Abstract
Maintaining active growth and effective immune responses is often costly for a living organism to survive. Fine-tuning the shared cross-regulators is crucial for metazoans and plants to make a trade-off between growth and immunity. The Arabidopsis regulatory receptor-like kinase BAK1 complexes with the receptor kinases FLS2 in bacterial flagellin-triggered immunity and BRI1 in brassinosteroid (BR)-mediated growth. BR homeostasis and signaling unidirectionally modulate FLS2-mediated immune responses at multiple levels. We have shown previously that BIK1, a receptor-like cytoplasmic kinase, is directly phosphorylated by BAK1 and associates with FLS2/BAK1 complex in transducing flagellin signaling. In contrast to its positive role in plant immunity, we report here that BIK1 acts as a negative regulator in BR signaling. The bik1 mutant displays various BR hypersensitive phenotypes accompanied with increased accumulation of de-phosphorylated BES1 proteins and transcriptional regulation of BZR1 and BES1 target genes. BIK1 associates with BRI1, and is released from BRI1 receptor upon BR treatment, which is reminiscent of FLS2-BIK1 complex dynamics in flagellin signaling. The ligand-induced release of BIK1 from receptor complexes is associated with BIK1 phosphorylation. However, in contrast to BAK1-dependent FLS2-BIK1 dissociation, BAK1 is dispensable for BRI1-BIK1 dissociation. Unlike FLS2 signaling which depends on BAK1 to phosphorylate BIK1, BRI1 directly phosphorylates BIK1 to transduce BR signaling. Thus, BIK1 relays the signaling in plant immunity and BR-mediated growth via distinct phosphorylation by BAK1 and BRI1, respectively. Our studies indicate that BIK1 mediates inverse functions in plant immunity and development via dynamic association with specific receptor complexes and differential phosphorylation events.
Keywords: brassinosteroid insensitive 1, BRI1-associated receptor kinase, flagellin sensing 2, botrytis-induced kinase 1, bri1-Ems-Suppressor 1
Metazoans and plants have evolved complex mechanisms to cope with the constant challenges of environmental stresses while maintaining their growth and development. Being sessile and lacking a sophisticated adaptive immune system, plants possess a large number of receptor-like kinases (RLKs) and receptor-like cytoplasmic kinases (RLCKs) that modulate growth, development, and innate immunity (1). RLKs sense different extrinsic and intrinsic cues through the extracellular domain and mediate diverse signaling events via the kinase domain. Arabidopsis Brassinosteroid Insensitive 1 (BRI1), a leucine-rich repeat (LRR)-receptor kinase perceives the polyhydroxylated growth hormones brassinosteroids (BR) to regulate plant growth and development (2). Despite structural similarity with BRI1, Flagellin Sensing 2 (FLS2) and EF-Tu Receptor (EFR) recognize microbe-associated molecular pattern (MAMP) flagellin and elongation factor Tu (EF-Tu), respectively, and initiate innate immune signaling to defend against pathogen attacks (3, 4). Apparently, signaling specificity is achieved by specific receptor–ligand interaction. Instead of ligand perception, RLCKs without an apparent extracellular domain often complex with RLKs and relay the signaling via phosphorylation.
In BR signaling, BRI1 receptor directly binds to brassinolide (BL), the most active form of BRs, via a surface pocket embedded in a 70-aa island of LRR ectodomain. Subsequent heterodimerization, reciprocal and sequential phosphorylation of BRI1 and BAK1 (BRI1-associated kinase 1), accompanied with release of the BRI1 C-terminal inhibitory effect and BRI1 Kinase Inhibitor protein, have been proposed to fully activate BRI1 (5-7). The activated BRI1 phosphorylates downstream RLCKs BR-Signaling Kinases (BSKs) and Constitutive Differential Growth 1 (CDG1), which further interact with and phosphorylate the phosphatase bri1 Suppressor 1 (BSU1) (8, 9). The phosphorylated BSU1 induces dephosphorylation and inactivation of Brassinosteroid Insensitive 2 (BIN2), a glycogen synthase kinase 3 (GSK3)-like kinase, leading to the nuclear accumulation of two dephosphorylated transcription factors Brassinazole-Resistant 1 (BRZ1) and bri1-Ems-Suppressor 1 (BES1) for the regulation of BR-responsive genes (9–11).
In immune signaling, the flagellin receptor FLS2 or EF-Tu receptor EFR instantaneously forms a ligand-induced complex with BAK1 and concomitant transphosphorylation events likely constitute key initial steps in signal transduction (12–14). The plasma membrane-associated RLCK Botrytis-Induced Kinase 1 (BIK1) associates with FLS2/EFR and BAK1 and is directly phosphorylated by BAK1 (15-16). BIK1 dissociates from FLS2 in a BAK1-dependent manner on flagellin perception. BIK1 positively regulates plant innate immunity, and the bik1 mutant was compromised in diverse flagellin-mediated responses and immunity to nonpathogenic bacterial infection. Activation of MAP kinases and calcium-dependent protein kinases, two independent intracellular signaling pathways downstream of the MAMP receptor complex, governs the expression of MAMP-responsive genes (17). In addition, MAMP perception leads to ion fluxes, production of reactive oxygen species (ROS), deposition of callose, and stomatal closure to prevent pathogen entry (18). Once activated, immune signaling is subjected to down-regulation to prevent excessive or prolonged activation of immune responses. Two plant E3 ubiquitin ligases, PUB12 and PUB13, directly ubiquitinate FLS2 and promote flagellin-induced FLS2 degradation, which, in turn, attenuates FLS2 signaling (19).
Despite distinct signaling outcomes, BAK1 is a shared component in BR and multiple immune signaling via heterodimerization with corresponding receptors BRI1, and immune receptors, such as FLS2 and EFR (12–14, 20, 21). BAK1 is also known as somatic embryogenesis receptor kinase (SERK) 3, belonging to a subfamily of RLKs with five members, SERK1 to SERK5 (22). In addition to BAK1/SERK3, BRI1 also associates with SERK1 and SERK4/BKK1 (BAK1-like 1) that play partially redundant roles with BAK1 in BR signaling (23–25). Similarly, BAK1 and SERK4 also exhibit redundant functions in plant innate immunity via association with multiple MAMP receptors (14). Recent studies have shown that BR homeostasis and signaling unidirectionally modulate FLS2-mediated immune responses (26, 27). The essential role of BAK1 in both BR and flagellin signaling pathways suggests that it may function as a rate-limiting factor to make a tradeoff between growth and immunity. Interestingly, BR antagonizes FLS2 signaling in both BAK1-dependent and BAK1-independent manners. To date, signaling components downstream of BAK1 appear to be divergent in FLS2 and BRI1 pathways. Here, we found that BIK1, a positive regulator in plant immunity, acts as a negative regulator in BR signaling. In contrast to its compromised immune responses, the bik1 mutant displays various BR hypersensitive phenotypes, including enhanced hypocotyl length of dark-grown seedlings and root growth inhibition upon BL treatment. The bik1 mutant also exhibits increased accumulation of dephosphorylated BES1 and expression of BZR1 and BES1 target genes. BIK1 interacts with BRI1 at low BR concentration and releases from BRI1 upon exogenous BL treatment in a kinase-dependent manner. Unlike BIK1-FLS2 dissociation, which is BAK1 dependent, the BIK1-BRI1 dissociation is independent of BAK1. Apparently, release of BIK1 from receptor complexes is a result of BIK1 phosphorylation upon signal perception. In FLS2 signaling, BAK1 is crucial for flagellin-mediated BIK1 phosphorylation and complex dissociation. However, in BR signaling, BRI1 is able to phosphorylate BIK1 directly and the phosphorylation event was further enhanced upon BL treatment.
Results
The bik1 Mutant Confers Hypersensitivity to Brassinolide.
Given the observation that the bik1 mutant plants exhibit slightly reduced primary root elongation, early flowering, and reduced fertility (28), it is likely that BIK1 is involved in plant growth and development. In addition, the bik1 mutant plants have moderately elongated and curling petioles, which were often observed in BRI1 overexpressing plants (Fig. S1) or mutants with constitutive activation of BR signaling (29). BIK1 is a direct phosphorylation target of BAK1, an important component in BR signaling, and BIK1 transphosphorylates BAK1 to enhance BAK1 kinase activity (15). These observations prompted us to examine the potential involvement of BIK1 in BR signaling.
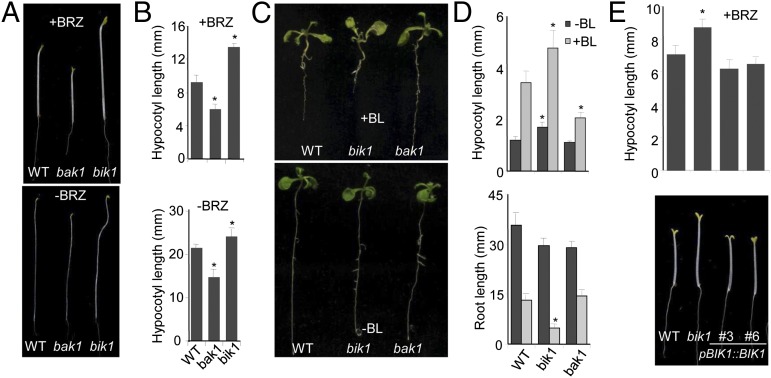
BAK1 positively regulates BR signaling by transphosphorylation of the BRI1 receptor (5, 20, 21). Surprisingly, in contrast to bak1-4 mutant, which is partially insensitive to BL treatment, bik1 mutant displays constitutive BR responses and are hypersensitive to BL treatment. When grown in the dark, the hypocotyls of bik1 mutant elongated slightly, but were significantly longer than those of wild-type (WT) plants, whereas bak1-4 mutant exhibited relatively short hypocotyls (Fig. 1 A and B, Lower). In the presence of brassinazole (BRZ), an inhibitor of BR biosynthesis, the bik1 mutant displayed much more pronounced hypocotyl elongation than WT plants (Fig. 1 A and B, Upper), suggesting that the bik1 mutant was less sensitive to BRZ treatment. The treatment of BL increases hypocotyl elongation and inhibits root growth of WT plant seedlings grown under the light. Consistent with its positive role in BR signaling, bak1-4 mutant was partially insensitive to BL treatment (Fig. 1 C and D). However, the bik1 mutant was hypersensitive to BL treatment with elevated root growth inhibition and hypocotyl elongation compared with WT seedlings (Fig. 1 C and D).
Fig. 1.
Elevated BR responses in bik1 mutant plants. (A) The bik1 mutant is partially insensitive to BRZ treatment. The seedlings of WT (Col-0), bak1-4, and bik1 mutants were grown in the dark for 8 d on 1/2 MS plates with or without 2 μM BRZ. (B) Quantification of hypocotyl length shown in A. (C) The bik1 mutant is hypersensitive to BL treatment. The seedlings were grown on 1/2 MS plates with or without 100 nM BL under the constant light for 14 d. (D) Quantification of root and hypocotyl length shown in C. The data are shown as mean ± SE from at least 25 seedlings. Asterisk indicates a significant difference with P < 0.05 compared with data from WT seedlings. (E) BIK1 complementation lines restore the BRZ insensitivity of the bik1 mutant. The above experiments were repeated three to four times with similar results.
To confirm that the phenotypes observed in the bik1 mutant were attributed to the mutation in BIK1, we complemented the bik1 mutant with the BIK1 cDNA under the control of its native promoter. The BIK1 transgene rescued bik1 mutant phenotypes in response to BRZ treatment (Fig. 1E). Interestingly, we observed that the bik1 mutant exhibited twisted hypocotyls when grown in the dark (Fig. S2). This phenotype is likely due to the constitutive activation of BR signaling in the bik1 mutant because the treatment of BL induced bik1-like hypocotyl twisting in the WT, but not bri1-5 seedlings (Fig. S2). The BL treatment further accelerated the hypocotyl twisting in the bik1 mutant, consistent with our observation that the bik1 mutant was hypersensitive to BL treatment. As expected, the BR biosynthesis mutant de-etiolated2 (det2) still responded to BL treatment to induce hypocotyl twisting (Fig. S2). In addition, the exogenously prolonged application of BRZ in the growth medium retarded the seedling growth and development likely due to the largely reduced BR biosynthesis (Fig. S3). Compared with WT plants, the bik1 mutant substantially ameliorated the BRZ-mediated growth inhibition (Fig. S3). Together, the data suggest that the loss of BIK1 activated BR signaling.
The bik1 mutant plants have elevated salicylic acid (SA) accumulation compared with WT plants (28). To investigate whether the high level of SA attributes to the observed BR phenotypes in the bik1 mutant, we examined the BR responses in the bik1sid2 double mutant, in which the high SA level is diminished by a SA biosynthesis mutant sid2 (30). Similar to the bik1 mutant, the bik1sid2 mutant exhibited elongated hypocotyls in the absence or presence of BRZ when grown in the dark and displayed hypersensitivity to BL treatment with elevated hypocotyl elongation and root inhibition when grown under the light (Fig. S4). The dark grown bik1sid2 double mutant also showed twisted hypocotyls in the absence of BL treatment and BL treatment exacerbated the phenotype (Fig. S2). The data suggest that BR hypersensitivity in the bik1 mutant was unlikely caused by the high level of SA.
BIK1 Negatively Regulates BR Signaling.
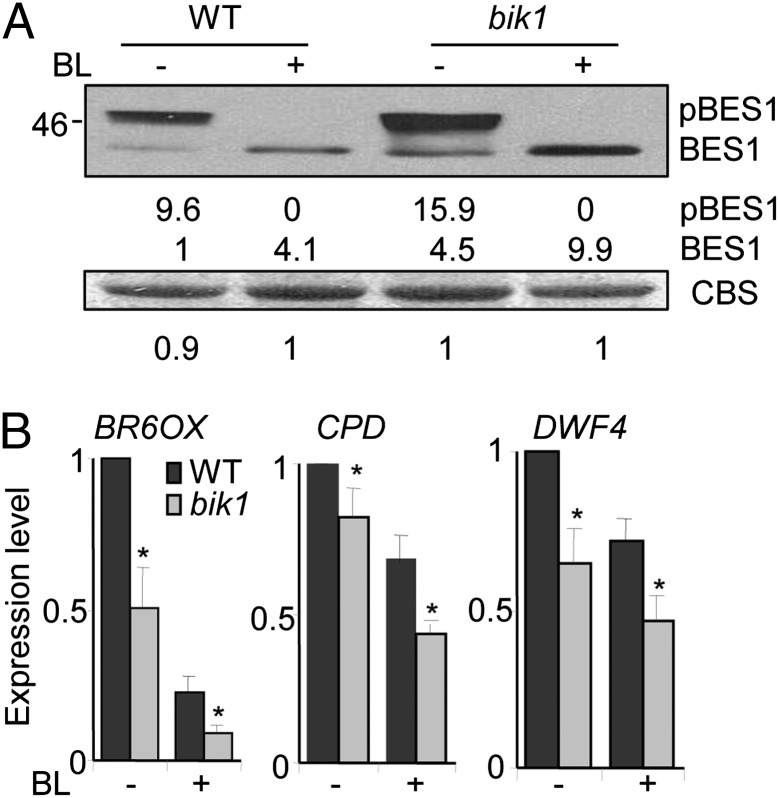
The elicitation of BR signaling induces dephosphorylation of two closely related transcription factors, BES1 and BZR1, which in turn regulate the expression of BR target genes including brassinosteroid-6-oxidase (BR6OX), constitutive photomorphogenic dwarf (CPD), and dwarf 4 (DWF) (31, 32). We examined the phosphorylation status of endogenous BES1 proteins with a specific α-BES1 antibody. In WT seedlings, the BL treatment induced BES1 dephosphorylation as indicated with the mobility shift of BES1 proteins from high molecular weight to low molecular weight in Western blot. Compared with WT plants, the bik1 mutant exhibited an elevated amount of dephosphorylated BES1 protein either with or without BL treatment (Fig. 2A). Apparently, both phosphorylated and dephosphorylated forms of BES1 proteins accumulated more in the bik1 mutant than those in WT plants (Fig. 2A).
Fig. 2.
BIK1 negatively regulates BR signaling. (A) BES1 phosphorylation in WT and bik1 mutant plants. The phosphorylated (pBES1) and dephosphorylated BES1 proteins were detected with an α-BES1 antibody (Upper). Equal loading was ensured by total protein quantification before loading and by Coomassie brilliant blue staining (CBS) of the membrane (Lower). The band intensity was quantified by ImageJ software and labeled under the gel. (B) Expression of BR responsive genes with qRT-PCR analysis. Ten-day-old seedlings were treated with 2 μM BL or H2O for 2 h. The expression of BR6OX, CPD, or DWF4 was normalized to the expression of UBQ10. The data are shown as mean ± SE from three independent biology repeats. Asterisk indicates a significant difference with P < 0.05 compared with data from WT seedlings. The above experiments were repeated three times with similar results.
Down-regulation of BR biosynthesis genes BR6OX, CPD, and DWF4 constitutes a negative feedback regulation mechanism in response to BR treatment or situations of enhanced BR signaling. Consistent with this notion, the expression of BR6OX, CPD, and DWF4 was significantly lower in the 10-d-old bik1 seedlings than that in WT before BL treatment, and the expression was further reduced in the bik1 mutant compared with WT plants upon BL treatment (Fig. 2B). We also investigated whether the activation of BR signaling in the bik1 mutant was specific to a plant developmental stage. Similar reduction of BR6OX, CPD, and DWF4 gene expression was observed in the 4-wk-old bik1 mutant plants compared with WT plants (Fig. S5). Thus, BIK1 negatively regulates BR signaling upstream of BES1 phosphorylation in both seedlings and mature plants.
BIK1 Associates with BRI1.
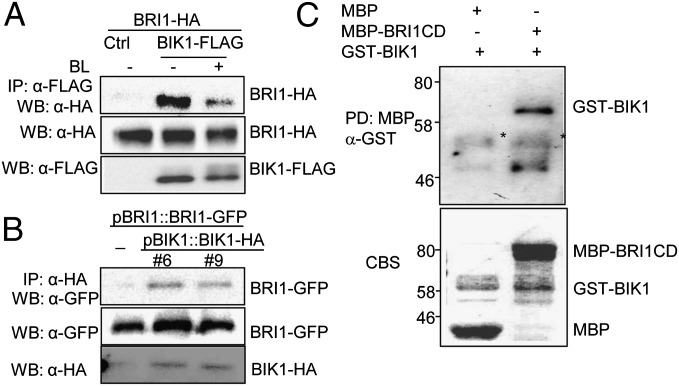
BIK1 is a plasma membrane localized protein with a putative myristoylation motif (28). In flagellin signaling, BIK1 associates with both FLS2 and BAK1 (15, 16). To examine whether BIK1 forms a complex with BRI1, we performed coimmunoprecipitation (Co-IP) assays by coexpressing FLAG epitope-tagged BIK1 and HA epitope-tagged BRI1 in protoplasts. As shown in Fig. 3A, BIK1 coimmunoprecipitated BRI1 in vivo (Fig. 3A). Interestingly, the association of BIK1 with BRI1 appears to be reduced upon BL treatment (Fig. 3A), suggesting that BIK1 might be released from the receptor complex upon BL perception. We further confirmed BIK1 and BRI1 association in the pBIK1::BIK1-HA/pBRI1::BRI1-GFP transgenic plants. As shown in Fig. 3B, BIK1-HA coimmunoprecipitated BRI1-GFP as detected with α-GFP antibody upon α-HA antibody immunoprecipitation. Consistently, bimolecular fluorescence complementation (BiFC) assay also indicated that BRI1 associates with BIK1 with cotransfection of BIK1 fused to the carboxyl-terminal half of YFP (yellow fluorescence protein) (BIK1-cYFP) and BRI1 fused to the amino-terminal half of YFP (BRI1-nYFP) in protoplasts (Fig. S6A). BIK1-BRI1 association and BL-induced dissociation were also confirmed with Nicotiana benthamiana transient assay (Fig. S6B). To test whether BRI1 directly interacts with BIK1 through the cytosolic kinase domain, we performed an in vitro pull-down assay with the BRI1 cytosolic domain (BRI1CD) fused to maltose binding protein (MBP) immobilized on amylose-agarose beads as bait against GST-BIK1 fusion proteins. As shown in Fig. 3C, GST-BIK1 could be pulled down by MBP-BRI1CD, not MBP itself. Similarly, GST-BRI1CD could be pulled down by MBP-BIK1 (Fig. S6C). Taken together, the data demonstrate that BIK1 functions in BR signaling by direct interaction with BRI1 cytosolic kinase domain.
Fig. 3.
BIK1 associates with BRI1. (A) BIK1 associates with BRI1 in protoplasts. BIK1-FLAG was coexpressed with BRI1-HA in Arabidopsis protoplasts. Co-IP was carried out with an α-FLAG antibody (IP: α-FLAG), and the proteins were analyzed by using Western blot with α-HA antibody. Top shows that BIK1-FLAG coimmunoprecipitated with BRI1-HA (IP: α-FLAG; WB: α-HA). Middle and Bottom show the expression of BRI1-HA and BIK1-FLAG proteins (WB: α-HA or α-FLAG for input control). Protoplasts were treated with 2 μM BL for 2 h. (B) BIK1 associates with BRI1 in transgenic plants. The membrane proteins from 4-wk-old pBIK1::BIK1-HA/pBRI1::BRI1-GFP (#6 and #9) or pBRI1::BRI1-GFP plants were immunoprecipitated with α-HA antibody and analyzed with Western blot using α-GFP antibody (Top). The expression of BRI1-GFP and BIK1-HA in transgenic plants are shown (Middle and Bottom). (C) BIK1 interacts with BRI1 cytosolic domain in vitro. GST-BIK1 proteins were incubated with MBP or MBP-BRI1CD beads (PD:MBP), and the beads were collected and washed for Western blot of immunoprecipitated proteins with α-GST antibody. Asterisk indicates nonspecific bands. The above experiments were repeated three times with similar results.
BL-Induced BRI1 Phosphorylation on BIK1.
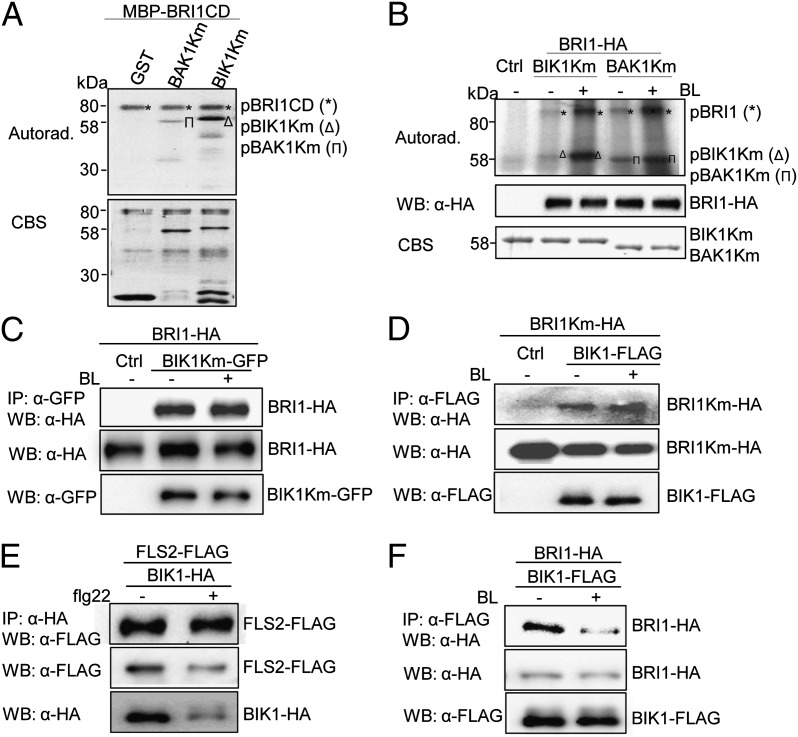
Because BIK1 interacts with BRI1, we tested whether BRI1 could directly phosphorylate BIK1 to transduce BR signaling. An in vitro kinase assay with GST-BIK1Km (the kinase-inactive mutant) as a substrate indicates that MBP-BRI1CD directly phosphorylated GST-BIK1Km (Fig. 4A). Interestingly, it appears that MBP-BRI1CD exhibited stronger kinase activity toward BIK1 than that to BAK1 (Fig. 4A). Phosphorylation of BIK1 by BRI1 was also observed with an immunocomplex kinase assay in which HA epitope-tagged full-length BRI1 was expressed in protoplasts, and BRI1 was pulled down with an α-HA antibody and used in an in vitro kinase assay by using GST-BIK1Km or GST-BAK1Km as a substrate. The immunoprecipitated BRI1 phosphorylated BIK1Km (Fig. 4B). Importantly, BIK1 phosphorylation by BRI1 was enhanced upon BRI1 activation with BL treatment (Fig. 4B). BL treatment did not induce BIK1 phosphorylation by BRI1Km (Fig. S6D). The BL treatment also enhanced BRI1 phosphorylation on BAK1Km, which was used as a control (Fig. 4B). BL-induced BIK1 phosphorylation by BRI1 was further detected by an α-pThr antibody with coexpressing BRI1 and BIK1 in protoplasts (Fig. S6E). The data indicate that BIK1 is a direct substrate of BRI1 and BL induces BRI1 phosphorylation on BIK1.
Fig. 4.
BL-induced BIK1 phosphorylation by BRI1. (A) BRI1 phosphorylates BIK1 in vitro. An in vitro kinase assay was performed by incubating MBP-BRI1CD with GST, GST-BIK1Km, or GST-BAK1Km proteins. Proteins were separated by 10% SDS/PAGE and analyzed by autoradiography (Upper), and the protein loading control was shown by CBS (Lower). (B) BL treatment enhances BRI1 phosphorylation on BIK1. BRI1-HA was expressed in WT protoplasts for 10 h followed by 2 μM BL treatment for 2 h. BRI1-HA proteins were immunoprecipitated with α-HA antibody and subjected to an in vitro kinase assay with GST-BIK1Km or GST-BAK1Km proteins as substrates (Top). Middle shows the BRI1-HA expression, and Bottom shows GST-BIK1Km and GST-BAK1Km proteins. (C) BIK1Km-BRI1 association in protoplasts. Protoplasts were treated with 2 μM BL for 2 h. (D) BIK1-BRI1Km association in protoplasts. (E) BAK1 is required for flg22-induced BIK1-FLS2 dissociation. The BIK1–FLS2 interaction was performed with bak1-4 protoplasts. Protoplasts were treated with 1 μM flg22 for 15 min. (F) BAK1 is not required for BL-induced BIK1-BRI1 dissociation. The BIK1–BRI1 interaction was performed with bak1-4 protoplasts. Protoplasts were treated with 2 μM BL for 2 h. The above experiments were repeated three times with similar results.
The 22-aa peptide of flagellin (flg22) treatment induces rapid BIK1 phosphorylation as indicated by a mobility shift of the protein on SDS/PAGE (Fig. S7A) (15, 16). We did not observe the clear and reproducible mobility shift of BIK1 upon BL treatment although we modified the SDS/PAGE gels with various ratios of bisacrylamide to acrylamide, which offers better separation of different phosphorylation states for phosphoproteins (Fig. S7A). The data suggest that the phosphorylation change of BIK1 mediated by BR might be distinct from that triggered by flagellin. However, when coexpressing with BRI1, the mobility shift of BIK1 was observed upon BL treatment (Fig. S7B), suggesting BRI1 phosphorylates BIK1 in vivo in BR signaling.
Differential Requirement of BAK1 for BIK1-FLS2 and BIK1-BRI1 Dissociation.
Because BRI1 interacts with and phosphorylates BIK1, we tested whether the kinase activity is required for their interaction. We coexpressed BIK1Km-GFP with BRI1-HA in protoplasts for Co-IP assay. As shown in Fig. 4C, BIK1Km-GFP coimmunoprecipitated BRI1-HA in vivo. However, the BL-induced BIK1-BRI1 dissociation was no longer observed with BIK1Km (Fig. 4C). Similarly, BIK1 still interacts with BRI1Km, and this interaction was not reduced upon BL treatment (Fig. 4D). Thus, the kinase activity of BIK1 and BRI1 is not required for their interaction, whereas it is indispensable for BL-induced BIK1-BRI1 dissociation. Apparently, release of BIK1 from BRI1 receptor complex upon BL-induced phosphorylation is one of early steps in transducing BR signaling.
BL-induced BIK1-BRI1 dissociation is strikingly similar with the dynamics of BIK1–FLS2 complex formation upon flagellin perception (15, 16). BIK1 constitutively interacts with FLS2 in the absence of flagellin, whereas the interaction was reduced upon treatment by flg22 (Fig. S7C). In FLS2 signaling, BAK1 is essential for flg22-induced BIK1 phosphorylation and BAK1 directly phosphorylates BIK1 in vitro, whereas FLS2 has little kinase activity (15, 16). Consistently, BAK1 is indispensible for flg22-induced BIK1-FLS2 dissociation because flg22 treatment no longer reduces BIK1-FLS2 association in bak1-4 mutant (Fig. 4E). These results support that phosphorylation of BIK1 by BAK1 upon flg22 perception leads to its dissociation from FLS2 receptor complex and, thereby, transducing intracellular FLS2 signaling. In contrast, BL-induced BIK1-BRI1 dissociation still occurred in the bak1-4 mutant (Fig. 4F), indicating that BAK1 is unlikely essential for BIK1 release from the BRI1 receptor in BR signaling. It is also possible that other SERK family members have redundant function with BAK1 for BIK1 release from the receptor complex (25). Consistent with the direct phosphorylation of BIK1 by BRI1, the BRI1 immunoprecipitated from bak1-4 mutant was still able to phosphorylate BIK1 in vitro (Fig. S7D). Considering that BIK1-BRI1 dissociation is a result of BIK1 phosphorylation and BRI1 phosphorylates BIK1, it is likely that BAK1 plays little direct role in BIK1 phosphorylation in BR signaling. Thus, BRI1 is able to directly phosphorylate BIK1 to transduce BR signaling, whereas BAK1 is essential to phosphorylate BIK1 in transducing flg22 signaling.
BIK1 Acts Downstream of BRI1 in BR Signaling.
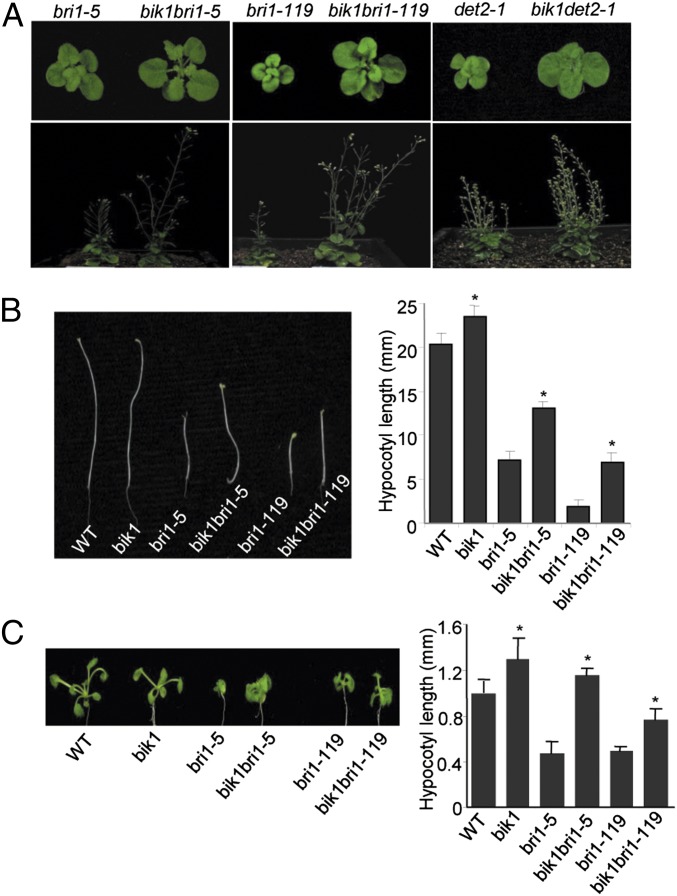
Our phenotypic, molecular, and biochemical data suggested that BIK1 negatively regulates BR signaling via association and phosphorylation by BRI1. We further investigated whether BRI1 functions genetically upstream of BIK1 by crossing the bik1 mutant with bri1-5 or bri1-119 mutant to generate the bik1bri1-5 and bik1bri1-119 double mutants. As shown in Fig. 5A, the bik1bri1-5 and bik1bri1-119 double mutants partially rescued the growth deficiency in the bri1 mutant. The double mutants displayed reduced growth dwarfism, enlarged leave size, and elongated inflorescence stems and branches compared with the bri1-5 or bri1-119 single mutant (Fig. 5A). Notably, the double mutants still exhibited the defects in silique development and had reduced seed yield, likely contributed by the mutation in BIK1 (Fig. S8) (28). The double mutant phenotype was unlikely caused by crossing of two different Arabidopsis ecotypes (24). The bik1det2-1 double mutant also partially suppressed the dwarf phenotype of det2 (Fig. 5A). Consistently, the hypocotyls of dark-grown bik1bri1-5 and bik1bri1-119 seedlings were longer than those of corresponding single mutants (Fig. 5B). Under the light, bik1bri1-5 and bik1bri1-119 seedlings also partially restored the leaf growth and hypocotyl length of the single mutants (Fig. 5C). These results suggest that BIK1 genetically interacts with BRI1 and inhibits BR signaling in vivo.
Fig. 5.
BIK1 acts downstream of BRI1 in BR signaling. (A) The bik1bri1-5, bik1bri1-119, and bik1det2-1 double mutants partially rescued the growth deficiency of single mutants. The phenotypes of 4-wk-old (Upper) and 8-wk-old (Lower) soil-grown plants are shown. (B) The hypocotyls of dark-grown bik1bri1-5 and bik1bri1-119 seedlings were longer than those of single mutants. Representatives of 8-d-old seedlings grown in the dark are shown on Left, and the quantification of hypocotyl length is shown on Right. The data are shown as mean ± SE from at least 25 seedlings. Asterisk indicates a significant difference with P < 0.05 compared with data from the corresponding single mutants. (C) The bik1bri1-5 and bik1bri1-119 seedlings partially restored the leave growth and hypocotyl length of single mutants grown under the light. Representatives of 14-d-old seedlings under the constant light are shown on Left, and the quantification of hypocotyl length is shown on Right.
Discussion
BIK1 was originally identified as Botrytis-induced kinase and plays critical roles in mediating plant resistance to necrotrophic fungal pathogens (28). We and others have shown that BIK1 is rapidly phosphorylated upon bacterial flagellin perception and associates with flagellin receptor complex in transducing plant immune signaling (15, 16). BIK1 is a direct substrate of BAK1, a shared regulatory component of multiple MAMP receptors and plant hormone BR receptor BRI1. Consistent with this notion, BIK1 is involved in immune signaling triggered by multiple MAMPs likely through association with the corresponding receptors. BIK1 is also phosphorylated upon ethylene treatment and required for responses to ethylene (30). Surprisingly, in this study, our extensive phenotypic, genetic, and biochemical examination of bik1 mutant revealed that in contrast to its positive roles in plant immune and ethylene signaling, BIK1 negatively regulates BR-mediated responses and signaling. BIK1 complexes with BRI1 and dissociates from the BRI1 complex upon BL perception, likely as a result of BIK1 phosphorylation by BRI1. Intriguingly, BAK1 is required for flagellin-induced BIK1 release from FLS2, but not for BL-induced BIK1 release from BRI1. Consistent with the data that FLS2 is a non–arginine-aspartate (RD) kinase with little kinase activity whereas BRI1 is an RD kinase with strong kinase activity (5), we observed a direct phosphorylation of BIK1 by BRI1, not by FLS2. Thus, BIK1 relays flagellin signaling via BAK1-mediated phosphorylation, whereas BIK1 is directly phosphorylated by BRI1 in transducing BR signaling (Fig. S9). Differential phosphorylation of BIK1 by BAK1 and BRI1 may determine the substrate specificity and signaling outcomes. Identification of the in vivo phosphorylation sites of BIK1 and characterization of their functional requirement in BR- and flagellin-mediated responses will shed light on how BIK1 positively regulates flagellin signaling but negatively controls BR signaling.
Several other members of the RLCKs, including BSKs from subfamily XII and CDG1 from subfamily VIIc have been identified as signaling components in transducing BR signaling via association with BR receptor BRI1 (8, 9). Similar to BIK1, BSKs associate with BRI1 and are released from the BRI1 complex upon BR perception. However, unlike BIK1, BSKs and CDG1 play positive roles in BR signaling. CDG1 directly phosphorylates BSU1, a phosphatase that dephosphorylates the negative regulator BIN2 (9). Phosphorylation of BSK1 by BRI1 promotes BSK1 binding to BSU1, which inactivates BIN2 kinase activity (33). Moreover, BSKs and CDG1 differ from BIK1 in that BSKs and CDG1 associate only with BRI1 and are phosphorylated by BRI1, not by BAK1. In contrast, BIK1 associates with both BRI1 and BAK1 and is phosphorylated by BRI1 and BAK1. Apparently, BIK1–BAK1 interaction and phosphorylation are more involved in flagellin signaling than that in BR signaling. Similar with BSKs and CDG1, BR-mediated BIK1 phosphorylation and BRI1-BIK1 dissociation are independent of BAK1. This result suggests the similarity and distinction of individual RLCKs in mediating different plant signaling pathways.
Our data indicate that the bik1 mutant possesses enhanced BR signaling. It has been shown that BR homeostasis and signaling antagonize flg22-induced responses (26, 27). One question is whether the compromised immune responses in the bik1 mutant are attributed to the elevated BR signaling. Accumulating evidence argues against this possibility. First, although exogenous application of BL inhibited certain FLS2- and EFR-mediated responses (26), it has not been reported that BR could promote disease symptom development or bacterial multiplication in Arabidopsis. In contrast, tobacco plants treated with BL exhibited enhanced resistance to multiple pathogens, including Pseudomonas bacteria (34). Besides compromised flg22-induced ROS production and defense gene activation, the bik1 mutant plants are also deficient in resistance to nonpathogenic bacterial infection and flg22-mediated restriction of bacterial growth (15, 16). Second, elevated BR signaling is not directly correlated with compromised immune responses. Although plants overexpressing BRI1 blocked FLS2- and EFR-mediated responses, BRI1sud1 plants with normal BRI1 protein level, but increased BRI1 signaling, display enhanced flg22-induced signaling (27). It has been proposed that the increased levels of BRI1, not increased BR signaling in BRI1 overexpressing plants, contribute to the antagonistic effects on MAMP signaling. We did not detect the reproducible difference of BRI1 protein level in WT and bik1 mutant, suggesting that enhanced BR signaling in bik1 mutant is not caused by overproduction of BRI1 receptor. Third, BR treatment did not affect flg22-mediated BIK1 phosphorylation (26). BIK1 phosphorylation is one of the earliest steps in flagellin signaling. Thus, the antagonistic effects on MAMP signaling by BL treatment are not due to the reduced BIK1 phosphorylation and activity. Together, the functions of BIK1 in flg22 and BR signaling are mechanistically uncoupled and the compromised immune responses in the bik1 mutant are not simply due to the elevated BR signaling.
Experimental Procedures
Plant Materials and Growth Conditions.
The bik1, sid2, bak1-4, and bik1sid2 mutants were reported (15, 30). The bri1-119 and det2-1 were obtained from the Arabidopsis Biological Resource Center. The bri1-5 mutant was obtained from Y. Yin (Ames, IA). The bik1bri1-5, bik1bri1-119, bik1det2-1 double mutants were generated by genetic crosses and confirmed by genotyping. Arabidopsis plants were grown in soil (Metro Mix 360) in a growth room at 23 °C, 60% relative humidity, and 75 μE m−2⋅s−1 light with a 12-h photoperiod for approximate 4 wk before protoplast isolations or RNA isolations. To grow Arabidopsis seedlings, the seeds were surface sterilized with 50% (vol/vol) bleach for 10 min, and then placed on the plates with 1/2 MS medium containing 0.5% sucrose, 0.8% agar, and 2.5 mM MES at pH 5.7. For various BR response assays, 100 nM BL (Chemiclones) or 2 μM BRZ (TCI America) were added in the medium. At least 25 seedlings were measured for each genotype and each treatment. All experiments were repeated three to four times, and the representative data were shown in the figures.
Plasmid Construction and Generation of Transgenic Plants.
Arabidopsis FLS2, BAK1, BAK1km, BIK1, and BIK1km constructs in plant expression vector or protein expression vector were reported (15). Full-length BRI1 was amplified by PCR from Col-0 cDNA and cloned into a plant expression vector. Cytosolic domain of BRI1 was cloned into the modified GST fusion protein expression vector pGEX4T-1 (Pharmacia) or pMAL-c2 (New England Biolabs). The BIK1 promoter up to 2.5 kb was amplified by PCR from Col-0 genomic DNA and introduced into pCB302 binary vector carrying BIK1-HA. All of the constructs were fully sequenced to verify mutations in the gene coding and promoter region. Stable transgenic lines were generated by using the standard Agrobacterium tumefaciens-mediated transformation in the pBRI1::BRI1-GFP transgenic plants or the bik1 mutant plants.
Protein Extraction, Co-IP, Phosphorylation, in Vitro Pull-Down, and Real-Time RT-PCR Assays.
The detailed procedures were described in SI Experimental Procedures. Briefly, 10-d-old seedlings were used for BES1 protein expression and 2 × 105 protoplasts or 15 g of leaf samples were used for Co-IP assays.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center, Drs. J. Li and Y. Yin for various Arabidopsis mutants and transgenic plants, Dr. Y. Yin for generous sharing of α-BES1 antibody, Dr. F. Rolland for protoplast BiFC vectors, and Dr. T. Devarenne for critical reading of the manuscript. The work was supported National Institute of Health Grants R01GM092893, USDA National Institute of Food and Agriculture Grant 2012-67013-19433 (to P.H.), NIH Grant R01GM097247, National Science Foundation Grant IOS-1030250, The Robert A. Welch Foundation (A-1795) (to L.S.), and National Science Foundation of China Grant 30800713 (to Z.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302154110/-/DCSupplemental.
References
- 1.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98(19):10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90(5):929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 3.Gómez-Gómez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5(6):1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 4.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125(4):749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15(2):220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313(5790):1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, et al. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell. 2005;8(6):855–865. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321(5888):557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim TW, Guan S, Burlingame AL, Wang ZY. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell. 2011;43(4):561–571. doi: 10.1016/j.molcel.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295(5558):1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 11.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109(2):181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 12.Heese A, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104(29):12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chinchilla D, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448(7152):497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 14.Roux M, et al. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell. 2011;23(6):2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu D, et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA. 2010;107(1):496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7(4):290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Boudsocq M, et al. Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature. 2010;464(7287):418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwessinger B, Ronald PC. Plant innate immunity: Perception of conserved microbial signatures. Annu Rev Plant Biol. 2012;63:451–482. doi: 10.1146/annurev-arplant-042811-105518. [DOI] [PubMed] [Google Scholar]
- 19.Lu D, et al. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science. 2011;332(6036):1439–1442. doi: 10.1126/science.1204903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110(2):203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 21.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110(2):213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 22.Chinchilla D, Shan L, He P, de Vries S, Kemmerling B. One for all: The receptor-associated kinase BAK1. Trends Plant Sci. 2009;14(10):535–541. doi: 10.1016/j.tplants.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He K, et al. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr Biol. 2007;17(13):1109–1115. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 24.Karlova R, et al. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell. 2006;18(3):626–638. doi: 10.1105/tpc.105.039412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gou X, et al. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 2012;8(1):e1002452. doi: 10.1371/journal.pgen.1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albrecht C, et al. Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci USA. 2012;109(1):303–308. doi: 10.1073/pnas.1109921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belkhadir Y, et al. Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci USA. 2012;109(1):297–302. doi: 10.1073/pnas.1112840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veronese P, et al. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell. 2006;18(1):257–273. doi: 10.1105/tpc.105.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan Z, Zhao J, Peng P, Chihara RK, Li J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 2009;150(2):710–721. doi: 10.1104/pp.109.138099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laluk K, et al. Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell. 2011;23(8):2831–2849. doi: 10.1105/tpc.111.087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19(5):765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu X, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65(4):634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 33.Kim TW, et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol. 2009;11(10):1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakashita H, et al. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 2003;33(5):887–898. doi: 10.1046/j.1365-313x.2003.01675.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.