Abstract
Insects are constantly adapting to human-driven landscape changes; however, the roles of their gut microbiota in these processes remain largely unknown. The western corn rootworm (WCR, Diabrotica virgifera virgifera LeConte) (Coleoptera: Chrysomelidae) is a major corn pest that has been controlled via annual rotation between corn (Zea mays) and nonhost soybean (Glycine max) in the United States. This practice selected for a “rotation-resistant” variant (RR-WCR) with reduced ovipositional fidelity to cornfields. When in soybean fields, RR-WCRs also exhibit an elevated tolerance of antiherbivory defenses (i.e., cysteine protease inhibitors) expressed in soybean foliage. Here we show that gut bacterial microbiota is an important factor facilitating this corn specialist’s (WCR's) physiological adaptation to brief soybean herbivory. Comparisons of gut microbiota between RR- and wild-type WCR (WT-WCR) revealed concomitant shifts in bacterial community structure with host adaptation to soybean diets. Antibiotic suppression of gut bacteria significantly reduced RR-WCR tolerance of soybean herbivory to the level of WT-WCR, whereas WT-WCR were unaffected. Our findings demonstrate that gut bacteria help to facilitate rapid adaptation of insects in managed ecosystems.
Keywords: anthropogenic disturbance, host–microbe interaction, contemporary evolution, digestive enzymes, dietary stress
Metagenomic studies are accelerating our understanding of host–microbe associations in various organisms; many microbial symbionts contribute directly to host evolution (1–4). However, unlike associations involving individual primary or secondary symbionts (2, 5), efforts to unravel interactions between the environment, host, and gut bacteria at the community scale—the bacterial microbiota—are often hindered by the intrinsic complexity of these interactions. Even if gut microbial species and their potential functions are identified through sequencing, in situ crosstalk among microbes and the host could prevent accurate inference regarding microbiota influences on host fitness and evolution (4). Here we find that rapid adaptation of the western corn rootworm (WCR, Diabrotica virgifera virgifera LeConte) (Coleoptera: Chrysomelidae) to human-mediated landscape changes (e.g., annual crop rotation) provides opportunities for exploring these interactions. Extensive knowledge of WCR biology, along with functional measurements of the microbial contribution to host fitness, make WCR a powerful model for studying the role of gut microbiota in insect adaptations to changing environments.
Throughout the US Corn Belt, annual crop rotation between corn (Zea mays) and soybean (Glycine max) is practiced to control the WCR (6, 7). The subterranean larvae of the WCR feed on corn roots, which may cause severe root injury and yield loss. After adult emergence, WCR beetles continue to feed on corn foliage, pollen, or immature ears and oviposit in the weeks after mating (7, 8). The life cycle of the univoltine WCR depends on the nearby availability of corn roots for newly hatched larvae. Close proximity to host tissues was historically assured by strong adult ovipositional fidelity to cornfields (8). As a pest management strategy, crop rotation disrupts the year-after-year availability of corn to which adult and larval WCR biology is adapted; larvae that emerge from eggs in soybean fields cannot survive. The strong selection pressures imposed by broad-scale adoption of crop rotation has resulted in the emergence of a “rotation-resistant” (RR) variant with reduced ovipositional fidelity to cornfields and greater mobility (6, 9). These behavioral changes increase the opportunity for RR-WCR females to lay eggs in nearby soybean fields, which may hatch in rotated cornfields in the following year and allow RR-WCR to circumvent crop rotation. Robust differences in movement patterns and oviposition in nonhost soybean plots suggest that rotation resistance could have a genetic basis (10, 11) and that its subsequent spread is correlated with landscape-level crop diversity (12). Aside from behavioral differences between the RR- and wild-type WCR (WT-WCR) (6, 9, 13), a recent study documented enhanced RR-WCR tolerance of soybean defenses (i.e., cysteine protease inhibitors, or CystPIs) (14), which are induced when soybean foliage is attacked by beetles (15). Compared with WT-WCR adults, the RR-WCR survived longer on soybean plant tissues and displayed more herbivory (14), which can prolong their residence time and opportunities to lay eggs in soybean fields. Moreover, the activity of gut cysteine proteases—the main group of enzymes responsible for WCR protein digestion (15, 16)—was also higher in RR-WCR guts, suggesting a physiological adaptation to CystPIs via enhanced proteolysis, similar to examples in other herbivorous species (14, 17, 18). However, because the expression of WCR protease genes in the two phenotypes could not fully explain differences in enzyme activity, contributions from microbial factors have been suggested (14).
Gut-associated bacteria can influence insect dietary preferences (4, 19) and are reciprocally regulated by immune genes and enzymes of the hosts (20–22). Interestingly, microarray studies of RR- and WT-WCR adult heads have identified differentially expressed sequence tags showing sequence similarity with immune proteins (23). Given the nature of the systemic and local insect immune response, their correlation with gut microbe colonization (24), and the repercussions for gut microbiota on host biology (4, 25, 26), we hypothesized that imposition of annual crop rotation selected for distinctive gut microbiota that contribute to the rapid adaptation of RR-WCR to crop rotation.
Understanding the roles of gut microbiota in insect adaptation to ecological disturbances could unveil mechanisms facilitating insect evolution and overlooked consequences of human activities. Here we used field-collected populations to test whether host–microbiota interactions in the WCR digestive tract were selected by crop rotation. We compared gut-associated bacterial community structures (designated as microbiota structures) of WT- and RR-WCR by incorporating the relative abundance of bacterial operative taxonomic units (OTUs) using automated ribosomal intergenic spacer analysis (ARISA). The functions of WT- and RR-WCR gut microbiotas were also compared by measuring the WCR tolerance of the soybean herbivory and the gut cysteine protease activity of WT- and RR-WCR after antibiotic suppression of gut microbes.
Results
Gut Microbiotas of Field-Collected WCR Populations and Their Correlation with RR/WT Phenotypes.
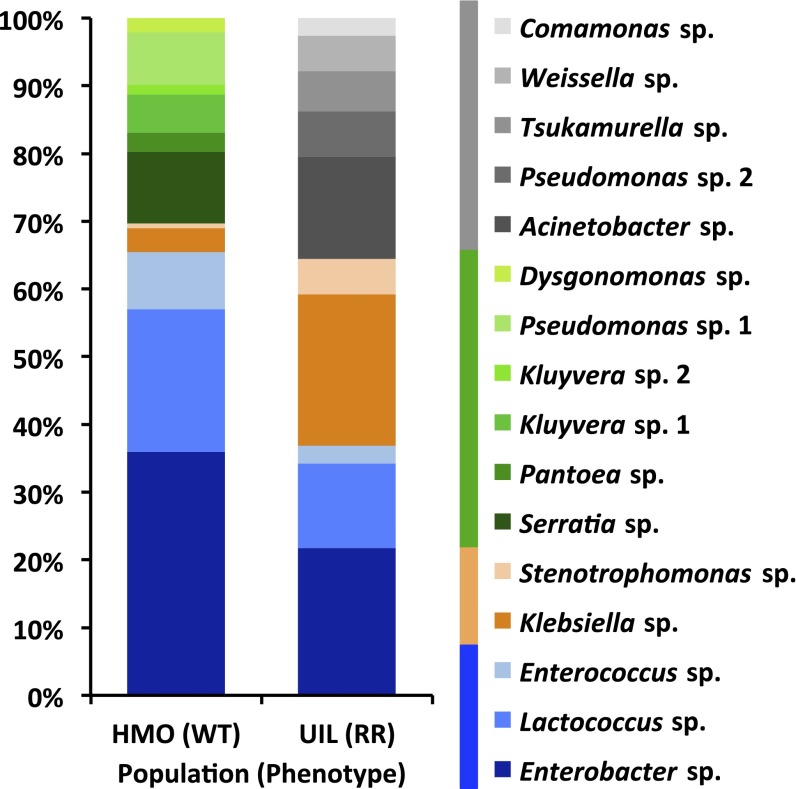
Gut microbiotas are closely linked to insect dietary preferences and trophic interactions (4, 19). To study potential differences between gut microbiotas of WT- and RR-WCR, 16S rDNA clone libraries of gut bacteria of phenotypically representative WT- and RR-WCR [based on their mobility (Fig. S1) and gut physiology (14)] from Higginsville, Missouri (WT) and Urbana, Illinois (RR) were compared. We identified significant proportional changes among five conserved bacterial taxa from Enterobacteriales, Lactobacillales, and Xanthomonadales (χ2 test, between populations, P < 0.0001), which together represent 70% and 65% of the microbiotas in the WT and RR population, respectively (Fig. 1). In WT-WCR, Enterobacter sp., Lactococcus sp., and Enterococcus sp. together make up 66% of the microbiota, whereas in RR-WCR they represent only 37% of the gut bacteria. In contrast, Klebsiella sp. and Stenotrophomonas sp. represent only 4% of the gut bacteria in WT-WCR, whereas in RR-WCR, the two taxa account for 28% of the gut microbiota. In addition, each population carried unique taxa that are known to exist in the environment (Fig. 1). Substantial compositional/proportional differences between microbiotas of the two populations suggest changes at the community/structural level that may influence gut physiology.
Fig. 1.
16S rDNA clone libraries constructed from WT-WCR (HMO, Higginsville, MO) and RR-WCR (UIL, Urbana, IL) gut microbiotas. A total of 154 and 142 clones were screened for the UIL and HMO population, respectively, until saturation of their collector’s curves (Fig. S3). The 100% stacked bar chart depicts proportional/compositional differences between microbiotas of WCRs from HMO and UIL. Blue fragments represent taxa that are more abundant in HMO; orange fragments represent taxa that are more abundant in UIL; green fragments represent taxa that exist only in HMO; gray fragments represent taxa that exist only in UIL. Top BLAST hits for each taxon (binned at 98% sequence similarity) are listed to the genus level.
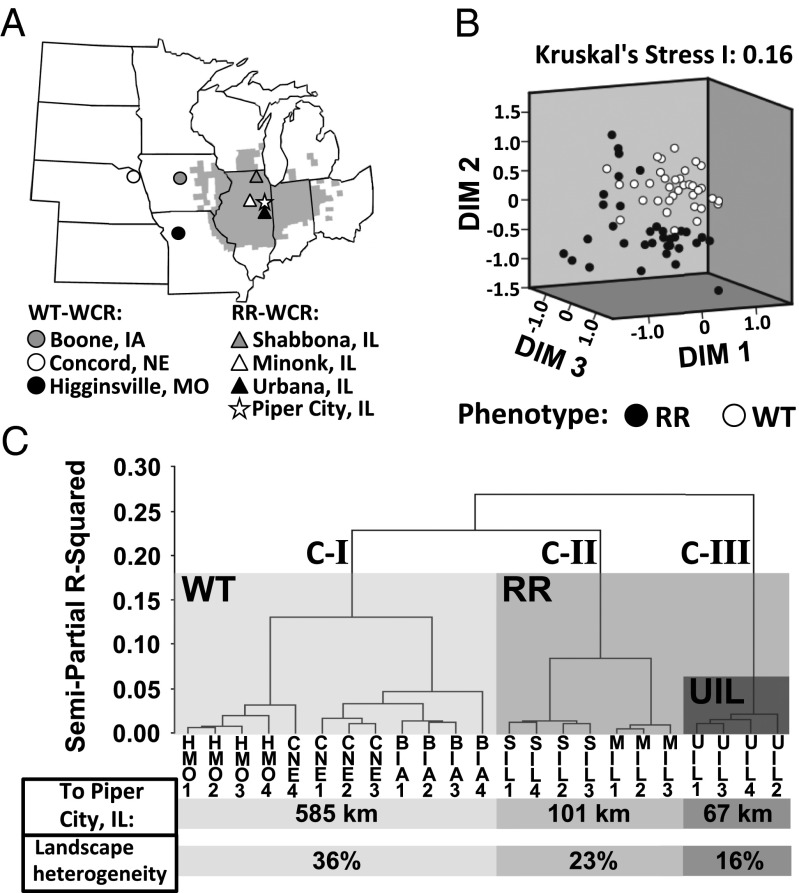
Given the prominent differences between bacterial community structures of the WT- and RR-WCR populations (Fig. 1), we tested whether gut microbiota structures are consistently different between the two phenotypes with multiple WCR populations using ARISA (Fig. 2 A and B). Collected beetles were experimentally kept under dietary conditions that they would encounter in the field (corn, soybean, or starvation) and sampled for their total gut DNA for ARISA (28). The ARISA profiles were compared within a Bray–Curtis dissimilarity matrix incorporating relative abundance of different OTUs (Fig. 2B). Results demonstrated that gut microbiota structures are different between the RR- and WT-WCR (P = 0.0001; Fig. 2B); the effect of dietary treatment (P = 0.0001) and the phenotype–diet interaction were also significant [P = 0.0147; two-way permutational multivariate analysis of variance (PERMANOVA) with Monte Carlo]. There was significantly greater heterogeneity in RR-WCR microbiota structures than in those of WT-WCR (permutational analysis of multivariate dispersions, or PERMDISP, P = 0.0078). When adding “population” as a random factor nested within “phenotype” (three-way nested PERMANOVA), a considerable amount of microbiota structural variation was explained (P = 0.0001). These data indicate correlations of microbiota structures with the RR and WT phenotypes and that there is a high level of heterogeneity in the gut bacterial communities at the population level, especially in RR-WCR.
Fig. 2.
Sampling and comparison of gut microbiota structures of WT- and RR-WCR populations on different diets and their correlation with host phenotype. (A) Locations of adult WCR collection. The gray zone indicates the range where RR-WCRs have been reported (7). Collection sites for WT (circles) and RR-WCRs (triangles) are illustrated with their corresponding symbols shown below. The star indicates Piper City (Ford County), Illinois, where the RR phenotype was first reported. (B) Nonmetric multidimensional scaling ordination depicting associations between WCR gut microbiota structures and the RR phenotype. Corresponding phenotypes are indicated below. (C) Hierarchical cluster analysis (Ward’s method) based on gut microbiota profiles of soybean-fed WCR. Populations included are the following: HMO, Higginsville, Missouri; CNE, Concord, Nebraska; BIA, Boone, Iowa; MIL, Minonk, Illinois; SIL, Shabbona, Illinois; and UIL, Urbana, Illinois. Numbers associated with each population designate different biological replicates (1–4). Each cluster (C-I–C-III) is labeled with its corresponding phenotype in light (WT) or darker (RR) boxes. The darkest box on C-III indicates the RR-WCR sample collected from the location closest to the reported epicenter (Piper City, IL) of the RR phenotype (27). Distances of each population from the epicenter and the county-scale landscape heterogeneity of each sampling site are separately averaged within clusters and labeled below.
To investigate correlations of microbiota structures and the RR phenotype at the population level, we extracted ARISA profiles of soybean-fed RR- and WT-WCR for further analysis (soybean was the diet of most interest). Pair-wise PERMANOVA tests showed significant differences in microbiota structures across nearly all populations fed on soybean (Holm–Bonferroni corrected, P < 0.05; with Monte Carlo), with the exception of WT-WCR from Concord, Nebraska, and Boone, Iowa. When analyzed with hierarchical cluster analysis using Ward’s method (29), the samples were divided into three, rather than two, large clusters (Fig. 2C). Cluster I (C-I) is composed of three WT populations, whereas clusters III and II (C-III and -II) are composed of one and two RR populations, respectively. Calculating the geographic distance between Piper City, Illinois—the historical origin and “epicenter” of the RR variant (27)—and each of the collection sites (Table S1) revealed a clustering of microbiota structures that followed the order of each location’s relative distance from the epicenter (Fig. 2C). Crop diversity in the areas where WCR were collected decreased toward the epicenter (Fig. 2C; Table S1). Moreover, the clustering results correlated with the mobility (measured as time spent to escape a cylindrical arena) of populations from each cluster, with the highest in C-III, followed by C-II, with the lowest mobility in C-I (Fig. S1). Also, there was greater microbial community heterogeneity among geographically clustered RR populations than among relatively dispersed WT populations (Fig. 2), suggesting that the patterns are not artifacts of environmental gradients.
Correlation of WCR Gut Microbiota Structures with WCR Survival on Soybean and Their Gut Cysteine Protease Activity.
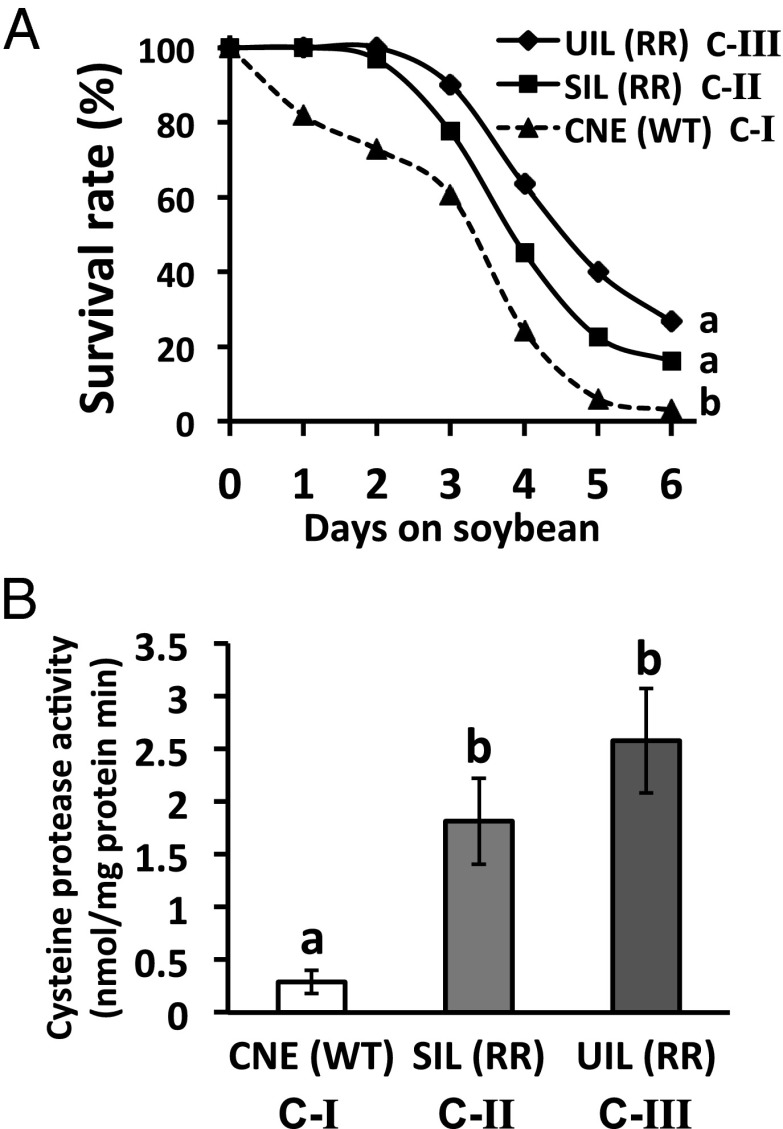
To test whether the observed gradient in microbiota community dissimilarity corresponded with WCR tolerance of soybean defense, we compared WCR survival (on soybean; Kaplan–Meier survival curves) and gut cysteine protease activities across three representative WCR populations (from each cluster in Fig. 2C). The results confirmed that RR-WCR from clusters II and III experience significantly greater survival on soybean and have higher gut cysteine protease activity than WT-WCR from cluster I (P < 0.05; Figs. 2C and 3), a pattern concordant with previous studies (14). Both phenotypic measurements of each population followed the same order as microbiota structure dissimilarity clustering among soybean-fed WCR (Figs. 2C and 3) and overall population mobility (Fig. S1).
Fig. 3.
(A) Survival of WCR populations representative of each microbiota cluster (in Fig 2C, C-I–C-III) on soybean. Survival of WCR is illustrated by the proportional change in surviving insects over time. Significant differences (log-rank test) between survival curves are declared at P < 0.05 (letters next to curves). (B) Gut cysteine protease activity while on corn diets. Activity is expressed as nanomoles of pNA released per milligram of gut protein per minute. Different letters above bars indicate significant difference in activity levels [Fisher’s Least Significant Difference (LSD), P < 0.05].
Contribution of RR and WT-WCR Gut Microbiotas to WCR Survival on Soybean and Their Gut Cysteine Protease Activity.
Using phenotypically well-characterized RR- and WT-WCR populations from Shabbona, Illinois, and Higginsville, Missouri, we compared the survival curves of RR- and WT-WCR adults feeding on soybean foliage following different antibiotic dosages (mixtures of erythromycin, gentamicin, kanamycin, and tetracycline at 0, 50, or 400 mg/L, Fig. 4 A and B). After control (0 mg/L) and 50-mg/L antibiotic treatments, the RR-WCR had greater survival on soybeans than WT-WCR (P < 0.05). After 400 mg/L treatments, however, there were no significant differences between survivorship of the two phenotypes. Compared with control groups, significant decreases in survivorship (P < 0.05) following high-dosage treatments occurred only in RR-WCR and not in WT-WCR (Fig. 4B). In contrast, survival of RR- and WT-WCR on corn diets after the same time period was unaffected by antibiotic treatments compared with control groups (Fig. S2A).
Fig. 4.
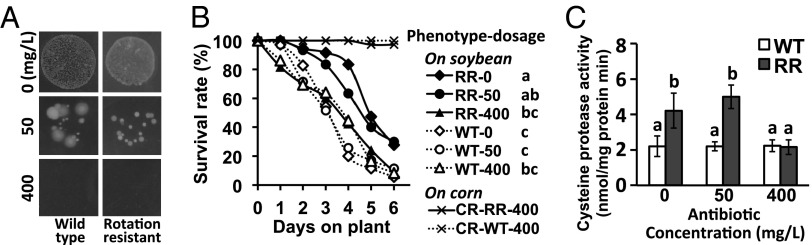
Effects of different antibiotic dosage treatments (0, 50, or 400 mg/L) on gut microbial growth and soybean tolerance in WT- and RR-WCR. (A) Antibiotic treatments (dosage indicated on the left) suppressed growth of culturable bacteria in WT- and RR-WCR guts. Pulverized gut-tissue suspensions (104-fold diluted, 100 µL) were dropped on nutrient agar plates and cultured for 48 h at 30 °C. (B) Survival of WT-WCR (Higginsville, MO, population) and RR-WCR (Shabbona, IL, population) treated with different antibiotic concentrations (mg/L) before feeding on soybean plants. Significant differences (log-rank test, Tukey–Kramer adjusted) between survival curves were declared at P < 0.05 (letters next to the key). Crosses represent WCRs treated with 400 mg/L of antibiotics and fed with corn (CR) during the same experiment. (C) Effects of antibiotic treatments on gut cysteine protease activities in WT-WCR (Higginsville, MO, population) and RR-WCR (Urbana, IL, population). Protease activities are expressed as nanomoles of pNA released per milligram of gut protein per minute. Different letters indicate significant differences between treatments (Fisher’s LSD; P < 0.05).
Because high activity levels of cysteine proteases in RR-WCR guts were previously demonstrated to explain their prolonged survival on soybean (14), we measured those levels in the two WCR phenotypes (populations from Urbana, IL, and Higginsville, MO) following the antibiotic treatments described above. Treatment with 400 mg/L of antibiotics significantly lowered the protease activity of RR-WCR to the level of WT-WCR (Fig. 4C). In the WT-WCR, protease activities were unaffected by antibiotic treatment (Fig. 4C).
To test whether reduced protease activity in RR-WCR after the 400-mg/L antibiotic treatments was related to antibiotic effects on WCR protease gene expression, the expression of cysteine protease gene DvRS5 (GenBank accession no. AJ583508) (30)—the most highly expressed protease gene in WCR when feeding on soybean foliage (14)—was measured in RR- and WT-WCR treated with 400 mg/L of antibiotics or water (control groups; Fig. S2B). Antibiotic treatments had virtually no effect on DvRS5 expression in both WCR phenotypes (F = 0.076, df = 1, P = 0.79), supporting the idea that gut bacteria are the main cause of the phenotypic differences among RR-WCR that received different antibiotic treatments (Fig. 4C).
Discussion
Human-mediated landscape changes are inducing insect adaptation to ecological disturbances at an unprecedented scale and pace (31). We have demonstrated that within few decades crop rotation has selected for a distinctly altered microbiota in RR-WCR, which provides digestive advantages endowing RR-WCR with enhanced tolerance of soybean defenses. Although it is unclear whether tolerance of soybean defense enabled the RR-WCR to reduce their fidelity to corn or was a subsequent adaptation following relaxation of host fidelity, improved RR-WCR performance in a nonhost environment led to greater reproductive success in rotated corn and soybean ecosystems. These changes present a mechanism facilitating WCR adaptation to cultural control that could lead to further ecological divergence if human-driven selection continues.
The gut bacterial community structures of RR-WCR populations were different from those of WT-WCR populations (Fig. 2B). The scale of dissimilarities between the microbiota structures of soybean-fed WCR paralleled their distances from the historical epicenter of rotation resistance, their mobility (Fig. S1) and capability to tolerate soybean diets, and the landscape heterogeneity of the area where they were collected (Figs. 2C and 3). Mobility assays showed proportional differences in adult mobility among and within RR-WCR populations (Fig. S1). In addition, previous studies indicated that movement into soybean fields and subsequent soybean herbivory, although rare, occurs in WT-WCR populations. It is the proportion of beetles exhibiting these behaviors that have greatly increased in RR-WCR populations (6). Also, a role for landscape heterogeneity in the selection of higher tolerance of soybean diets in RR-WCR has been proposed (14). Our data therefore suggest that the proportion of RR-WCR harboring distinctive gut microbiota within populations is distributed in a gradient reflecting the penetration of RR phenotypes into various parts of the Corn Belt. Gut microbiotas are known to regulate or contribute to insect digestive enzyme activities (25, 26). We demonstrated that the RR-WCR microbiota contribute to the proteolysis and survival of the WCR on soybeans (Fig. 4). These results, together with the RR-WCR’s digestive adaptation to soybean CystPIs, (14) suggest that the functionally distinctive RR-WCR gut microbiota could act as an adaptive trait that persists among WCR in rotated corn and soybean agro-ecosystems.
Our study of gut bacterial clone libraries revealed substantial differences between conserved/unique bacterial taxa in WT- and RR-WCR guts (Fig. 1). Various bacterial species could produce intra/extracellular proteases (32–35), regulate host gene expression in the gut (36, 37), or modify biochemical properties of their surrounding environments (such as Enterococcus species in termites) (38). Moreover, species like Lactococcus sp. are known to regulate the growth of other gut bacteria (39, 40), which suggests correlations between their relative abundance and the gut bacterial community structure. These interactions involve complex mechanisms that are difficult to dissect based on the identity of individual taxa that are mostly environmental bacteria. The RR phenomenon itself is also intrinsically obscured because genetic diagnostic markers differentiating RR individuals from WT are lacking (41). Moreover, modeling studies showed that the spread of the RR phenotype could be explained by the expansion of an adaptive allele across populations rather than by displacement of WT-WCR by a “RR strain” (42), indicating that heterogeneity in allele frequency of the gene(s) responsible can exist in any RR-WCR population. Given these challenges, we considered the host (WCR), gut bacterial microbiota, and environment as entities and studied their interconnections. By comparing quantitative measurements representing these components at the population level, we demonstrated that the microbiotas are not merely passive players influenced by the host, but functional components of an insect mechanism to confront dietary stress. This adaptation could affect other aspects of insect biology, such as distorting the outcomes of a pest’s reduced fidelity to optimal diets. Recognizing host–microbiota interactions as potent ecological factors facilitating insect resistance evolution may provide avenues for pest resistance management and for developing pest control strategies.
Materials and Methods
Insect and Plant Materials.
Insects were collected and bioassayed from July to August in 2010–2012. WT-WCR adults were collected as individuals from cornfields in Concord, Nebraska; Higginsville, Missouri; and Boone, Iowa, whereas RR-WCRs were collected from Urbana, Illinois; Minonk, Illinois; and Shabbona, Illinois (Fig. 2A and Table S1). Sampling sites are separated by over 100 km from any other site so that confounding effects of local adaptation did not restrict the analyses. Populations were separated into different phenotypes based on the documented RR-WCR distribution (7). Four populations (all except Boone, IA, and Shabbona, IL) have been phenotypically characterized in previous work (14). Moreover, four of the most abundant populations (all except Boone, IA, and Minonk, IL) were tested for their mobility; each population included some individuals exhibiting characteristics of WT-WCR (longer escape time = lower mobility) and RR-WCR populations (shorter escape time = greater mobility) (Fig. S1). Collection of all field populations was completed within a week to reduce insect phenology effects. To minimize laboratory effects on gut physiology and microbiota composition, WCR were maintained on corn ears from their field of collection; WCR were used in experiments soon after collection. Correlation of gut microbiota structures with the RR phenotype was first tested across all populations. Thereafter, because of limited insect availability, phenotypically representative populations of WT- and RR-WCR were used to conduct subsequent bioassays and antibiotic treatments.
Soybean plants (G. max “Williams 82”) were grown in a greenhouse under light intensities of 1,200–1,500 µmol⋅m−2⋅s−1 at the University of Illinois at Urbana–Champaign for 28 d. For dietary treatments, corn ears (Z. mays “Sugar Buns”) grown in an experimental plot at the University of Illinois were washed and confirmed to be free of transgenes using test strips (QuickStix, Envirologix Inc.). Ears used for each experiment were obtained and prepared from the same batch of hand-harvested corn.
Constructing and Sequencing 16S rDNA Libraries of WCR Gut Microbiotas.
Gut DNA from Urbana, Illinois, and Higginsville, Missouri, WCR beetles feeding on a soybean diet (Williams 82) for 48 h were separately used as templates (100 ng) for PCR using bacterial universal primers (Table S1). For each population, total gut DNA was extracted from a pool of 20 WCR guts (10 per sex). Amplified products including bacterial 16S rDNA were cloned into the pGEM-T Easy vector (Promega Inc.). Inserts were sequenced using primers 27f and 1525r (43). Details of clone screening (Fig. S3) and data processing are described in SI Materials and Methods.
Comparison of WCR Gut Microbiota Structures Under Different Dietary Treatments.
Field-collected WCR populations were kept separately on soybean foliage, corn, or a starvation treatment for 48 h and aseptically dissected to collect their complete digestive tracts (SI Materials and Methods). Bacterial community profiles incorporating relative abundance of bacterial OTUs were analyzed and compared across populations and treatments.
For DNA sampling, 20 WCR guts (10 per sex) were pooled as one biological replicate and subjected to DNA extraction using the FastDNA SPIN kit for soil (MP Biomedicals). Three to four samples were collected for each population × diet combination. In microbial community analyses using ARISA, 69 gut DNA samples (from a total of 1,380 WCR adults) were used as templates for amplification using the primers ITSF and ITSReub (44), with the former 5′-labeled with the fluorescent dye 6-carboxyfluorescein (SI Materials and Methods). Amplified products were analyzed using the ABI 3730xl genetic analyzers (Applied Biosystems Inc.). Details of ARISA and data processing are presented in SI Materials and Methods.
Multivariate Statistical Methods and Environmental Data.
Using Bray–Curtis dissimilarity measures calculated from analyzed ARISA profiles, nonmetric multidimensional scaling, PERMANOVA, PERMDISP (PRIMER6, PRIMER-E Ltd.), and hierarchical cluster analysis (SAS 9.2, SAS Institute Inc.) were conducted to determine data structures and associations of WCR microbiota with their phenotype, population, and diets (SI Materials and Methods).
To estimate the cropping diversity of each WCR sampling site, county-scale landscape heterogeneity values (percentage of land area that is neither corn nor soybean, obtained from online databases) were calculated for each site (Table S1).
WCR Survival Test.
Tests of field-collected WCR survival on soybean plants were conducted as previously described (14) with slight changes (SI Materials and Methods). Before the tests, 30–35 female WCR from different treatments were starved for 36 h (with water) to facilitate soybean herbivory. Soybean plants were pretreated with 250 µM of methyl-jasmonate 4 d before the test as described in a previous study for induction of soybean CystPIs (15). Survival data were analyzed using the Kaplan–Meier method (45). Survival distribution curves of all treatment groups (phenotype × antibiotic dosage) on soybean were compared using the log-rank test in SAS (LIFETEST procedure, Tukey–Kramer adjusted). For the antibiotic treatment tests described below, survival tests were repeated three times to confirm reproducibility.
Determination of Gut Cysteine Protease Activities in WCR.
Before gut sampling, WCR were either standardized on identical corn diets for 48 h (for comparisons across populations) or subjected to antibiotic treatments on the same commercial diets (described below). Independent triplicates of gut samples pooled from multiple insects of equal sex ratios (10 for antibiotic treatment tests and 6 for comparison across populations) were extracted for their gut proteases (SI Materials and Methods). After inhibition of inducible cathepsin B activity by the inhibitor CA-074 [l-3-trans(propylcarbamyl) oxirane-2-carbonyl)-l-isoleucyl-l-proline] (14), constitutive cysteine protease activities of each sample were determined by monitoring substrate [L-pyroglutamyl-L-phenylalanyl-L-leucine-p-nitroanilide (p-Glu-Phe-Leu-pNA)] cleavage under 405 nm using a spectrophotometer (SI Materials and Methods). The Bradford method (Bio-Rad Laboratories Inc.) was used to determine protein concentrations of all protease samples. Protease activity measurements were then calculated into units of nanomoles of pNA released per milligram of protein per minute and compared across treatments using one-way ANOVA.
Antibiotic Treatment of WCR adults.
To suppress gut microbes, antibiotic mixtures of erythromycin, gentamicin, kanamycin, and tetracycline were added to sterile water and flash-autoclaved commercial diets (BioServ; F9766B) to achieve low (50 mg/L) or high (400 mg/L) concentrations and fed to the insects. In control groups, antibiotic solutions were replaced with sterile water. Before treatments, WCR were starved for 12 h to promote diet ingestion. Each treatment was replicated in three containers and continued for 5 d. To confirm treatment effectiveness, guts from three treated insects were pooled and pulverized in 100 µL of sterile water. After 104-fold dilution, 100 µL of these suspensions was dropped on nutrient agar and grown for 48 h at 30 °C (Fig. 4A). To rule out detrimental effects (to the WCR) caused by antibiotics, survival of WT-WCR (Higginsville, MO) and RR-WCR (Shabbona, IL) treated with 400 mg/L of antibiotics were also compared with those from control groups (water only) on corn diets for 6 d. All antibiotic-treated insects were then subjected to survival and protease activity tests described above. Antibiotic-treated samples were also used for total RNA extraction and determination of DvRS5 expression using the primers DvRS5-rtF/R and EF-rtF/R (internal control; Table S2).
Supplementary Material
Acknowledgments
We thank D. Huckla, B. Hibbard, M. Dunbar, D. Lindgren, and T. Hunt for their assistance in WCR collection and supply; J. Juvik and R. Nelson for plant materials; Y. Hanzawa, M. Band, and A. Yannarell for equipment and technical support; and M. Berenbaum, J. Cheeseman, G. Caetano-Anollés, H. Lim, W. Anthonysamy, M. Davis, and R. Weigel for their useful comments. This research was funded by US Department of Agriculture-National Institute of Food and Agriculture Grant 2009-35505-06012 (to M.J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KC865711–KC865726).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301886110/-/DCSupplemental.
References
- 1.Russell JA, et al. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc Natl Acad Sci USA. 2009;106(50):21236–21241. doi: 10.1073/pnas.0907926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 3.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dillon RJ, Dillon VM. The gut bacteria of insects: Nonpathogenic interactions. Annu Rev Entomol. 2004;49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 5.Brownlie JC, Johnson KN. Symbiont-mediated protection in insect hosts. Trends Microbiol. 2009;17(8):348–354. doi: 10.1016/j.tim.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Levine E, Spencer JL, Isard SA, Onstad DW, Gray ME. Adaptation of the western corn rootworm to crop rotation: Evolution of a new strain in response to a management practice. Am Entomol. 2002;48(2):94–117. [Google Scholar]
- 7.Gray ME, Sappington TW, Miller NJ, Moeser J, Bohn MO. Adaptation and invasiveness of western corn rootworm: Intensifying research on a worsening pest. Annu Rev Entomol. 2009;54:303–321. doi: 10.1146/annurev.ento.54.110807.090434. [DOI] [PubMed] [Google Scholar]
- 8.Spencer JL, Hibbard BE, Moeser J, Onstad DW. Behaviour and ecology of the western corn rootworm (Diabrotica virgifera virgifera LeConte) Agric For Entomol. 2009;11:9–27. [Google Scholar]
- 9.Knolhoff LM, Onstad DW, Spencer JL, Levine E. Behavioral differences between rotation-resistant and wild-type Diabrotica virgifera virgifera (Coleoptera: Chrysomelidae) Environ Entomol. 2006;35(4):1049–1057. [Google Scholar]
- 10.Pierce CMF, Gray ME. Western corn rootworm, Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae), oviposition: A variant’s response to maize phenology. Environ Entomol. 2006;35:423–434. [Google Scholar]
- 11.Pierce CMF, Gray ME. Seasonal oviposition of a western corn rootworm, Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae), variant in east central Illinois commercial maize and soybean fields. Environ Entomol. 2006;35(3):676–683. [Google Scholar]
- 12.Onstad DW, et al. Does landscape diversity slow the spread of rotation-resistant western corn rootworm? Environ Entomol. 2003;32(5):992–1001. [Google Scholar]
- 13.Mabry TR, Spencer JL. Survival and oviposition of a western corn rootworm variant feeding on soybean. Entomol Exp Appl. 2003;109(2):113–121. [Google Scholar]
- 14.Curzi MJ, Zavala JA, Spencer JL, Seufferheld MJ. Abnormally high digestive enzyme activity and gene expression explain the contemporary evolution of a Diabrotica biotype able to feed on soybeans. Ecol Evol. 2012;2(8):2005–2017. doi: 10.1002/ece3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zavala JA, Casteel CL, Delucia EH, Berenbaum MR. Anthropogenic increase in carbon dioxide compromises plant defense against invasive insects. Proc Natl Acad Sci USA. 2008;105(13):5129–5133. doi: 10.1073/pnas.0800568105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koiwa H, et al. A plant defensive cystatin (soyacystatin) targets cathepsin L-like digestive cysteine proteinases (DvCALs) in the larval midgut of western corn rootworm (Diabrotica virgifera virgifera) FEBS Lett. 2000;471(1):67–70. doi: 10.1016/s0014-5793(00)01368-5. [DOI] [PubMed] [Google Scholar]
- 17.Moon J, Salzman RA, Ahn JE, Koiwa H, Zhu-Salzman K. Transcriptional regulation in cowpea bruchid guts during adaptation to a plant defence protease inhibitor. Insect Mol Biol. 2004;13(3):283–291. doi: 10.1111/j.0962-1075.2004.00485.x. [DOI] [PubMed] [Google Scholar]
- 18.Gruden K, et al. Molecular basis of Colorado potato beetle adaptation to potato plant defence at the level of digestive cysteine proteinases. Insect Biochem Mol Biol. 2004;34(4):365–375. doi: 10.1016/j.ibmb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Berenbaum MR. In: Novel Aspects of Insect-Plant Interactions. Barbosa P, Letourneau DK, editors. New York: Wiley; 1988. pp. 91–123. [Google Scholar]
- 20.Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16(2):228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verberkmoes NC, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3(2):179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 22.Shao Q, et al. Hindgut innate immunity and regulation of fecal microbiota through melanization in insects. J Biol Chem. 2012;287(17):14270–14279. doi: 10.1074/jbc.M112.354548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knolhoff LM, Walden KKO, Ratcliffe ST, Onstad DW, Robertson HM. Microarray analysis yields candidate markers for rotation resistance in the western corn rootworm beetle, Diabrotica virgifera virgifera. Evol Appl. 2011;3(1):17–27. doi: 10.1111/j.1752-4571.2009.00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 25.Visôtto LE, Oliveira MG, Guedes RN, Ribon AO, Good-God PI. Contribution of gut bacteria to digestion and development of the velvetbean caterpillar, Anticarsia gemmatalis. J Insect Physiol. 2009;55(3):185–191. doi: 10.1016/j.jinsphys.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 26.Engel P, Martinson VG, Moran NA. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci USA. 2012;109(27):11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine E, Oloumi-Sadeghi H. Western corn rootworm (Coleoptera: Chrysomelidae) larval injury to corn grown for seed production following soybeans grown for seed production. J Econ Entomol. 1996;89(4):1010–1016. [Google Scholar]
- 28.Fisher MM, Triplett EW. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl Environ Microbiol. 1999;65(10):4630–4636. doi: 10.1128/aem.65.10.4630-4636.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58(301):236–244. [Google Scholar]
- 30.Bown DP, Wilkinson HS, Jongsma MA, Gatehouse JA. Characterisation of cysteine proteinases responsible for digestive proteolysis in guts of larval western corn rootworm (Diabrotica virgifera) by expression in the yeast Pichia pastoris. Insect Biochem Mol Biol. 2004;34(4):305–320. doi: 10.1016/j.ibmb.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Carroll SP, Loye JE. Soapberry bug (Hemiptera: Rhopalidae: Serinethinae) native and introduced host plants: Biogeographic background of anthropogenic evolution. Ann Entomol Soc Am. 2012;105(5):671–684. [Google Scholar]
- 32.Hotson A, Mudgett MB. Cysteine proteases in phytopathogenic bacteria: Identification of plant targets and activation of innate immunity. Curr Opin Plant Biol. 2004;7(4):384–390. doi: 10.1016/j.pbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Chapot-Chartier MP, Nardi M, Chopin MC, Chopin A, Gripon JC. Cloning and sequencing of pepC, a cysteine aminopeptidase gene from Lactococcus lactis subsp. cremoris AM2. Appl Environ Microbiol. 1993;59(1):330–333. doi: 10.1128/aem.59.1.330-333.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukomski S, et al. Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect Immun. 1999;67(4):1779–1788. doi: 10.1128/iai.67.4.1779-1788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao MB, Tanksale AM, Ghatge MS, Deshpande VV. Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev. 1998;62(3):597–635. doi: 10.1128/mmbr.62.3.597-635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalmasso G, et al. Microbiota modulate host gene expression via microRNAs. PLoS ONE. 2011;6(4):e19293. doi: 10.1371/journal.pone.0019293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292(5519):1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 38.Tholen A, Schink B, Brune A. The gut microflora of Reticulitermes flavipes, its relation to oxygen, and evidence for oxygen-dependent acetogenesis by the most abundant Enterococcus sp. FEMS Microbiol Ecol. 2006;24(2):137–149. [Google Scholar]
- 39.Pérez T, et al. Host-microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol. 2010;3(4):355–360. doi: 10.1038/mi.2010.12. [DOI] [PubMed] [Google Scholar]
- 40.Fooks LJ, Gibson GR. Probiotics as modulators of the gut flora. Br J Nutr. 2002;88(Suppl 1):S39–S49. doi: 10.1079/BJN2002628. [DOI] [PubMed] [Google Scholar]
- 41.Miller NJ, et al. Absence of genetic divergence between western corn rootworms (Coleoptera: Chrysomelidae) resistant and susceptible to control by crop rotation. J Econ Entomol. 2006;99(3):685–690. doi: 10.1603/0022-0493-99.3.685. [DOI] [PubMed] [Google Scholar]
- 42.Onstad DW, Spencer JL, Guse CA, Levine E, Isard SA. Modeling evolution of behavioral resistance by an insect to crop rotation. Entomol Exp Appl. 2001;100(2):195–201. [Google Scholar]
- 43. Lane DJ (1991) in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt, M. D. Goodfellow (John Wiley & Sons, New York), pp. 115–175.
- 44.Cardinale M, et al. Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl Environ Microbiol. 2004;70(10):6147–6156. doi: 10.1128/AEM.70.10.6147-6156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee ET. Statistical Methods for Survival Data Analysis. 2nd Ed. New York: Wiley; 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.