Abstract
Complex microbial ecosystems contain large reservoirs of unexplored biosynthetic diversity. Here we provide an experimental framework and data analysis tool to facilitate the targeted discovery of natural-product biosynthetic gene clusters from the environment. Multiplex sequencing of barcoded PCR amplicons is followed by sequence similarity directed data parsing to identify sequences bearing close resemblance to biosynthetically or biomedically interesting gene clusters. Amplicons are then mapped onto arrayed metagenomic libraries to guide the recovery of targeted gene clusters. When applied to adenylation- and ketosynthase-domain amplicons derived from saturating soil DNA libraries, our analysis pipeline led to the recovery of biosynthetic clusters predicted to encode for previously uncharacterized glycopeptide- and lipopeptide-like antibiotics; thiocoraline-, azinomycin-, and bleomycin-like antitumor agents; and a rapamycin-like immunosuppressant. The utility of the approach is demonstrated by using recovered eDNA sequences to generate glycopeptide derivatives. The experiments described here constitute a systematic interrogation of a soil metagenome for gene clusters capable of encoding naturally occurring derivatives of biomedically relevant natural products. Our results show that previously undetected biosynthetic gene clusters with potential biomedical relevance are very common in the environment. This general process should permit the routine screening of environmental samples for gene clusters capable of encoding the systematic expansion of the structural diversity seen in biomedically relevant families of natural products.
Keywords: nonribosomal peptide synthetase, polyketide synthase, secondary metabolite, drug discovery, uncultured bacteria
Microbial secondary metabolites produced by nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) biosynthetic machinery have been an enormously valuable source of pharmacologically relevant chemical entities (1, 2). A common theme within these natural compounds is the existence of families of molecules whose similar structural features and biological activities derive from evolutionarily related biosynthetic pathways (2, 3). Structural variations within natural-product families often lead to differences in potency, solubility, toxicity, bioavailability, and even target specificity. These changes can dramatically affect the potential of a natural product to successfully transition from a lead structure to a therapeutic agent (4–6). The development of a facile method for the routine identification of gene clusters capable of encoding natural variants of biomedically relevant natural-product families or natural-product lead structures (“variant clusters”) would therefore be a powerful addition to current drug-development pipelines.
At the beginning of this study, the extent to which undiscovered variant gene clusters existed in nature was unclear. Based on recent metagenomic evidence (7–9), we hypothesized that hidden reservoirs of biosynthetic diversity might be common throughout nature and that their discovery would require the development of an experimental framework that would: (i) faithfully replicate the immense genomic diversity found within complex microbial ecosystems; (ii) facilitate the cloning of complete natural-product biosynthetic gene clusters; and (iii) correctly identify gene clusters of interest from within the much larger pool of closely related but undesired biosynthetic sequences.
It is now well established that most environments contain several orders of magnitude more microbial species than have traditionally been examined using pure culture methods (10, 11). Soils are particularly rich niches of microbial species diversity, with potentially in excess of 10,000 unique bacterial species present in a single gram (12–14). Based on these extraordinary diversity estimates and the historical importance of soil-dwelling microbes as a source of bioactive natural products (15), we elected to focus on soil bacteria as potential sources of biosynthetic gene clusters capable of encoding for metabolites that are structurally and functionally related to clinically relevant natural-product families. Here we show that previously undetected variant biosynthetic gene clusters are likely to be common in nature and present a data-generation pipeline to permit the discovery of these gene clusters from diverse environmental samples.
Results and Discussion
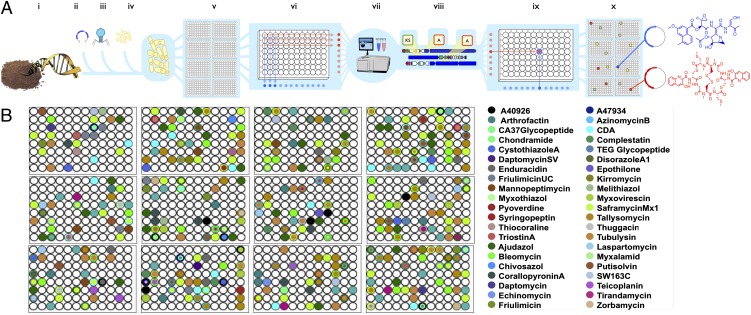
For our initial systematic screening study, total community DNA was extracted directly from a randomly selected soil sample obtained from the Chihuahuan Desert (New Mexico). The resulting high-molecular-weight DNA was then used to construct an environmental DNA (eDNA) cosmid library containing >1.5 × 107 unique clones, hereafter referred to as the New Mexico megalibrary. To facilitate sequence mapping and downstream clone recovery efforts (Fig. 1A), the library was arrayed into 96-well plates at a density of 4–5 × 103 clones per well. Soil DNA megalibraries of this size have been shown to approach saturation of the genetic diversity present in soils, allowing for the recovery of even the largest natural-product biosynthetic gene clusters on sets of overlapping cosmid clones (16).
Fig. 1.
Overview of library construction, clone arraying, amplicon mapping, and targeted gene cluster discovery pipeline. Total community DNA is extracted from a soil (A, i), ligated into a cosmid vector (A, ii), packaged into lambda phage (A, iii), and transfected into E. coli (A, iv). Cosmid clones are then arrayed in 96-well plates at a density of ∼4–5 × 103 clones per well (A, v). Cosmid DNA is prepared from each row and column in a plate (A, vi) and used as template in PCR reactions targeting A and KS domains. Forward primers contain barcodes to allow subsequent position assignment of all amplicons. Tagged PCR amplicons are pooled, 454-pyrosequenced (A, vii), and submitted to a “curated” BLAST analysis to identify amplicons (natural-product sequence tags) indicative of the presence of pathways of interest (A, viii). Amplicons returning a top hit to a gene cluster of interest are positionally located within the library using the primer-encoded row and column information (A, ix), and the resulting map (A, x) is used to guide the systematic recovery of specific gene clusters of interest. (B) A-domain amplicon hit positions mapped onto individual wells of 96-well plates from the arrayed New Mexico eDNA megalibrary. Although amplicons related to known biosynthetic systems are common, they only represent a small fraction of the thousands of diverse sequences found in a library.
With the exception of rare cases of convergent evolution, gene clusters encoding structurally related metabolites will share common ancestry and are therefore likely to exhibit high sequence identity (3). We reasoned that the identification of variant clusters could be accomplished by comparing appropriately selected eDNA sequences (natural-product sequence tags) to a reference database of equivalent sequences derived from characterized biosynthetic systems. Conserved regions within both NRPS adenylation (A) domain and PKS ketosynthase (KS) domain sequences were selected to be the target sequences in the first application of our experimental framework. In addition to their ubiquitous presence in NRPS and PKS biosynthetic gene clusters (17), the choice of these domains as targets was dictated by two factors. First, although they encompass a diverse set of sequences, conserved motifs within both domains have allowed for the design of robust degenerate primers targeted toward each group (18, 19), Second, these domains are typically present multiple times in a given gene cluster (17), thereby increasing the likelihood of successfully amplifying at least one tag from any given cluster. The primer sets we used in this study are known to provide robust amplification of a wide variety of A/KS domains, particularly those found in the genomes of GC-rich soil bacteria traditionally associated with the production of medicinally relevant secondary metabolites (18, 19). The approach we outline is equally applicable to the survey of fungal or AT-rich (adenine/thymine-rich) bacterial sequences by using alternative degenerate primer sets.
To generate PKS and NRPS sequence tags from the partially arrayed New Mexico mega-library, aliquots of DNA representing the separately pooled rows and columns from each of the 96-well plates making up this library were used as templates in degenerate PCR reactions targeting these sequences. Each of the forward primers used in these amplification reactions was designed to contain a unique eight base-pair barcode sequence immediately downstream of a 454 sequencing adapter to permit tracking of the sequence tags to individual rows and columns within the megalibrary. The resulting row and column PCR amplicons were subsequently pooled and 454-pyrosequenced (SI Appendix, Table S1). The collection of raw sequencing reads was first quality filtered and trimmed, and the resulting pool of clean sequences was then clustered at 95% identity to compensate for potential sequencing error and natural-sequence polymorphisms (20). Following these steps, 16,949 unique A-domain clusters and 4,167 unique KS-domain clusters remained, the latter number representing 30-fold more unique KS domains than were identified in a previous analysis of the largest available shotgun metagenomic sequencing datasets (21). The resulting consensus sequence from each 95% cluster was taken to be representative of a unique A- or KS-domain sequence within the library, with position information assigned based on the associated row and column barcodes.
Library-derived A/KS-domain sequence tags were compared using BLAST (22) to a curated database containing A/KS-domain sequences from either completed genomes or individually sequenced biosynthetic gene clusters found in GenBank. Within this reference set, domains derived from >250 biosynthetically and/or biomedically interesting NRPS/PKS biosynthetic gene clusters (SI Appendix, Table S3) were marked to indicate their respective associations with specific natural products of interest. Library-derived A/KS-domain amplicons that returned one of these biosynthetic systems as their top BLAST match were classified as hits. The net effect of this curated BLAST analysis was to identify environmental sequences that are more closely related to biosynthetic sequences of interest than to any other biosynthetic sequences in NCBI (National Center for Biotechnology Information), an attribute we believe to be a good indicator of a functional relationship between an environmental gene cluster yielding an A/KS-domain sequence tag and a curated gene cluster. The analysis of the A/KS-domain sequence tags in this fashion led to the identification of 1,026 unique library sequences that could be classified as hits (Fig. 1B, SI Appendix, Tables S1 and S3). Collectively, these hits recognize 281 different domains derived from 124 of the >250 gene clusters found in our database. This analysis served to pare down the large initial number of A/KS-domain consensus sequences into a smaller number of sequences from which gene clusters of interest (i.e., clones containing natural variants of biosynthetically and/or biomedically interesting NRPS/PKS biosynthetic gene clusters) might be identified. The subset of “hit” amplicon sequences were then mapped onto our arrayed library to guide the recovery of clones containing biosynthetic gene clusters that might encode for new members of biomedically interesting families of secondary metabolites (Fig. 1B).
To validate this screening process as a means of correctly targeting gene clusters of interest, amplicon maps derived from three arrayed soil megalibraries were used to guide recovery of cosmids comprising 26 different biosynthetic systems. Recovered gene clusters were sequenced and bioinformatically analyzed to determine their relationship to the targeted gene-cluster families of interest. A detailed summary of the analyses carried out for each gene cluster is presented in SI Appendix, Figs. S1–11. The closest previously sequenced relative of each recovered gene cluster was determined using the Cluster-BLAST function of AntiSMASH (Antibiotics and Secondary Metabolite Analysis Shell) (23) and confirmed through extensive manual gene-cluster comparisons. In general, the higher the identity observed between a library amplicon sequence and the corresponding domain from a characterized gene cluster, the higher the likelihood of there being a true functional relationship between the recovered gene cluster and the previously characterized homologous gene cluster. Seventeen of the twenty-one (∼81%) KS/A-domain sequence tags returning expectation values below 10−45 in the initial BLAST analysis led to the recovery of gene clusters resembling the targeted gene cluster of interest (“on target” eDNA gene clusters), and only one of the five (20%) hits returning expectation values above 10−45 proved to be on target. In future analyses, productive expectation value cutoffs may vary depending on the specific family of gene clusters being examined. All 18 on-target eDNA gene clusters were subsequently examined for changes in core NRPS/PKS biosynthetic machinery [e.g., number, type, arrangement, identity, or predicted substrate specificity of megasynth(et)ase domains] and for changes in the complement of tailoring enzymes (SI Appendix, Figs. S1–10).
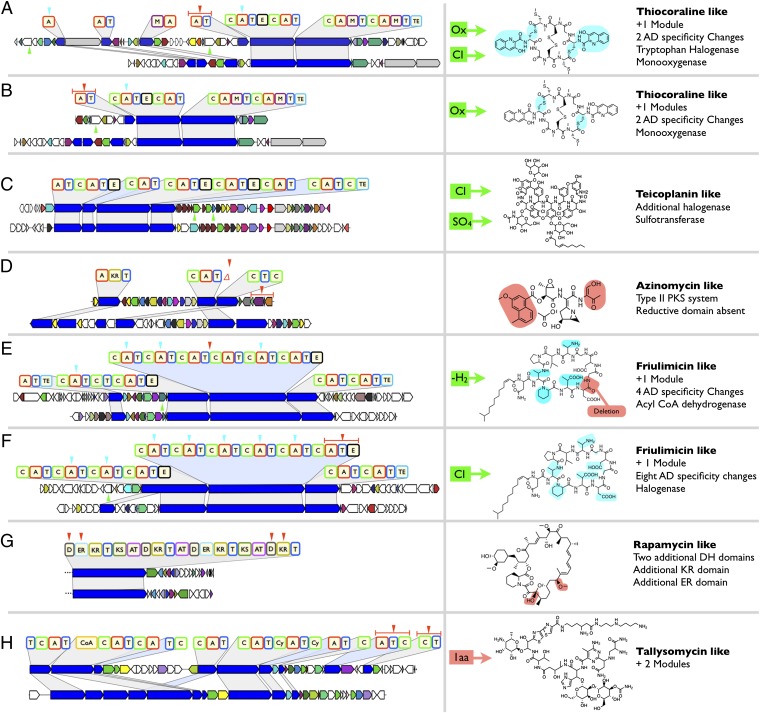
Half (9/18) of the on-target gene clusters we recovered contain changes that are predicted to confer onto them the ability to encode for natural products that differ in structure from any previously reported metabolites (Fig. 2, SI Appendix, Figs. S1–10). Targeting the recovery of clones associated with lipo- and glycopeptide antibiotic related A-domain amplicons led to the identification of four gene clusters that are predicted to encode new members of these families. Two of these are lipopeptide antibiotic-like gene clusters (Fig. 2 E and F, SI Appendix, Figs. S1 and S2) most closely related to the friulimicin (24) biosynthetic gene cluster but with differences in both the megasynthetase biosynthetic machinery and the tailoring enzyme content. The two remaining antibiotic gene clusters are predicted to encode molecules structurally related to the glycopeptides teicoplanin and A47934 (8, 25). In each case, the combination of tailoring enzymes seen in these clusters is predicted to result in the production of a previously unknown glycopeptide derivative (Figs. 2C and 3, SI Appendix, Figs. S7 and S10). Five gene clusters predicted to encode the production of DNA-damaging agents, with potential applications toward the development of anticancer therapeutics, were also recovered. These were related to known gene clusters encoding thiocoraline (26) (Fig. 2 A and B, SI Appendix, Figs. S4–S6), bleomycin (27) (Fig. 2H, SI Appendix, Fig. S8), and azinomycin (28) (Fig. 2D, SI Appendix, Fig. S3). Each of these eDNA-derived gene clusters is predicted to encode a metabolite that differs from known structures within these respective families due to changes in core megasynthetase biosynthetic machinery and/or tailoring enzyme content. Finally, by targeting a gene cluster associated with a rapamycin-like (29) KS amplicon, we recovered a partial gene cluster (Fig. 2G, SI Appendix, Fig. S9) that is closely related to the rapamycin biosynthetic system but with predicted changes in megasynthase domain content that suggest it belongs to a biosynthetic system encoding a previously uncharacterized rapamycin-like structure.
Fig. 2.
Biosynthetic pathways (A–H) encoding new derivatives of medicinally relevant bacterial natural products. eDNA pathways (Upper) are presented with their closest characterized relatives (Lower). A red triangle indicates a module addition or deletion. A blue triangle indicates a change in domain substrate specificity. A green triangle indicates the presence of a new tailoring enzyme. When possible, sites of predicted substructure changes are indicated on the known structures shown at Right. In instances where the regiospecificity could not be predicted, colored boxes with arrows indicate the functional group additions or residue changes that are predicted to occur. ORFs are color coded to indicate conserved functions between eDNA and characterized pathway.
Fig. 3.
Production of glycopeptide derivatives. (A) eDNA-derived glycopeptide gene cluster and scheme showing the creation of the ermE* promoter-driven tailoring enzyme construct. (B) Bioinformatically predicted functional group modifications encoded by this glycopeptide gene cluster. (C) Conjugation of the ermE* promoter-driven tailoring construct into S. toyocaensis:ΔStaL resulted in the production of three new glycopeptide derivatives (peaks 1–3) (A = dsA47934). (D) Structure for compound 1. Functional groups incorporated by eDNA-encoded tailoring enzymes are colored to match ORFs in A and predictions in B.
Taken together, the on-target clusters we identified provide strong evidence supporting our original hypothesis that the genomes of environmental bacteria contain a large hidden reservoir of variant gene clusters that can be systematically identified using sequence-similarity-guided search strategies. Moreover, gene clusters predicted to encode new variants appear to be as common in the environment as gene clusters that are functionally identical to known clusters. The gene clusters described here are all relatives of well-characterized clinically relevant biosynthetic systems. This experimental framework is equally applicable to the diversification of newly discovered lead structures found in high-throughput screening programs or (meta)genome mining studies. The only prerequisite for identifying variant gene clusters using this method is the knowledge of a single A or KS domain associated with the biosynthesis of a lead structure of interest.
To demonstrate the utility of this approach for generating derivatives of medicinally relevant natural products, heterologous expression studies were conducted using one of the recovered glycopeptide gene clusters. When using derivative clusters to generate secondary metabolites, it is possible to envision using either de novo heterologous biosynthesis or mixed heterologous biosynthesis methods. In mixed biosynthesis studies, the unique genetic elements found in a newly discovered cluster would ideally be introduced into a strain that produces a minimally functionalized conserved core structure that can serve as a generic substrate for new gene-cluster-encoded enzymatic activities. For this study we elected to use a mixed biosynthesis strategy, in which the glycopeptide producer Streptomyces toyocaensis:ΔStaL was used as the host. S. toyocaensis:ΔStaL is genetically tractable and produces high titers of desulfo-A47934 (dsA47934), a glycopeptide core structure devoid of tailoring enzyme functionality (30).
In the eDNA-derived glycopeptide gene cluster selected for functional expression studies, all of the tailoring enzymes (a sulfotransferase, a glycosyltransferase, and an N-methyltransferase) are encoded by a single operon. To ensure the constitutive expression of this operon in our mixed biosynthesis study, an ermE* promoter cassette was introduced upstream of the first ORF (Fig. 3A). The introduction of the cosmid containing the now constitutively expressed tailoring operon into S. toyocaensis:ΔStaL, resulted in the production of three new compounds (Fig. 3C). The molecular formulas of these compounds were determined by high-resolution mass spectrometry (SI Appendix, Text S2), and in each case they were found to be consistent with the functionalization of dsA47934 by one or more eDNA-encoded tailoring enzymes. The molecular formula deduced for compound 1 (C65H56Cl3N7O26S) suggested it was the product of the combined action of all three eDNA-tailoring enzymes. This compound was purified and its structure elucidated by NMR (Fig. 3D, SI Appendix, Fig. S12). The structure of compound 1 represents a glycopeptide derivative, resulting from sulfation, methylation, and glycosylation of dsA47934, a functionalization pattern that is completely consistent with our bioinformatics predictions for this gene cluster (Fig. 3B, SI Appendix, Fig. S10). The masses of the two remaining compounds suggest they are intermediates in the production of 1. The deduced molecular formula for 2 (C59H46Cl3N7O21S) is consistent with the addition of a methyl and sulfate to dsA47934, and that for 3 (C59H46Cl3N7O18) corresponds to the addition of a methyl group only.
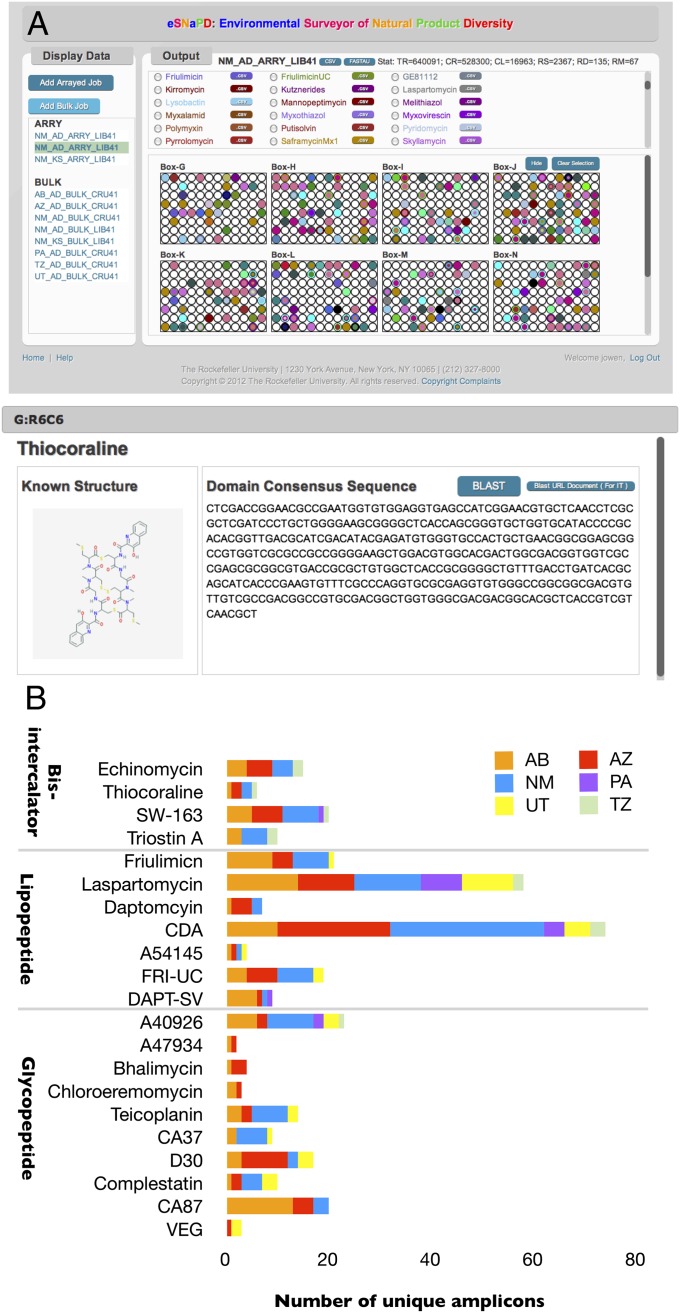
To facilitate the analysis of crude eDNA samples in a similar manner, all of the steps performed in the course of the analytical process described above have been integrated into a web-based sequence analysis platform (environmental Surveyor of Natural Product Diversity [eSNaPD]). The analysis package can be accessed at http://esnapd2.rockefeller.edu. eSNaPD is an interactive tool designed to perform an automated analysis of raw amplicon sequence data, identify key biosynthetic markers, and use bar-coded primer data, if available, to map the location of these markers onto a library map (Fig. 4A). The graphical user interface facilitates the storing, management, and exploration of large, natural-product-associated amplicon data generated from multiple environmental samples. All output data are available in a click-and-see format and in downloadable .csv format to facilitate subsequent user-specific analyses. The reference databases used in the analysis can be easily modified, allowing searches to be customized for the analysis of amplicon sequences arising from the use of degenerate primer pairs designed to recognize any biosynthetic gene sequences of interest.
Fig. 4.
eSNaPD software and direct analysis of crude soil eDNA for clinically relevant families of secondary metabolites. (A) Screen shot from eSNaPD user interface showing interactive color-coded map of locations for biosynthetic markers within the New Mexico soil library arrayed in 96-well plates. The list of NPRS related A-domain molecule hits from the New Mexico megalibrary are shown in Upper. By scrolling down this page the list of PKS-related KS-domain molecule hits is also available. Each radio button links to the specific location within the library where similar sequences reside and to downloadable files containing information related to each respective natural product. The second screen shot shows archived data that is available by clicking on an individual well hit. This includes molecule structure, amplicon sequence, BLAST results, library location statistics, and associated links to outside databases. (Left) Additional archived environmental data sets that are accessible by clicking on these entries. (B) The number of unique amplicon hits found to resemble bisintercalator, lipopeptide, and glycopeptide gene-cluster-associated A domains in eSNaPD analyses of crude eDNA samples isolated from six different soils is shown. The gene cluster to which each amplicon sequence most closely matches is indicated immediately below the bar. Fri-UC = Friulimicn cluster from eDNA (16), Dapt-SV = daptomycin-like cluster from Saccharomonospora viridis (31), CA37, D30, CA87 and VEG = glycopeptide like clusters previously cloned from eDNA (7, 8).
To evaluate the suitability of eSNaPD for the purpose of prescreening environmental samples, we carried out the same general analysis described above directly on eDNA derived from six geographically diverse soil samples (Fig. 4B). A-domain PCR amplicons were generated from each sample and the resulting sequences were analyzed using the eSNaPD amplicon analysis tool (SI Appendix, Table S2). Natural-product sequence tags corresponding to glycopeptide-, lipopeptide-, and bisintercalator-like gene clusters were then quantified and compared, revealing widely differing biosynthetic profiles across these six soil environments (Fig. 4B). This suggests that environmental samples can vary considerably in their gene-cluster content and, as such, sample selection should be an important consideration for future efforts to target the recovery of biosynthetic pathways encoding new members of biomedically important families of natural products.
The eDNA sample used in the construction of the New Mexico megalibrary was among the six samples subjected to direct analysis. Even at greatly reduced sequence depth (∼10% the number of reads) the data from the direct analysis of this sample was found to be in good agreement with that generated from the library. Of the 58 different NRPS gene clusters identified in the library, 45 of these were also detected in the direct soil analysis. The significant overlap (>75%) seen between these two analyses indicates that the assessment of crude soil eDNA samples, even at limited sequencing depth, is likely to provide a good representation of the gene-cluster diversity that can be cloned from these samples. The workup required for direct soil analysis can be achieved using commercially available soil DNA isolation kits which, when coupled with multiplexed parallel sequencing, will allow for large numbers of environmental samples to be analyzed rapidly in parallel. Although we have described the application of this method to culture independent studies, it is equally applicable to the prescreening of large culture collections, or of environmental samples before attempts at strain isolation.
In conclusion, we have presented a data-generation pipeline and an associated analysis software tool that allows for the rapid screening of crude eDNA and saturating eDNA megalibraries for biosynthetic pathways encoding variants of bacterial natural products of interest. The general approach we have described provides a blueprint for the large-scale screening of diverse environmental samples for natural-product targets of interest, which, when coupled to library construction and gene-cluster recovery, should allow for the identification of biosynthetic pathways capable of expanding upon the structural diversity seen in biomedically relevant families of natural products.
Materials and Methods
Library Construction.
Library construction methods have been described in detail previously (32). Briefly: Approximately 1 kg of soil was collected from the Chihuahuan Desert in southwestern New Mexico. Soil was sifted to remove large particulates, and then heated (70 °C) in lysis buffer [100 mM Tris-HCl, 100 mM EDTA, 1.5 M NaCl, 1% (wt/vol) CTAB (cetyltrimethylammonium bromide), 2% (wt/vol) SDS, pH 8.0] for 2 h. Soil particulates were removed from the crude lysate by centrifugation, and eDNA was precipitated from the resulting supernatant with the addition of 0.7 volume isopropanol. Crude eDNA was collected by centrifugation, washed with 70% (vol/vol) ethanol, and resuspended in TE (Tris/EDTA). Gel-purified (1% agarose) high-molecular-weight eDNA was blunt ended (Epicentre, End-It), ligated into pWEB-TNC (Epicentre) or the broad host range vector pWEB436, packaged into lambda phage, and transfected into Escherichia coli EC100. Following recovery, transfected cells were inoculated into 5 mL LB with selective antibiotic (100 μg/mL ampicillin or 50 μg/mL apramycin) in 48-well plates at a density of ∼4–5 × 103 clones per well and grown overnight. Matching glycerol stocks and cosmid DNA minipreps were prepared from each well and arrayed into 96-well plates for screening.
Amplicon Generation and Sequencing.
Cosmid DNA was pooled for each row and column of all library plates and these individual pools were used as template for PCR reactions with degenerate primers targeting NRPS adenylation (A) domains and PKS ketosynthase (KS) domains. A-domain fragments (∼795 bp) were amplified using primers A3F (5′-GCSTACSYSATSTACACSTCSGG) and A7R (5′-SASGTCVCCSGTSCGGTA) (18, 19). These primers are designed to recognize the conserved A3 and A7 regions in NRPS A domains. KS-domain fragments (∼760 bp) were amplified using primers degKS2F (5′-GCIATGGAYCCICARCARMGIVT) and degKS2R (5′-GTICCIGTICCRTGISCYTCIAC) (18, 19). These primers are designed to amplify the most conserved regions of PKSI KS-domains, including the active site residues. The 5′ ends of forward primers were augmented with 454 sequencing adapters followed by unique 8-bp barcode sequences identifying the template pool to which they were assigned. PCR reactions: 25 μL reaction, 1× G buffer (Epicentre), 50 pmol of each primer, 2.5 U Taq polymerase and 100 ng cosmid DNA. Cycle conditions for A-domain amplification: 94 °C, 4 min, (94 °C, 30 s; 67.5 °C, 30 s; 72 °C, 60 s) × 35 cycles, 72 °C, 5 min. Cycle conditions for KS-domain amplification: 94 °C, 4 min, (95 °C, 40 s; 56.3 °C, 40 s; 72 °C, 75 s) × 35 cycles, 72 °C, 5 min. Before sequencing, all PCR products were quantified by gel electrophoresis and mixed in an equal molar ratio. Fluorometrically quantified (PicoGreen Quant-iT; Invitrogen, DNA 7500; Agilent Technologies) gel purified (Qiagen MinElute, crystal violet staining) PCR amplicon pools were sequenced using 454 GS-FLX Titanium pyrosequencing technologies.
Amplicon Sequence Data Processing.
Reads with any ambiguous calls and those <200 bp in length were removed. All remaining reads were trimmed to ≤ 400 bp, sorted by decreasing length and then clustered at 95% identity using UCLUST (20). The cluster consensus sequence (CCS) generated for each 95% identity cluster was taken to represent a unique A or KS sequence in the library. Each unique A/KS-domain sequence was located in the arrayed library using the 8-bp PCR primer barcodes uniquely associated with each row and column, with one position assigned for each library plate in which a sequence was detected. For example: a 95% identity cluster found to contain sequences from row G and column 6 of plate M is assigned the library position MG6 (plate M, well G6). Location information for each sequence in a 95% identity cluster was subsequently assigned to the CCS generated from this cluster. A/KS-domain CCSs were searched using Blast against our NCBI-NT-AD and NCBI-NT-KS datasets (a description of these datasets appears in the additional material and methods located in the SI Appendix), respectively. Blast hits with e-value greater than 10−20 were discarded. CCS that return A or KS domains from functionally characterized gene clusters of interest were considered hits. The New Mexico library was used as the primary source of clones in this study. A similar analysis was performed on previously archived libraries created from California and Arizona soils, and clones recovered from these libraries were also included in this study.
Additional material and methods outlining recovery of target clones, sequencing and in silico analysis of recovered gene clusters, the development of eSNaPD software, production of compound 1 by mixed biosynthesis, purification, and structure determination of compound 1 are located in SI Appendix.
Supplementary Material
Acknowledgments
We thank Jerry Wright for S. toyocaensis:ΔStaL. This work was supported by National Institutes of Health Grant GM077516. S.F.B. is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this manuscript have been deposited in GenBank [accession nos. KF264537–KF264565 (29 entries)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222159110/-/DCSupplemental.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh CT. The chemical versatility of natural-product assembly lines. Acc Chem Res. 2008;41(1):4–10. doi: 10.1021/ar7000414. [DOI] [PubMed] [Google Scholar]
- 3.Fischbach MA, Walsh CT, Clardy J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc Natl Acad Sci USA. 2008;105(12):4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kahne D, Leimkuhler C, Lu W, Walsh C. Glycopeptide and lipoglycopeptide antibiotics. Chem Rev. 2005;105(2):425–448. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]
- 5.Strieker M, Marahiel MA. The structural diversity of acidic lipopeptide antibiotics. ChemBioChem. 2009;10(4):607–616. doi: 10.1002/cbic.200800546. [DOI] [PubMed] [Google Scholar]
- 6.Zolova OE, Mady AS, Garneau-Tsodikova S. Recent developments in bisintercalator natural products. Biopolymers. 2010;93(9):777–790. doi: 10.1002/bip.21489. [DOI] [PubMed] [Google Scholar]
- 7.Banik JJ, Brady SF. Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc Natl Acad Sci USA. 2008;105(45):17273–17277. doi: 10.1073/pnas.0807564105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banik JJ, Craig JW, Calle PY, Brady SF. Tailoring enzyme-rich environmental DNA clones: a source of enzymes for generating libraries of unnatural natural products. J Am Chem Soc. 2010;132(44):15661–15670. doi: 10.1021/ja105825a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang FY, Brady SF. Cloning and characterization of an environmental DNA-derived gene cluster that encodes the biosynthesis of the antitumor substance BE-54017. J Am Chem Soc. 2011;133(26):9996–9999. doi: 10.1021/ja2022653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 11.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308(5721):554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 12.Roesch LF, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1(4):283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniel R. The metagenomics of soil. Nat Rev Microbiol. 2005;3(6):470–478. doi: 10.1038/nrmicro1160. [DOI] [PubMed] [Google Scholar]
- 14.Fierer N, et al. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl Environ Microbiol. 2007;73(21):7059–7066. doi: 10.1128/AEM.00358-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baltz RH. Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol. 2008;8(5):557–563. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Kim JH, et al. Cloning large natural product gene clusters from the environment: piecing environmental DNA gene clusters back together with TAR. Biopolymers. 2010;93(9):833–844. doi: 10.1002/bip.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cane DE, Walsh CT, Khosla C. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science. 1998;282(5386):63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 18.Ayuso-Sacido A, Genilloud O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb Ecol. 2005;49(1):10–24. doi: 10.1007/s00248-004-0249-6. [DOI] [PubMed] [Google Scholar]
- 19.Reddy BV, et al. Natural product biosynthetic gene diversity in geographically distinct soil microbiomes. Appl Environ Microbiol. 2012;78(10):3744–3752. doi: 10.1128/AEM.00102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 21.Foerstner KU, Doerks T, Creevey CJ, Doerks A, Bork P. A computational screen for type I polyketide synthases in metagenomics shotgun data. PLoS ONE. 2008;3(10):e3515. doi: 10.1371/journal.pone.0003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Medema MH, et al. 2011. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39(Web Server issue):W339–346.
- 24.Müller C, et al. Sequencing and analysis of the biosynthetic gene cluster of the lipopeptide antibiotic Friulimicin in Actinoplanes friuliensis. Antimicrob Agents Chemother. 2007;51(3):1028–1037. doi: 10.1128/AAC.00942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pootoolal J, et al. Assembling the glycopeptide antibiotic scaffold: The biosynthesis of A47934 from Streptomyces toyocaensis NRRL15009. Proc Natl Acad Sci USA. 2002;99(13):8962–8967. doi: 10.1073/pnas.102285099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lombó F, et al. Deciphering the biosynthesis pathway of the antitumor thiocoraline from a marine actinomycete and its expression in two streptomyces species. ChemBioChem. 2006;7(2):366–376. doi: 10.1002/cbic.200500325. [DOI] [PubMed] [Google Scholar]
- 27.Galm U, et al. Comparative analysis of the biosynthetic gene clusters and pathways for three structurally related antitumor antibiotics: bleomycin, tallysomycin, and zorbamycin. J Nat Prod. 2011;74(3):526–536. doi: 10.1021/np1008152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Q, et al. Characterization of the azinomycin B biosynthetic gene cluster revealing a different iterative type I polyketide synthase for naphthoate biosynthesis. Chem Biol. 2008;15(7):693–705. doi: 10.1016/j.chembiol.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 29.Schwecke T, et al. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci USA. 1995;92(17):7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamb SS, Patel T, Koteva KP, Wright GD. Biosynthesis of sulfated glycopeptide antibiotics by using the sulfotransferase StaL. Chem Biol. 2006;13(2):171–181. doi: 10.1016/j.chembiol.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Baltz RH. Genomics and the ancient origins of the daptomycin biosynthetic gene cluster. J Antibiot (Tokyo) 2010;63(8):506–511. doi: 10.1038/ja.2010.82. [DOI] [PubMed] [Google Scholar]
- 32.Brady SF. Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nat Protoc. 2007;2(5):1297–1305. doi: 10.1038/nprot.2007.195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.