progression of myopic retinopathy in an older population. Ophthalmology. 2002;109:704–711. doi: 10.1016/s0161-6420(01)01024-7. [DOI] [PubMed] [Google Scholar]
- 49.Pierro L, Camesasca FI, Mischi M, Brancato R. Peripheral retinal changes and axial myopia. Retina. 1992;12:12–17. doi: 10.1097/00006982-199212010-00003. [DOI] [PubMed] [Google Scholar]
- 50.Spitznas M, Boker T. Idiopathic posterior subretinal neovascularization (IPSN) is related to myopia. Graefes Arch Clin Exp Ophthalmol. 1991;229:536–538. doi: 10.1007/BF00203317. [DOI] [PubMed] [Google Scholar]
- 51.Lin LL, Hung PT, Ko LS, Hou PK. Study of myopia among aboriginal school children in Taiwan. Acta Ophthalmol Suppl. 1988;185:34–36. doi: 10.1111/j.1755-3768.1988.tb02658.x. [DOI] [PubMed] [Google Scholar]
- 52.Young FA. Myopia and personality. Am J Optom Arch Am Acad Optom. 1967;44:192–201. doi: 10.1097/00006324-196703000-00005. [DOI] [PubMed] [Google Scholar]
- 53.Young FA. Reading, measures of intelligence and refractive errors. Am J Optom Arch Am Acad Optom. 1963;40:257–264. doi: 10.1097/00006324-196305000-00002. [DOI] [PubMed] [Google Scholar]
- 54.Rosner M, Belkin M. Intelligence, education, and myopia in males. Arch Ophthalmol. 1987;105:1508–1511. doi: 10.1001/archopht.1987.01060110054030. [DOI] [PubMed] [Google Scholar]
- 55.Wallman J. Parental history and myopia: taking the long view. JAMA. 1994;272:1255–1256. doi: 10.1001/jama.1994.03520160039037. [DOI] [PubMed] [Google Scholar]
- 56.Mutti DO, Mitchell GL, Moeschberger ML, Jones LA, Zadnik K. Parental myopia, near work, school achievement, and children's refractive error. Invest Ophthalmol Vis Sci. 2002;43:3633–3640. [PubMed] [Google Scholar]
- 57.Goss DA, Jackson TW. Clinical findings before the onset of myopia in youth: 4. Parental history of myopia. Optom Vis Sci. 1996;73:279–282. doi: 10.1097/00006324-199604000-00010. [DOI] [PubMed] [Google Scholar]
- 58.Pacella R, McLellan J, Grice K, Del Bono EA, Wiggs JL, Gwiazda JE. Role of genetic factors in the etiology of juvenile-onset myopia based on a longitudinal study of refractive error. Optom Vis Sci. 1999;76:381–386. doi: 10.1097/00006324-199906000-00017. [DOI] [PubMed] [Google Scholar]
- 59.Zadnik K. The Glenn A. Fry Award Lecture (1995). Myopia development in childhood. Optom Vis Sci. 1997;74:603–608. [PubMed] [Google Scholar]
- 60.Zadnik K, Satariano WA, Mutti DO, Sholtz RI, Adams AJ. The effect of parental history of myopia on children's eye size. JAMA. 1994;271:1323–1327. [PubMed] [Google Scholar]
- 61.Yap M, Wu M, Liu ZM, Lee FL, Wang SH. Role of heredity in the genesis of myopia. Ophthalmic Physiol Opt. 1993;13:316–319. doi: 10.1111/j.1475-1313.1993.tb00479.x. [DOI] [PubMed] [Google Scholar]
- 62.Goldschmidt E. [On the etiology of myopia. An epidemiological study] Acta Ophthalmol (Copenh) 1968;1(Suppl):11–172. [PubMed] [Google Scholar]
- 63.Teikari J, O'Donnell JJ, Kaprio J, Koskenvuo M. Genetic and environmental effects on oculometric traits. Optom Vis Sci. 1989;66:594–599. doi: 10.1097/00006324-198909000-00005. [DOI] [PubMed] [Google Scholar]
- 64.Ashton GC. Segregation analysis of ocular refraction and myopia. Hum Hered. 1985;35:232–239. doi: 10.1159/000153551. [DOI] [PubMed] [Google Scholar]
- 65.Goss DA, Hampton MJ, Wickham MG. Selected review on genetic factors in myopia. J Am Optom Assoc. 1988;59:875–884. [PubMed] [Google Scholar]
- 66.Naiglin L, Clayton J, Gazagne C, Dallongeville F, Malecaze F, Calvas P. Familial high myopia: evidence of an autosomal dominant mode of inheritance and genetic heterogeneity. Ann Genet. 1999;42:140–146. [PubMed] [Google Scholar]
- 67.Guggenheim JA, Kirov G, Hodson SA. The heritability of high myopia: a reanalysis of Goldschmidt's data. J Med Genet. 2000;37:227–231. doi: 10.1136/jmg.37.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farbrother JE, Kirov G, Owen MJ, Guggenheim JA. Family aggregation of high myopia: estimation of the sibling recurrence risk ratio. Invest Ophthalmol Vis Sci. 2004;45:2873–2878. doi: 10.1167/iovs.03-1155. [DOI] [PubMed] [Google Scholar]
- 69.Klein AP, Duggal P, Lee KE, Klein R, Bailey-Wilson JE, Klein BE. Support for polygenic influences on ocular refractive error. Invest Ophthalmol Vis Sci. 2005;46:442–446. doi: 10.1167/iovs.04-0794. [DOI] [PubMed] [Google Scholar]
- 70.Teikari JM, O'Donnell J, Kaprio J, Koskenvuo M. Impact of heredity in myopia. Hum Hered. 1991;41:151–156. doi: 10.1159/000153994. [DOI] [PubMed] [Google Scholar]
- 71.Sorsby A, Sheridan M, Leary GA. Refraction and Its Components in Twins. Medical Research Council Special Report. London: Her Majesty's Stationery Office, Medical Research Counsil; 1962. [Google Scholar]
- 72.Sorsby A, Leary GA, Fraser GR. Family studies on ocular refraction and its components. J Med Genet. 1966;3:269–273. doi: 10.1136/jmg.3.4.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lyhne N, Sjolie AK, Kyvik KO, Green A. The importance of genes and environment for ocular refraction and its determiners: a population based study among 20–45 year old twins. Br J Ophthalmol. 2001;85:1470–1476. doi: 10.1136/bjo.85.12.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hammond CJ, Snieder H, Gilbert CE, Spector TD. Genes and environment in refractive error: the twin eye study. Invest Ophthalmol Vis Sci. 2001;42:1232–1236. [PubMed] [Google Scholar]
- 75.Hammond CJ, Andrew T, Mak YT, Spector TD. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am J Hum Genet. 2004;75:294–304. doi: 10.1086/423148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carney LG, Mainstone JC, Henderson BA. Corneal topography and myopia. A cross-sectional study. Invest Ophthalmol Vis Sci. 1997;38:311–320. [PubMed] [Google Scholar]
- 77.Grosvenor T, Scott R. Role of the axial length/corneal radius ratio in determining the refractive state of the eye. Optom Vis Sci. 1994;71:573–579. doi: 10.1097/00006324-199409000-00005. [DOI] [PubMed] [Google Scholar]
- 78.Mainstone JC, Carney LG, Anderson CR, Clem PM, Stephensen AL, Wilson MD. Corneal shape in hyperopia. Clin Exp Optom. 1998;81:131–137. doi: 10.1111/j.1444-0938.1998.tb06731.x. [DOI] [PubMed] [Google Scholar]
- 79.Strang NC, Schmid KL, Carney LG. Hyperopia is predominantly axial in nature. Curr Eye Res. 1998;17:380–383. doi: 10.1080/02713689808951218. [DOI] [PubMed] [Google Scholar]
- 80.Wong TY, Foster PJ, Ng TP, Tielsch JM, Johnson GJ, Seah SK. Variations in ocular biometry in an adult Chinese population in Singapore: the Tanjong Pagar Survey. Invest Ophthalmol Vis Sci. 2001;42:73–80. [PubMed] [Google Scholar]
- 81.Touzeau O, Allouch C, Borderie V, Kopito R, Laroche L. [Correlation between refraction and ocular biometry] J Fr Ophtalmol. 2003;26:355–363. [PubMed] [Google Scholar]
- 82.Olsen T, Arnarsson A, Sasaki H, Sasaki K, Jonasson F. On the ocular refractive components: the Reykjavik Eye Study. Acta Ophthalmol Scand. 2007;85:361–366. doi: 10.1111/j.1600-0420.2006.00847.x. [DOI] [PubMed] [Google Scholar]
- 83.Biino G, Palmas MA, Corona C, Prodi D, Fanciulli M, Sulis R, Serra A, Fossarello M, Pirastu M. Ocular refraction: heritability and genome-wide search for eye morphometry traits in an isolated Sardinian population. Hum Genet. 2005;116:152–159. doi: 10.1007/s00439-004-1231-6. [DOI] [PubMed] [Google Scholar]
- 84.Zhu G, Hewitt AW, Ruddle JB, Kearns LS, Brown SA, Mackinnon JR, Chen CY, Hammond CJ, Craig JE, Montgomery GW, Martin NG, Mackey DA. Genetic dissection of myopia: evidence for linkage of ocular axial length to chromosome 5q. Ophthalmology. 2008;115:1053–1057. doi: 10.1016/j.ophtha.2007.08.013. e2. [DOI] [PubMed] [Google Scholar]
- 85.Grosvenor T, Goss DA. Role of the cornea in emmetropia and myopia. Optom Vis Sci. 1998;75:132–145. doi: 10.1097/00006324-199802000-00017. [DOI] [PubMed] [Google Scholar]
- 86.Schwartz M, Haim M, Skarsholm D. X-linked myopia: Bornholm eye disease. Linkage to DNA markers on the distal part of Xq. Clin Genet. 1990;38:281–286. [PubMed] [Google Scholar]
- 87.Young TL, Deeb SS, Ronan SM, Dewan AT, Alvear AB, Scavello GS, Paluru PC, Brott MS, Hayashi T, Holleschau AM, Benegas N, Schwartz M, Atwood LD, Oetting WS, Rosenberg T, Motulsky AG, King RA. X-linked high myopia associated with cone dysfunction. Arch Ophthalmol. 2004;122:897–908. doi: 10.1001/archopht.122.6.897. [DOI] [PubMed] [Google Scholar]
- 88.Michaelides M, Johnson S, Bradshaw K, Holder GE, Simunovic MP, Mollon JD, Moore AT, Hunt DM. X-linked cone dysfunction syndrome with myopia and protanopia. Ophthalmology. 2005;112:1448–1454. doi: 10.1016/j.ophtha.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 89.Young TL, Ronan SM, Alvear AB, Wildenberg SC, Oetting WS, Atwood LD, Wilkin DJ, King RA. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet. 1998;63:1419–1424. doi: 10.1086/302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lam DS, Lee WS, Leung YF, Tam PO, Fan DS, Fan BJ, Pang CP. TGFbeta-induced factor: a candidate gene for high myopia. Invest Ophthalmol Vis Sci. 2003;44:1012–1015. doi: 10.1167/iovs.02-0058. [DOI] [PubMed] [Google Scholar]
- 91.Heath S, Robledo R, Beggs W, Feola G, Parodo C, Rinaldi A, Contu L, Dana D, Stambolian D, Siniscalco M. A novel approach to search for identity by descent in small samples of patients and controls from the same mendelian breeding unit: a pilot study on myopia. Hum Hered. 2001;52:183–190. doi: 10.1159/000053375. [DOI] [PubMed] [Google Scholar]
- 92.Scavello GS, Paluru PC, Ganter WR, Young TL. Sequence variants in the transforming growth beta-induced factor (TGIF) gene are not associated with high myopia. Invest Ophthalmol Vis Sci. 2004;45:2091–2097. doi: 10.1167/iovs.03-0933. [DOI] [PubMed] [Google Scholar]
- 93.Pertile KK, Schache M, Islam FM, Chen CY, Dirani M, Mitchell P, Baird PN. Assessment of TGIF as a candidate gene for myopia. Invest Ophthalmol Vis Sci. 2008;49:49–54. doi: 10.1167/iovs.07-0896. [DOI] [PubMed] [Google Scholar]
- 94.Li J, Zhang QJ, Xiao XS, Li JZ, Zhang FS, Li SQ, Li W, Li T, Jia XY, Guo L, Guo XM. [The SNPs analysis of encoding sequence of interacting factor gene in Chinese population] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2003;20:454–456. [PubMed] [Google Scholar]
- 95.Hasumi Y, Inoko H, Mano S, Ota M, Okada E, Kulski JK, Nishizaki R, Mok J, Oka A, Kumagai N, Nishida T, Ohno S, Mizuki N. Analysis of single nucleotide polymorphisms at 13 loci within the transforming growth factor-induced factor gene shows no association with high myopia in Japanese subjects. Immunogenetics. 2006;58:947–953. doi: 10.1007/s00251-006-0155-9. [DOI] [PubMed] [Google Scholar]
- 96.Young TL, Ronan SM, Drahozal LA, Wildenberg SC, Alvear AB, Oetting WS, Atwood LD, Wilkin DJ, King RA. Evidence that a locus for familial high myopia maps to chromosome 18p. Am J Hum Genet. 1998;63:109–119. doi: 10.1086/301907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Farbrother JE, Kirov G, Owen MJ, Pong-Wong R, Haley CS, Guggenheim JA. Linkage analysis of the genetic loci for high myopia on 18p, 12q, and 17q in 51 U.K. families. Invest Ophthalmol Vis Sci. 2004;45:2879–2885. doi: 10.1167/iovs.03-1156. [DOI] [PubMed] [Google Scholar]
- 98.Nurnberg G, Jacobi FK, Broghammer M, Becker C, Blin N, Nurnberg P, Stephani U, Pusch CM. Refinement of the MYP3 locus on human chromosome 12 in a German family with Mendelian autosomal dominant high-grade myopia by SNP array mapping. Int J Mol Med. 2008;21:429–438. [PubMed] [Google Scholar]
- 99.Naiglin L, Gazagne C, Dallongeville F, Thalamas C, Idder A, Rascol O, Malecaze F, Calvas P. A genome wide scan for familial high myopia suggests a novel locus on chromosome 7q36. J Med Genet. 2002;39:118–124. doi: 10.1136/jmg.39.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Paluru P, Ronan SM, Heon E, Devoto M, Wildenberg SC, Scavello G, Holleschau A, Makitie O, Cole WG, King RA, Young TL. New locus for autosomal dominant high myopia maps to the long arm of chromosome 17. Invest Ophthalmol Vis Sci. 2003;44:1830–1836. doi: 10.1167/iovs.02-0697. [DOI] [PubMed] [Google Scholar]
- 101.Paluru PC, Nallasamy S, Devoto M, Rappaport EF, Young TL. Identification of a novel locus on 2q for autosomal dominant high-grade myopia. Invest Ophthalmol Vis Sci. 2005;46:2300–2307. doi: 10.1167/iovs.04-1423. [DOI] [PubMed] [Google Scholar]
- 102.Chen CY, Stankovich J, Scurrah KJ, Garoufalis P, Dirani M, Pertile KK, Richardson AJ, Baird PN. Linkage replication of the MYP12 locus in common myopia. Invest Ophthalmol Vis Sci. 2007;48:4433–4439. doi: 10.1167/iovs.06-1188. [DOI] [PubMed] [Google Scholar]
- 103.Nallasamy S, Paluru PC, Devoto M, Wasserman NF, Zhou J, Young TL. Genetic linkage study of high-grade myopia in a Hutterite population from South Dakota. Mol Vis. 2007;13:229–236. [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. A new locus for autosomal dominant high myopia maps to 4q22-q27 between D4S1578 and D4S1612. Mol Vis. 2005;11:554–560. [PubMed] [Google Scholar]
- 105.Zhang Q, Li S, Xiao X, Jia X, Guo X. Confirmation of a genetic locus for X-linked recessive high myopia outside MYP1. J Hum Genet. 2007;52:469–472. doi: 10.1007/s10038-007-0130-9. [DOI] [PubMed] [Google Scholar]
- 106.Mutti DO, Semina E, Marazita M, Cooper M, Murray JC, Zadnik K. Genetic loci for pathological myopia are not associated with juvenile myopia. Am J Med Genet. 2002;112:355–360. doi: 10.1002/ajmg.10683. [DOI] [PubMed] [Google Scholar]
- 107.Ibay G, Doan B, Reider L, Dana D, Schlifka M, Hu H, Holmes T, O'Neill J, Owens R, Ciner E, Bailey-Wilson JE, Stambolian D. Candidate high myopia loci on chromosomes 18p and 12q do not play a major role in susceptibility to common myopia. BMC Med Genet. 2004;5:20. doi: 10.1186/1471-2350-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stambolian D, Ibay G, Reider L, Dana D, Moy C, Schlifka M, Holmes T, Ciner E, Bailey-Wilson JE. Genomewide linkage scan for myopia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 22q12. Am J Hum Genet. 2004;75:448–459. doi: 10.1086/423789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Simpson CL, Hysi P, Bhattacharya SS, Hammond CJ, Webster A, Peckham CS, Sham PC, Rahi JS. The Roles of PAX6 and SOX2 in Myopia: lessons from the 1958 British Birth Cohort. Invest Ophthalmol Vis Sci. 2007;48:4421–4425. doi: 10.1167/iovs.07-0231. [DOI] [PubMed] [Google Scholar]
- 110.Stambolian D, Ciner EB, Reider LC, Moy C, Dana D, Owens R, Schlifka M, Holmes T, Ibay G, Bailey-Wilson JE. Genome-wide scan for myopia in the Old Order Amish. Am J Ophthalmol. 2005;140:469–476. doi: 10.1016/j.ajo.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 111.Wallman J. Retinal factors in myopia and emmetropization: clues from research on chicks. In: Grosvenor TP, Flom MC, editors. Refractive Anomalies: Research and Clinical Applications. Boston: Butterworth-Heinemann; 1991. pp. 268–286. [Google Scholar]
- 112.Wallman J, McFadden S. Monkey eyes grow into focus. Nat Med. 1995;1:737–739. doi: 10.1038/nm0895-737. [DOI] [PubMed] [Google Scholar]
- 113.Wallman J, Wildsoet C, Xu A, Gottlieb MD, Nickla DL, Marran L, Krebs W, Christensen AM. Moving the retina: choroidal modulation of refractive state. Vision Res. 1995;35:37–50. doi: 10.1016/0042-6989(94)e0049-q. [DOI] [PubMed] [Google Scholar]
- 114.Wildsoet CF. Active emmetropization—evidence for its existence and ramifications for clinical practice. Ophthalmic Physiol Opt. 1997;17:279–290. [PubMed] [Google Scholar]
- 115.Norton TT. Animal models of myopia: learning how vision controls the size of the eye. ILAR J. 1999;40:59–77. doi: 10.1093/ilar.40.2.59. [DOI] [PubMed] [Google Scholar]
- 116.Fischer AJ, McGuire JJ, Schaeffel F, Stell WK. Light- and focus-dependent expression of the transcription factor ZENK in the chick retina. Nat Neurosci. 1999;2:706–712. doi: 10.1038/11167. [DOI] [PubMed] [Google Scholar]
- 117.Mertz JR, Wallman J. Choroidal retinoic acid synthesis: a possible mediator between refractive error and compensatory eye growth. Exp Eye Res. 2000;70:519–527. doi: 10.1006/exer.1999.0813. [DOI] [PubMed] [Google Scholar]
- 118.Siegwart JT, Jr, Norton TT. Regulation of the mechanical properties of tree shrew sclera by the visual environment. Vision Res. 1999;39:387–407. doi: 10.1016/s0042-6989(98)00150-3. [DOI] [PubMed] [Google Scholar]
- 119.Siegwart JT, Jr, Norton TT. The time course of changes in mRNA levels in tree shrew sclera during induced myopia and recovery. Invest Ophthalmol Vis Sci. 2002;43:2067–2075. [PMC free article] [PubMed] [Google Scholar]
- 120.Gentle A, Liu Y, Martin JE, Conti GL, McBrien NA. Collagen gene expression and the altered accumulation of scleral collagen during the development of high myopia. J Biol Chem. 2003;278:16587–16594. doi: 10.1074/jbc.M300970200. [DOI] [PubMed] [Google Scholar]
- 121.Guggenheim JA, McBrien NA. Form-deprivation myopia induces activation of scleral matrix metalloproteinase-2 in tree shrew. Invest Ophthalmol Vis Sci. 1996;37:1380–1395. [PubMed] [Google Scholar]
- 122.Norton TT, Rada JA. Reduced extracellular matrix in mammalian sclera with induced myopia. Vision Res. 1995;35:1271–1281. doi: 10.1016/0042-6989(94)00243-f. [DOI] [PubMed] [Google Scholar]
- 123.Wallman J. Retinal control of eye growth and refraction. Prog Retin Res. 1993;12:133–153. [Google Scholar]
- 124.Sohocki MM, Malone KA, Sullivan LS, Daiger SP. Localization of retina/pineal-expressed sequences: identification of novel candidate genes for inherited retinal disorders. Genomics. 1999;58:29–33. doi: 10.1006/geno.1999.5810. [DOI] [PubMed] [Google Scholar]
- 125.den Hollander AI, van Driel MA, de Kok YJ, van de Pol DJ, Hoyng CB, Brunner HG, Deutman AF, Cremers FP. Isolation and mapping of novel candidate genes for retinal disorders using suppression subtractive hybridization. Genomics. 1999;58:240–249. doi: 10.1006/geno.1999.5823. [DOI] [PubMed] [Google Scholar]
- 126.Bortoluzzi S, d'Alessi F, Danieli GA. A novel resource for the study of genes expressed in the adult human retina. Invest Ophthalmol Vis Sci. 2000;41:3305–3308. [PubMed] [Google Scholar]
- 127.Bowes Rickman C, Ebright JN, Zavodni ZJ, Yu L, Wang T, Daiger SP, Wistow G, Boon K, Hauser MA. Defining the human macula transcriptome and candidate retinal disease genes using EyeSAGE. Invest Ophthalmol Vis Sci. 2006;47:2305–2316. doi: 10.1167/iovs.05-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhou G, Williams RW. Mouse models for the analysis of myopia: an analysis of variation in eye size of adult mice. Optom Vis Sci. 1999;76:408–418. doi: 10.1097/00006324-199906000-00021. [DOI] [PubMed] [Google Scholar]
- 129.Zhou G, Williams RW. Eye1 and Eye2: gene loci that modulate eye size, lens weight, and retinal area in the mouse. Invest Ophthalmol Vis Sci. 1999;40:817–825. [PubMed] [Google Scholar]
- 130.Zhou G, Strom RC, Giguere V, Williams RW. Modulation of retinal cell populations and eye size in retinoic acid receptor knockout mice. Mol Vis. 2001;7:253–260. [PubMed] [Google Scholar]
- 131.Schippert R, Burkhardt E, Feldkaemper M, Schaeffel F. Relative axial myopia in Egr-1 (ZENK) knockout mice. Invest Ophthalmol Vis Sci. 2007;48:11–17. doi: 10.1167/iovs.06-0851. [DOI] [PubMed] [Google Scholar]
- 132.Bitzer M, Schaeffel F. Defocus-induced changes in ZENK expression in the chicken retina. Invest Ophthalmol Vis Sci. 2002;43:246–252. [PubMed] [Google Scholar]
- 133.Brand C, Burkhardt E, Schaeffel F, Choi JW, Feldkaemper MP. Regulation of Egr-1, VIP, and Shh mRNA and Egr-1 protein in the mouse retina by light and image quality. Mol Vis. 2005;11:309–320. [PubMed] [Google Scholar]
- 134.Zhong X, Ge J, Smith EL, 3rd, Stell WK. Image defocus modulates activity of bipolar and amacrine cells in macaque retina. Invest Ophthalmol Vis Sci. 2004;45:2065–2074. doi: 10.1167/iovs.03-1046. [DOI] [PubMed] [Google Scholar]
- 135.Spielman RS, Ewens WJ. The TDT and other family-based tests for linkage disequilibrium and association. Am J Hum Genet. 1996;59:983–989. [PMC free article] [PubMed] [Google Scholar]
- 136.Han W, Yap MK, Wang J, Yip SP. Family-based association analysis of hepatocyte growth factor (HGF) gene polymorphisms in high myopia. Invest Ophthalmol Vis Sci. 2006;47:2291–2299. doi: 10.1167/iovs.05-1344. [DOI] [PubMed] [Google Scholar]
- 137.Tang WC, Yip SP, Lo KK, Ng PW, Choi PS, Lee SY, Yap MK. Linkage and association of myocilin (MYOC) polymorphisms with high myopia in a Chinese population. Mol Vis. 2007;13:534–544. [PMC free article] [PubMed] [Google Scholar]
- 138.Inamori Y, Ota M, Inoko H, Okada E, Nishizaki R, Shiota T, Mok J, Oka A, Ohno S, Mizuki N. The COL1A1 gene and high myopia susceptibility in Japanese. Hum Genet. 2007;122:151–157. doi: 10.1007/s00439-007-0388-1. [DOI] [PubMed] [Google Scholar]
- 139.Mutti DO, Cooper ME, O'Brien S, Jones LA, Marazita ML, Murray JC, Zadnik K. Candidate gene and locus analysis of myopia. Mol Vis. 2007;13:1012–1019. [PMC free article] [PubMed] [Google Scholar]
- 140.Young TL, Guo XD, King RA, Johnson JM, Rada JA. Identification of genes expressed in a human scleral cDNA library. Mol Vis. 2003;9:508–514. [PubMed] [Google Scholar]
- 141.Young TL, Scavello GS, Paluru PC, Choi JD, Rappaport EF, Rada JA. Microarray analysis of gene expression in human donor sclera. Mol Vis. 2004;10:163–176. [PubMed] [Google Scholar]
- 142.Zhou J, Rappaport EF, Tobias JW, Young TL. Differential gene expression in mouse sclera during ocular development. Invest Ophthalmol Vis Sci. 2006;47:1794–1802. doi: 10.1167/iovs.05-0759. [DOI] [PubMed] [Google Scholar]
- 143.Boehnke M, Cox NJ. Accurate inference of relationships in sib-pair linkage studies. Am J Hum Genet. 1997;61:423–429. doi: 10.1086/514862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Epstein MP, Duren WL, Boehnke M. Improved inference of relationship for pairs of individuals. Am J Hum Genet. 2000;67:1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 148.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 149.Boyles AL, Scott WK, Martin ER, Schmidt S, Li YJ, Ashley-Koch A, Bass MP, Schmidt M, Pericak-Vance MA, Speer MC, Hauser ER. Linkage disequilibrium inflates type I error rates in multipoint linkage analysis when parental genotypes are missing. Hum Hered. 2005;59:220–227. doi: 10.1159/000087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li Y-J, Bulusu A, Metlapally R, Malecaze F, Calvas P, Guggenheim J, Mackey D, Rosenberg T, Paget S, Young TL. An international collaborative family-based whole genome linkage screen for high-grade myopia. Invest Ophthalmol Vis Sci. 2008;49 doi: 10.1167/iovs.08-2781. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 152.Arnold N, Gross E, Schwarz-Boeger U, Pfisterer J, Jonat W, Kiechle M. A highly sensitive, fast, and economical technique for mutation analysis in hereditary breast and ovarian cancers. Hum Mutat. 1999;14:333–339. doi: 10.1002/(SICI)1098-1004(199910)14:4<333::AID-HUMU9>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 153.Zou Z, Eibl C, Koop HU. The stem-loop region of the tobacco psbA 5'UTR is an important determinant of mRNA stability and translation efficiency. Mol Genet Genomics. 2003;269:340–349. doi: 10.1007/s00438-003-0842-2. [DOI] [PubMed] [Google Scholar]
- 154.Ohata T, Yoshida K, Sakai H, Hamanoue H, Mizuguchi T, Shimizu Y, Okano T, Takada F, Ishikawa K, Mizusawa H, Yoshiura K, Fukushima Y, Ikeda S, Matsumoto N. A −16C>T substitution in the 5' UTR of the puratrophin-1 gene is prevalent in autosomal dominant cerebellar ataxia in Nagano. J Hum Genet. 2006;51:461–466. doi: 10.1007/s10038-006-0385-6. [DOI] [PubMed] [Google Scholar]
- 155.Hamai Y, Matsumura S, Matsusaki K, Kitadai Y, Yoshida K, Yamaguchi Y, Imai K, Nakachi K, Toge T, Yasui W. A single nucleotide polymorphism in the 5' untranslated region of the EGF gene is associated with occurrence and malignant progression of gastric cancer. Pathobiology. 2005;72:133–138. doi: 10.1159/000084116. [DOI] [PubMed] [Google Scholar]
- 156.Hanna MC, Platts JT, Kirkness EF. Identification of a gene within the tandem array of red and green color pigment genes. Genomics. 1997;43:384–386. doi: 10.1006/geno.1997.4830. [DOI] [PubMed] [Google Scholar]
- 157.Ueyama H, Torii R, Tanabe S, Oda S, Yamade S. An insertion/deletion TEX28 polymorphism and its application to analysis of red/green visual pigment gene arrays. J Hum Genet. 2004;49:548–557. doi: 10.1007/s10038-004-0189-5. [DOI] [PubMed] [Google Scholar]
- 158.Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:627–633. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Metlapally R, Moore AT, Michealides M, Schwartz M, Rosenberg T, Hunt D, Bowes-Rickman C, Young TL. Evaluation of the X-linked high grade myopia locus (MYP1) with cone dysfunction and color vision deficiencies. Invest Ophthalmol Vis Sci. 2008;49 doi: 10.1167/iovs.08-2455. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Scavello GS, Jr, Paluru PC, Zhou J, White PS, Rappaport EF, Young TL. Genomic structure and organization of the high grade Myopia-2 locus (MYP2) critical region: mutation screening of 9 positional candidate genes. Mol Vis. 2005;11:97–110. [PubMed] [Google Scholar]
- 161.Ghosh S, Watanabe RM, Valle TT, Hauser ER, Magnuson VL, Langefeld CD, Ally DS, Mohlke KL, Silander K, Kohtamaki K, Chines P, Balow J, Jr, Birznieks G, Chang J, Eldridge W, Erdos MR, Karanjawala ZE, Knapp JI, Kudelko K, Martin C, Morales-Mena A, Musick A, Musick T, Pfahl C, Porter R, Rayman JB. JB. The Finland-United States investigation of non-insulin-dependent diabetes mellitus genetics (FUSION) study. I. An autosomal genome scan for genes that predispose to type 2 diabetes. Am J Hum Genet. 2000;67:1174–1185. [PMC free article] [PubMed] [Google Scholar]
- 162.Silander K, Scott LJ, Valle TT, Mohlke KL, Stringham HM, Wiles KR, Duren WL, Doheny KF, Pugh EW, Chines P, Narisu N, White PP, Fingerlin TE, Jackson AU, Li C, Ghosh S, Magnuson VL, Colby K, Erdos MR, Hill JE, Hollstein P, Humphreys KM, Kasad RA, Lambert J, Lazaridis KN, Lin G, Morales-Mena A, Patzkowski K, Pfahl C, Porter R, Rha D, Segal L, Suh YD, Tovar J, Unni A, Welch C, Douglas JA, Epstein MP, Hauser ER, Hagopian W, Buchanan TA, Watanabe RM, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A large set of Finnish affected sibling pair families with type 2 diabetes suggests susceptibility loci on chromosomes 6, 11, and 14. Diabetes. 2004;53:821–829. doi: 10.2337/diabetes.53.3.821. [DOI] [PubMed] [Google Scholar]
- 163.Saw SM, Chua WH, Hong CY, Wu HM, Chan WY, Chia KS, Stone RA, Tan D. Nearwork in early-onset myopia. Invest Ophthalmol Vis Sci. 2002;43:332–339. [PubMed] [Google Scholar]
- 164.Gwiazda J, Hyman L, Hussein M, Everett D, Norton TT, Kurtz D, Leske MC, Manny R, Marsh-Tootle W, Scheiman M. A randomized clinical trial of progressive addition lenses versus single vision lenses on the progression of myopia in children. Invest Ophthalmol Vis Sci. 2003;44:1492–1500. doi: 10.1167/iovs.02-0816. [DOI] [PubMed] [Google Scholar]
- 165.Martin ER, Bass MP, Hauser ER, Kaplan NL. Accounting for linkage in family-based tests of association with missing parental genotypes. Am J Hum Genet. 2003;73:1016–1026. doi: 10.1086/378779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]