Significance
Neurofilaments, which are the intermediate filaments of neurons, are major components of nerve cells, but their assembly dynamics have not been determined. Here we demonstrate efficient end-to-end annealing of neurofilaments in nerve cells, and we show that there also is a mechanism that severs these polymers. Our efforts thus identify severing of neuronal intermediate filaments in vivo and suggest a mechanism for regulating intermediate filament length involving a dynamic cycle of severing and end-to-end annealing.
Abstract
We have shown previously that neurofilaments and vimentin filaments expressed in nonneuronal cell lines can lengthen by joining ends in a process known as “end-to-end annealing.” To test if this also occurs for neurofilaments in neurons, we transfected cultured rat cortical neurons with fluorescent neurofilament fusion proteins and then used photoconversion or photoactivation strategies to create distinct populations of red and green fluorescent filaments. Within several hours we observed the appearance of chimeric filaments consisting of alternating red and green segments, which is indicative of end-to-end annealing of red and green filaments. However, the appearance of these chimeric filaments was accompanied by a gradual fragmentation of the red and green filament segments, which is indicative of severing. Over time we observed a progressive increase in the number of red–green junctions along the filaments accompanied by a progressive decrease in the average length of the alternating red and green fluorescent segments that comprised those filaments, suggesting a dynamic cycle of severing and end-to-end-annealing. Time-lapse imaging of the axonal transport of chimeric filaments demonstrated that the red and green segments moved together, confirming that they were indeed part of the same filament. Moreover, in several instances, we also were able to capture annealing and severing events live in time-lapse movies. We propose that the length of intermediate filaments in cells is regulated by the opposing actions of severing and end-to-end annealing, and we speculate that this regulatory mechanism may influence neurofilament transport within axons.
The assembly dynamics of actin filaments and microtubules are well understood, but relatively little is known about the assembly dynamics of intermediate filaments. Several studies over the past two decades have suggested that intermediate filaments lengthen by fusion of preexisting filaments (1, 2), known as “end-to-end annealing,” and this idea is supported by mathematical modeling and electron microscopy of intermediate filament assembly in vitro (3–6). Several other studies also have suggested that the addition and loss of intermediate filament subunits is not confined to the filament ends and can occur along the entire length of intermediate filaments (7–9), which we refer to as “intercalary subunit exchange.” Collectively, these studies suggest that intermediate filaments exhibit novel dynamic properties that are distinct from those of microtubules and actin filaments.
In a previous study on neurofilaments and vimentin filaments in adrenal carcinoma and immortalized fibroblast cell lines, we reported experimental evidence for end-to-end annealing and intercalary subunit exchange in vivo (10). However, those cell lines are heterologous systems for studying neurofilament assembly because they do not naturally express neurofilament proteins. Moreover, the neurofilaments formed in those cells were simple heteropolymers composed of neurofilament L (NFL) and either neurofilament M (NFM) or neurofilament H (NFH), whereas neurofilaments in neurons are more complex heteropolymers composed of NFL, NFM, NFH, internexin, and, in peripheral neurons, peripherin (11, 12). Thus, it is important to confirm these assembly dynamics in neurons. Here we demonstrate robust and efficient end-to-end annealing of neurofilaments in neurons, and we also report evidence for a neurofilament-severing activity.
Results
To test for end-to-end annealing of neurofilaments, we transfected primary cultures of rat cortical neurons with fluorescent fusions of neurofilament proteins. After waiting 7–10 d for the fusion proteins to express and incorporate throughout the neurofilament array, we used photobleaching in conjunction with photoconversion or photoactivation to create distinct populations of red and green fluorescent neurofilaments in the cell bodies or axons. We then allowed the red and green filaments to intermix through their motor-driven movement and looked for the appearance of chimeric filaments composed of alternating red and green fluorescent segments.
Photoconversion Strategy.
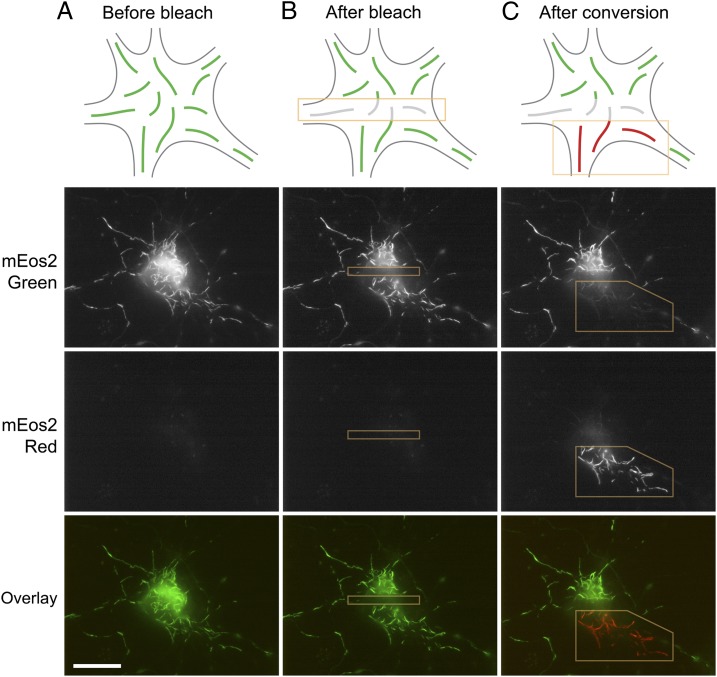
For the photoconversion strategy, we transfected cultured neurons with NFM fused to the monomeric photoconvertible fluorescent protein mEos2 (mEos2-NFM) (Fig. 1). mEos2 is a monomeric variant of the Eos fluorescent protein which switches from green to red fluorescence emission upon illumination with violet light (13). Before photoconversion, all of the mEos2-tagged neurofilaments were green (Fig. 1A). To create a population of red fluorescent filaments in these cells, we photoconverted the mEos2-tagged neurofilaments in a region of the cell body or axon by excitation with violet light (Fig. 1C). To ensure that we did not create any red–green junctions at the borders of the photoconverted region, we bleached the green filaments in the border region by excitation with blue light before photoconversion (Fig. 1B).
Fig. 1.
Photoconversion strategy. Neurons were transfected with mEos2-NFM and were observed 7–10 d later to allow sufficient time for the fusion proteins to incorporate throughout the neurofilament array. Shown here is a nerve cell body from which several processes emanate. (A) Initially the neurofilaments are all green. (B) Blue light is used to photobleach the green fluorescence (mEos2 Green) in a stripe across the cell body (yellow box), effectively bisecting the green filaments into two separate populations. The purpose of this step is to ensure that the subsequent photoconversion step does not create red–green junctions. Depending on the density and distribution of the neurofilaments in the processes emerging from the cell body, it sometimes was necessary to bleach around the entire perimeter of the region to be activated. (C) Violet light is used to photoconvert a subpopulation of the green neurofilaments on one side of the bleached zone (yellow box) to make them red (mEos2 Red). Note that some green fluorescence remains because of incomplete photoconversion of the mEos2 fluorescence. The result is that one half of the cell body now contains red filaments, and the other half contains green filaments, with a bleached region between them. The schematic drawings represent a generalized nerve cell body and are not meant to depict the specific cell shown in this figure. (Scale bar, 20 µm.)
Photoactivation Strategy.
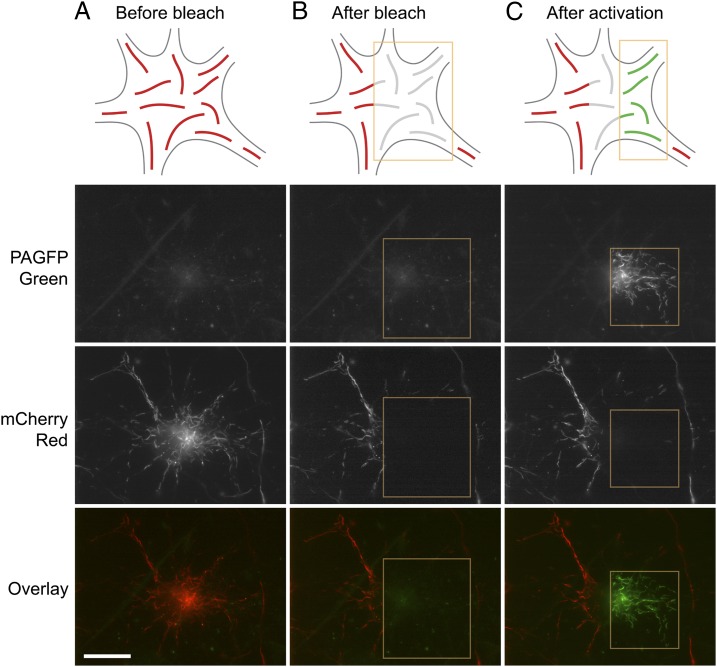
For the photoactivation strategy, we cotransfected the neurons with NFM fused to photoactivatable green fluorescent protein (PAGFP-NFM) and with NFL fused to the monomeric red fluorescent protein mCherry (NFL-mCherry) (Fig. 2). Using this strategy, the neurofilaments were tagged with PAGFP and mCherry but initially were all red because the PAGFP had not been converted (Fig. 2A). To create a subpopulation of green fluorescent neurofilaments in these cells, we bleached the mCherry fluorescence in a region of the cell by excitation with green light (Fig. 2B) and then activated the PAGFP fluorescence in a subregion of this bleached region using violet light (Fig. 2C). By making the activated region smaller than the bleached region, we were able to ensure that we did not create any red–green junctions artificially.
Fig. 2.
Photoactivation strategy. Neurons were cotransfected with PAGFP-NFM and NFL-mCherry and were observed 7–10 d later to allow sufficient time for the fusion proteins to incorporate throughout the neurofilament array. Shown here is a nerve cell body from which several processes emanate. (A) The neurofilaments initially are all red. (B) Green light is used to photobleach the red mCherry-NFL fluorescence in one half of the cell body (yellow box). (C) Violet light is used to photoactivate the PAGFP-NFM in the photobleached filaments (yellow box), making them green. Note that the activated region is smaller than the bleached region to ensure that the photoactivation does not create red–green junctions. The result is that one half of the cell body now contains red filaments, and the other half contains green filaments, with a bleached region between them. The schematic drawings represent a generalized nerve cell body and are not meant to depict the specific cell shown in this figure. (Scale bar, 20 µm.)
End-to-End Annealing of Red and Green Filaments.
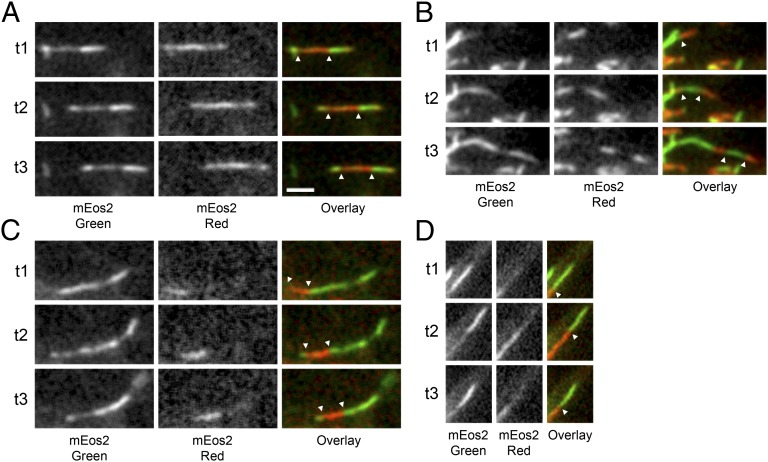
After creating distinct populations of red and green filaments using each of the above strategies, we imaged the cells at intervals over a period of up to 24 h. We have shown previously that neurofilaments in nerve cells move in a rapid, intermittent manner, pausing most of the time (14–17). Over time this motile behavior caused the red and green filaments to become intermixed, and this intermixing was accompanied by the appearance of increasing numbers of chimeric filaments composed of alternating red and green segments (Fig. 3). To document these observations, we acquired images of chimeric filaments that were isolated from their neighbors (Fig. 4). In each case, the filaments appeared smooth and continuous across the red–green junctions with no overlap of the red and green fluorescence. Thus, the red and green segments of these filaments abut each other directly, and the chimeric appearance cannot be explained by overlap of separate red and green filaments.
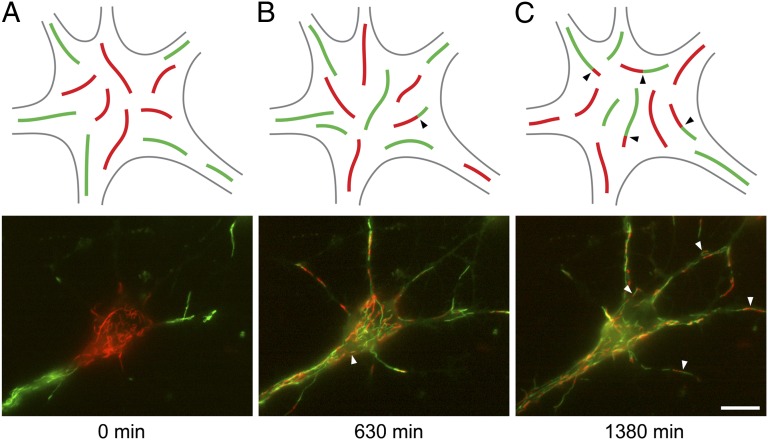
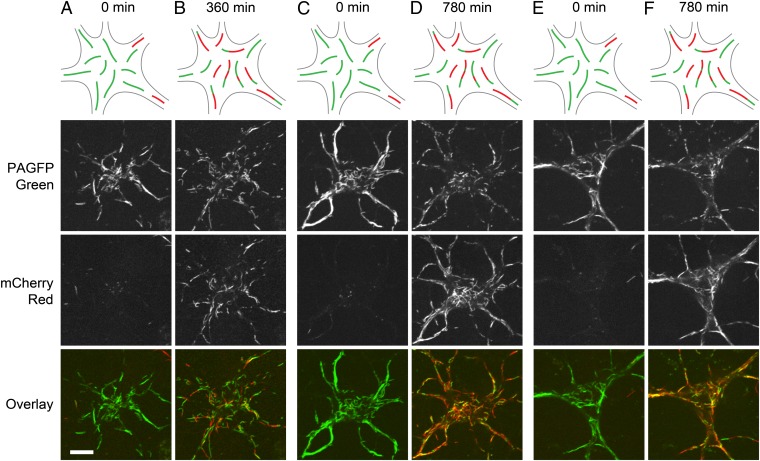
Fig. 3.
The red and green filaments intermix over time. (A) The cell body of a nerve cell immediately after using the photoconversion strategy to create distinct populations of red and green filaments. (B and C) The same cell 630 (B) and 1,380 min (C) after photoconversion. The red and green filaments intermixed over time via neurofilament movement, and this intermixing was accompanied by the appearance of chimeric filaments composed of alternating red and green segments (arrowheads). The number of chimeric filaments increased with time. The schematic drawings represent a generalized nerve cell body and are not meant to depict the specific cell shown in this figure. (Scale bar, 10 µm.)
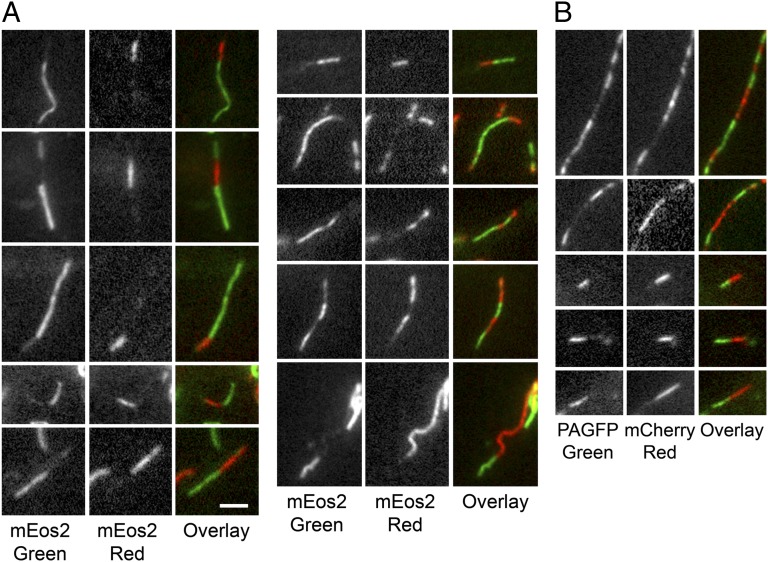
Fig. 4.
Examples of chimeric filaments imaged 300–900 min after photoconversion or photoactivation. (A) Photoconversion strategy. (B) Photoactivation strategy. In all cases the filaments appeared smooth and continuous across the red–green junctions with no overlap of the red and green fluorescence, indicative of end-to-end annealing. Often we observed several alternating red and green segments along a single filament, indicative of multiple end-to-end annealing events. Note that there was weak green fluorescence on some of the red fluorescent segments and weak red fluorescence on some of the green fluorescent segments. Although not the focus of this article, this weak fluorescence may have been caused by intercalary subunit exchange (10). In the photoconversion strategy, we also expected some residual green fluorescence on the red fluorescent segments because of incomplete conversion of the mEos2 fluorescence. (Scale bar, 2 µm.)
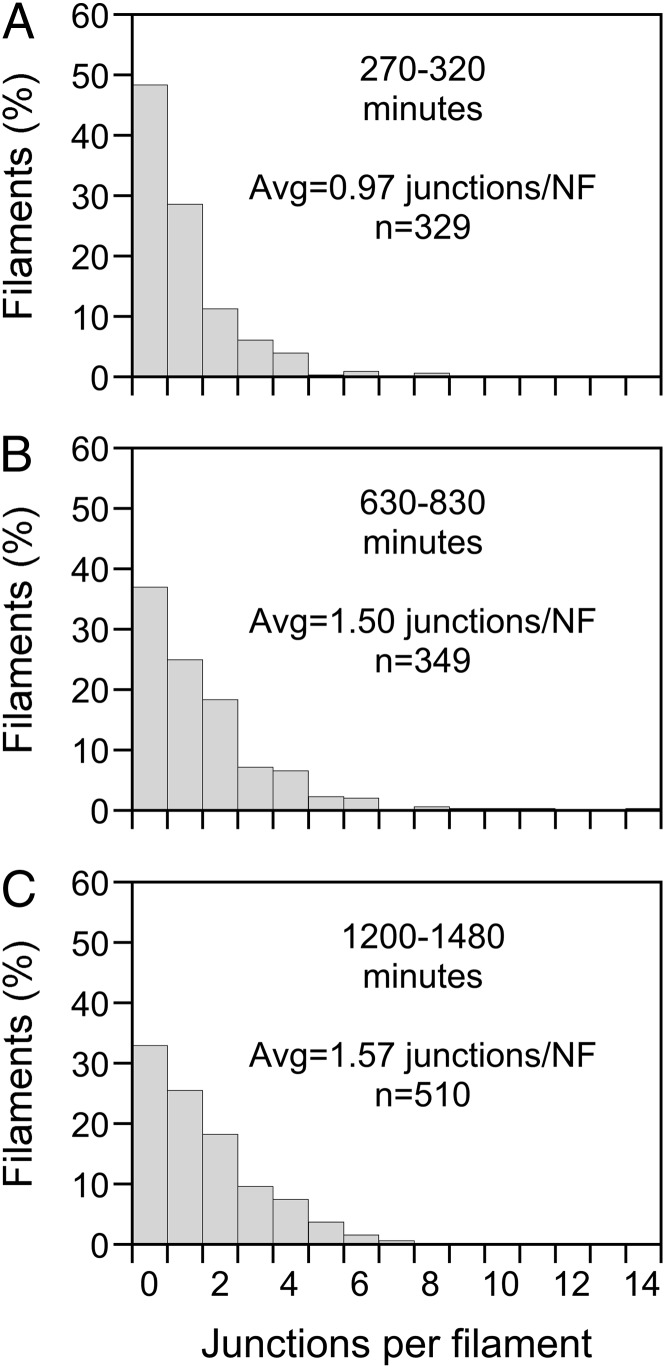
Some chimeric filaments contained one red and one green fluorescent segment, suggesting a single end-to-end annealing event, whereas others contained multiple alternating red and green fluorescent segments, suggesting multiple end-to-end annealing events. To quantify the extent of annealing, we used the photoconversion strategy in cell bodies to create distinct populations of red and green fluorescent filaments, and then we scored the number of red–green junctions along the filaments over time. We included all neurofilaments or portions of neurofilaments that were isolated from their neighbors and that could be traced without ambiguity. These measurements underestimate the extent of end-to-end annealing, because we could detect only junctions created by annealing between filaments of different color. In addition, they underestimate the total number of red–green junctions per neurofilament because overlap with other neurofilaments prevented the tracing of many neurofilaments along their entire length. Therefore these measurements should be considered a minimum estimate. Nevertheless, they revealed evidence of extensive annealing. On average, 52% of the neurofilaments were chimeric after 270–320 min (average of 0.97 red–green junctions per neurofilament; n = 329 filaments), increasing to 63% of the filaments after 630–830 min (average of 1.50 junctions per neurofilament; n = 349 filaments). By 1,200–1,480 min, 67% of the neurofilaments contained at least one red–green junction (average of 1.57 junctions per neurofilament; n = 510 filaments), with as many as 14 red–green junctions along a single filament (Fig. 5).
Fig. 5.
Quantification of annealing junctions along neurofilaments. We used the photoconversion strategy to create distinct populations of red and green fluorescent filaments in neuronal cell bodies, and then we tracked the filaments over time. Data are from six cells imaged at multiple time points extending up to 1,480 min after photoconversion. The graphs are frequency histograms of the number of red–green junctions per neurofilament. Because the cells were imaged at different times, we grouped the time points into three time windows: (A) 270–320 min (n = 329 filaments); (B) 630–830 min (n = 349 filaments); and (C) 1,200–1,480 min (n = 510 filaments). By 1,200–1,480 min after photoconversion, approximately two-thirds of the filaments were chimeric.
To confirm that the red and green segments were indeed part of the same filament, we performed simultaneous two-color time-lapse imaging of the movement of chimeric filaments. Fig. 6 shows four examples, all obtained using the photoconversion strategy. In each case, the red and green segments of the chimeric filaments can be seen to move together with no overlap, confirming that they are indeed part of the same filament (also see Movies S1, S2, and S3). Thus, end-to-end annealing of neurofilament polymers is an efficient and robust process in neurons.
Fig. 6.
Movement of chimeric filaments. Dual-color time-lapse imaging of chimeric filaments obtained using the photoconversion strategy. The filaments shown here were all imaged 360–1,320 min after photoconversion. The white arrowheads mark the red–green junctions. Note that the red and green segments of the chimeric filaments moved together, confirming that they were part of the same filament. The filaments in A, B, and C all moved unidirectionally, whereas the filament in D moved forward and then reversed direction. (Scale bar, 2 µm.)
Fragmentation of Red and Green Filaments.
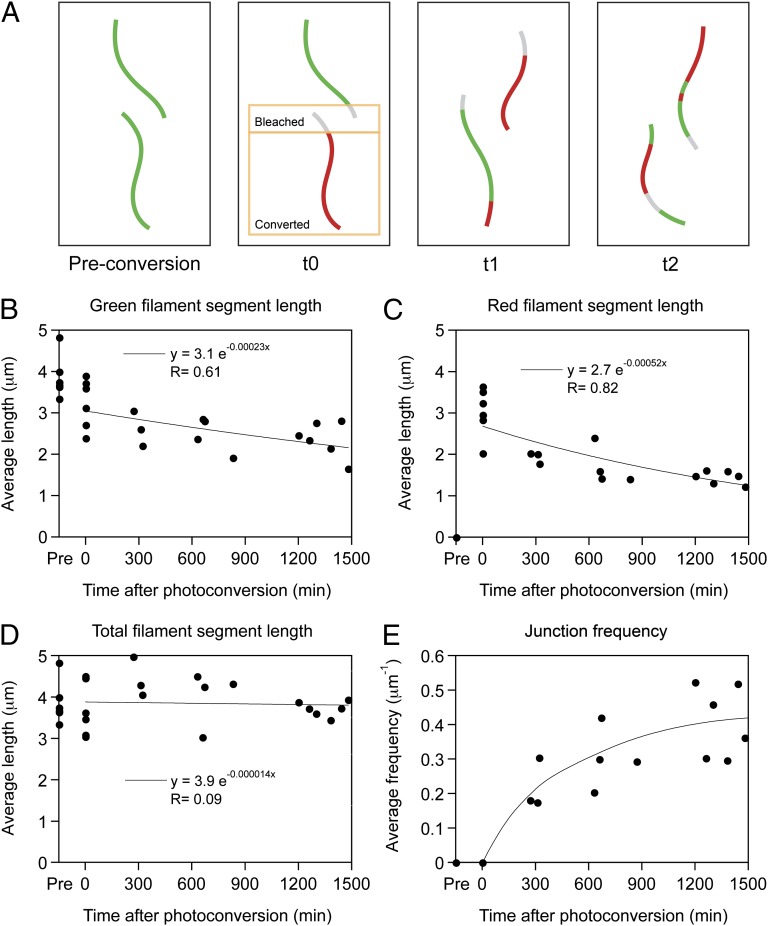
During the course of the experiments described above, we noticed that the increase in the number of chimeric junctions along the filaments over time was accompanied by a decrease in the average length of the red and green filament segments (Fig. 7). To quantify these observations, we measured the lengths of the filaments at multiple time points in the same cells that were analyzed for Fig. 5. We observed a progressive decrease in the average lengths of both the red and green fluorescent segments over a 24-h period (Fig. 8 B and C), but the average overall length of the filaments (including contiguous red and green segments along the same filament) remained constant (Fig. 8D). Consistent with these observations, we also observed a progressive increase in the average frequency of red–green junctions along the filaments (Fig. 8E). The filaments became increasingly fragmented over time, but this fragmentation was counterbalanced by end-to-end annealing of the fragments, resulting in chimeric filaments composed of an increasing number of progressively shorter alternating red and green fluorescent segments (Fig. 8A). Thus, there are mechanisms in these cells that can sever neurofilaments in addition to the mechanisms that can join them together, resulting in a constant and dynamic rearrangement of filament composition and length.
Fig. 7.
The fluorescent segments appear to fragment over time. We used the photoactivation strategy to make all the neurofilaments in neuronal cell bodies green, and then we tracked the cells over time. Immediately after photoactivation, the cells contained green filaments in their cell bodies and red filaments in their processes. Over time, the green and red filaments intermixed via their bidirectional movement. (A, C, and E) Three neurons immediately after photoactivation. Note the green filaments in the cell bodies. Some red filaments are visible in the processes. (B, D, and F) The same cells 360 or 780 min later. Note the many red and green filaments in the cell bodies, including many chimeric filaments. Note also that the green filaments are shorter in B, D, and F than in A, C, and E, indicating neurofilament severing. The schematic drawings represent a generalized nerve cell body and are not meant to depict the specific cells shown in this figure. (Scale bar, 10 µm.)
Fig. 8.
Quantification of neurofilament fragmentation and annealing. Data were obtained from the cells analyzed in Fig. 5. (A) Schematic diagram of a photoconversion experiment showing the effect of breakage and annealing over time. Before photoconversion (“Pre-conversion”), the filaments are green. After bleaching and photoconversion (t0), the cell contains a population of noncontiguous red and green filaments. Over time, these filaments break and anneal to form chimeric filaments comprised of alternating red and green segments (t1). As time progresses, repeated breakage and annealing events result in filaments that are composed of increasing numbers of red and green fluorescent segments that are progressively shorter in length (t2). If the kinetics of breakage and annealing are comparable, the overall filament length remains unchanged. (B and C) The average length of the green and red filament segments decreased steadily over time, indicating filament severing. Note that the average segment length decreased by about 20% immediately after photoconversion (t0). The explanation for this decrease is that we used photobleaching to ensure that we did not introduce red–green junctions artificially, resulting in shorter lengths for filaments that entered or traversed the bleached region, as shown in A. Note also that the extent of shortening is less for the green filaments than for the red filaments. There are several possible reasons for this difference. First, there is new synthesis and assembly of green fluorescent neurofilament protein throughout these experiments, whereas the number of red fluorescent neurofilaments is fixed at the time of photoconversion. Second, the green filaments may have been more abundant because we did not photoconvert filaments in the axonal and dendritic processes, so one would expect a higher probability of green–green annealing events that would go undetected in our analyses. (D) The total filament length (measured in green–red overlay images) did not change over time, indicating that neurofilament severing was accompanied by end-to-end annealing. (E) The frequency of annealing junctions (average number of red–green junctions per micrometer of neurofilament polymer) increased with time, indicating that neurofilament annealing was accompanied by severing. Each data point in B–D represents the average filament segment length for one cell at one time point (average of 218 filament segments measured per data point). Each data point in E represents the average number of red–green junctions per micrometer of neurofilament (average of 126 junctions counted per data point). The trend lines in B–D represent least squares exponential curve fits. The trend line in E is a manual interpolation.
Annealing and Severing Events Captured on Camera.
To confirm the existence of end-to-end annealing and severing, we acquired time-lapse movies of neurofilaments in cortical neurons expressing GFP-tagged neurofilament proteins. To be confident of annealing and severing events in these movies, we set stringent criteria. For annealing events, we looked for instances in which the adjoined filaments moved together as one after the putative annealing event, and we measured the filaments before and after the putative annealing event to ensure conservation of length. For severing events, we looked for instances in which the filaments moved before breaking (to be sure that they were indeed single filaments) and in which there was clear separation between the resulting fragments (e.g., because of the movement of one or both of the fragments). In total we identified three confirmed annealing events and three confirmed severing events. Fig. 9 shows an end-to-end annealing event, and Fig. 10 shows a severing event. Movies S4 and S5 show these two events, respectively, and Movies S6 and S7 show one additional end-to-end annealing and severing event, respectively. These observations confirm that end-to-end annealing and severing of neurofilaments occur in neurons and are not artifacts of our photoactivation or photoconversion strategies.
Fig. 9.
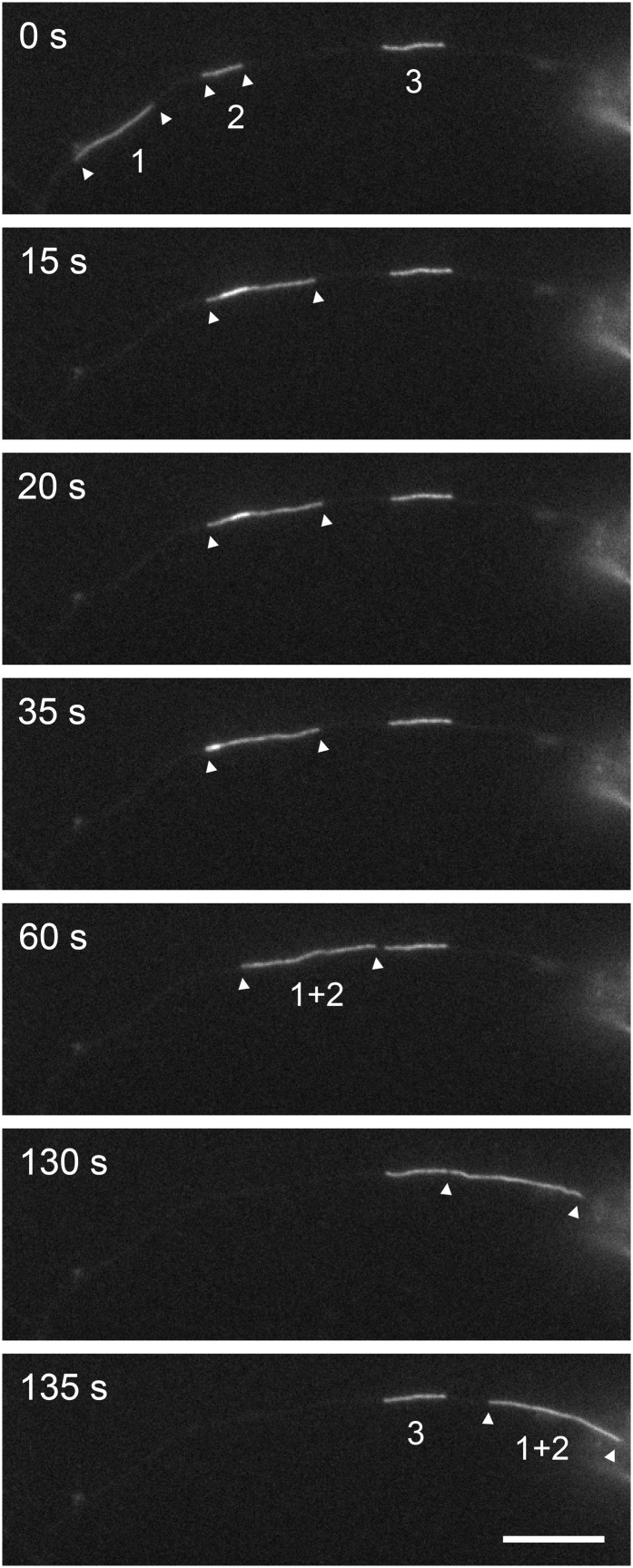
An annealing event captured on camera. Time-lapse imaging of neurofilaments in an axon of a cultured mouse cortical neuron expressing GFP-NFM. This portion of axon contained three filaments, labeled 1, 2, and 3. Filament 1 moved into the field of view and then paused, overlapping with filament 2 (t = 15 s). Note that the region of overlap is evident by the local increase in the fluorescence intensity, although the two filaments cannot be resolved. Subsequently the two filaments joined end to end (t = 35 s) and moved as a single longer filament (1+2; t = 60–135 s). The length of this longer filament (13.5 µm) was close to the sum of the lengths of filaments 1 and 2 (4.2 µm + 9.4 µm = 13.6 µm), consistent with end-to-end annealing. Filament 3 remained stationary throughout this sequence. See Movie S4. (Scale bar, 10 µm.)
Fig. 10.
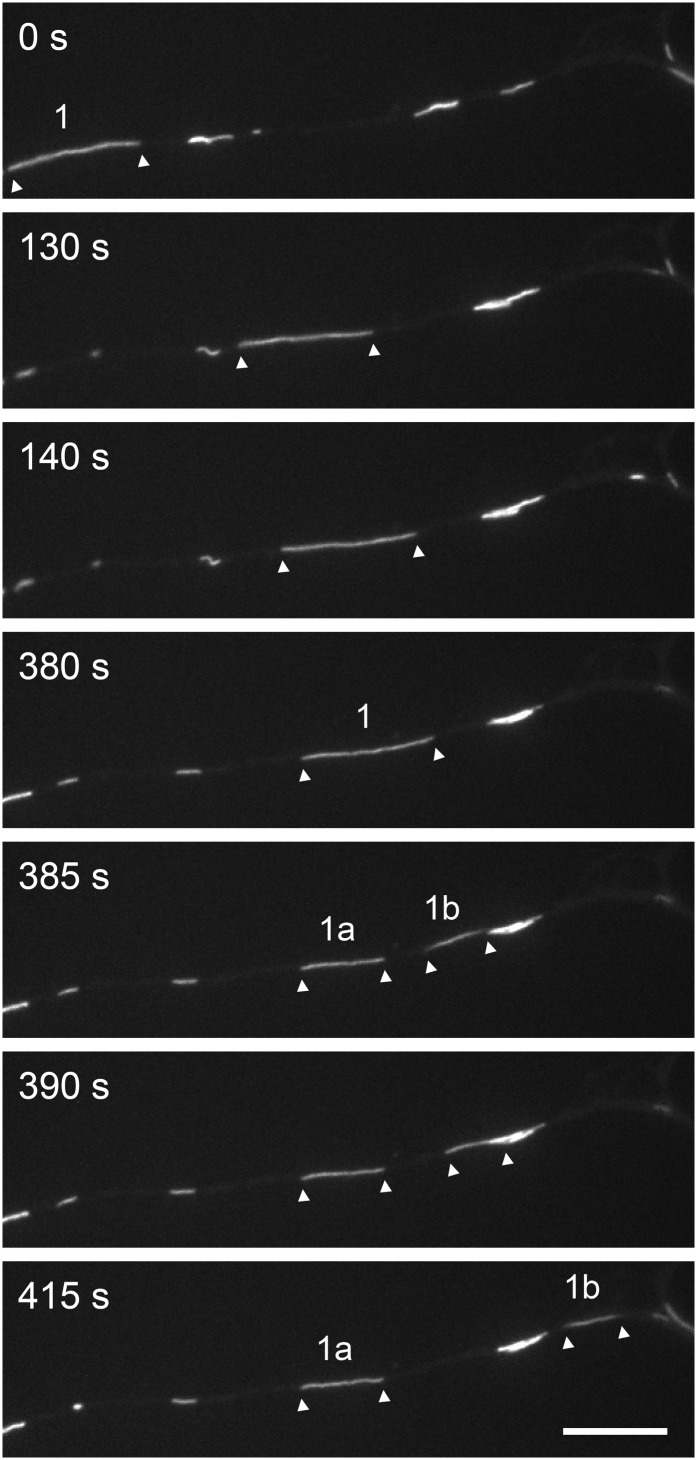
A severing event captured on camera. Time-lapse imaging of neurofilaments in an axon of a cultured mouse cortical neuron expressing GFP-NFM. This portion of axon contained multiple filaments, some isolated and some overlapping. Filament 1 moved into the field of view and paused and then broke at t = 380–385 s, leaving two fragments of unequal length (designated 1a and 1b; t = 385 s). Filament 1a remained stationary, but filament 1b continued to move (t = 390–415 s). The length of the parent filament (13.2 μm) was very close to the sum of the lengths of filaments 1a and 1b (8.3 μm + 4.8 μm = 13.1 μm). See Movie S5. (Scale bar, 10 μm.)
Discussion
Neurofilament Polymers Can Lengthen by End-to-End Annealing.
We have used photoconversion and photoactivation strategies to create distinct populations of red and green fluorescent neurofilaments in cultured neurons. Over time, we found that these filaments joined together end to end, resulting in chimeric filaments composed of alternating red and green fluorescent segments. After 24 h, approximately two-thirds of the filaments exhibited red–green junctions, with an average frequency of ∼0.4 junctions per micrometer. The smooth and continuous appearance of these chimeric filaments and the absence of any overlap of the red and green fluorescence at the red–green junctions suggest that these junctions represented a true end-to-end fusion. To confirm this notion, we performed live-cell imaging of moving chimeric filaments in axons and demonstrated that the red and green segments did indeed move as one. Finally, we also captured end-to-end annealing events live in time-lapse movies of GFP-tagged neurofilaments. Thus, we have shown that neurofilament polymers can lengthen by end-to-end annealing in neurons and that this annealing is a robust and efficient process in these cells. In conjunction with our previous report of end-to-end annealing of neurofilaments and vimentin filaments in adrenal carcinoma and immortalized fibroblast cell lines (10), these data suggest that end-to-end annealing may be a general mechanism by which intermediate filaments lengthen in cells.
Mechanism of Intermediate Filament Assembly and End-to-End Annealing.
Studies on intermediate filaments in vitro have established that the subunit of assembly is a tetramer (18, 19). Each tetramer is composed of two rod-shaped dimers that associate in a staggered antiparallel configuration, giving rise to 12-nm-long dimer overhangs at either end (20–23). The tetramers associate laterally to form unit-length filaments (ULFs) that measure 60 nm in length and ∼16 nm in width; the ULFs elongate by joining end to end to form thick filaments (3, 24, 25). The end-to-end joining of these ULFs is stabilized by interdigitation of the dimer overhangs of the tetramer subunits at the ends of the ULFs (5). Subsequently the thick filaments undergo a radial compaction to form mature intermediate filament polymers, which measure ∼11 nm in width (3, 18, 19, 24).
Although it is not possible to measure neurofilament diameter by light microscopy, the long length of the filaments that annealed end to end in our experiments suggests they are mature, compact intermediate filament polymers. Thus, it would appear that end-to-end annealing is not unique to the concatenation of ULFs and that it also contributes to the elongation of mature compact filaments. This interpretation is consistent with the recent demonstration of end-to-end annealing of vimentin filaments in vitro (26) as well as with predictions based on computational modeling of intermediate filament assembly in vitro (5, 6). Presumably the end-to-end joining of these mature polymers is stabilized by the same interactions between the dimer overhangs that stabilize the end-to-end joining of ULFs described above (10). The fact that vimentin filaments can anneal end-to-end in vitro in the absence of other proteins indicates that this annealing is an intrinsic property of intermediate filaments (26). However, we cannot exclude the possibility that there may also be proteins or other factors that catalyze or regulate this process in cells.
Efficiency of End-to-End Annealing in Neurons.
For intermediate filament polymers to fuse end to end, there must be a diffusional encounter between two filament ends, and those ends also must be in the right spatial configuration to permit a molecular integration. Given these requirements, it is remarkable that preformed neurofilament polymers can join end to end in neuronal cytoplasm with the efficiency that we have reported in this study. This efficiency may explain the remarkable length of neurofilaments in axons. For example, we have observed neurofilaments as long as 183 µm in axons of cultured neurons (27), and the one published estimate of neurofilament length in vivo, which was obtained by electron microscopy of serial sections of bullfrog olfactory nerve, has yielded an average length of 118 µm (28). We propose that neurofilaments attain these lengths progressively over time by concatenation of shorter filaments to form longer ones. In support of this idea, we observed as many as 14 red–green junctions along a single neurofilament, indicating that filaments indeed can be formed by multiple end-to-end annealing events.
Severing Mechanism for Neurofilaments.
In the course of our experiments we noticed that the increase in the number of chimeric filaments over time was accompanied by a decrease in the average lengths of the photoactivated or photoconverted fluorescent filament segments. Because the number of photoactivated or photoconverted filaments was fixed at the start of our experiments in our experimental strategies, this decrease cannot be explained by de novo synthesis and assembly of new polymer. Thus, we conclude that there are mechanisms in these cells that sever neurofilaments. In support of this conclusion, we were able to capture severing events live in time-lapse movies of GFP-tagged neurofilaments. Based on these observations, we propose that the lengthening of neurofilaments caused by end-to-end annealing in nerve cells is opposed by shortening caused by severing, implying that the length of these polymers is regulated by a dynamic balance of severing and annealing mechanisms. In our photoconversion and photoactivation experiments these mechanisms result in a gradual interchange of the red and green filament populations, effectively merging them into a single population of patchwork filaments.
Our study is a direct demonstration of the intermediate filament severing that was proposed as mechanism of keratin filament disassembly in mitotic oocytes by Klymkowsky et al. (29) more than 20 y ago based on density gradient sedimentation of cell lysates. Severing is well established for microtubules and microfilaments, and it is known to be catalyzed by severing proteins, including gelsolin and cofilin for microfilaments (30, 31) and katanin and spastin for microtubules (32, 33). These proteins bind to the sides of preexisting polymers and destabilize the interactions between the subunits, resulting in fragmentation. Klymkowsky et al. (29) proposed that keratin filaments could be severed by hyperphosphorylation, either by destabilizing the filament or by targeting the filament for the action of a putative severing factor. Our data do not address the severing mechanism, but, given the extraordinary flexibility and mechanical strength of intermediate filament polymers (34–38), it seems unlikely that the breakage could be passive or spontaneous.
Are End-to-End Annealing and Severing Artifacts of Fluorescent Fusion Proteins?
One potential criticism of these data are that they were all obtained with fluorescent fusion proteins. Unfortunately this is a general limitation in the field at present, because it is not possible to perform live imaging of proteins in cells without tagging them. We do screen all our neurofilament fusion proteins for their ability to coassemble with other neurofilament proteins, and the low level of diffuse fluorescence in our movies (e.g., Movies S4, S5, S6, and S7) confirms that the fusion proteins are fully assembled. Thus, the fusion proteins do not interfere with neurofilament assembly. Moreover, we were able to demonstrate end-to-end annealing of untagged neurofilaments in our experiments in heterologous cell lines (10). For those experiments we created two populations of cells, one expressing neurofilaments composed of NFL and NFM and the other expressing neurofilaments composed of NFL and NFH. We then induced those cells to fuse by using polyethylene glycol and looked for evidence of annealing of NFM-containing and NFH-containing neurofilaments in the resulting hybrid cells by using immunofluorescence microscopy with specific antibodies. Such experiments are not possible in neurons, because neurons do not tolerate cell fusion, but they give us added confidence that end-to-end annealing is not an artifact of our experimental strategy. However, different experimental strategies will be required to confirm severing without the use of fluorescent fusion proteins.
Do Neurofilaments “Cut and Run”?
Neurofilament polymers are transported along axons in a rapid, intermittent, and bidirectional manner (14, 15, 39). The filaments move along microtubule tracks powered by microtubule motor proteins (40, 41). Kinetic analysis of this movement has shown that the filaments alternate between two distinct kinetic states, a mobile state in which they exhibit short bouts of rapid motion interrupted by short pauses (seconds or minutes) and a stationary state in which they pause for prolonged periods of time (hours) without movement (42, 43). These kinetic states, which we have termed “on-track” and “off-track,” can be resolved by live-cell imaging on a time scale of minutes or hours, but on a time scale of days or weeks this behavior averages out, and the filaments behave like a single population (42, 44–46).
It is important to note that these on-track and off-track states are defined kinetically and that at present we do not know what distinguishes them mechanistically. As their names suggest, we have speculated that they may differ in the proximity of the neurofilaments to their microtubule tracks (43). However, they also may differ in some other respect. Although speculative, one possibility raised by our present data is that on-track and off-track filaments differ in their length. For example, we noted above that neurofilaments in axons can be very long, with the average length in bullfrog olfactory nerve estimated to be greater than 100 µm. However, in our live-cell imaging studies on neurofilament transport in cultured nerve cells, the average length of the moving filaments was much shorter, ranging from 4.1 to 8.6 µm (14, 16, 41, 47–49). The longest moving neurofilament that we observed was 44.8 µm in length (41), which certainly is remarkable, but it is hard to conceive of the movement of neurofilaments in excess of 100 µm.
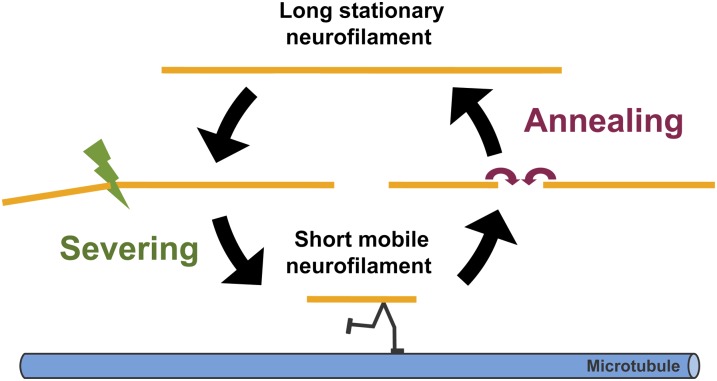
Based on these considerations, we speculate that the pausing behavior of neurofilaments may be regulated, at least in part, by the length of these polymers. According to this hypothesis, short and long neurofilaments coexist in axons, but the short ones are more likely to move, and the long ones are more likely to pause. Severing of long stationary neurofilaments may liberate short neurofilaments for axonal transport, and end-to-end annealing of these short mobile neurofilaments may sequester them in a temporarily immobile state. In this way, neurofilaments may be reversibly mobilized and immobilized during their transport along axons by a dynamic cycle of severing and annealing (Fig. 11). The severing part of this model is analogous to the “cut and run” hypothesis of microtubule transport (50–52) in which short mobile microtubules are thought to be liberated from longer stationary microtubules by the action of severing proteins such as katanin and spastin. An attractive feature of this model for neurofilaments is that it can explain how these polymers become so long in axons without being permanently immobilized (45, 46).
Fig. 11.
We hypothesize that neurofilament length is regulated by a dynamic cycle of severing and annealing. We know from prior studies that there are distinct populations of moving and stationary neurofilaments and that the average length of the moving filaments is relatively short. We speculate that long and short neurofilaments coexist in axons and that the short ones move more readily than the long ones. Severing of long filaments may liberate short neurofilaments for axonal transport by allowing them to move along microtubule tracks. Subsequently, these short mobile filaments may pause and anneal end to end, effectively sequestering them temporarily in an immobile state. Over time, all neurofilaments move; there is no significant number of permanently stationary neurofilaments in axons (47, 48).
Materials and Methods
Molecular Cloning.
The pPAGFP-NFM fusion construct encoded PAGFP connected to the N terminus of rat NFM by a 25-aa linker, as described previously (43). To make the pNFL-PAGFP fusion construct, we obtained the mouse NFL cDNA by PCR using the pNFL vector of Yan et al. (53) as a template and then subcloned it into the pPAGFP-N1 vector of Patterson and Lippincott-Schwartz (54) between the NheI and AgeI sites. To make the pNFL-mCherry fusion construct, we excised the PAGFP-coding sequence from the pNFL-PAGFP vector using AgeI and BsrGI and then replaced it with the mCherry sequence which was obtained by PCR using the pRSET-B-mCherry vector of Shaner et al. (55) as a template. To make the pmEos2-NFM fusion construct, we excised the EGFP sequence from the pEGFP-rat NFM vector of Wang et al. (14) using NheI and BspEI and then replaced it with the mEos2 sequence which was obtained by PCR using the pRSETa-mEos2 vector of McKinney et al. (13) as template (plasmid 20341; Addgene). All constructs were confirmed by sequencing.
Cell Culture and Transfection.
Cortical neurons were cultured using the glial sandwich technique of Kaech & Banker (56), essentially as described by Wang & Brown (49). To prepare glial cultures, the cerebral cortices of 4–6 postpartum day 1 (P1) rat pups were dissected and chopped in Ca2+/Mg2+-free HBSS; (Invitrogen). The tissue pieces were dissociated in PBS containing 0.25% (wt/vol) trypsin (Worthington Biochemical Corp.), 1% (wt/vol) DNase-I (Sigma), and 0.27 mM EDTA (Sigma), and the resulting cell suspension was plated in a 75-cm3 flask at 37 °C/5% CO2 in glial medium, which consisted of Minimum Essential Medium (Invitrogen) supplemented with 10% (vol/vol) horse serum (Invitrogen), 0.7% (wt/vol) glucose (Sigma), and 5 μg/mL gentamicin (Invitrogen). After 2 h the flask was swirled to remove loosely attached cells, and then the remaining attached cells were fed with fresh glial medium. The cells typically were passaged onto glass coverslips 1 wk before the intended day of use, and the culture medium was replaced with neuronal cell-plating medium (see below) 1 d before the intended day of use. To prepare neuronal cultures, the cerebral cortices of two P0 rats were dissociated in PBS containing 0.25% (wt/vol) Trypsin, 1% (wt/vol) DNase-I, and 0.27 mM EDTA (Sigma).
The dissociated cortical neurons were transfected by electroporation before plating using an Amaxa Nucleofector (Lonza) with the rat neuron nucleofection kit (VPG-1003) and program O-03. The volume of the cell suspension was 100 μL, and the cell density ranged from 4–6 × 106 cells/mL. For the photoactivation and photoconversion experiments, we used either 8 μg pmEos2-NFM construct or 8 μg pPAGFP-NFM construct and 8 μg pNFL-mCherry construct. For time-lapse imaging of GFP-tagged neurofilaments we used 2 µg pEGFP-NFM (49). The nucleofected cells were plated onto glass-bottomed dishes that had been coated with poly-d-lysine (Sigma). Glass coverslips bearing glia (>80% confluency) were suspended over the neurons using dots of paraffin wax as spacers, and the resulting sandwich cultures were maintained initially at 37 °C/5% CO2 in neuronal cell-plating medium, which consisted of NbActiv4 (BrainBits) supplemented with 5% (vol/vol) FBS (Thermo Scientific) and 5 μg/mL gentamicin. The day after plating, the medium was replaced with neuronal cell-culturing medium, which was identical to the plating medium except that it lacked serum and also contained 5 μM cytosine arabinoside (AraC; Sigma). Every 2 d, half the medium was removed and replaced with fresh cell-culturing medium lacking AraC.
Microscopy and Imaging.
For imaging, the neuronal cell-culture medium was replaced with Hibernate-E low fluorescence medium (BrainBits). A layer of silicone fluid (polydimethylsiloxane, 5 centipoise; Sigma) was floated onto the medium to prevent evaporation during observation on the microscope stage. For most of the photoactivation and photoconversion experiments, the cells were observed with a Nikon TiE inverted widefield epifluorescence microscope (Nikon) using a Nikon Intensilight light source and a 100×/1.4NA Plan-Apochromat VC or a 100/1.4NA Plan-Apochromat DM oil immersion objective. The temperature and humidity on the microscope stage was maintained using a stage-top incubating chamber (Okolab). The exciting light was attenuated with a 25% transmission neutral-density filter, and images were acquired with no pixel binning using a CoolSNAP HQ cooled CCD camera (Photometrics) and MetaMorph software (Molecular Devices). For dual-wavelength time-lapse imaging, we acquired green and red fluorescent images simultaneously at 5-s intervals using 1-s exposures and a DualView two-channel imager (Photometrics) in combination with a custom ET-GFP/mCherry dual-bandpass excitation filter and dichroic (Chroma). Photobleaching and photoconversion were performed using a Mosaic Digital Diaphragm (Andor Technology). mCherry and photoconverted mEos2 were observed with an ET-mCherry filter cube (model 49008; Chroma Technology Corp.). mEos2, EGFP, and photoactivated PAGFP were observed with an ET-GFP filter cube (model 49002; Chroma). mEos2 was photoconverted by illumination for 10–20 s using a violet filter cube (model 11005v2; Chroma). PAGFP was photoactivated by illumination for 10 s using the same violet filter cube. mCherry was photobleached by excitation for 1–2 min using the ET-mCherry filter cube. For figure and movie preparation, all images acquired using the DualView were subjected to low-pass 2 × 2 spatial filtering to remove high-frequency noise. For time-lapse imaging of GFP-tagged neurofilaments, the exciting light was attenuated with an 8% transmission neutral-density filter, and images were acquired at 4-s intervals with 1-s exposures using a Micromax 512BFT cooled CCD camera (Princeton Instruments) as described previously (49). For the images in Fig. 7, we used an Andor Revolution WD spinning disk confocal microscope (Andor Technology) attached to a Nikon TiE inverted microscope, and a 100×/1.4NA Plan-Apochromat VC oil immersion objective. Photoactivation, photobleaching, and photoconversion on this system were performed with the 405, 488, and 561 nm laser lines and a FRAPPA dual galvo laser scanner introduced through a back port of the microscope. Images of the green and red fluorescence were acquired sequentially using the 488- and 561-nm laser lines and an Andor Ultra 897 cooled EMCCD camera.
Quantification of End-to-End Annealing.
To quantify end-to-end annealing, we traced the red and green fluorescent segments of neurofilaments using the line-tracing tools in the MetaMorph software and then we analyzed the number and frequency of the red–green junctions. We traced all neurofilaments or portions of neurofilaments that were isolated from their neighbors and that could be identified without ambiguity. To avoid the possibility of user bias, the images were renamed, and the measurements were performed by an individual who did not know their identity.
Supplementary Material
Acknowledgments
We thank George Patterson and Jennifer Lippincott-Schwartz for the PAGFP vector and Roger Tsien for the mCherry vector. This work was funded by National Institutes of Health (NIH) Grant R01-NS38526 (to A.B.) and by a grant from the Takeda Science Foundation (to A.U.) with additional support from the Ohio State University Neuroscience Core funded by NIH Grant P30-NS045758.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221835110/-/DCSupplemental.
References
- 1.Prahlad V, Yoon M, Moir RD, Vale RD, Goldman RD. Rapid movements of vimentin on microtubule tracks: Kinesin-dependent assembly of intermediate filament networks. J Cell Biol. 1998;143(1):159–170. doi: 10.1083/jcb.143.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wöll S, Windoffer R, Leube RE. Dissection of keratin dynamics: Different contributions of the actin and microtubule systems. Eur J Cell Biol. 2005;84(2-3):311–328. doi: 10.1016/j.ejcb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Herrmann H, Häner M, Brettel M, Ku NO, Aebi U. Characterization of distinct early assembly units of different intermediate filament proteins. J Mol Biol. 1999;286(5):1403–1420. doi: 10.1006/jmbi.1999.2528. [DOI] [PubMed] [Google Scholar]
- 4.Wickert U, et al. Characterization of the in vitro co-assembly process of the intermediate filament proteins vimentin and desmin: Mixed polymers at all stages of assembly. Eur J Cell Biol. 2005;84(2-3):379–391. doi: 10.1016/j.ejcb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Kirmse R, et al. A quantitative kinetic model for the in vitro assembly of intermediate filaments from tetrameric vimentin. J Biol Chem. 2007;282(25):18563–18572. doi: 10.1074/jbc.M701063200. [DOI] [PubMed] [Google Scholar]
- 6.Portet S, et al. Vimentin intermediate filament formation: In vitro measurement and mathematical modeling of the filament length distribution during assembly. Langmuir. 2009;25(15):8817–8823. doi: 10.1021/la900509r. [DOI] [PubMed] [Google Scholar]
- 7.Ngai J, Coleman TR, Lazarides E. Localization of newly synthesized vimentin subunits reveals a novel mechanism of intermediate filament assembly. Cell. 1990;60(3):415–427. doi: 10.1016/0092-8674(90)90593-4. [DOI] [PubMed] [Google Scholar]
- 8.Coleman TR, Lazarides E. Continuous growth of vimentin filaments in mouse fibroblasts. J Cell Sci. 1992;103(Pt 3):689–698. doi: 10.1242/jcs.103.3.689. [DOI] [PubMed] [Google Scholar]
- 9.Vikstrom KL, Lim S-S, Goldman RD, Borisy GG. Steady state dynamics of intermediate filament networks. J Cell Biol. 1992;118(1):121–129. doi: 10.1083/jcb.118.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colakoğlu G, Brown A. Intermediate filaments exchange subunits along their length and elongate by end-to-end annealing. J Cell Biol. 2009;185(5):769–777. doi: 10.1083/jcb.200809166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan A, et al. Alpha-internexin is structurally and functionally associated with the neurofilament triplet proteins in the mature CNS. J Neurosci. 2006;26(39):10006–10019. doi: 10.1523/JNEUROSCI.2580-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan A, et al. Peripherin is a subunit of peripheral nerve neurofilaments: Implications for differential vulnerability of CNS and peripheral nervous system axons. J Neurosci. 2012;32(25):8501–8508. doi: 10.1523/JNEUROSCI.1081-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6(2):131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Ho C-L, Sun D, Liem RKH, Brown A. Rapid movement of axonal neurofilaments interrupted by prolonged pauses. Nat Cell Biol. 2000;2(3):137–141. doi: 10.1038/35004008. [DOI] [PubMed] [Google Scholar]
- 15.Brown A. Slow axonal transport: Stop and go traffic in the axon. Nat Rev Mol Cell Biol. 2000;1(2):153–156. doi: 10.1038/35040102. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Brown A. Rapid intermittent movement of axonal neurofilaments observed by fluorescence photobleaching. Mol Biol Cell. 2001;12(10):3257–3267. doi: 10.1091/mbc.12.10.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan Y, Brown A. Neurofilament polymer transport in axons. J Neurosci. 2005;25(30):7014–7021. doi: 10.1523/JNEUROSCI.2001-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrmann H, Aebi U. Structure, assembly, and dynamics of intermediate filaments. Subcell Biochem. 1998;31:319–362. [PubMed] [Google Scholar]
- 19.Herrmann H, Aebi U. Intermediate filaments: Molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- 20.Sokolova AV, et al. Monitoring intermediate filament assembly by small-angle x-ray scattering reveals the molecular architecture of assembly intermediates. Proc Natl Acad Sci USA. 2006;103(44):16206–16211. doi: 10.1073/pnas.0603629103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parry DA, Strelkov SV, Burkhard P, Aebi U, Herrmann H. Towards a molecular description of intermediate filament structure and assembly. Exp Cell Res. 2007;313(10):2204–2216. doi: 10.1016/j.yexcr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Qin Z, Buehler MJ. Structure and dynamics of human vimentin intermediate filament dimer and tetramer in explicit and implicit solvent models. J Mol Model. 2011;17(1):37–48. doi: 10.1007/s00894-010-0696-6. [DOI] [PubMed] [Google Scholar]
- 23.Chernyatina AA, Nicolet S, Aebi U, Herrmann H, Strelkov SV. Atomic structure of the vimentin central α-helical domain and its implications for intermediate filament assembly. Proc Natl Acad Sci USA. 2012;109(34):13620–13625. doi: 10.1073/pnas.1206836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrmann H, et al. Structure and assembly properties of the intermediate filament protein vimentin: The role of its head, rod and tail domains. J Mol Biol. 1996;264(5):933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- 25.Herrmann H, Aebi U. Intermediate filament assembly: Fibrillogenesis is driven by decisive dimer-dimer interactions. Curr Opin Struct Biol. 1998;8(2):177–185. doi: 10.1016/s0959-440x(98)80035-3. [DOI] [PubMed] [Google Scholar]
- 26.Winheim S, et al. Deconstructing the late phase of vimentin assembly by total internal reflection fluorescence microscopy (TIRFM) PLoS ONE. 2011;6(4):e19202. doi: 10.1371/journal.pone.0019202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown A. Visualization of single neurofilaments by immunofluorescence microscopy of splayed axonal cytoskeletons. Cell Motil Cytoskeleton. 1997;38(2):133–145. doi: 10.1002/(SICI)1097-0169(1997)38:2<133::AID-CM3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 28.Burton PR, Wentz MA. Neurofilaments are prominent in bullfrog olfactory axons but are rarely seen in those of the tiger salamander, Ambystoma tigrinum. J Comp Neurol. 1992;317(4):396–406. doi: 10.1002/cne.903170406. [DOI] [PubMed] [Google Scholar]
- 29.Klymkowsky MW, Maynell LA, Nislow C. Cytokeratin phosphorylation, cytokeratin filament severing and the solubilization of the maternal mRNA Vg1. J Cell Biol. 1991;114(4):787–797. doi: 10.1083/jcb.114.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLaughlin PJ, Gooch JT, Mannherz HG, Weeds AG. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature. 1993;364(6439):685–692. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- 31.Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int Rev Cytol. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- 32.Roll-Mecak A, McNally FJ. Microtubule-severing enzymes. Curr Opin Cell Biol. 2010;22(1):96–103. doi: 10.1016/j.ceb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharp DJ, Ross JL. Microtubule-severing enzymes at the cutting edge. J Cell Sci. 2012;125(Pt 11):2561–2569. doi: 10.1242/jcs.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mücke N, et al. Assessing the flexibility of intermediate filaments by atomic force microscopy. J Mol Biol. 2004;335(5):1241–1250. doi: 10.1016/j.jmb.2003.11.038. [DOI] [PubMed] [Google Scholar]
- 35.Kreplak L, Bär H, Leterrier JF, Herrmann H, Aebi U. Exploring the mechanical behavior of single intermediate filaments. J Mol Biol. 2005;354(3):569–577. doi: 10.1016/j.jmb.2005.09.092. [DOI] [PubMed] [Google Scholar]
- 36.Guzmán C, et al. Exploring the mechanical properties of single vimentin intermediate filaments by atomic force microscopy. J Mol Biol. 2006;360(3):623–630. doi: 10.1016/j.jmb.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 37.Kreplak L, Herrmann H, Aebi U. Tensile properties of single desmin intermediate filaments. Biophys J. 2008;94(7):2790–2799. doi: 10.1529/biophysj.107.119826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin Z, Buehler MJ, Kreplak L. A multi-scale approach to understand the mechanobiology of intermediate filaments. J Biomech. 2010;43(1):15–22. doi: 10.1016/j.jbiomech.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 39.Brown A. Slow axonal transport. In: Squire LR, editor. Encyclopedia of Neuroscience. Vol 9. Oxford, UK: Academic; 2009. pp. 1–9. [Google Scholar]
- 40.Francis F, Roy S, Brady ST, Black MM. Transport of neurofilaments in growing axons requires microtubules but not actin filaments. J Neurosci Res. 2005;79(4):442–450. doi: 10.1002/jnr.20399. [DOI] [PubMed] [Google Scholar]
- 41.Uchida A, Alami NH, Brown A. Tight functional coupling of kinesin-1A and dynein motors in the bidirectional transport of neurofilaments. Mol Biol Cell. 2009;20(23):4997–5006. doi: 10.1091/mbc.E09-04-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown A, Wang L, Jung P. Stochastic simulation of neurofilament transport in axons: The “stop-and-go” hypothesis. Mol Biol Cell. 2005;16(9):4243–4255. doi: 10.1091/mbc.E05-02-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trivedi N, Jung P, Brown A. Neurofilaments switch between distinct mobile and stationary states during their transport along axons. J Neurosci. 2007;27(3):507–516. doi: 10.1523/JNEUROSCI.4227-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jung P, Brown A. Modeling the slowing of neurofilament transport along the mouse sciatic nerve. Phys Biol. 2009;6(4):046002. doi: 10.1088/1478-3975/6/4/046002. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Jung P, Brown A. Axonal transport of neurofilaments: A single population of intermittently moving polymers. J Neurosci. 2012;32(2):746–758. doi: 10.1523/JNEUROSCI.4926-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown A, Jung P. A critical reevaluation of the stationary axonal cytoskeleton hypothesis. Cytoskeleton (Hoboken) 2013;70(1):1–11. doi: 10.1002/cm.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uchida A, Brown A. Arrival, reversal, and departure of neurofilaments at the tips of growing axons. Mol Biol Cell. 2004;15(9):4215–4225. doi: 10.1091/mbc.E04-05-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alami NH, Jung P, Brown A. Myosin Va increases the efficiency of neurofilament transport by decreasing the duration of long-term pauses. J Neurosci. 2009;29(20):6625–6634. doi: 10.1523/JNEUROSCI.3829-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, Brown A. A hereditary spastic paraplegia mutation in kinesin-1A/KIF5A disrupts neurofilament transport. Mol Neurodegener. 2010;5:52. doi: 10.1186/1750-1326-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Brown A. Rapid movement of microtubules in axons. Curr Biol. 2002;12(17):1496–1501. doi: 10.1016/s0960-9822(02)01078-3. [DOI] [PubMed] [Google Scholar]
- 51.Baas PW, Karabay A, Qiang L. Microtubules cut and run. Trends Cell Biol. 2005;15(10):518–524. doi: 10.1016/j.tcb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 52.Baas PW, Vidya Nadar C, Myers KA. Axonal transport of microtubules: The long and short of it. Traffic. 2006;7(5):490–498. doi: 10.1111/j.1600-0854.2006.00392.x. [DOI] [PubMed] [Google Scholar]
- 53.Yan Y, Jensen K, Brown A. The polypeptide composition of moving and stationary neurofilaments in cultured sympathetic neurons. Cell Motil Cytoskeleton. 2007;64(4):299–309. doi: 10.1002/cm.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297(5588):1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 55.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22(12):1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 56.Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1(5):2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.