Abstract
Placental trophoblasts form the interface between the fetal and maternal environments and serve to limit the maternal–fetal spread of viruses. Here we show that cultured primary human placental trophoblasts are highly resistant to infection by a number of viruses and, importantly, confer this resistance to nonplacental recipient cells by exosome-mediated delivery of specific microRNAs (miRNAs). We show that miRNA members of the chromosome 19 miRNA cluster, which are almost exclusively expressed in the human placenta, are packaged within trophoblast-derived exosomes and attenuate viral replication in recipient cells by the induction of autophagy. Together, our findings identify an unprecedented paracrine and/or systemic function of placental trophoblasts that uses exosome-mediated transfer of a unique set of placental-specific effector miRNAs to directly communicate with placental or maternal target cells and regulate their immunity to viral infections.
Keywords: C19MC, primary human trophoblasts, miR-517-3p
Strategies to reduce maternal–fetal infections are essential during pregnancy, where maternal infections can compromise the host, and maternal to fetal transmission of microbes can adversely impact the developing embryo (1–3). In eutherian organisms, the placenta shields the fetus from the hematogenous spread of diverse pathogens, including viruses, by providing a physical barrier interfacing the maternal and fetal blood systems. Located at the placental villous surface and in direct contact with the maternal blood is the syncytiotrophoblast, which comprises multinucleated, terminally differentiated cells that mediate the crucial exchange of gases, nutrients, and waste products between the mother and fetus. Once thought to function as a passive barrier to pathogens, it is now clear that trophoblasts actively orchestrate an extensive repertoire of signals designed to optimize placental transport functions, produce crucial hormones, and immunologically defend the developing fetus.
Placental trophoblasts are ideally positioned to govern the critical cross-talk between the maternal and fetal microenvironments and provide a mechanical and immunological defensive barrier to restrict virus access to the fetus. Antiviral innate immune signaling is crucial at the maternal–fetal interface, where vertical transmission of viruses from the maternal blood to the fetus can have profound pathological outcomes, ranging from neurodevelopment dysfunction to birth defects and fetal death. Additionally, viruses that compromise maternal health jeopardize the pregnancy even in the absence of vertical transmission to the fetus. Thus, elucidating the defense mechanism(s) used by placental trophoblasts to combat viral infections, as well as the possible mechanisms of viral counter measures, is paramount for designing therapeutic strategies aimed at preventing fetal disease.
Here we show that cultured primary human trophoblasts (PHTs) are highly resistant to infection by diverse and unrelated viruses and that conditioned PHT culture medium confers resistance to viral infections in nonplacental recipient cells. We find that a unique group of primate-specific microRNAs (miRNAs), which are highly expressed in human trophoblasts from the chromosome 19 miRNA cluster (C19MC) (4, 5), are packaged within PHT-derived exosomes and confer viral resistance to recipient cells. Complementing these findings, we show that PHT-derived exosomes and select C19MC miRNAs robustly induce autophagy in nonplacental recipient cells to facilitate enhanced antiviral defenses. Our findings illuminate a pathway used by human trophoblasts to suppress viral infections systemically by conferring viral resistance to nonplacental cells, suggesting a unique mechanism for shielding placental and maternal cells against viral infections during pregnancy.
Results
PHT-Derived Exosomes Protect Recipient Cells from Viral Infection.
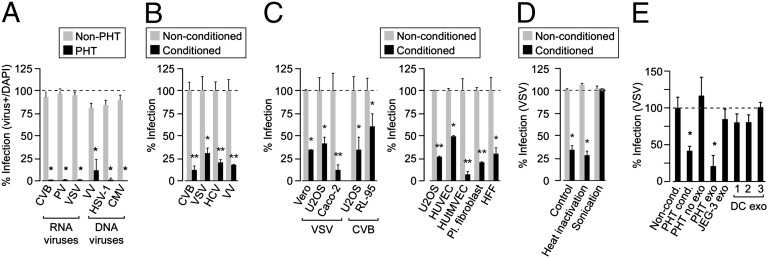
We found that PHT cells were resistant to infection by a panel of viruses, including coxsackievirus B3 (CVB), poliovirus (PV), vesicular stomatitis virus (VSV), vaccinia virus (VV), herpes simplex virus-1 (HSV-1), and human cytomegalovirus (CMV) compared with non-PHT cells (Fig. 1A; Fig. S1 A–D). This lack of viral replication was not due to inefficient viral binding and/or entry or to defects in common endocytic pathways used by viruses for their entry such as clathrin- or caveolar-mediated pathways (Fig. S1 E and F). Strikingly, we found that exposure of diverse non-PHT recipient cells for 24 h before infection to PHT conditioned medium (isolated from naïve PHT cells 48–72 h after plating) decreased the replication of CVB, VSV, hepatitis C virus (HCV), and VV (Fig. 1 B and C; Fig. S2A). We also observed the antiviral effect of conditioned PHT medium in several physiologically relevant fetal and/or maternal primary cells, including human umbilical vein endothelial cells (HUVECs), human uterine microvascular endothelial cells (HUtMVECs), human placental fibroblasts, and human foreskin fibroblasts (HFFs; Fig. 1C, Right). In contrast, conditioned medium from other cell types such as immortalized trophoblast BeWo cells had no effect (Fig. S2B). This effect was not the result of direct neutralization of the virus or induction of cell death, as conditioned medium had no direct effect on viral titers or cell viability (Fig. S2 C and D). Furthermore, we observed antiviral effects across multiple conditioned medium samples isolated from independent and unrelated PHT preparations (Fig. S2 E–G). Together, our data indicated that PHT cells release specific components to the medium, which are capable of conferring viral resistance to nonplacental recipient cells.
Fig. 1.
Conditioned PHT medium and exosomes confer viral resistance to recipient cells. (A) PHT or non-PHT cells were infected with a panel of viruses, including coxsackievirus B (CVB), poliovirus (PV), vesicular stomatitis virus (VSV), vaccinia virus (VV), herpes simplex virus-1 (HSV-1), or cytomegalovirus (CMV). Non-PHT cells were as follows: HeLa (CVB, PV), U2OS (VSV, HSV-1, and VV), and human foreskin fibroblasts (HFF; CMV). Shown are the percent infected cells [assessed by immunofluorescence (IF); *P < 0.0001]. (B) Non-PHT recipient cells were exposed for 24 h to nonconditioned or conditioned PHT medium and then infected with CVB, VSV, HCV, or VV. Non-PHT cells were as follows: HFF (CVB), U2OS (VSV, VV), and Huh 7.5 (HCV). Shown are the percent of infected cells [assessed by IF (CVB, VSV), luciferase assay (HCV), or qRT-PCR (VV); *P < 0.05, **P < 0.005]. (C) (Left) Cells were exposed to nonconditioned or conditioned PHT medium for 24 h and then infected with VSV or CVB. (Right) Cells were infected with VSV following exposure to nonconditioned or conditioned PHT medium (*P < 0.05, **P < 0.005). (D) Conditioned PHT medium was subjected to heat inactivation or sonication before 24-h exposure to Vero cells and then infected with VSV. Percent infection assessed as in A (*P < 0.0001). (E) U2OS cells were exposed for 24 h to nonconditioned, conditioned, exosome-depleted conditioned medium, exosomes purified from PHT, JEG-3, or from three preparations of murine dendritic cells (DCs) and then infected with VSV. Percent infection assessed as in A (*P < 0.0005); each PHT exosome preparation was derived from a different placental preparation.
To better define the component in conditioned medium of PHT cells that is responsible for conferring viral resistance, we exposed the conditioned medium to heat inactivation but found no effect (Fig. 1D). In contrast, sonication completely abolished the antiviral effect of PHT conditioned medium (Fig. 1D). Because exosomes, which function as cargo nanovesicles (6, 7), are characteristically released from trophoblasts and are sensitive to sonication (5, 8–11), we examined their role in PHT-mediated transfer of viral resistance. We found that exosomes purified from PHT conditioned medium attenuated VSV infection in recipient cells (Fig. 1E). Importantly, the antiviral effect was abrogated using exosome-depleted PHT conditioned medium (Fig. 1E). In addition, exosomes isolated from other cell types such as an immortalized human placental choriocarcinoma cell line (JEG-3) or primary murine dendritic cells had no effect on viral infection (Fig. 1E). Taken together, these data point to a direct role for PHT-derived exosomes in the transfer of viral resistance to nonplacental recipient cells.
C19MC-Associated miRNAs Confer Viral Resistance.
The transfer of RNA and/or miRNAs via exosomes may play an important role in exosome-based intercellular communication (6, 11, 12). The human C19MC is the largest known miRNA cluster, comprising 46 miRNAs that are highly expressed almost exclusively in the human placenta. Moreover, as a group, C19MC miRNAs are also the most abundant miRNA species in trophoblastic exosomes, with a strong correlation between C19MC miRNA levels in PHT cells and in PHT-derived exosomes (4, 5, 8, 13). To date, the function of these miRNAs has remained elusive. To test whether the expression of C19MC miRNAs could induce viral resistance in non-PHT cells, which do not naturally express these miRNAs, we stably transfected U2OS cells with a bacterial artificial chromosome (BAC) that contained the entire human C19MC cluster. Compared with U2OS cells transfected with a control BAC (that is deficient for the C19MC expression sequence), cells stably expressing C19MC BAC or cells exposed to PHT conditioned media expressed a higher level of C19MC miRNAs, as confirmed by RNA sequencing (RNAseq) and quantitative RT-PCR (qRT-PCR; Fig. S3 A and B; Table S1), and exhibited resistance to VSV infection (Fig. 2A). Likewise, transient transfection of U2OS cells with miRNA mimics of 16 C19MC-associated miRNAs [representing highly expressed miRNAs, or the two subfamilies of the C19MC (14)] markedly reduced VSV infection (Fig. 2B; Table S2). We also found that transfection of mimics of the highest six expressed C19MCs (5, 13) attenuated VSV infection, whereas transfection with mimics of the lowest expressed seven had no significant effect (Fig. 2B). To define the impact of individual miRNAs, we expressed individual mimics from among the highest expressed C19MC miRNAs and detected a significant inhibition of VSV infection with mimics of miR-517-3p, -516b-5p, and -512-3p, but not with mimics of several non–C19MC-associated miRNAs (miR-1, -424, -630, and -720; Fig. 2 C and D). Likewise, we found that a mimic of miR-517-3p also attenuated infection by the DNA viruses VV and HSV-1 (Fig. 2E).
Fig. 2.
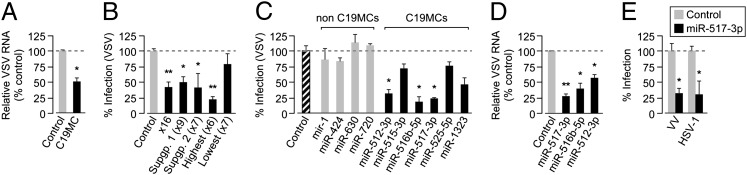
PHT and exosomal C19MC miRNAs confer viral resistance to recipient cells. (A) U2OS cells stably expressing control or C19MC BAC were infected with VSV (infection levels assessed by qRT-PCR; *P < 0.0001). (B) U2OS cells were transfected with C19MC miRNA mimics that represent the miRNA subgroups detailed in Table S2 or control mimics and then infected with VSV (shown as percent infected cells assessed by IF; *P < 0.05, **P < 0.001). (C) U2OS cells, transfected with mimics of the six highest expressed C19MC miRNAs, scrambled control, or non-C19MC (miR-1, -424, -630, -720) miRNA mimics, were infected with VSV (infection level assessed by IF or qRT-PCR; *P < 0.0005). (D) U2OS cells, transfected with mimics of the top three antiviral C19MC miRNAs or with scrambled control mimics, were infected with VSV (infection assessed by qRT-PCR; *P < 0.05, **P < 0.0001). (E) U2OS cells, transfected with scrambled control or miR-517b mimic, were infected with VV or HSV-1 (infection assessed as in D; *P < 0.0001).
PHT-Derived Exosomes and C19MC-Associated miRNAs Up-Regulate Autophagy.
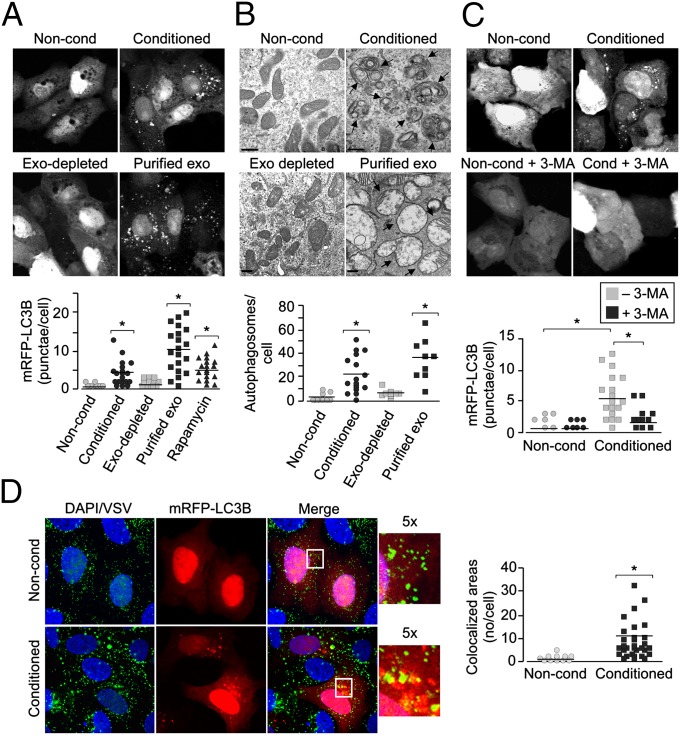
Mammalian cells use diverse defense mechanisms to combat microbial pathogens. One crucial mechanism is the induction of autophagy, an evolutionarily conserved lysosomal degradation pathway that has been associated with an array of cellular functions. Autophagy also degrades intracellular foreign microbial invaders (a process sometimes referred to as xenophagy) and thus serves as an important cellular response to suppress microbial infections. We found that exposure of U2OS cells to PHT conditioned medium or to purified PHT-derived exosomes markedly stimulated autophagy, as assessed by the formation of mRFP-LC3b–containing punctae and by electron microscopy, whereas conditioned medium depleted of PHT exosomes had no effect (Fig. 3 A and B; Fig. S4A). In contrast, we observed no effect of PHT conditioned medium or C19MC miRNAs on cell death induction or type I IFN signaling in recipient cells (Figs. S2D, S4E, and S5) and observed antiviral activity of conditioned PHT medium in cells that fail to respond to type I IFNs (Fig. S5E). In addition, PHT cells themselves also do not exhibit enhanced type I IFN signaling (Fig. S5A) but do exhibit high levels of autophagy (Fig. 4A; Fig. S6A–C). Autophagy induction was observed in diverse cell types (Fig. 3 A and B; Fig. S4B) and was absent in cells exposed to exosome-depleted PHT conditioned medium (Fig. 3 A and B). In addition, PHT conditioned medium induced the up-regulation (greater than threefold) of several key proautophagy transcripts (e.g., ATG4C, UVRAG, and LC3A) while having no effect on other innate immune pathway components (e.g., Toll-like receptors, IFN regulatory factors, cytokine-mediated signaling) in U2OS cells exposed to conditioned PHT medium (Fig. S4C; Table S3), further supporting the induction of autophagy. We found that 3-methyladenine (3-MA), an inhibitor of autophagosome biogenesis, inhibited autophagosome formation in recipient cells exposed to conditioned PHT medium (Fig. 3C). Last, we found that incoming VSV particles were trafficked to LC3b-positive punctae formed following exposure of cells to conditioned PHT medium, suggesting that the mislocalization or targeting of incoming viral particles to autophagosomes and/or autolysosomes might impact viral replication (Fig. 3D).
Fig. 3.
PHT-derived exosomes induce autophagy in recipient cells. (A) U2OS cells transfected with mRFP-LC3b were exposed to nonconditioned, PHT conditioned, or exosome-depleted conditioned PHT medium or purified PHT exosomes for 24 h, and LC3b punctae formation was assessed by confocal microscopy. (Upper) Confocal micrographs. (Lower) Quantification of mRFP-LC3b punctae per cell (*P < 0.0001). (B) (Upper) Electron micrographs of cells exposed to nonconditioned or conditioned PHT medium (Vero), exosome-depleted conditioned PHT medium (Vero), or purified PHT exosomes (U2OS). Arrows denote autophagosomes. (Scale bar, 500 nm.) (Lower) Quantification of electron micrographs of cells exposed to nonconditioned (Vero and U2OS), conditioned PHT media samples (Vero and U2OS), exosome-depleted conditioned medium (Vero), or purified PHT exosomes (U2OS) (*P < 0.0001). (C) U2OS cells transfected with mRFP-LC3b were exposed to nonconditioned or conditioned PHT medium in the absence or presence of 3-MA for 8 h, and LC3b punctae formation was assessed by confocal microscopy. (Upper) Confocal micrographs. (Lower) Quantification of mRFP-LC3b punctae (*P < 0.0005). (D) (Left) Confocal images of VSV entry into U2OS cells transiently transfected with mRFP-LC3b exposed to nonconditioned (Upper) or conditioned (Lower) PHT medium. VSV particles are shown in green and DAPI-stained nuclei are shown in blue (Inset, magnification: 5×). Areas of colocalization appear as yellow. (Right) Quantification of the extent of colocalization between VSV particles and mRFP-LC3B positive punctae (*P < 0.0001).
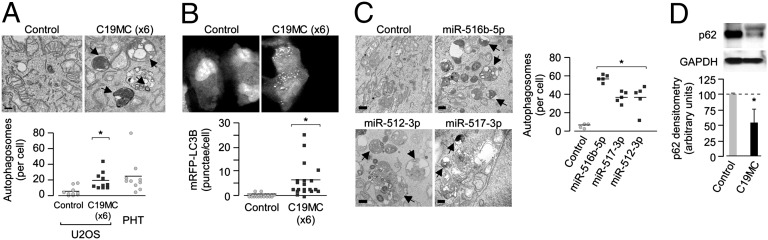
Fig. 4.
C19MC miRNAs induce autophagy. (A) (Upper) Electron micrographs of U2OS cells transfected with scrambled control or the six highest expressed C19MC miRNA mimics (Table S2). Black arrows denote autophagosomes and/or autolysosomes. (Scale bar, 500 nm.) (Lower) Quantification of electron micrographs shown at top (*P < 0.005) or in PHT cells. (B) U2OS cells were transfected with mRFP-LC3b and either scrambled control or the six highest expressed C19MC miRNA mimics. (Upper) Confocal micrographs. (Lower) Quantification of mRFP-LC3b punctae per cell (*P = 0.0005). (C) (Left) Electron micrographs of U2OS cells transfected with scrambled control or the most potent antiviral miRNA mimics. Black arrows denote autophagosomes and/or autolysosomes. (Scale bar, 500 nm.) (Right) Quantification of adjacent electron micrographs (*P < 0.005). (D) (Upper) A representative immunoblot for p62 or GAPDH in U2OS cells stably transfected with either control Del or C19MC BAC. (Lower) Densitometry of p62 levels (normalized to GAPDH) from three independent immunoblots as described above (*P < 0.05).
Because we observed a role for C19MC-associated miRNAs in the induction of viral resistance, we assessed whether these miRNAs could induce autophagy. We found that transfection of cells with mimics of six of the highest expressed C19MC miRNAs (Fig. 4 A and B), the entire C19MC (Fig. S4D), or mimics of individual C19MC miRNAs that attenuated viral infection (Fig. 4C) also stimulated autophagy, as observed by mRFP-LC3b punctate formation or by electron microscopy, and did not induce cell death (Fig. S4E). Furthermore, we found that C19MC-associated induction of autophagy occurred via the up-regulation of autophagic flux, as supported by a decrease in p62 protein levels in cells expressing the entire C19MC (Fig. 4D) without a corresponding reduction in p62 mRNA levels (Fig. S4F).
Antiviral Effects of C19MC-Associated miRNAs Require Autophagy.
We observed an inhibition of viral replication and a pronounced up-regulation of autophagy in cells exposed to PHT conditioned medium and in cells expressing C19MC-associated miRNAs. To determine whether the antiviral effects of these conditions involved autophagy, we suppressed autophagy by treatment of cells with 3-MA or by RNAi-mediated silencing of beclin-1, a key factor in autophagic induction (15). We found that inhibition of autophagy using 3-MA or by RNAi-mediated silencing of beclin-1 expression significantly restored the level of VSV infection in U2OS cells expressing the entire C19MC (Fig. 5 A and B) or in cells exposed to PHT conditioned medium (Fig. S4G). Interestingly, we also found that addition of 3-MA to PHT cells also enhanced VSV infection (Fig. 5C). These data show that the induction of autophagy is critical for the antiviral effect of C19MC miRNAs.
Fig. 5.
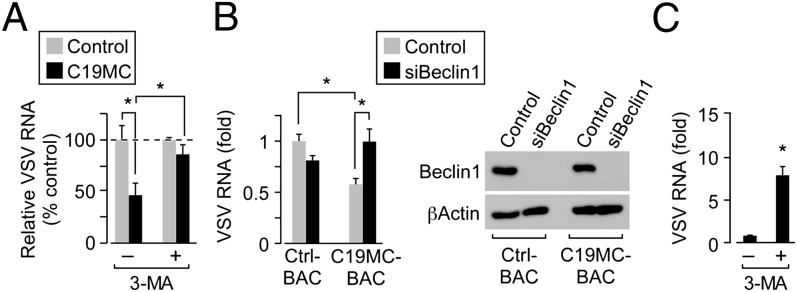
Suppression of autophagy restores C19MC-medated antiviral effects. (A) U2OS cells transfected with scrambled control or miRNA mimics of the six most prevalent C19MC miRNA mimics. Cells were exposed to 3-MA before and during VSV infection. Relative VSV RNA was analyzed by qRT-PCR (*P < 0.0005). (B) (Left) U2OS cells stably expressing control or C19MC BAC transfected with scrambled control siRNA or beclin-1 siRNA for 72 h were infected with VSV, and relative infection was determined by qRT-PCR. (*P < 0.05, determined using ANOVA with Boneferroni correction). (Right) Immunoblots for beclin-1 or actin in cells transfected as described above. (C) PHT cells were treated with 3-MA before infection with GFP-VSV (in the presence of 3-MA). Relative VSV RNA was analyzed by qRT-PCR (*P < 0.005).
Discussion
The placenta shields the embryo from the spread of pathogens. Here we report on the striking finding that human placental trophoblasts are resistant to viral infection and are capable of conferring viral resistance to nonplacental cells. Viral resistance is transferrable via trophoblastic conditioned medium, trophoblastic exosomes, or miRNA members of the C19MC primate-specific cluster, which are packaged within exosomes (5). Together, this pathway may constitute a powerful evolutionary adaptation to enhance the protection of the developing fetus against viral invaders.
We and others have shown that PHT cells produce robust levels of miRNAs throughout pregnancy, as well as other small RNAs [piwi-interacting RNAs (piRNAs), small nuclear (snRNAs), small nucleolar (snoRNAs)] (10, 13, 17, 18). Interestingly, many of these miRNAs, including members of the C19MC, are found in the maternal blood throughout pregnancy and rapidly decline in the first 24 h postpartum (19, 20), suggesting a miRNA-based mechanism for fetal–maternal communication (13, 21). Our data thus provide evidence for a unique paracrine and/or systemic function of placental trophoblasts, using exosome-mediated transport of a unique set of primate-specific effector miRNAs to directly communicate with maternal cells and possibly neighboring placental cells and regulate their immunity to viral infections. It is possible that PHT-derived C19MC miRNA–containing exosomes specifically target their cargo to a discrete subpopulation of maternal cells, or may aid in the selectively eliciting antiviral responses and up-regulating autophagy.
Our data show that conditioned media from PHT cells, purified PHT-derived exosomes, and miRNA mimics of several members of the C19MC potently induce autophagy, an important component of host antiviral signaling (22). The degradation of microbes via the fusion of autophagosomes with lysosomes is a key component in the antimicrobial effects of autophagy, yet autophagy can also direct MHC class II presentation (23), the production of antiviral type I IFNs downstream of Toll-like receptor 7 engagement (24), or even altered T-cell signaling (25). Strikingly, we show that blocking autophagy using either a pharmacological or molecular approach at least partially restored viral susceptibility of recipient cells exposed to PHT conditioned medium or expressing C19MC-associated miRNAs, supporting a role for this cellular pathway in the induction of viral resistance by these conditions. Whereas some of the viruses used in our study (e.g., CVB and HCV) may benefit from the formation of autophagic vesicles during their replication (26, 27), these viruses were also sensitive to the antiviral effects of C19MC miRNAs. There may be several explanations for this, and it is important to point out that the pathway(s) by which C19MC-associated miRNAs induce autophagy may differ from the pathways that have been described previously, thus complicating the direct comparison of our results using C19MC-associated miRNAs and the work of others who used different mechanisms to stimulate autophagy (most commonly by rapamycin- or starvation-induced autophagy). In addition, many studies focus specifically on autophagy induced in response to viral replication (likely via the induction of innate immune signaling downstream of pattern recognition receptor activation). In contrast, our findings specifically focus on the induction of autophagy before the initiation of viral replication, which could profoundly impact the ability of incoming viral particles to properly traffic and/or release their genomes, consistent with our findings with VSV (Fig. 3D). Similarly, we found that expression of a VV gene rpo35 (encoding a subunit of a DNA-directed RNA polymerase) that is expressed within 2 h postentry (28, 29) is inhibited in PHT cells and by conditioned PHT medium in recipient cells (Figs. S1D and S2G), suggesting that the inhibition of viral replication in PHT cells and non-PHT cells exposed to conditioned medium occurs early in the virus life cycle. Interestingly, we found that VSV infection was enhanced in PHT cells by treatment with 3-MA (Fig. 5C), suggesting that autophagy may be at least one mechanism by which these cells resist viral infections. Notably, the overall level of infection in PHT remained quite low (∼10% overall infection), suggesting additional mechanisms may protect these cells from viral infections. Our results do not rule out other functions for autophagy in placental trophoblasts, such as maintenance of cellular homeostasis in terminally differentiated syncytiotrophoblasts. Likewise, although our data point to a prominent role for autophagy in the antiviral effects of conditioned PHT medium, PHT-derived exosomes, and C19MC-associated miRNAs, they do not exclude other possible cellular pathways that may be affected or converge on the autophagic pathway, such as other innate immune pathways (30), although our data indicate that type I IFN signaling plays no role in C19MC-induced viral resistance (Fig. S5).
Surprisingly, we found that, unlike the other viruses tested in our study, conditioned PHT medium and expression of C19MC miRNAs significantly enhanced hCMV infection (Fig. S2 H and I), indicating that, although C19MC miRNAs attenuate the replication of many viruses, they may function in a proviral manner to enhance the infection of CMV and possibly other viruses. Our findings (Fig. 1A) and the work of others (31) suggest that PHT cells are resistant to CMV infection, and studies of CMV-infected placentas suggest that CMV specifically targets invasive and endovascular cytotrophoblasts as a means of entry into the fetal compartment (31–33). CMV is known to inhibit host autophagosome biogenesis during its infection, even when autophagy is stimulated (34, 35). However, the specific mechanism(s) underlying the up-regulation of CMV infection by C19MC miRNAs are likely complex, involving diverse viral and/or cellular strategies.
Our work defines an unexpected role for C19MC miRNAs in transferrable, autophagy-mediated antiviral responses (Fig. S7). Whereas our data demonstrate the influence of human miR-512-3p, -516b-5p, and -517-3p on viral infection and autophagy, the identification of relevant molecular targets and autophagic pathways that are impacted by these and other members of the C19MC is ongoing (36). Importantly, our data suggest that key members of the C19MC miRNAs identified in our studies mount an antiviral response through synergistic interactions with other C19MC members and possibly even through up-regulation of gene expression (37). It is also likely that other components within PHT-derived exosomes or within conditioned PHT medium potentiate the antiviral effects of C19MC-associated miRNAs. These components may interact with a network of C19MC miRNA targets to mount an antiviral response, spanning diverse and possibly redundant pathways (38). Thus, exosomal C19MC miRNAs may direct a pathogen-specific response, facilitating the deployment of a selective repertoire of defense mechanisms designed to protect the developing feto-placental unit and maternal tissues against viral infections.
Materials and Methods
Cell and Exosome Purification.
PHT cells were isolated from healthy singleton term placentas using the trypsin-DNase-dispase/Percoll method as described by Kliman et al., with previously published modifications (39, 40) under an exempt protocol approved by the Institutional Review Board at the University of Pittsburgh. Under the protocol, patients provided written consent for the use of de-identified discarded tissues for research on admittance to the hospital. Cells were maintained in DMEM (Sigma) containing 10% (vol/vol) FBS (HyClone) and antibiotics at 37 °C in a 5% CO2 air atmosphere. Cells were maintained 72 h after plating, with cell quality monitored both morphologically (by microscopy) and by medium human chorionic gonadotropin (hCG) levels (ELISA; DRG International), showing a characteristic increase in medium hCG as cytotrophoblasts differentiate into syncytiotrophoblasts (40, 41).
For isolation of PHT (or JEG-3) exosomes, PHT cells were maintained for 48 h in DMEM containing 10% FBS that was ultracentrifuged at 108,000 × g for 10 h to deplete preexisting FBS exosomes. Exosomes were isolated as described previously (5, 9). Briefly, supernatants from 200 million PHT cells were centrifuged at 300 × g for 5 min, 1,200 × g for 10 min, and 10,000 × g for 30 min. Exosomes were concentrated by centrifugation at 2,500 × g for 25 min using a Vivacell 100 filter (BioExpress) and then ultracentrifuged at 108,000 × g for 1 h, and the pellet was subsequently ultracentrifuged on top of a 30% sucrose/D2O density cushion at 108,000 × g for 1 h (42). The exosomal phase was collected and resuspended in PBS. Dendritic cell (DC)-derived exosomes were purified as previously described (43) from culture supernatants of C57BL/6 mouse DCs generated from bone marrow precursors cultured in medium supplemented 10% FCS, GM-CSF (1,000 U/mL), and IL-4 (500 U/mL).
Conditioned media from PHT or other cells were harvested between 48 and 72 h after plating. Nonconditioned medium was complete PHT medium (described in detail above) that had not been incubated with PHT cells. Conditioned media were subjected to sonication or heat inactivation for 30 min at 65 °C. Recipient cells were exposed to conditioned medium for ∼24 h before assays.
Viruses.
Experiments were performed with VSV, GFP-tagged VSV, recombinant yellow fluorescent protein (YFP)-tagged VV as described (44), coxsackievirus B3-RD isolate (CVB3-RD) as described (45), PV as described (46), CMV (hCMV Towne strain, obtained from William Goins, University of Pittsburgh), cell culture grown HCV expressing firefly luciferase (HCVcc-luc), or GFP-tagged HSV-1 (strain KOS, provided by Prashant Desai, The Johns Hopkins University) as described (47). Viral titers were determined by plaque assays as described (48). HCVcc-luc propagation was performed as described (49).
For experiments assessing productive viral infection, PHT cells were infected with CVB, PV, VSV, VV, or HSV-1 for 14–15 h [multiplicity of infection (MOI) = 5] or CMV for 24 h for immunofluorescence. For 3-MA experiments assessed by qRT-PCR, PHT cells were infected with GFP-VSV for 15 h at MOI = 5. For experiments analyzing immediate early viral gene expression measured by qRT-PCR, PHT cells were infected with CVB, VSV, VV, or HSV-1 for 6–7 h at MOI = 1. HeLa cells were infected with CVB or PV at an MOI = 5 for 8 h. HFF cells were infected with CMV for 24 h and VSV or CVB (MOI = 5) for 15 h. Vero cells were infected with VSV for 6 h (MOI = 5). Caco-2 cells were infected with VSV or CVB for 7 h (MOI = 5). RL-95 cells were infected with CVB for 15 h (MOI = 5). For immunofluorescence, U2OS cells were infected with CVB for 7 h (MOI = 5) or VSV (MOI = 5), VV, or HSV-1 (MOI = 1) for 15 h. For qRT-PCR, U2OS cells were infected with CMV, VSV, HSV-1, or VV for 5–6 h (MOI = 1). Huh7.5 cells were infected with HCVcc as described previously (49).
miRNA Mimics, Plasmids, and Transfections.
Mimics for C19MC miRNAs (miRIDIAN), as well as a nontargeting control miRNA mimic, were obtained from Thermo-Fisher Scientific (Dharmacon) as described (5). U2OS cells were reverse transfected with one or multiple miRNA mimics or miRNA mimic control (final concentration 6 nM for each miRNA mimic) using DharmaFECT-1 transfection reagent (Thermo-Fisher Scientific) or Hiperfect (Qiagen) according to the manufacturers’ instructions. Cells were assayed 48 h posttransfection. The total concentration of nontargeting control miRNA mimics was adjusted to that of all active miRNA mimics. For siRNA transfections, U2OS cells were reverse transfected using DharmaFECT-1 transfection reagent. For silencing of beclin-1, 40 nM per well of scrambled nontargeting siRNA (siControl) or Beclin-1 siRNA (Cell Signaling, #6222S) was transfected. Plasmid transfections were performed using X-tremeGENE 9 (Roche) according to the manufacturer’s protocol. The mRFP-LC3b expression construct was purchased from Addgene (plasmid 21075) and originally constructed by Tamotsu Yoshimori (50). For experiments with conditioned media and purified exosomes, cells were exposed to media 24 h posttransfection and fixed 48 h posttransfection. For all other experiments, cells were assayed 48 h posttransfection.
General Statistical Analysis.
Experiments were performed at least three times as indicated in the figure legends or as detailed. Data are presented as mean ± SD. Except where specified, Student t test was used to determine statistical significance for virus infection and autophagy assays when two sets were compared, and one-way ANOVA with Bonferroni’s correction for post hoc multiple comparisons was used to determine the statistical significance for reporter gene assays. P < 0.05 was determined significant.
Additional methods can be found in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jon Boyle and William Goins (University of Pittsburgh) for generously providing reagents, Prashant Desai (The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins) for GFP-HSV-1, and Carl Hubel (Magee Womens Research Institute) for HUVECs and HUtMVECs. We thank Elena Sadovsky, William Shufesky, Magdalena Jennings, Stefanie Morosky, Jana Jacobs, Katharine Harris, and Judy Ziegler for technical assistance and Lori Rideout for assistance in manuscript preparation. E.D.A. was supported by a teaching fellowship obtained through the University of Pittsburgh School of Medicine and R.B.D was supported by a postdoctoral research fellowship from Magee-Womens Research Institute. The generation of the C19MC BAC, as described, was also made possible by National Institutes of Health (NIH) Grants UL1-RR024153 and UL1-TR000005. The project was supported by the Pennsylvania Department of Health Research Formula Funds (to J.-F.M. and T.C.), NIH Grants R01-HD065893 and R21-HD071707 (to Y.S.), NIH Grant R01-AI081759 (C.B.C.), and NIH Grant R01-HD075665 (to C.B.C. and Y.S.). In addition, C.B.C. is a recipient of the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award.
Footnotes
Conflict of interest statement: C.B.C. and Y.S. are named inventors on a pending patent application describing the use of C19MC microRNAs as therapeutics.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304718110/-/DCSupplemental.
References
- 1.Ornoy A, Tenenbaum A. Pregnancy outcome following infections by coxsackie, echo, measles, mumps, hepatitis, polio and encephalitis viruses. Reprod Toxicol. 2006;21(4):446–457. doi: 10.1016/j.reprotox.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Euscher E, Davis J, Holzman I, Nuovo GJ. Coxsackie virus infection of the placenta associated with neurodevelopmental delays in the newborn. Obstet Gynecol. 2001;98(6):1019–1026. doi: 10.1016/s0029-7844(01)01625-8. [DOI] [PubMed] [Google Scholar]
- 3.Horstmann DM. Viral infections in pregnancy. Yale J Biol Med. 1969;42(2):99–112. [PMC free article] [PubMed] [Google Scholar]
- 4.Noguer-Dance M, et al. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet. 2010;19(18):3566–3582. doi: 10.1093/hmg/ddq272. [DOI] [PubMed] [Google Scholar]
- 5.Donker RB, et al. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod. 2012;18(8):417–424. doi: 10.1093/molehr/gas1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 7.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor DD, Akyol S, Gercel-Taylor C. Pregnancy-associated exosomes and their modulation of T cell signaling. J Immunol. 2006;176(3):1534–1542. doi: 10.4049/jimmunol.176.3.1534. [DOI] [PubMed] [Google Scholar]
- 9.Montecalvo A, et al. Exosomes as a short-range mechanism to spread alloantigen between dendritic cells during T cell allorecognition. J Immunol. 2008;180(5):3081–3090. doi: 10.4049/jimmunol.180.5.3081. [DOI] [PubMed] [Google Scholar]
- 10.Luo SS, et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81(4):717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 11.Pegtel DM, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107(14):6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39(1):133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Mouillet JF, et al. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta. 2010;31(9):781–784. doi: 10.1016/j.placenta.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin S, et al. Computational identification and characterization of primate-specific microRNAs in human genome. Comput Biol Chem. 2010;34(4):232–241. doi: 10.1016/j.compbiolchem.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 16.Mouillet JF, Chu T, Sadovsky Y. Expression patterns of placental microRNAs. Birth Defects Res A Clin Mol Teratol. 2011;91(8):737–743. doi: 10.1002/bdra.20782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barad O, et al. MicroRNA expression detected by oligonucleotide microarrays: System establishment and expression profiling in human tissues. Genome Res. 2004;14(12):2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pineles BL, et al. 2007. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol 196(3):e261–e266.
- 19.Ng EK, et al. mRNA of placental origin is readily detectable in maternal plasma. Proc Natl Acad Sci USA. 2003;100(8):4748–4753. doi: 10.1073/pnas.0637450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilad S, et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chim SS, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54(3):482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 22.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.English L, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10(5):480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315(5817):1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 25.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455(7211):396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 26.Jackson WT, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3(5):e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106(33):14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amegadzie BY, Ahn BY, Moss B. Identification, sequence, and expression of the gene encoding a Mr 35,000 subunit of the vaccinia virus DNA-dependent RNA polymerase. J Biol Chem. 1991;266(21):13712–13718. [PubMed] [Google Scholar]
- 29.Wang Z, et al. Skin mast cells protect mice against vaccinia virus by triggering mast cell receptor S1PR2 and releasing antimicrobial peptides. J Immunol. 2012;188(1):345–357. doi: 10.4049/jimmunol.1101703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jounai N, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci USA. 2007;104(35):14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan G, Hemmings DG, Yurochko AD, Guilbert LJ. Human cytomegalovirus-caused damage to placental trophoblasts mediated by immediate-early gene-induced tumor necrosis factor-alpha. Am J Pathol. 2002;161(4):1371–1381. doi: 10.1016/s0002-9440(10)64413-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maidji E, Percivalle E, Gerna G, Fisher S, Pereira L. Transmission of human cytomegalovirus from infected uterine microvascular endothelial cells to differentiating/invasive placental cytotrophoblasts. Virology. 2002;304(1):53–69. doi: 10.1006/viro.2002.1661. [DOI] [PubMed] [Google Scholar]
- 33.Maidji E, Genbacev O, Chang HT, Pereira L. Developmental regulation of human cytomegalovirus receptors in cytotrophoblasts correlates with distinct replication sites in the placenta. J Virol. 2007;81(9):4701–4712. doi: 10.1128/JVI.02748-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaumorcel M, et al. The human cytomegalovirus protein TRS1 inhibits autophagy via its interaction with Beclin 1. J Virol. 2012;86(5):2571–2584. doi: 10.1128/JVI.05746-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaumorcel M, Souquère S, Pierron G, Codogno P, Esclatine A. Human cytomegalovirus controls a new autophagy-dependent cellular antiviral defense mechanism. Autophagy. 2008;4(1):46–53. doi: 10.4161/auto.5184. [DOI] [PubMed] [Google Scholar]
- 36.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: MicroRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 38.Labialle S, Cavaillé J. Do repeated arrays of regulatory small-RNA genes elicit genomic imprinting?: Concurrent emergence of large clusters of small non-coding RNAs and genomic imprinting at four evolutionarily distinct eutherian chromosomal loci. Bioessays. 2011;33(8):565–573. doi: 10.1002/bies.201100032. [DOI] [PubMed] [Google Scholar]
- 39.Kliman HJ, Nestler JE, Sermasi E, Sanger JM, Strauss JF., 3rd Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology. 1986;118(4):1567–1582. doi: 10.1210/endo-118-4-1567. [DOI] [PubMed] [Google Scholar]
- 40.Nelson DM, Johnson RD, Smith SD, Anteby EY, Sadovsky Y. Hypoxia limits differentiation and up-regulates expression and activity of prostaglandin H synthase 2 in cultured trophoblast from term human placenta. Am J Obstet Gynecol. 1999;180(4):896–902. doi: 10.1016/s0002-9378(99)70661-7. [DOI] [PubMed] [Google Scholar]
- 41.Chen B, Nelson DM, Sadovsky Y. N-myc down-regulated gene 1 modulates the response of term human trophoblasts to hypoxic injury. J Biol Chem. 2006;281(5):2764–2772. doi: 10.1074/jbc.M507330200. [DOI] [PubMed] [Google Scholar]
- 42.Lamparski HG, et al. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods. 2002;270(2):211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 43.Montecalvo A, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119(3):756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moser TS, Jones RG, Thompson CB, Coyne CB, Cherry S. A kinome RNAi screen identified AMPK as promoting poxvirus entry through the control of actin dynamics. PLoS Pathog. 2010;6(6):e1000954. doi: 10.1371/journal.ppat.1000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coyne CB, Bergelson JM. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell. 2006;124(1):119–131. doi: 10.1016/j.cell.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 46.Coyne CB, Kim KS, Bergelson JM. Poliovirus entry into human brain microvascular cells requires receptor-induced activation of SHP-2. EMBO J. 2007;26(17):4016–4028. doi: 10.1038/sj.emboj.7601831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72(9):7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bozym RA, et al. Focal adhesion kinase is a component of antiviral RIG-I-like receptor signaling. Cell Host Microbe. 2012;11(2):153–166. doi: 10.1016/j.chom.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu S, et al. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83(4):2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.