Abstract
The ataxia telangiectasia mutated (ATM) checkpoint is the central surveillance system that maintains genome integrity. We found that in the context of childhood sarcoma, mammalian target of rapamycin (mTOR) signaling suppresses ATM by up-regulating miRNAs targeting ATM. Pharmacological inhibition or genetic down-regulation of the mTOR pathway resulted in increase of ATM mRNA and protein both in mouse sarcoma xenografts and cultured cells. mTOR Complex 1 (mTORC1) suppresses ATM via S6K1/2 signaling pathways. microRNA-18a and microRNA-421, both of which target ATM, are positively controlled by mTOR signaling. Our findings have identified a negative feedback loop for the signaling between ATM and mTOR pathways and suggest that oncogenic growth signals may promote tumorigenesis by dampening the ATM checkpoint.
Keywords: childhood cancer, kinase inhibitor
Genome instability is an enabling hallmark of cancer cells (1, 2). The ataxia telangiectasia mutated (ATM) checkpoint is the central genome caretaker to promote cell cycle arrest, DNA damage repair and cell death via apoptosis in response to genotoxins (3, 4). One of the mechanisms by which the ATM checkpoint prevents tumorigenesis is to inhibit mTOR signaling at the convergence of RAS-mitogen-activated protein kinase (RAS-MAPK) and phosphatidylinositol-3′-kinase and protein kinase B, also named AKT signaling pathways (5). mTORC1 is one of the complexes formed by mTOR kinase and the central integrator and processor of intracellular and extracellular signals (6). The tumor suppressor TP53 plays an essential role between mTORC1 signaling and the ATM checkpoint (5, 7). Following activation by ATM-checkpoint kinase 2 (ATM-CHK2) and/or ATM and Rad3-related protein-checkpoint kinase 1 (ATR-CHK1), p53 suppresses mTORC1 signaling through inducing the expression of negative regulators upstream of mTORC1 that include phosphatase and tensin homolog (PTEN), 5′-AMP-activated protein kinase catalytic subunit beta-1 (AMPK1β), and tuberin (TSC2) (8, 9). Activated p53 also promotes the phosphorylation and activation of 5′-AMP-activated protein kinase catalytic subunit alpha (AMPKα) via SESTRIN1/2 (10). Hypoxia-activated ATM phosphorylates hypoxia inducible factor 1 alpha (HIF-1α), which in turn leads to TSC1/2 activation and hence inhibition of mTORC1 through regulated in DNA damage and development 1 and 2 (REDD1/2) signaling (11–13). Thus, ATM negatively regulates mTORC1 signaling in response to cellular stress such as DNA damage and hypoxia (5, 13).
Most childhood solid tumors, including rhabdomyosarcoma, neuroblastoma, brain tumors, and osteosarcoma, are nonfamilial cancers. The Cancer Genome Project showed that there are an average of 100 genetic changes in a solid tumor and it is widely agreed that it takes several decades for a sporadic tumor to happen (14, 15). The early occurrence of childhood cancers suggests a higher mutator phenotype of pediatric tumors, although the drivers are unknown. Previously, we showed that the levels of ATM mRNA expression and protein were significantly suppressed in most pediatric solid tumor xenografts compared with xenografts derived from acute lymphoblastic leukemia (ALL) (13). We hypothesized that there may be attenuated ATM-mediated genome surveillance in these pediatric tumors, and that maintained signaling by mTORC1 may provide feedback to suppress ATM, potentially enhancing progression. To test this hypothesis, we have manipulated TOR signaling using pharmacologic and genetic approaches in a sarcoma cell line and xenograft models of childhood sarcoma. Our results suggest that mTORC1 signaling negatively regulates ATM levels in these models.
Results
Inhibition of mTOR Signaling Results in Up-Regulation of ATM Protein Levels.
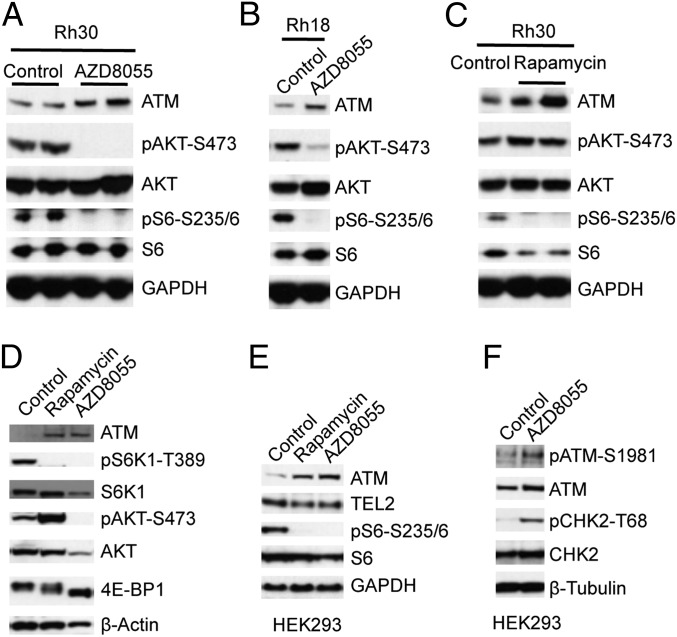
Previously we demonstrated that levels of ATM mRNA and protein were lower in xenografts derived from childhood solid tumors compared with xenografts derived from pediatric ALL (13). ATM protein levels were recently found to be controlled by TEL2 at the posttranslational level (16), but we did not detect an apparent correlation between ATM and telomere maintenance 2 (TEL2) (Fig. S1). Because mTOR signaling is up-regulated in most cancers and accumulating evidence demonstrates that oncogenic alteration of this pathway leads to cancer (17–19), we hypothesized that the decreased ATM may be due to the increased activity of the mTOR pathway in childhood solid cancers. To test this, we determined ATM protein levels in rhabdomyosarcoma (Rh) xenograft models Rh30 and Rh18 that were treated with an mTOR kinase inhibitor Astrazeneca (AZD)8055 (20). AZD8055 efficiently inhibited both mTORC1 and mTORC2 complexes as demonstrated by the disappearance of pS6-S235/6 and pAKT-S473 signals, markers for the activity of mTORC1 and mTORC2, respectively. Intriguingly, AZD8055 increased ATM both in Rh30 and Rh18 models (Fig. 1 A and B). We next examined ATM levels in Rh30 xenografts treated with rapamycin. Rapamycin treatment abrogated pS6-S235/6, indicating inhibition of mTORC1, whereas slightly increasing levels of pAKT-S473 by suppressing the S6K1-IRS–negative feedback loop. Similarly to AZD8055, rapamycin increased ATM levels (Fig. 1C). These data suggest that mTOR signaling suppresses ATM in vivo.
Fig. 1.
Inhibition of mTOR signaling results in increase of ATM protein levels in sarcoma xenografts. (A) Rh30 tumor xenografts were propagated s.c. in SCID mice and were treated with the mTOR kinase inhibitor AZD8055 at 20 mg/kg per day. Twenty-four hours after treatment on day 4, tumors were excised, rapidly frozen in liquid N2, and pulverized under liquid N2. Total proteins were extracted for immunoblotting. Anti-pS6-S235/6, S6, pAKT-S473, AKT, ATM, and GAPDH antibodies were used to analyze mTORC1 inhibition, mTORC2 inhibition, and ATM protein levels. Tumors were randomly picked for each group. Control, mouse treated with drug vehicle only. (B) Rh18 rhabdomyosarcoma xenografts were processed for immunoblotting as in A. (C) Rh30 xenografts were treated with rapamycin at 5 mg/kg per day. Twenty-four hours posttreatment on day 4, tumors were pulverized under liquid N2 and processed for immunoblotting as in A. (D) Rh30 cells were treated with rapamycin (100 ng/mL) or AZD8055 (2 μM) for 24 h. Total proteins were extracted for immunoblotting for ATM, pS6K1-T389, S6K1, pAKT-S473, AKT, pGSK3β-S9, and GSK3β. (E) HEK293 cells were treated with rapamycin (100 ng/mL) or AZD8055 (2 μM) for 24 h. Total proteins were extracted for immunoblotting of ATM, TEL2, S6, and pS6-S235/6. (F) HEK293 cells were treated with AZD8055 (2 μM) for 48 h. Total proteins were extracted and ATM, pATM-S1981, CHK2, and pCHK2-T68 were determined by immunoblotting. β-Actin, GAPDH, or β-tubulin served as loading controls.
To extend these observations, we treated cultured Rh30 cells with rapamycin and AZD8055 and determined levels of ATM. Rapamycin decreased pS6K1-T389 but increased pAKT-S473 and pGSK3β-S9 signals. AZD8055 down-regulated the signals of pS6K1-T389, pAKT-S473 and pGSK3β-S9. However, both rapamycin and AZD8055 increased ATM levels (Fig. 1D). Similar results were observed for Rh30 cells treated with rapamycin or AZD8055 for 24, 48 and 72 h, whereas TEL2 levels were not affected (Fig. S2A). In addition, treatment of lymphoblast cells with AZD8055 also resulted in up-regulation of ATM (Fig. S2B). In Rh18 cells, rapamycin slightly increased, whereas AZD8055 more markedly induced ATM (Fig. S2C). Similar to Rh30 cells, both rapamycin- and AZD8055-treated HEK293 cells displayed a marked increase of ATM associated with a slight decrease in TEL2 levels (Fig. 1E). Moreover, the increase of ATM in HEK293 cells induced by AZD8055 was accompanied by enhanced signals of pATM-S1981 and pCHK2-T68, indicating that increase of ATM protein level leads to ATM-CHK2 checkpoint activation (Fig. 1F). These results support the conjecture that mTOR signaling controls ATM independently of TEL2 both in tumors and in cultured cells.
ATM mRNA Levels Are Controlled by mTOR Signaling.
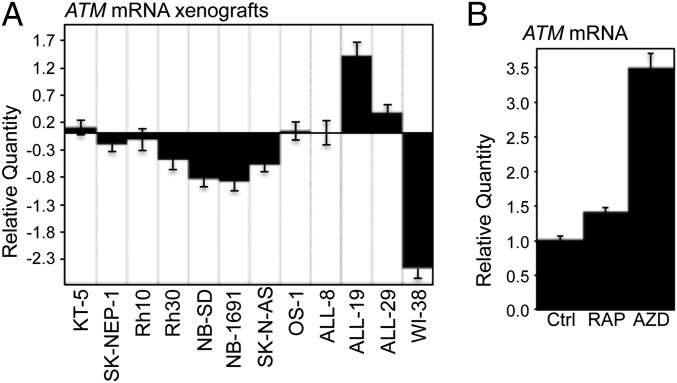
The data presented here demonstrate that inhibition of mTOR leads to an increase of ATM protein both in vivo and in vitro. A survey of the Pediatric Preclinical Testing Program (PPTP) tumor panels showed a general decrease of ATM expression in solid childhood xenografts in comparison with ALL xenografts, which is similar to the pattern of ATM protein (13) (Fig. S1), suggesting that the decreased ATM protein results, from at least in part, the down-regulation of ATM mRNA. This led us to investigate the regulation of ATM mRNA by mTOR signaling. To validate the data from the Affymetrix gene expression profile of PPTP xenografts, we determined the mRNA levels of ATM of randomly picked solid tumors and ALL models from the PPTP, cultured HEK293, and WI-38 cells (ATCC CCL75). Six of the eight solid tumors demonstrated decreased ATM mRNA by real-time quantitative RT-PCR compared with that of the three ALL. Consistent with the undetectable ATM protein (Fig. S1), WI-38 showed significant down-regulation of ATM mRNA (Fig. 2A). In cultured Rh18 cells, rapamycin slightly, whereas AZD8055 robustly, increased ATM mRNA (Fig. 2B). Moreover, Rh30 cells treated with rapamycin or AZD8055 for either 12 or 24 h displayed increased ATM mRNA (Fig. S2D). These results indicate that the down-regulation of ATM in the solid tumors of the PPTP panel, at least in part, results from the suppression of ATM mRNA mediated by mTOR signaling.
Fig. 2.
The mTOR pathway regulates mRNA levels of ATM. (A) Total RNA of randomly picked pediatric cancers (from the PPTP panel) propagated s.c. in SCID mice, and cultured WI-38 were extracted, and subjected to real-time RT-PCR with GAPDH as internal control. Relative quantity of ATM mRNA was plotted using ALL-8 as calibrator. Ctrl, control; GMB2, glioblastoma; KT-5, Wilms tumor; NB-SD, NB-1691, and SK-N-AS, neuroblastoma; OS-1, osteosarcoma; SK-NEP-1, Ewing tumor; WI-38, human fibroblast cells derived from normal embryonic lung tissue. (B) Rh18 cells were treated with rapamycin (RAP, 100 ng/mL) or AZD8055 (AZD, 2 μM) for 24 h. Total RNA was extracted to detect ATM mRNA by real-time RT-PCR with GAPDH as the internal control. Relative quantity of ATM mRNA was plotted. Ctrl, DMSO. Error bars, mean ± SD (n = 3).
mTOR forms both the Raptor-containing complex mTORC1 and Rictor-containing complex mTORC2. mTORC2 phosphorylates AKT at S473, essential for the full activation of AKT kinase (21), and AKT activates mTORC1 signaling (6). To further test our results obtained from pharmacologic inhibition of mTOR signaling, we down-regulated Raptor, Rictor, or mTOR by siRNAs, and determined the protein and mRNA levels of ATM. Partial down-regulation of Raptor, Rictor, or mTOR resulted in a decrease of the pS6-S235/6 signal, demonstrating the suppression of mTORC1 signaling. Each siRNA increased ATM levels (Fig. S2E). Raptor siRNA slightly increased ATM mRNA, whereas siRNA of Rictor or mTOR induced a more increase of ATM mRNA (Fig. S2F). These data support the conjecture that mTOR signaling negatively regulates ATM mRNA.
mTOR Signaling Suppresses ATM mRNA via the S6K Pathway.
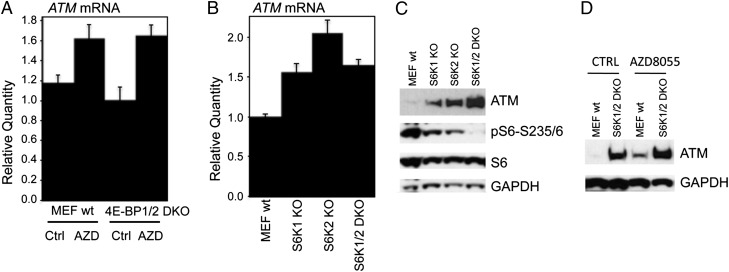
We next examined which pathways downstream of mTORC1, S6K1, and 4E-BP1 regulate ATM mRNA by determining ATM mRNA and protein levels in 4E-BP1/2 double knockout (4E-BP1/2 DKO), S6K1 knockout (S6K1 KO), S6K2 knockout (S6K2 KO), and S6K1/2 double knockout (S6K1/2 DKO) mouse embryonic fibroblasts (MEFs). Compared with that of WT MEFs, 4E-BP1/2 DKO MEFs displayed slightly decreased ATM mRNA, but AZD8055 still increased ATM mRNA to a level comparable to that of WT MEF cells (Fig. 3A). In sharp contrast, ATM mRNAs were robustly elevated in S6K1 KO, S6K2 KO, or S6K1/2 DKO MEFs (Fig. 3B) and KO of either S6K1 or S6K2 led to increase of ATM proteins (Fig. 3C). Impressively, S6K1/2 DKO MEFs showed robust up-regulation of ATM protein, accompanied by disappearance of pS6-S235/6 signal (Fig. 3C). Furthermore, AZD8055 increased ATM in WT MEF cells and only slightly in S6K1/2 DKO MEFs (Fig. 3D). These data demonstrate that mTOR suppresses ATM protein and mRNA primarily through S6K-mediated signaling pathways.
Fig. 3.
The mTOR pathway suppresses ATM through S6K-dependent pathways. (A) WT MEFs and 4E-BP1/2 DKO MEFs were treated with AZD8055 (2 μM) for 12 h; total RNA was extracted to detect murine ATM mRNA by real-time RT-PCR with GAPDH as the internal control. Relative quantity of ATM mRNA was plotted. AZD, AZD8055; Ctrl, DMSO. (B) Total RNA from WT (MEF wt), S6K1 KO, S6K2 KO, and S6K1/2 DKO MEF cells were extracted to detect murine ATM mRNA by real-time RT-PCR with GAPDH as the internal control. Relative quantity of ATM mRNA was plotted. (C) Total proteins from WT MEF, S6K1 KO, S6K2 KO, and S6K1/2 DKO MEFs were immunoblotted for ATM, pS6-S235/6, S6, and GAPDH. (D) WT (MEF wt) and S6K1/2 DKO MEFs were treated with AZD8055 (2 μM) for 24 h. Total proteins were extracted to detect ATM and GAPDH. Error bars, mean ± SD (n = 3).
The mTOR Pathway Controls miRNAs Targeting ATM.
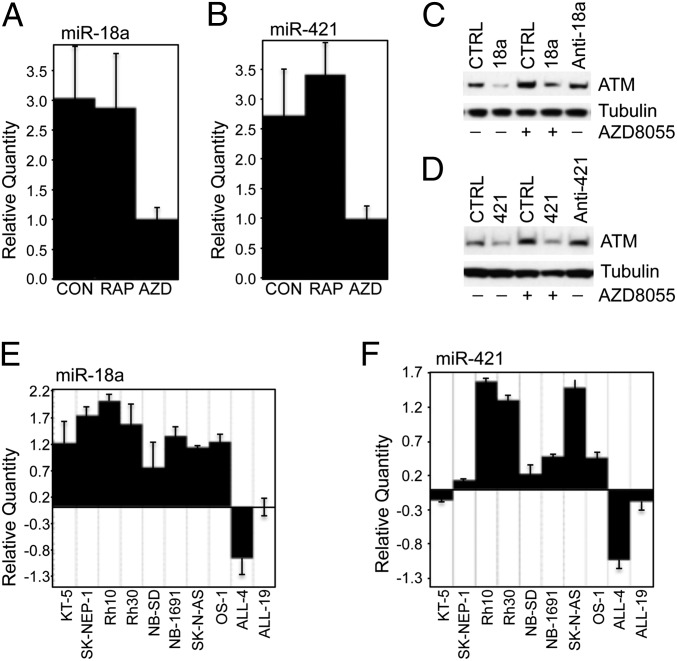
Deregulation of miRNAs has been found in most cancers (22, 23), and several miRNAs have been shown to regulate ATM (24–27). Oncogenic signaling, such as RAS- and MYC-mediated enhancement of cell growth signaling pathways, regulates miRNAs, especially the oncogenic microRNA-17-92 (miR-17-92) cluster (28, 29). These observations led us to postulate that the mTOR pathway might control ATM via regulating miRNAs. In Rh18 cells, AZD8055 apparently decreased miR-18a (Fig. 4A) and miR-421 (Fig. 4B), two miRNAs that have been well-validated in the regulation of ATM (24, 27). In Rh30 cells, AZD8055 also decreased miR-18a and miR-421 (Fig. S3A), whereas the effect of rapamycin was less robust. Moreover, knockdown of mTOR by siRNA resulted in decrease of both miR-18a and miR-421 (Fig. S3B). Consistent with these results, mimics of miR-18a reduced ATM and abolished the induction of ATM by AZD8055 in Rh18 cells. In contrast, inhibitor of miR-18a increased ATM (Fig. 4C). miR-421 down-regulated ATM and abolished the up-regulation of ATM by AZD8055. Inhibitor of miR-421 also increased ATM (Fig. 4D). Similarly, in Rh30 cells, both miR-18a (Fig. S3C) and miR-421 (Fig. S3D) reduced ATM for up to 120 h (Fig. S3E) and abolished the induction of ATM by AZD8055, whereas inhibitors of either miR-18a or miR-421 increased ATM. Moreover, cotransfection of mimics of miR-18a or miR-421 with a Renilla luciferase reporter construct with WT 3′UTR of ATM reduced the luciferase activity (Fig. S3F), whereas miR-421 did not decrease the luciferase activity of the construct with a mutation of the seed sequence of miR-421–binding site of ATM 3′UTR (Fig. S3G). These results indicate that miR-18a and miR-421 are positively controlled by mTOR-mediated signaling pathways.
Fig. 4.
mTOR signaling controls miRNAs that suppress ATM. (A and B) Rh18 cells were treated with RAP (100 ng/mL) or AZD8055 (AZD, 2 μM) for 24 h. Total small RNA was extracted to detect miR-18a (A) and miR-421 (B) by real-time RT-PCR with small nuclear RNA RNU66 as the internal control. Relative quantity of miRNAs was plotted. CON or CTRL, DMSO. (C) Rh18 cells at 60% confluence were transfected with CTRL, miR-18a (18a), or anti-miR-18a (anti-18a) using lipofectamine 2000. Forty-eight hours later, AZD8055 (AZD, 2 μM) was added as indicated for an additional 48 h. Total proteins were extracted for immunoblotting of ATM and Tubulin. (D) Rh18 cells at 60% confluence were transfected with CTRL, miR-421 (421), or anti-miR-421 (anti-421) using lipofectamine 2000. Forty-eight hours later, AZD (2 μM) was added as indicated for an additional 48 h. Total proteins were extracted for immunoblotting of ATM and Tubulin. (E) Total small RNA of randomly picked pediatric solid tumor xenografts as presented in Fig. 2A; ALL4 and ALL19 leukemia xenografts were extracted and subjected to detection of miR-18a by real-time RT-PCR with RNU66 as the internal control. Relative quantity of miR-18a was plotted. (F) Total small RNA of the samples from E was subjected to detection of miR-421 by real-time RT-PCR with RNU66 as the internal control. Relative quantity of miR-421 was plotted. Error bars, mean ± SD (n = 3).
miR-18a and miR-421 Are Generally Increased in Pediatric Solid Tumors.
Our data suggest that the general down-regulation of ATM in solid pediatric tumors may be due to the up-regulation of miRNAs targeting ATM and mTOR signaling might suppress ATM by up-regulating these miRNAs. Indeed, the levels of miR-18a (Fig. 4E) and miR-421 (Fig. 4F) were in general inversely correlated with the levels of ATM mRNA in solid pediatric tumors (Fig. 2A). In agreement, our previous miRNA microarray profile of the PPTP tumor panels demonstrated that the miR-17–92 cluster, containing miR-18a, is overexpressed in pediatric rhabdomyosarcoma and neuroblastoma (30). To validate the miRNA microarray data and test if miR-421 is also up-regulated in these tumors, we compared miR-18a and miR-421 levels in rhabdomyosarcoma and neuroblastoma xenograft models of the PPTP panel with two acute lymphoblastic leukemia models (ALL-4 and ALL-19). In comparison with the leukemias, all of the eight rhabdomyosarcoma overexpressed both miR-18a (Fig. S4A) and miR-421 (Fig. S4B). Five of the eight neuroblastoma displayed increased expression of miR-18a (Fig. S4C) and six of eight neuroblastoma showed increased levels of miR-421 (Fig. S4D). Thus, the up-regulation of miR-18a and miR-421 might contribute to, at least in part, the down-regulation of ATM in solid pediatric tumor xenografts.
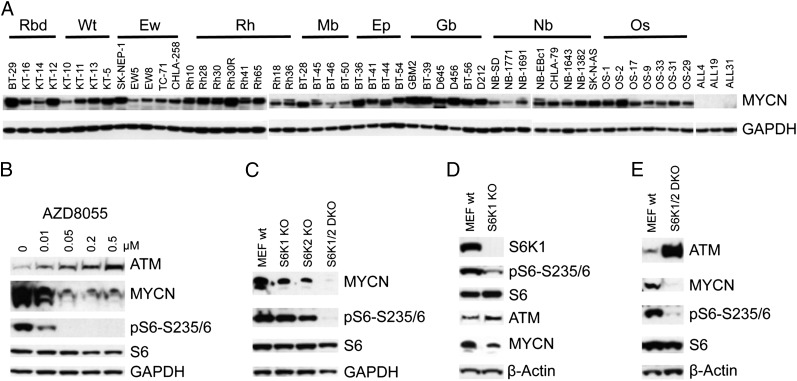
MYCN Is Under Control of the mTOR-S6K Pathway.
It was recently demonstrated that both miR-18a and miR-421 are regulated by the MYCN (v-myc myelocytomatosis viral related oncogene, neuroblastoma derived) transcription factor (27, 29). In vitro, down-regulation of MYCN by siRNA led to a slight increase of ATM (Fig. S5A), but a decrease of miR-18a and miR-421 (Fig. S5B). In contrast, ectopic overexpression of MYCN resulted in a decrease of ATM (Fig. S5C), but an increase of miR-18a (Fig. S5D). In vivo, most pediatric solid tumors of PPTP panels displayed overexpression of MYCN protein (Fig. 5A). It has been well documented that MYCN protein is stabilized by the PI3K-AKT-mTOR pathway via multiple mechanisms (31, 32). Thus, it is possible that enhanced mTOR activity in pediatric solid tumors may up-regulate MYCN protein, which in turn leads to increase of miR-18a and miR-421 and concomitant down-regulation of ATM. To test this hypothesis, we treated Rh30 cells with different concentrations of AZD8055 for 24 h and checked ATM and MYCN. The decrease of MYCN and increase of ATM was correlated with the inhibition of pS6-S235/6 signal by AZD8055 in a concentration-dependent manner (Fig. 5B), supporting the contention that MYCN is under tight control of PI3K-AKT-mTOR signaling (32). Similar inverse correlation of ATM and MYCN was observed in Rh18 cells treated with rapamycin and AZD8055 (Fig. S2A). Furthermore, depletion of either S6K1 or S6K2 led to partial decrease of MYCN in MEFs, whereas knockout of both S6K1 and S6K2 resulted in loss of MYCN detection (Fig. 5C), which is consistent with the robust induction of ATM in S6K1/2 DKO MEFs (Fig. 3 C and D). Moreover, the protein levels of ATM and MYCN were inversely correlated in both S6K1 KO (Fig. 5D) and S6K1/2 DKO MEFs (Fig. 5E). These data suggest that one of the mechanisms by which mTORC1 suppresses ATM is through sustaining MYCN via S6K1/2 signaling.
Fig. 5.
The mTOR pathway controls MYCN. (A) Protein profile of MYCN in PPTP tumor xenograft models. Protein extracts were used for immunoblots to detect the protein levels of MYCN. Ep, ependymoma; Ew, Ewing sarcoma; Gb, glioblastoma; Mb, medulloblastoma; Nb, neuroblastoma; Os, osteosarcoma; Rbd, rhabdoid tumor; Rh; rhabdomyosarcoma; Wt, Wilms tumor. (B) Rh30 cells were treated with AZD8055 at the concentrations indicated for 24 h. Total proteins were extracted for immunoblotting of ATM, MYCN, pS6-S235/6, S6, and GAPDH. (C) Total proteins from WT MEFs, S6K1 KO, S6K2 KO, and S6K1/2 DKO MEFs were immunoblotted for MYCN, pS6-S235/6, S6, and GAPDH. (D) Total proteins from WT MEF or S6K1 KO MEF cells were immunoblotted for S6K1, ATM, pS6-S235/6, S6, MYCN, and GAPDH. (E) Total proteins from WT MEF or S6K1/2 DKO MEF cells were immunoblotted for ATM, pS6-S235/6, S6, MYCN, and GAPDH.
Discussion
Cellular stress, such as DNA damage, nutritional deprivation, and hypoxia, results in inhibition of mTORC1 signaling. For DNA damage and hypoxia, ATM plays a critical role in such regulation and has been postulated to be one mechanism by which ATM functions as a tumor suppressor. In our previous study, we noticed that levels of ATM were generally far lower in xenografts of pediatric solid tumors compared with ALL xenografts and speculated that suppression of ATM may provide a proliferative or survival advantage for cells under hypoxic conditions through maintenance of mTORC1 signaling (13). However, the mechanism by which ATM is suppressed was not addressed.
Using an mTOR kinase inhibitor or genetic ablation of mTORC1 signaling, we discovered that the mTORC1 pathway negatively controls ATM by S6K signaling, and that mTORC1-S6K1/2 suppresses ATM via MYCN-mediated up-regulation of miR-18a and miR-421 (Fig. S6A). The discovery of this negative control of ATM by mTORC1 signaling adds a feedback loop for the relationship between mTOR and ATM, at least in the context of pediatric solid tumors (Fig. S6B).
Generally, cancer cells are deficient in ATM-CHK2-p53 circuitry (33), while demonstrating enhanced mTORC1 signaling (34). Thus, in most cancer cells, the mTOR and ATM-CHK2-p53 pathways are uncoupled. Suppression of ATM might be one of the mechanisms by which mTORC1 signaling promotes and maintains tumorigenesis (34). This uncoupling may provide a growth advantage for premalignant cells during the multistep processes of tumorigenesis. First, increased mTORC1 activity directly leads to enhanced macromolecule synthesis, the increase of which is the prerequisite for cell-cycle progression and hence cell proliferation (5). Second, up-regulated mTOR activity results in unchecked cell survival and proliferation (17, 35). Moreover, without ATM-CHK2-p53 function, unrestrained cell proliferation may tolerate environmental and intracellular genotoxins (36, 37). This tolerance in turn would result in karyotypic chaos: gene mutations, chromosome translocations, and aneuploidy (38). In rare conditions, via sequential clonal expansion, a cell (probably a cancer-initiating cell) will stabilize this karyotypic chaos with augmentation of other cancer-promoting capabilities such as reactivation of telomerase and sustained angiogenesis, leading to malignant progression (39).
Overexpression of MYCN has been reported in a high frequency of rhabdomyosarcomas (40); MYCN amplification also occurs frequently (41). MYCN protein levels are elevated in many xenografts derived from rhabdomyosarcoma and neuroblastoma, but also in atypical teratoid rhabdoid tumors (ATRT), glioblastoma, and ependymoma, and to a lesser extent in osteosarcoma and Ewing sarcoma; therefore, overexpression of MYCN and suppression of ATM expression may have relevance to many childhood solid tumors. The relationship between MYCN overexpression and ATM is not absolute, suggesting that factors other than miR-18a and miR-421 may be involved, such as promoter hypermethylation (42, 43). The role of ATM in the initiation or progression of childhood solid tumors has not been studied extensively; however undetectable levels of ATM were reported in 7 of 17 (41%) rhabdomyosarcoma samples (44). Persistent MYCN signaling in paravertebral ganglia cells initiates tumorigenesis by altering the physiologic process of neural crest cell deletion (45), suggesting that mTORC1-mediated up-regulation of MYCN may initiate transformation in other embryonal cancers. Our hypothesis is that maintained mTORC1 signaling leads to persistent MYCN overexpression and, in some tumors, down-regulation of ATM, further enhancing genomic instability and leading to tumor initiation and progression.
Our results indicate that ATM is suppressed, at least in part, by mTORC1-S6K1/2 regulation of MYCN and miRNAs controlled by MYCN. The identification of this reciprocal inhibition of mTOR and ATM may provide insight for several characteristics of childhood cancer: early onset of disease and hypersensitivity to drugs or ionizing radiation that damage DNA. Standard curative therapy for most childhood solid tumors incorporates drugs that damage DNA including topoisomerase poisons (topotecan, irinotecan, etoposide, doxorubicin), bifunctional alkylating agents (cyclophosphamide, ifosfamide), cisplatin, and ionizing radiation. This uncoupling of mTOR and ATM-CHK2-p53 may not only contribute to primary tumorigenesis and initial chemosensitivity, but also play an important role in chemoradiation resistance and genesis of secondary tumors, two of the major problems of the current cancer chemotherapies (46). Our data provide further support for the proposal that down-regulation of the DNA damage checkpoint is required for cancer cells to bypass DNA replication of stress-induced DNA damage response (47), which is believed to be a barrier for tumorigenesis (48, 49).
Experimental Procedures
Chemicals.
AZD8055 was provided by AstraZeneca. Rapamycin was from the National Cancer Institute drug repository. Mimics and inhibitors of miR-421 and miR-18a were from Applied Biosystems.
Solid Tumor Xenografts Studies.
CB17SC SCID−/− female mice (Taconic Farms) were used to propagate s.c. implanted tumors. All mice were maintained under barrier conditions, and experiments were conducted using protocols and conditions approved by the Institutional Animal Care and Use Committee of Nationwide Children's Hospital. AZD8055 was administered orally daily at 20 mg/kg per d (20). Rh18 and Rh30 rhabdomyosarcomas were harvested 24 h posttreatment on day 4. Rapamycin was administered I.P. daily at a dose of 5 mg/kg (50). Rh30 rhabdomyosarcomas were harvested 24 h posttreatment on day 4.
Supplementary Material
Acknowledgments
We thank Said Sebti for plasmid pCMV-AKT1-wt; Elizabeth Beierle for plasmid pcDNA3.1D/V5-His-N-MYC; Hailiang Hu for plasmids pRL-ATM 3′UTR WT and pRL-ATM 3′UTR Δ6; Nahum Sonenberg for 4E-BP1/2 double knockout mouse embryonic fibroblast (MEF) cells; and George Thomas for S6K1, S6K2, and S6K1/2 knock-out MEFs. Purified acute lymphoblastic leukemia cells from xenografts were provided by Richard Lock (Children’s Cancer Institute of Australia). This work was supported by US Public Health Service Grant CA77776 and National Cancer Institute Grant NO1-CM42216.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220898110/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: Oncogene and non-oncogene addiction. Cell. 2009;136(5):823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432(7015):316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 4.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine AJ, Feng Z, Mak TW, You H, Jin S. Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev. 2006;20(3):267–275. doi: 10.1101/gad.1363206. [DOI] [PubMed] [Google Scholar]
- 6.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20(7):427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Z, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: Stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67(7):3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 9.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102(23):8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134(3):451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brugarolas J, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18(23):2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22(2):239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1α. Mol Cell. 2010;40(4):509–520. doi: 10.1016/j.molcel.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salk JJ, Fox EJ, Loeb LA. Mutational heterogeneity in human cancers: Origin and consequences. Annu Rev Pathol. 2010;5:51–75. doi: 10.1146/annurev-pathol-121808-102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takai H, Wang RC, Takai KK, Yang H, de Lange T. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131(7):1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 17.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441(7092):424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 18.Bjornsti MA, Houghton PJ. The TOR pathway: A target for cancer therapy. Nat Rev Cancer. 2004;4(5):335–348. doi: 10.1038/nrc1362. [DOI] [PubMed] [Google Scholar]
- 19.Menon S, et al. Chronic activation of mTOR complex 1 is sufficient to cause hepatocellular carcinoma in mice. Sci Signal. 2012;5(217):ra24. doi: 10.1126/scisignal.2002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houghton PJ, et al. Initial testing (stage 1) of the mTOR kinase inhibitor AZD8055 by the pediatric preclinical testing program. Pediatr Blood Cancer. 2012;58(2):191–199. doi: 10.1002/pbc.22935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 22.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33(6):1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song L, et al. miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS ONE. 2011;6(9):e25454. doi: 10.1371/journal.pone.0025454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng WL, Yan D, Zhang X, Mo YY, Wang Y. Over-expression of miR-100 is responsible for the low-expression of ATM in the human glioma cell line: M059J. DNA Repair (Amst) 2010;9(11):1170–1175. doi: 10.1016/j.dnarep.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 26.Yan D, et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS ONE. 2010;5(7):e11397. doi: 10.1371/journal.pone.0011397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci USA. 2010;107(4):1506–1511. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dews M, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38(9):1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terrile M, et al. miRNA expression profiling of the murine TH-MYCN neuroblastoma model reveals similarities with human tumors and identifies novel candidate miRNAs. PLoS ONE. 2011;6(12):e28356. doi: 10.1371/journal.pone.0028356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei JS, et al. microRNA profiling identifies cancer-specific and prognostic signatures in pediatric malignancies. Clin Cancer Res. 2009;15(17):5560–5568. doi: 10.1158/1078-0432.CCR-08-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnsen JI, et al. Inhibitors of mammalian target of rapamycin downregulate MYCN protein expression and inhibit neuroblastoma growth in vitro and in vivo. Oncogene. 2008;27(20):2910–2922. doi: 10.1038/sj.onc.1210938. [DOI] [PubMed] [Google Scholar]
- 32.Gustafson WC, Weiss WA. Myc proteins as therapeutic targets. Oncogene. 2010;29(9):1249–1259. doi: 10.1038/onc.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine AJ, Oren M. The first 30 years of p53: Growing ever more complex. Nat Rev Cancer. 2009;9(10):749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7(8):606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 36.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 37.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 38.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9(4):138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 39.Holland AJ, Cleveland DW. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10(7):478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toffolatti L, et al. MYCN expression in human rhabdomyosarcoma cell lines and tumour samples. J Pathol. 2002;196(4):450–458. doi: 10.1002/path.1068. [DOI] [PubMed] [Google Scholar]
- 41.Dias P, et al. N-myc gene is amplified in alveolar rhabdomyosarcomas (RMS) but not in embryonal RMS. Int J Cancer. 1990;45(4):593–596. doi: 10.1002/ijc.2910450403. [DOI] [PubMed] [Google Scholar]
- 42.Ai L, et al. Ataxia-telangiectasia-mutated (ATM) gene in head and neck squamous cell carcinoma: Promoter hypermethylation with clinical correlation in 100 cases. Cancer Epidemiol Biomarkers Prev. 2004;13(1):150–156. doi: 10.1158/1055-9965.epi-082-3. [DOI] [PubMed] [Google Scholar]
- 43.Bolt J, et al. The ATM/p53 pathway is commonly targeted for inactivation in squamous cell carcinoma of the head and neck (SCCHN) by multiple molecular mechanisms. Oral Oncol. 2005;41(10):1013–1020. doi: 10.1016/j.oraloncology.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 44.Zhang P, et al. Association of ataxia telangiectasia mutated (ATM) gene mutation/deletion with rhabdomyosarcoma. Cancer Biol Ther. 2003;2(1):87–91. doi: 10.4161/cbt.231. [DOI] [PubMed] [Google Scholar]
- 45.Hansford LM, et al. Mechanisms of embryonal tumor initiation: Distinct roles for MycN expression and MYCN amplification. Proc Natl Acad Sci USA. 2004;101(34):12664–12669. doi: 10.1073/pnas.0401083101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varmus H. The new era in cancer research. Science. 2006;312(5777):1162–1165. doi: 10.1126/science.1126758. [DOI] [PubMed] [Google Scholar]
- 47.Diffley JF. The many faces of redundancy in DNA replication control. Cold Spring Harb Symp Quant Biol. 2010;75:135–142. doi: 10.1101/sqb.2010.75.062. [DOI] [PubMed] [Google Scholar]
- 48.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 49.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 50.Houghton PJ, et al. Initial testing (stage 1) of the mTOR inhibitor rapamycin by the pediatric preclinical testing program. Pediatr Blood Cancer. 2008;50(4):799–805. doi: 10.1002/pbc.21296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.