Abstract
Peptidylarginine deiminase 4 (PAD4) is a Ca2+-dependent enzyme that converts arginine and methylarginine residues to citrulline, with histone proteins being among its best-described substrates to date. However, the biological function of this posttranslational modification, either in histones or in nonhistone proteins, is poorly understood. Here, we show that PAD4 recognizes, binds, and citrullinates glycogen synthase kinase-3β (GSK3β), both in vitro and in vivo. Among other functions, GSK3β is a key regulator of transcription factors involved in tumor progression, and its dysregulation has been associated with progression of human cancers. We demonstrate that silencing of PAD4 in breast cancer cells leads to a striking reduction of nuclear GSK3β protein levels, increased TGF-β signaling, induction of epithelial-to-mesenchymal transition, and production of more invasive tumors in xenograft assays. Moreover, in breast cancer patients, reduction of PAD4 and nuclear GSK3β is associated with increased tumor invasiveness. We propose that PAD4-mediated citrullination of GSK3β is a unique posttranslational modification that regulates its nuclear localization and thereby plays a critical role in maintaining an epithelial phenotype. We demonstrate a dynamic and previously unappreciated interplay between histone-modifying enzymes, citrullination of nonhistone proteins, and epithelial-to-mesenchymal transition.
Breast cancer is the most common cancer in women worldwide; major obstacles to successful treatment of this disease are tumor recurrence and metastasis. Metastatic progression is a complex and multistep process initiated via an epithelial-to-mesenchymal transition (EMT) (1). Typically, EMT involves loss of epithelial polarity, adhesive properties, and acquisition of a fibroblastoid phenotype with increased cell motility. Collectively, these changes result in dispersed and isolated cells, capable of invading the surrounding stroma, intravasating into the bloodstream, and eventually populating distant sites as micrometastases (2). Numerous signaling pathways have been implicated in this process, such as TGF-β, RAS, PI3K/AKT, and Wnt, and several important downstream transcription factor targets have been identified, including Snail, Slug, Smad, Twist1, and Zeb1 (1, 3, 4).
Protein citrullination is a poorly understood posttranslational modification (PTM) but has recently gained increased attention because of its potential role in human disease, including cancer (5–7). Citrullination, also referred to as deimination, involves conversion of positively charged arginine residues to uncharged, nonribosomally encoded citrulline residues (8). The resulting biochemical change can lead to alterations in protein structure and protein interactions (9). Citrullination is mediated by peptidylarginine deiminases (PADs), a family of Ca2+-dependent sulfhydryl enzymes consisting of PAD1–PAD4 and PAD6 (10). PADs display extensive sequence homologies, but each PAD isoform has its own characteristic subcellular localization, tissue distribution, and substrate specificity (8). PAD4, for example, has been found in a variety of cells, such as embryonic stem cells, leukocytes, and lung and breast cancer cells. PAD4 is the only PAD family member that contains a distinct nuclear localization sequence (11). We and others have reported that PAD4 can convert arginine and methylarginine to citrulline in the histones H2A, H3, and H4. Citrullination has been linked to either transcriptional repression or activation, depending on the context (12–16). Recently PAD4 was also shown to citrullinate nonhistone proteins, raising important issues as to what other substrates may be targets of citrullination in different biological contexts (16–19).
Because PAD4 is expressed in breast cancer cells, we aimed to investigate whether PAD4 activity might play a role in breast cancer initiation or progression. We show that knockdown of PAD4 induces TGF-β signaling, EMT, and increases the invasive potential of breast cancer cells. Depletion of PAD4 causes a dramatic reduction in nuclear levels of glycogen synthase kinase-3β (GSK3β), a key signaling intermediate in pathways known to initiate and regulate EMT (20). Our data show that PAD4 specifically citrullinates arginine residues in the N-terminal domain of GSK3β that closely resemble its known target sites on histones H2A and H4. We conclude that this unique PTM of GSK3β is crucial for nuclear localization of the kinase, and that this, in turn, is necessary for maintaining an epithelial phenotype.
Results
Stable Knockdown of PAD4 Induces EMT and Increases the Invasive Potential of Noninvasive MCF7 Cells.
Using shRNAs, we first knocked down expression of PAD4 in the noninvasive, estrogen receptor (ER)α-positive human mammary adenocarcinoma cell line MCF7 (Fig. 1A and Fig. S1A). This resulted in substantial silencing of PADI4 but did not affect the mRNA expression levels of other relevant PADI family members (Fig. S1C). Using differential interference contrast imaging, we observed that PAD4-depleted MCF7 cells (shPAD4) showed active spreading, loss of cell–cell contact, and a fibroblast-like morphology in contrast to control MCF7 cells (MCF7ctr), which grew as densely packed clusters with limited cell spreading (Fig. 1B and Fig. S1B). Detection of epithelial markers E-cadherin and β-catenin at the plasma membrane and in whole-cell extracts was drastically reduced in shPAD4 cells, whereas immunofluorescence (IF) and Western blotting showed higher levels of the mesenchymal marker vimentin in shPAD4 cells (Fig. 1 C and D). To verify that depletion of PAD4 induced similar phenotypic changes in a different ERα-positive breast carcinoma cell line, we generated T47D cells with a stable PAD4 knockdown. These cells also showed a reduction of E-cadherin protein levels and elevated protein levels of vimentin (Fig. S1 D–F). We next evaluated the motility and invasiveness of these cells in vitro using reconstituted basement membrane invasion assays. PAD4-depleted MCF7 cells showed significantly more invasion than control cells (Fig. 1E and Fig. S1G), and comparable results were obtained in PAD4-depleted T47D cells (Fig. S1H). Our findings thus demonstrate that silencing of PAD4 leads to morphological and functional changes consistent with EMT, a process known as an initiating step in tumor invasion and metastasis.
Fig. 1.
Knockdown of PAD4 induces EMT in MCF7 cells. (A) Western blot of nuclear extracts from MCF7 control (ctr) and PAD4 knockdown (shPAD4) cells demonstrating silencing of PAD4 in shPAD4 cells. (B) Epithelial morphology of MCF7ctr and mesenchymal morphology of shPAD4 cells. (C) IF images of MCF7ctr and shPAD4 cells stained for E-cadherin (green), β-catenin (red), vimentin (red), and nuclei (blue). (Scale bar, 10 μm.) (D) Indicated cell lysates were analyzed by SDS/PAGE and probed for E-cadherin, vimentin, and α-tubulin. (E) Cell invasion assays using ECM Extract coated transwell filters (**P < 0.01 control vs. shPAD4). (F) MCF7ctr, shPAD4, and PAD4 rescue cells (shPAD4+PAD4) were processed for bright field (Upper) and IF imaging (Lower) and stained for E-cadherin (green) and nuclei (blue). (Scale bar, 10 µm.) (G) Western blot of whole-cell extracts from shPAD4, MCF7ctr, and PAD4 rescue cells for E-cadherin. PAD4 reexpression was detected by HA-tag. Bottom lane shows densitometry of E-cadherin levels normalized to Gapdh.
To confirm that the EMT phenotype was specifically due to the knockdown of PAD4, we performed rescue experiments in shPAD4 cells using a PAD4-lentiviral construct. This resulted in partial restoration of the epithelial phenotype, as observed by changes in cell morphology and increased E-cadherin expression levels in the rescued cells (Fig. 1 F and G). Additionally, overexpression of PAD4 in MCF7 cells (MCF7-PAD4) resulted in an even more pronounced epithelial phenotype, with flattened cells growing in tight clusters and increased expression of E-cadherin and β-catenin (Fig. S2 A–C). Thus, the consequences of PAD4 knockdown and subsequent rescue, as well as its overexpression in MCF7 cells, indicate that PAD4 is involved in maintenance of an epithelial cell phenotype.
PAD4 Citrullinates GSK3β in Vitro and in Vivo.
Given the phenotypic switch in shPAD4 cells, we sought to identify putative targets of PAD4 that might account for induction of the EMT phenotype. Crystallographic data suggest that PAD4 does not recognize specific arginine residues in a unique sequence context but rather recognizes five successive residues with the consensus sequence 1ϕXRXX5, in which ϕ denotes amino acids with small side-chain moieties, and X denotes any amino acid (21). The target sequence must also be accessible and flexible enough to fit into the active pocket of PAD4, much like the N-terminal sequence SGRGK of human histones H4 and H2A, whose arginine at position 3 (R3) is one of the better described targets of PAD4 (14, 21, 22). Because initiating methionines (M) often are removed during protein maturation, we searched the proteome for sequences beginning with an amino (N)-terminal MSGR motif using pBLAST and PredMod (23). We identified 12 nonhistone proteins with an N-terminal MSGR motif (Table S1). Interestingly, 1 of the 12 putative PAD4 target proteins was GSK3β (Fig. S2D). Because GSK3β is an important inhibitor in several signaling pathways that have been implicated in EMT-mediated breast cancer progression, we investigated the hypothesis that PAD4 might directly citrullinate GSK3β. We performed in vitro citrullination assays in which recombinant GSK3β was incubated with purified, recombinant wild-type PAD4 or with PAD4 carrying a mutation in its catalytic center (PAD4mut). Citrullination was evaluated by Western blotting with an anti-modified–citrulline antibody. In samples treated with wild-type PAD4, a strong signal for citrullination of GSK3β was readily detected, whereas mutant PAD4 or mock-treated samples produced either a weak signal or no signal at all (Fig. 2A). To map the sites of citrullination in GSK3β, we performed LC-MS/MS analysis, revealing that PAD4 preferentially citrullinates the N-terminal arginine residues R3 and R5 of GSK3β, and to a lesser extent other arginine residues throughout the protein (Fig. S3 A and B). In accordance, a citrullination assay using GSK3β peptides (amino acids 1–13) showed that PAD4 recognizes and citrullinates the N-terminal sequence of GSK3β containing the SGR motif (Fig. S3C).
Fig. 2.

PAD4 targets GSK3β for citrullination in MCF7 cells. (A) Recombinant GSK3β was incubated with recombinant GST-PAD4wt, GST-PAD4mut (inactive), or left untreated in the presence of calcium. Reaction products were analyzed by Western blot using modified-citrulline and GSK3β antibodies. (B) Interaction of PAD4 with GSK3β in MCF7 cells. Nuclear extracts of MCF7 cells stably overexpressing PAD4-FLAG were immunoprecipitated (IP) with antibodies against either FLAG-tag or GSK3β. Complexes were then immunoblotted (IB) using antibodies against the indicated proteins. (C) Citrullination of GSK3β by PAD4 in MCF7 cells. After treatment with calcium ionophore A23187, nuclear extracts of MCF7 cells overexpressing either PAD4wt-FLAG or PAD4mut-FLAG were immunoprecipitated (IP) with an antibody against GSK3β. Immunoprecipitation material was then immunoblotted with antibodies against GSK3β and modified citrulline.
To examine whether PAD4 can directly interact with GSK3β in vivo, we performed anti-FLAG immunoprecipitation assays with nuclear extracts from MCF7 cells, showing coprecipitation of endogenous nuclear GSK3β in cells stably expressing PAD4-FLAG but not in cells lacking PAD4-FLAG (control) (Fig. 2B). Reciprocally, when endogenous GSK3β was immunoprecipitated from PAD4-FLAG containing nuclear extracts, coimmunoprecipitated PAD4 proteins were readily detected with anti-PAD4 antibody but were not seen in IgG control precipitates (Fig. 2B). To determine whether endogenous, nuclear GSK3β is citrullinated by active PAD4 in vivo, we incubated MCF7 cells overexpressing either wild-type PAD4 (MCF7-PAD4) or catalytically inactive PAD4 (PAD4mut) in calcium-containing medium including calcium ionophore A23187 before generating nuclear extracts and immunoprecipitations of GSK3β. Western blotting using an anti-modified–citrulline antibody showed a band corresponding to citrullinated GSK3β in the nuclear extracts of MCF7-PAD4 cells but only a very faint band in MCF7-PAD4mut cells (Fig. 2C). Together, these results provide evidence that PAD4 citrullinates nuclear GSK3β both in vitro and in vivo.
PAD4 Regulates Nuclear Levels of GSK3β.
Next we sought to determine whether silencing of PAD4, and the consequent reduction in citrullination activity, might affect GSK3β levels in MCF7 cells. We observed a modest reduction of GSK3B mRNA in shPAD4 cells (43% reduction) (Fig. S4A), concordant with previously reported down-regulation of GSK3β levels in cells undergoing EMT. We then investigated GSK3β total protein levels in MCF7ctr and shPAD4 cells using flow cytometric analysis. shPAD4 cells showed a clear reduction in the intensity and percentage of GSK3β-positive cells (Fig. S4D). In contrast, both the PAD4-rescued knockdown cells and MCF7 cells stably overexpressing PAD4 showed higher levels of total GSK3β expression (Fig. S4 B, E, and F), whereas expression of catalytically inactive PAD4 had no effect on GSK3β total levels (Fig. S4F). GSK3β is regulated by a variety of mechanisms (24), including its inactivation by phosphorylation of serine 9. Therefore, we tested whether citrullination of GSK3β by PAD4 affects serine 9 phosphorylation, but no difference was observed between MCF7ctr and shPAD4 cells (Fig. S4C).
Although GSK3β is predominantly located in the cytosol, a proportion of the protein is constantly shuttled in and out of the nucleus, where it plays important roles in regulating apoptosis, ribosomal biogenesis, and gene expression (25–27). To investigate whether PAD4 equally affects both pools, we analyzed whole-cell lysates (Fig. 3A) and cytosolic and nuclear fractions. Although GSK3β protein was modestly reduced in the cytosolic fraction of shPAD4 cells, its abundance was greatly reduced in the nuclear fraction (Fig. 3B). Similar results were obtained with PAD4-depleted T47D cells (Fig. S5A). These data suggest that PAD4 is required for maintaining a pool of GSK3β protein in the nucleus of breast cancer cells.
Fig. 3.

Loss of PAD4 leads to reduced nuclear levels of GSK3β. (A) Whole-cell lysates and (B) cellular fractions from MCF7ctr and shPAD4 cells were analyzed with antibodies against GSK3β. Calnexin, Gapdh, and histone H3 were used as loading controls, respectively.
To investigate whether citrullination directly regulates the nuclear accumulation or retention of GSK3β, we transfected MCF7 cells with either wild-type PAD4 (MCF7-PAD4) or inactive PAD4 (MCF7-PAD4mut). Nuclear accumulation of GSK3β was observed only in the cells transfected with wild-type PAD4, indicating that this is dependent on the enzymatic activity of PAD4 (Fig. 4A). In addition, upon overexpression of GSK3β in MCF7ctr cells, GSK3β displayed mostly cytoplasmic localization, whereas in MCF7 cells overexpressing PAD4, GSK3β was readily detected in nuclei (Fig. S5B).
Fig. 4.
Nuclear accumulation of GSK3β is regulated by citrullination of its N terminus. (A) Catalytically active PAD4 leads to accumulation of GSK3β in the nucleus. MCF7 cells were transfected with (A) PAD4wt-FLAG or PAD4mut-FLAG and stained against the FLAG-tag (green) and GSK3β (red). Nuclei were stained with DAPI. Signals were quantified and expressed in a bar graph (**P < 0.01 PAD4wt vs. PAD4mut). (Scale bar, 40 μm.) (B) Arginine residues R3 and R5 of GSK3β are critical for nuclear accumulation in the presence of PAD4. HEK293 cells were cotransfected with PAD4-FLAG or empty control vector and either wild-type (wt) GSK3β-NLSmyc or GSK3β K3,K5-NLSmyc constructs and stained against the FLAG-tag (red), myc-tag (green). Quantification of nuclear GSK3β signal: bar graph depicts GSK3β-NLSmyc vs. GSK3β-NLSmyc+PAD4 (*P = 0.02) and GSK3β-NLSmyc+PAD4 vs. GSK3β K3,K5-NLSmyc+PAD4 (**P = 0.003). (Scale bar, 10 μm.) (C) MCF7ctr, shPAD4, and GSK3β rescue cells were processed for brightfield and IF imaging and stained for E-cadherin (green), and nuclei (blue). (Scale bar, 10 µm.) (D) Migration assay: shPAD4 and GSK3β rescue cells were observed by live microscopy for 15 h, and distance of migration was measured (**P < 0.01 shPAD4 vs. shPAD4+GSK3β).
Truncation of the first nine amino acids of GSK3β has been reported to reduce its nuclear abundance, but the mechanism has not been fully elucidated (26). An alignment of GSK3β protein sequences from five species demonstrated complete conservation of the first nine amino acids, suggesting that this domain is functionally important (Fig. S5C). Given that the first nine amino acids of GSK3β contain two arginine residues (R3 and R5), within a motif that resembles the N termini of histones H4 and H2A (Fig. S2D), we asked whether citrullination of these two arginines is required for the nuclear accumulation of GSK3β. To test this, we replaced arginines with lysines at positions 3 and 5 of a full-length GSK3β-NLS (nuclear localization sequence)-myc fusion protein, thus preserving the charge of these residues but precluding their possible citrullination. A triple NLS sequence was included in these constructs to ensure a basal level of nuclear import, even when the N-terminal residues were mutated (Fig. S5D). The GSK3βK3/K5NLS mutant and wild-type GSK3β (GSK3β-NLS) constructs were transfected into HEK293 cells with or without PAD4-FLAG. Expression of PAD4 caused a significant increase in the nuclear accumulation of wild-type GSK3β (Fig. 4B). In contrast, mutation of R3 and R5 resulted in significantly reduced nuclear accumulation of GSK3β (Fig. 4B), despite the triple NLS sequence. Thus, we conclude that citrullination of R3 and/or R5 of GSK3β plays a critical role in the nuclear accumulation of GSK3β.
To determine whether the EMT phenotype of shPAD4 cells was attributable to the accompanying loss of nuclear GSK3β, we restored nuclear GSK3β expression levels by a GSK3β-NLS lentiviral construct. Strikingly, expression of nuclear GSK3β in shPAD4 cells partially restored epithelial cell morphology and induced increased E-cadherin expression (Fig. 4C and Fig. S5E). Furthermore, GSK3β-rescued cells migrated slower than shPAD4 cells in a direct comparison by live microscopy (Fig. 4D). Collectively these data demonstrate the importance of PAD4-mediated citrullination for nuclear abundance of GSK3β and maintenance of an epithelial cell phenotype.
Knockdown of PAD4 Induces TGF-β Signaling in MCF7 Cells.
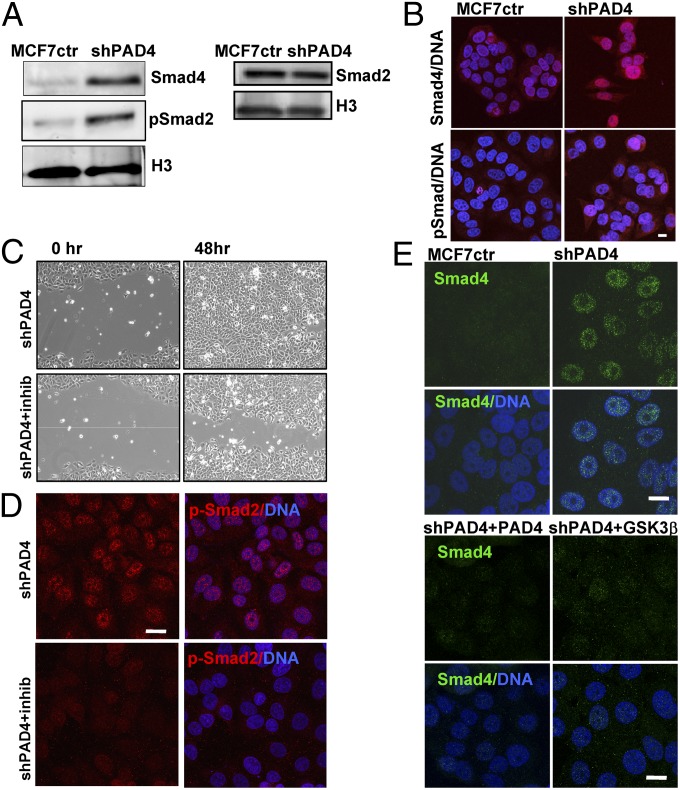
TGF-β transmitted regulation of Smad transcription factors is an important mediator of EMT (1, 28). Because GSK3β is a known direct regulator of Smads (24, 29), we next asked whether TGF-β signaling was altered in shPAD4 cells. Western blot analysis (Fig. 5A) and IF imaging (Fig. 5B) showed elevated levels of the TGF-β effector proteins Smad4 and pSmad2 in shPAD4 cells, consistent with activation of the TGF-β pathway. Total levels of Smad2 remained unchanged (Fig. 5A, Right). To test for functional TGF-β signaling, we treated MCF7ctr and shPAD4 cells with a specific TGF-β receptor inhibitor (SB431542) and analyzed known Smad downstream target genes (Fig. S6). Transcripts of all three target genes investigated, protooncogene serine/threonine-protein kinase, (PIM1), serpin peptidase inhibitor type 1 (SERPINE1), and Thrombospondin (THSB1), were elevated in shPAD4 cells and significantly reduced upon drug treatment, suggesting activated TGF-β signaling in shPAD4 cells. Because TGF-β signaling can induce cell migration accompanying EMT, we investigated whether treatment with TGF-β receptor inhibitor would change the migratory phenotype of shPAD4 cells in a wound-healing assay. shPAD4 cells closed the wound slower upon inhibitor treatment (Fig. 5C) and displayed reduced pSmad2 staining (Fig. 5D), consistent with active TGF-β signaling in the untreated shPAD4 cells. We next investigated Smad4 levels in shPAD4 cells transduced with either the PAD4 rescue construct or the GSK3β-NLS rescue construct. Lower Smad4 levels were observed when either the effector protein (i.e., PAD4) or the target protein (i.e., nuclear GSK3β) was reintroduced into shPAD4 cells (Fig. 5E).
Fig. 5.
Knockdown of PAD4 leads to activation of TGFβ signaling in MCF7 cells. (A) Nuclear extracts from MCF7ctr and shPAD4 cells were analyzed for Smad4, pSmad2, and Smad2. Histone H3 was used as control. (B) IF analysis of Smad4 and pSmad-2 (red) in MCF7ctr and shPAD4 cells. Nuclei were stained with DAPI (blue). (Scale bar, 10 μm.) (C) Scratch assay in shPAD4 cells in the presence or absence of the specific TGF-β inhibitor SB431542. Wound closure was observed for 48 h. (D) IF analysis of p-Smad2 (red) in shPAD4 cells treated with SB431542 for 48 h. (Scale bar, 10 μm.) (E) IF analysis of Smad4 (green) levels in MCF7ctr, shPAD4, PAD4 rescue, and GSK3β rescue cells. (Scale bar, 10 μm.)
PAD4 Knockdown Increases the Invasive Potential of Noninvasive MCF7 Cells in Vivo.
We next evaluated the potential of PAD4-depleted cells to form invasive tumors in vivo. To this end, shPAD4 and MCF7ctr cells were injected in the mammary fat pads of female NCr-nu/nu mice, and tumor growth was measured for 7 wk. No significant difference in mean tumor volume was observed (Fig. 6A). However, upon histological analysis, tumors derived from shPAD4 cells displayed significantly more evidence of invasiveness. Strikingly, 65% of analyzed shPAD4 tumor sections showed a characteristic invasive front at the tumor margin compared with 24% of MCF7ctr tumors (Fig. 6 B and C). shPAD4 tumor sections also revealed a reduction in the proportion of tumor cells expressing GSK3β, cytoplasmic and nuclear (Fig. 6 D and E). In contrast, the relative number of tumor cells with nuclear Smad4, and consequently active TGF-β signaling, a well-described activator of EMT, was increased (Fig. 6 D and E). These results reinforce the notion that down-regulation of PAD4 and subsequent loss of nuclear GSK3β induces a TGF-β signaling loop and increases the invasive potential of breast cancer cells in vivo.
Fig. 6.
Correlation between expression levels of PAD4 and invasiveness of breast cancer cells in vivo. Orthotopic tumors were generated in nude mice by injection of MCF7ctr or shPAD4 cells (n = 8 mice per group). (A) Tumor volume was measured at 7 wk after injection. (B) Quantification of 10× fields at primary tumor margins that display an invasive front (**P < 0.01 control vs. PAD4). (C) Representative images of tumor margins of MCF7ctr and shPAD4 orthotopic tumors (H&E staining). Arrows indicate the invasive front. (Magnification: 10×; scale bars, 100 μm.) (D) Representative images of MCF7ctr and shPAD4 orthotopic tumors stained with GSK3β and Smad4 (Magnification: 40×; scale bars, 25 μm.) (E) Quantification of histological images of MCF7ctr and shPAD4 orthotopic tumors stained with GSK3β and Smad4 antibodies. (F) Abundance of PAD4 and (G) GSK3β in human normal breast tissue and in breast tumor samples. (i) Normal breast epithelium. (ii) DCIS. (iii and iv) Invasive carcinoma. (Scale bars, 50 μm.)
Loss of PAD4 Is Associated with Invasive Breast Carcinomas.
To investigate the relevance of altered PAD4 expression in human breast cancer, we determined whether loss of PAD4 correlates with malignant progression in human breast cancer progression tissue arrays (NCI Frederick). These consisted of normal breast tissue, ductal carcinoma in situ (DCIS), and invasive breast cancer samples. Although PAD4 nuclear staining was not significantly associated with age, histological score, and hormonal receptor status, PAD4 nuclear staining was significantly less in invasive carcinomas compared with DCIS tumors or normal breast tissue using a semiquantitative approach (Fig. 6F, i–iii and Table S2). Additionally, we evaluated GSK3β staining on a matching tissue array (Fig. 6G, i–iv and Table S2). We observed a significant correlation between tumors that stained positive in the nucleus for PAD4 and tumors that stained positive for nuclear GSK3β (Table S2). Further evidence solidifying an in vivo connection between the two enzymes came from the intensity of stain and relative percentage of cells showing positive staining of nuclear PAD4 and GSK3β (Table S3). Together these data are consistent with our in vitro and in vivo findings, showing that loss of PAD4 correlates with reduced nuclear GSK3β and is associated with invasive breast cancer in human tumor samples.
Discussion
Citrullination of arginine and methylarginine residues by PAD4 mediates changes in protein–protein interactions and regulates gene transcription, thereby impacting various cellular processes. The importance of this modification in human pathologies remains largely unknown. In this study we show that loss of PAD4 from breast tumor cells initiates early steps of the metastatic process (i.e., migration and invasion). We further demonstrate that PAD4 is an important posttranslational modifier of GSK3β that regulates nuclear protein levels of GSK3β by citrullinating its N-terminal domain. Furthermore, PAD4-depleted MCF7 cells form invasive tumors in nude mice, and PAD4 expression is drastically reduced during human breast cancer progression, strongly correlating with the loss of nuclear GSK3β in these tissues.
Previously PAD4 has been shown to convert arginine and methylarginine residues in the histones H2A, H3, and H4 to citrulline in various human cancer cells (12–15, 18, 22), influencing the regulation of target genes, such as TFF1, p21, and OKL38 (12–16). PAD4 was recently also shown to citrullinate the nonhistone proteins p300, nucleophosmin (NPM1), ING4, Elk-1 and RPS2, thereby partially influencing the activity or localization of these proteins (16–19). Despite this progress, in-depth studies linking experiments that examine cell morphology, mouse xenograft, and human tissue array analyses to elucidate the role of PAD4 in breast cancer progression have largely been lacking.
To evaluate the role of PAD4 in breast cancer processes, we generated experimental cell models with stable knockdown of PAD4 in noninvasive human breast cancer cell lines. Our analysis demonstrated that silencing of PAD4 leads to morphological changes defined as EMT. Consistent with cells undergoing EMT, our results also show that down-regulation of PAD4 increases cell migration and invasion.
To identify putative targets of PAD4 that might account for the induction of EMT, we searched the proteome for proteins with an N-terminal MSGR motif and identified GSK3β as a potential target substrate of PAD4. This was of particular interest because the loss or down-regulation of GSK3β activity has previously been associated with EMT and cancer progression (3, 26, 30). GSK3β was originally identified as a kinase regulating conversion of glucose to glycogen. Subsequent studies have indicated that this kinase is directly involved in numerous human diseases, including neurological disorders, EMT, and cancer (20). Known nuclear targets of GSK3β in these processes include Smads, β-catenin, CREB, p53, c-myc, HIF1α, and cyclin D1. GSK3β controls these transcription factors by affecting their nuclear localization, protein stability, and/or DNA binding activity (31, 32). The protein itself is subject to regulation by a variety of mechanisms (24). Given that GSK3β is an important and diverse signaling mediator, we investigated whether PAD4 could citrullinate and regulate GSK3β directly. Our data indicate that PAD4 both interacts with, and citrullinates, GSK3β in vitro and in vivo.
Although PAD4 depletion resulted in decreased nuclear GSK3β levels, expression of wild-type PAD4 drastically increased nuclear GSK3β levels in MCF7 cells. Catalytically inactive mutant PAD4 had no such effect. These data support a model wherein citrullination by PAD4 promotes the nuclear accumulation of GSK3β, although the underlying mechanisms remain to be determined. We provide evidence that citrullination of the N terminus is involved in nuclear retention or stabilization of GSK3β because mutating R3 and R5 at the SGR motif lead to decreased levels of nuclear to GSK3β in the presence of active PAD4 despite an added 3XNLS motif.
These data fit well with a previous report showing that the N-terminal nine amino acids of GSK3β are involved in nuclear localization of the protein (26). The GSK3β nuclear pool has been shown to modulate several cellular functions (27, 33) and is regulated in a dynamic manner (26). Our study identifies citrullination as a unique PTM of GSK3β and demonstrates that this modification regulates the nuclear GSK3β pool. Loss of PAD4-mediated citrullination of GSK3β and the resulting EMT phenotype highlights the importance of nuclear GSK3β for the maintenance of an epithelial phenotype and, thus, its function as a suppressor of EMT. Our data are in line with a recent study describing the importance of H4R3 citrullination by PAD4 in carcinogenesis and indicating a tumor suppressor function (34). Our data support PAD4’s role as a tumor suppressor by ensuring adequate nuclear levels of GSK3β.
PAD4-depleted MCF7 cells also displayed activation of functional TGF-β signaling, a critical inducer of EMT (1, 28). Expression of TGF-β target genes was enhanced in shPAD4 cells and reduced to the levels of MCF7 control cells by a specific TGF-β receptor inhibitor. Interestingly, one of the genes induced in PAD4 depleted cells was THSB1, which has been shown to convert latent inactive TGF-β ligand to its active form (35), thus supporting the notion of a continuous TGF-β signaling loop in these cells. The possibility that additional signaling pathways contribute to the EMT phenotype of PAD4-depleted cells cannot be excluded. However, the observed stabilization of Smad4 and phosphorylated Smad2 in PAD4-depleted cells and the fact that overexpression of nuclear GSK3β reduces Smad4 levels suggests that this may occur as a direct consequence of altered nuclear GSK3β levels. Together with the reduced migratory properties of cells rescued with nuclear GSK3β, these data suggest that PAD4-GSK3β inhibits Smad stabilization and TGF-β signaling and therefore ensures an epithelial cell phenotype.
Citrullination neutralizes positively charged arginine residues and therefore has an impact on protein structure, folding, and interprotein binding capabilities (9). Thus, it is conceivable that citrullination of the N terminus of GSK3β may regulate the binding of additional proteins involved in destabilization or nuclear export of the kinase in much the same way that nearby or adjacent histone modifications, such as those embedded in relatively unstructured “tail domains,” may regulate histone function through PTM effector (“reader”) binding, drawing upon an emerging concept being described as “histone mimicry” in a wide range of biological processes (36–39). Citrullination of the N terminus is an entirely new concept in GSK3β regulation, and we look forward to future studies aimed at addressing the function(s) of N-terminal citrullination of GSK3β and the pathway being described here.
Materials and Methods
Reagents, cell lines, transfections, and viral transductions are listed in SI Materials and Methods.
Immunofluorescence, invasion assays, intracellular staining procedures, citrullination assays, protein purification, nuclear extract preparation, immunoprecipitation assays, and mass spectrometry were conducted using standard procedures detailed in SI Materials and Methods. Xenograft tumor studies, tissue arrays, and immunohistochemistry followed established procedures described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank G. Meares, R. Jope, and H. Dormann for providing plasmids, A. Moustakas, R. Burkhardt, and J. Zavadil for helpful discussions, and S. Mazel and A. North of the Rockefeller University Flow Cytometry and Bio-Imaging Resource Centers. We also thank L. Angelov for assistance with confocal microscopy. Tissue microarray slides were provided by the Cooperative Breast Cancer Tissue Resource, which is funded by the National Cancer Institute. This work was supported by an American Recovery and Reinvestment Act of 2009 grant and National Institutes of Health (NIH) Grant 5R01GM098870-03 (to C.D.A.); Department of Defense Breast Cancer Research Program, Era of Hope Scholar Award W871XWH-07-0372 (to S.A.C.), NIH Grant 5R01CA123238 and NYSTEM Grant C024286 (to A.B.); NIH Grant CA100324 (to A.P. and J.S.C.); an American Society for Mass Spectrometry research award sponsored by the Waters Corporation, a New Jersey Cancer Commission SEED grant, and National Science Foundation (NSF) Grant CBET-0941143 (to B.A.G.). B.M.Z. receives support from an NSF predoctoral fellowship. C.T.V. was supported by the Swedish Research Council, Georg and Eva Klein Visiting Junior Scientist Award, American Scandinavia Foundation, and a Vinnova Marie Curie international qualification grant. S.C.S. was supported by a research fellowship of the Deutsche Forschungsgemeinschaft (DFG Sta 1151/1-1). V.D.F. is supported by NIH Medical Scientist Training Program Grant GM07739.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308362110/-/DCSupplemental.
References
- 1.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 3.Vincent T, et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol. 2009;11(8):943–950. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuxe J, Vincent T, Garcia de Herreros A. Transcriptional crosstalk between TGF-β and stem cell pathways in tumor cell invasion: Role of EMT promoting Smad complexes. Cell Cycle. 2010;9(12):2363–2374. doi: 10.4161/cc.9.12.12050. [DOI] [PubMed] [Google Scholar]
- 5.Anzilotti C, Pratesi F, Tommasi C, Migliorini P. Peptidylarginine deiminase 4 and citrullination in health and disease. Autoimmun Rev. 2010;9(3):158–160. doi: 10.1016/j.autrev.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Chang X, et al. Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer. 2009;9:40. doi: 10.1186/1471-2407-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang X, Han J. Expression of peptidylarginine deiminase type 4 (PAD4) in various tumors. Mol Carcinog. 2006;45(3):183–196. doi: 10.1002/mc.20169. [DOI] [PubMed] [Google Scholar]
- 8.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: Genes, features and involvement in disease. Bioessays. 2003;25(11):1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 9.Tarcsa E, et al. Protein unfolding by peptidylarginine deiminase. Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J Biol Chem. 1996;271(48):30709–30716. doi: 10.1074/jbc.271.48.30709. [DOI] [PubMed] [Google Scholar]
- 10.Senshu T, et al. Detection of deiminated proteins in rat skin: probing with a monospecific antibody after modification of citrulline residues. J Invest Dermatol. 1995;105(2):163–169. doi: 10.1111/1523-1747.ep12317070. [DOI] [PubMed] [Google Scholar]
- 11.Nakashima K, Hagiwara T, Yamada M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J Biol Chem. 2002;277(51):49562–49568. doi: 10.1074/jbc.M208795200. [DOI] [PubMed] [Google Scholar]
- 12.Cuthbert GL, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118(5):545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 13.Li P, et al. Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol Cell Biol. 2008;28(15):4745–4758. doi: 10.1128/MCB.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306(5694):279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 15.Yao H, et al. Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J Biol Chem. 2008;283(29):20060–20068. doi: 10.1074/jbc.M802940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, et al. Genome-wide analysis reveals PADI4 cooperates with Elk-1 to activate c-Fos expression in breast cancer cells. PLoS Genet. 2011;7(6):e1002112. doi: 10.1371/journal.pgen.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci USA. 2005;102(10):3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanikawa C, et al. Regulation of protein Citrullination through p53/PADI4 network in DNA damage response. Cancer Res. 2009;69(22):8761–8769. doi: 10.1158/0008-5472.CAN-09-2280. [DOI] [PubMed] [Google Scholar]
- 19.Guo Q, Fast W. Citrullination of inhibitor of growth 4 (ING4) by peptidylarginine deminase 4 (PAD4) disrupts the interaction between ING4 and p53. J Biol Chem. 2011;286(19):17069–17078. doi: 10.1074/jbc.M111.230961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doble BW, Woodgett JR. Role of glycogen synthase kinase-3 in cell fate and epithelial-mesenchymal transitions. Cells Tissues Organs. 2007;185(1-3):73–84. doi: 10.1159/000101306. [DOI] [PubMed] [Google Scholar]
- 21.Arita K, et al. Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4. Proc Natl Acad Sci USA. 2006;103(14):5291–5296. doi: 10.1073/pnas.0509639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagiwara T, Nakashima K, Hirano H, Senshu T, Yamada M. Deimination of arginine residues in nucleophosmin/B23 and histones in HL-60 granulocytes. Biochem Biophys Res Commun. 2002;290(3):979–983. doi: 10.1006/bbrc.2001.6303. [DOI] [PubMed] [Google Scholar]
- 23.Basu A, et al. Proteome-wide prediction of acetylation substrates. Proc Natl Acad Sci USA. 2009;106(33):13785–13790. doi: 10.1073/pnas.0906801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taelman VF, et al. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143(7):1136–1148. doi: 10.1016/j.cell.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bijur GN, Jope RS. Glycogen synthase kinase-3 beta is highly activated in nuclei and mitochondria. Neuroreport. 2003;14(18):2415–2419. doi: 10.1097/00001756-200312190-00025. [DOI] [PubMed] [Google Scholar]
- 26.Meares GP, Jope RS. Resolution of the nuclear localization mechanism of glycogen synthase kinase-3: Functional effects in apoptosis. J Biol Chem. 2007;282(23):16989–17001. doi: 10.1074/jbc.M700610200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vincent T, Kukalev A, Andäng M, Pettersson R, Percipalle P. The glycogen synthase kinase (GSK) 3beta represses RNA polymerase I transcription. Oncogene. 2008;27(39):5254–5259. doi: 10.1038/onc.2008.152. [DOI] [PubMed] [Google Scholar]
- 28.Heldin CH, Landström M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21(2):166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 29.Guo X, et al. Axin and GSK3- control Smad3 protein stability and modulate TGF- signaling. Genes Dev. 2008;22(1):106–120. doi: 10.1101/gad.1590908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armanious H, et al. Clinical and biological significance of GSK-3β inactivation in breast cancer-an immunohistochemical study. Hum Pathol. 2010;41(12):1657–1663. doi: 10.1016/j.humpath.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 31.Xu C, Kim NG, Gumbiner BM. Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle. 2009;8(24):4032–4039. doi: 10.4161/cc.8.24.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou BP, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6(10):931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 33.Jope RS, Johnson GV. The glamour and gloom of glycogen synthase kinase-3. Trends Biochem Sci. 2004;29(2):95–102. doi: 10.1016/j.tibs.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Tanikawa C, et al. Regulation of histone modification and chromatin structure by the p53-PADI4 pathway. Nat Commun. 2012;3:676. doi: 10.1038/ncomms1676. [DOI] [PubMed] [Google Scholar]
- 35.Schultz-Cherry S, Murphy-Ullrich JE. Thrombospondin causes activation of latent transforming growth factor-beta secreted by endothelial cells by a novel mechanism. J Cell Biol. 1993;122(4):923–932. doi: 10.1083/jcb.122.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425(6957):475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 37.Zhao X, et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22(5):640–653. doi: 10.1101/gad.1632608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gamsjaeger R, et al. Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3. Mol Cell Biol. 2011;31(13):2632–2640. doi: 10.1128/MCB.05413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Y, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29(11):1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.