Significance
Rickettsia Sca2 mimics eukaryotic formins by promoting actin filament nucleation and elongation to assemble actin comet tails for Rickettsia motility. We show that unlike formins, Sca2 is monomeric, but has N- and C-terminal repeat domains (NRD and CRD) that interact with each other. The structure of NRD reveals a new crescent-like fold. CRD is predicted to share this fold, and might form together with NRD a doughnut-shaped formin-like structure for processive elongation. Between NRD and CRD, proline-rich sequences incorporate profilin-actin for elongation, and WASP-homology 2 (WH2) domains recruit actin monomers for nucleation. Rickettsia has therefore “rediscovered” formin-like actin nucleation and elongation.
Keywords: passenger domain, translocator domain, spotted fever
Abstract
Sca2 (surface cell antigen 2) is the only bacterial protein known to promote both actin filament nucleation and profilin-dependent elongation, mimicking eukaryotic formins to assemble actin comet tails for Rickettsia motility. We show that Sca2’s functional mimicry of formins is achieved through a unique mechanism. Unlike formins, Sca2 is monomeric, but has N- and C-terminal repeat domains (NRD and CRD) that interact with each other for processive barbed-end elongation. The crystal structure of NRD reveals a previously undescribed fold, consisting of helix–loop–helix repeats arranged into an overall crescent shape. CRD is predicted to share this fold and might form together with NRD, a doughnut-shaped formin-like structure. In between NRD and CRD, proline-rich sequences mediate the incorporation of profilin-actin for elongation, and WASP-homology 2 (WH2) domains recruit actin monomers for nucleation. Sca2’s α-helical fold is unusual among Gram-negative autotransporters, which overwhelmingly fold as β-solenoids. Rickettsia has therefore “rediscovered” formin-like actin nucleation and elongation.
Many bacterial pathogens use the actin cytoskeleton of host eukaryotic cells for invasion and motility (1, 2). In so doing, bacteria often resort to mimicry by expressing proteins that adopt core functions of key actin cytoskeletal components, particularly actin filament nucleation and elongation factors. However, bacterial proteins tend to bypass the elaborate regulatory networks characteristic of their eukaryotic counterparts, offering a rare opportunity to dissect their functions within a simplified system (2, 3), with implications for our understanding of pathogenicity and the eukaryotic actin cytoskeleton alike.
Rickettsiae are obligate intracellular Gram-negative pathogens that are transmitted to humans via arthropod vectors, such as ticks, fleas, and lice (4). Rickettsia species are responsible for a number of severe human diseases, including typhus and spotted fever (5). The spotted fever group, including Rickettsia parkeri, Rickettsia conorii, Rickettsia rickettsii, and over 20 other species throughout the world, uses the host-cell actin cytoskeleton to spread inter- and intracellularly. Similar to Listeria and Shigella, Rickettsia forms actin comet tails to propel its movement. However, the actin tails of Rickettsia consist of long and unbranched actin filaments, whereas those of Listeria and Shigella contain shorter and densely branched filaments (6, 7). These morphological differences stem from different molecular mechanisms for comet tail formation by these pathogens. Listeria and Shigella rely heavily on the activity of the host Arp2/3 complex that localizes uniformly along their tails (6). Although the Arp2/3 complex, activated by either host nucleation promoting factors (8) or the Rickettsia surface protein RickA (9, 10), is necessary for Rickettsia invasion (11), it is absent from Rickettsia tails (6). Another protein, Sca2 (surface cell antigen 2), has been implicated in comet tail formation by spotted fever group Rickettsia species (12, 13).
Sca2 is an autotransporter protein, comprising a short N-terminal signal sequence, a large passenger domain, and a C-terminal translocator domain (Fig. 1A). The signal peptide targets autotransporter proteins for passage across the inner bacterial membrane. This first step is followed by insertion of the translocator domain into the outer membrane, where it forms a pore through which the passenger domain is translocated and either cleaved or held tethered on the surface of the bacterium (14). By immunofluorescence microscopy, Sca2 has been localized to the actin tail-associated surface of R. parkeri (13), and disruption of the sca2 gene in R. rickettsii inhibits actin-tail formation and motility (12), which also results in reduced virulence. Sca2 may play a separate role in mediating the association of Rickettsia pathogens with mammalian host cells. Thus, Escherichia coli cells expressing R. conorii Sca2 gain the ability to adhere and invade mammalian cells (15). The minimal domain sufficient for association with mammalian cells was recently mapped to Sca2 residues 34–556 (16).
Fig. 1.
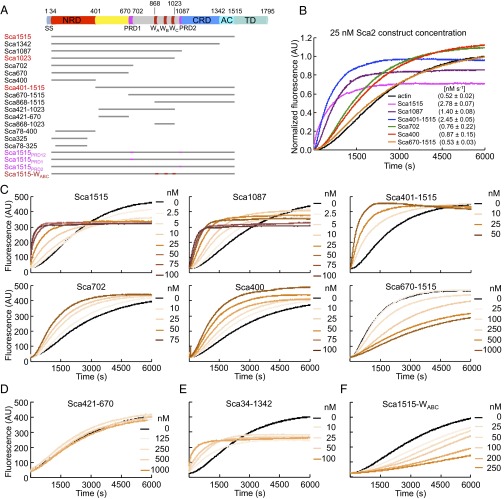
Actin polymerization activities of Sca2 constructs. (A) Domain organization of Sca2 and constructs used in this study (SS, signaling sequence; W, WH2 domains A, B and C; AC, autochaperone domain; TD, translocator domain. Constructs that were also made as Avi-tag fusions for Qdot labeling are highlighted in red. Constructs containing mutations in the proline-rich regions or WH2 domains are highlighted in magenta or maroon, respectively (the specific mutations are described in Fig. S2A). (B) Normalized time course of polymerization of 2 μM Mg-ATP-actin (6% pyrene-labeled) alone (black) or in the presence of 25 nM Sca2 constructs, as indicated. Initial polymerization rates were calculated between 0.1 and 0.3 of the normalized fluorescence and are shown in parenthesis. (C–F) Time course of polymerization of 2 μM Mg-ATP-actin (6% pyrene-labeled) alone (black) or with increasing concentrations of the Sca2 constructs shown in B (C), as well as constructs Sca421-670 (D), Sca34-1342 (E), and Sca1515-WABC (F), which are not shown in B.
In vitro, Sca2 displays actin assembly properties that resemble those of eukaryotic formins; it nucleates unbranched actin filaments, processively associates with growing barbed ends, requires profilin-actin for efficient elongation, and inhibits the activity of capping protein (13). Although these activities led to the suggestion that Sca2 functionally mimics eukaryotic formins, it is unknown how this is mechanistically achieved. Here, we show that Rickettsia Sca2 has evolved formin-like activity through an entirely unique structural and functional mechanism.
Results
N- and C-terminal Domains of Sca2 Cooperate for Optimal Polymerization Activity.
The polymerization activity of R. parkeri Sca2 decreased incrementally with C-terminal deletions, but the fragment 34-670 retained appreciable activity, leading to the suggestion that the N-terminal region of Sca2 structurally and functionally mimics the formin-homology 2 (FH2) domain (13). Here, we set out to test this hypothesis. Guided by the domain organization of R. conorii Sca2 (the species studied here, sharing 91.3% sequence identity with R. parkeri Sca2), we designed 20 protein constructs, including both N- and C-terminal deletions, as well as middle fragments and mutants (Fig. 1A, and Figs. S1 and S2A). Purification tags were removed (Fig. S2B), as they can potentially affect polymerization, particularly the GST tag that induces dimerization, a factor known to increase the polymerization activity of WH2 domain-based nucleators (17). Bulk polymerization was measured using the pyrene-actin assay with 2 μM Mg-ATP-actin (6% pyrene-labeled) (18). Although the results confirmed some of the previous observations (13), we made several new and unexpected findings that suggest cooperation between the N- and C-terminal domains of Sca2 for activity (Fig. 1 and Fig. S3).
Sca400 (residues 34–400) had nucleation activity that increased in a dose-dependent manner, and was nearly indistinguishable from that of Sca670, with polymerization rates at 25 nM of 0.87 and 0.82 nM⋅s−1 (the polymerization rate for actin alone was 0.5 nM⋅s−1). In contrast, Sca421-670 had no measurable activity at any concentration. Note that residues 400–420 were not included in the latter construct because this region was prone to degradation (Fig. S2B). Combined, these results suggested that Sca2 residues 34–400 accounted for all of the polymerization activity of Sca670, proposed previously to mimic the FH2 domain of eukaryotic formins. Sca702, including proline-rich domain (PRD)1, had similar activity to Sca400 and Sca670. In contrast, a marked increase in polymerization activity was observed with construct Sca1023 (polymerization rate 1.4 nM⋅s−1), comprising the WH2 domains, suggesting that the WH2 domains contribute significantly to the nucleation activity. The activity of Sca1087, including PRD2, was essentially indistinguishable from that of Sca1023. In contrast, every construct comprising the WH2 domains but lacking either the N- or C-terminal domains (421–1023, 670–1515, 868–1023, 868–1515) sequestered actin monomers and inhibited polymerization. Surprisingly, construct Sca401-1515 had very strong activity (polymerization rate 2.5 nM⋅s−1), approaching that of the full-length passenger domain Sca1515 (polymerization rate 2.8 nM⋅s−1). Sca1515-WABC, carrying four point mutations in each of the WH2 domains of residues implicated in actin binding (19), had no detectable nucleation activity (Fig. 1F) and additionally inhibited elongation (see below). It thus emerges that the constructs with the highest polymerization activity (Sca1515, Sca401-1515 and Sca1023/Sca1087, in that order) contained the WH2 domains and either the entire N-terminal domain or the entire C-terminal domain and at least part of the N-terminal domain. Therefore, contrary to previous results (13), we find that the nucleation activity of Sca2 decreases somewhat symmetrically with N- or C-terminal deletions.
Profilin-Dependent Barbed-End Elongation.
Constructs Sca1515, Sca401-1515, and Sca1023/Sca1087 were also set apart from the others in that they all lowered the steady-state level of actin polymer (Fig. 1 B and C and Fig. S3), suggesting that they remain bound at the barbed end and raise the critical concentration for actin polymerization. This effect was less pronounced for Sca401-1515, lacking the N-terminal Sca400 fragment, which was also the smallest fragment displaying polymerization activity (Fig. 1B). To investigate this point further, we monitored the elongation of phalloidin-stabilized F-actin seeds in the presence of 0.5 μM actin (6% pyrene-labeled) i.e., below the critical concentration for monomer addition at the pointed end, such that elongation could only occur at the barbed end. Sca400 and Sca670 had no effect on elongation at any concentration, whereas Sca1515, Sca1023, and Sca1087 all severely inhibited barbed-end elongation in a concentration-dependent manner (Fig. S4A). The WH2 domain mutant Sca1515-WABC also strongly inhibited barbed-end elongation, and this effect was not a result of monomer sequestration, as suggested by comparison with Sca421-1023, which inhibited polymerization by sequestration (Figs. S3 and S4B). Therefore, the WH2 domains are required for nucleation (Fig. 1F), but not for barbed-end binding. Sca401-1515 also inhibited barbed-end elongation but at higher concentrations. Therefore, removing the first 400 aa of Sca2 results in a construct with strong nucleation activity (Fig. 1B), but reduced barbed-end binding affinity.
To conclusively show the barbed-end tracking activity of Sca2 constructs, we used total internal reflection fluorescence (TIRF) microscopy to directly observe filament assembly in the presence of 1.5 μM Mg-ATP-actin (33% Oregon Green-labeled) and Sca2 constructs (Fig. 2). In control experiments (in the absence of Sca2 constructs), filaments grew at a rate of 10 sub⋅s−1. In the presence of 0.1 nM Sca1515, we observed two populations of filaments growing either at the control rate or at the much slower rate of 0.1 sub⋅s−1 (Fig. 2 A and C). The addition of 4 μM profilin accelerated 28-fold the elongation rate of the second population, to 2.8 sub⋅s−1. Profilin-actin accelerates the elongation rate of formins by binding to their PRDs (i.e., the FH1 domain) (20). Sca2 contains two PRDs spaced 370-aa apart. We asked whether one or both of the PRDs contributed to the binding of profilin-actin by mutating them individually and simultaneously within construct Sca1515. PRD1 and PRD2 contain three and two predicted profilin-actin binding sites, respectively. We substituted two consecutive proline residues in each of the predicted profilin-actin binding sites with serine residues (Fig. 1A and Fig. S2A). All three mutants (Sca1515PRD1, Sca1515PRD2, and Sca1515PRD12) inhibited elongation from profilin-actin to approximately the level observed in the absence of profilin (Fig. 2C). This result suggested that profilin-actin is incorporated through both PRD1 and PRD2, and that these two sites cooperate for efficient elongation. However, the PRD mutants displayed nucleation activities and steady-state polymer levels very similar to those of Sca1515 (Fig. S3), indicating that the PRD regions are not implicated in nucleation nor barbed-end binding.
Fig. 2.
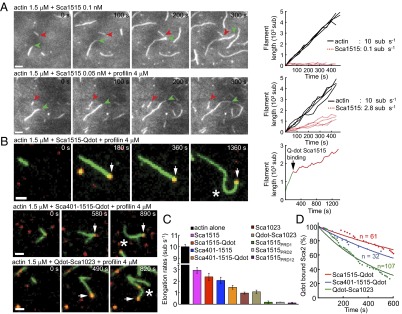
Actin filament elongation by Sca2. (A) Polymerization of 1.5 μM ATP-actin in the presence of 0.1 nM Sca1515 (Upper) or 0.05 nM Sca1515 and 4 μM profilin (Lower). Green and red arrowheads point to the barbed ends of actin filaments growing on their own or associated with Sca1515, respectively. (Scale bars, 2 μm.) The plots on the right illustrate the growth of individual actin filaments alone (black) or associated with Sca1515 (red dashed) versus time. (B) Polymerization of 1.5 μM ATP-actin in the presence of 4 μM profilin and Sca1515-Qdot, Sca401-1515-Qdot, and Qdot-Sca1023. Arrows point to Qdot-Sca2 constructs (red) bound to actin filament barbed ends (green). White asterisk indicates filament-buckling events. The plot on the right of Sca1515-Qdot illustrates the growth of the actin filament shown on the left, before (green) and after (red) its association with Sca1515-Qdot. (Scale bars, 1 μm.) (C) Elongation rates of actin filaments alone (black) or in the presence of Sca2 constructs and profilin, as indicated by color-coded labels. At least eight filaments were measured for each construct. (D) Exponential fits of the percent of Sca2 constructs (red, Sca1515-Qdot; blue, Sca401-1515-Qdot; green, Qdot-Sca1023) associated with filament-barbed ends over time (dots). The calculated half lifetimes and number of measurements are: Sca1515-Qdot (t1/2 = 1,050 ± 40 s, n = 61), Sca401-1515-Qdot (t1/2 = 707 ± 28 s, n = 32), Qdot-Sca1023 (t1/2 = 361 ± 8 s, n = 107).
To further clarify the mechanism of barbed-end binding and elongation by Sca2, we labeled the three constructs that lowered the steady-state level of actin polymer (Sca1515, Sca401-1515, and Sca1023) with quantum dots (Qdots) and tracked their activities by TIRF. These experiments allowed us to conclusively show that Sca2 was bound at the barbed end and further clarified the role of the N- and C-terminal domains for this activity. Indeed, the barbed-end elongation rate and association lifetime were reduced for the N- and C-terminal deletion constructs Sca401-1515 and Sca1023 compared with Sca1515 (Fig. 2 B and D and Movies S1, S2, and S3).
The N- and C-terminal Domains of Sca2 Interact and Cooperate for Filament Assembly.
The formin FH2 domain forms a doughnut-shaped dimer (21), a fold that appears optimally adapted for elongation of the two long-pitch helices of the actin filament (22). In contrast, we found here by analytical size-exclusion chromatography (SEC) that Sca2 is a monomer (Fig. S5), which is also consistent with the fact that Sca2 has a monomeric type of translocator domain (14). How does a monomeric protein track the two long-pitch helices of the filament? Up to this point, our data suggest cooperation and a certain level of pseudosymmetry between the N- and C-terminal domains of Sca2. Indeed, the polymerization activity of Sca2 decreases gradually with both N- or C-terminal deletions (Fig. 1B). Sca421-670 and Sca670-1515 had no activity or sequestered monomers, respectively, but construct Sca401-1515, in which these two fragments were combined, had strong polymerization activity (Fig. 1B). Optimal barbed-end processivity also required intact N- and C-terminal domains (Fig. 2 B–D and Fig. S4). Finally, the two distantly separated PRDs appeared to cooperate for elongation from profilin-actin, suggesting that they are both near the barbed end of the filament (Fig. 2C). We thus speculated that the N- and C-terminal domains might interact with each other. To test this possibility, we coexpressed constructs His-Sca670 and GST-Sca670-1515, which copurified through His- and GST-tag affinity columns and SEC, and remained as a complex after removal of the GST-tag, as confirmed by mass spectrometry (Fig. 3 A and B). What is more, the purified complex restored the activity of the full-length passenger domain in a pyrene-actin polymerization assay (Fig. 3C). Very similar results were obtained by coexpressing His-Sca670 and GST-Sca868-1515 (Fig. 3 D–F), showing that the ∼200 aa from 670 to 868 are not necessary for this interaction. Smaller C-terminal fragments could not be analyzed because of limited protein expression. We thus conclude that, at least functionally, Sca2 is not a formin; it displays formin-like nucleation and elongation activities through cooperation of its N- and C-terminal domains within a monomeric protein.
Fig. 3.
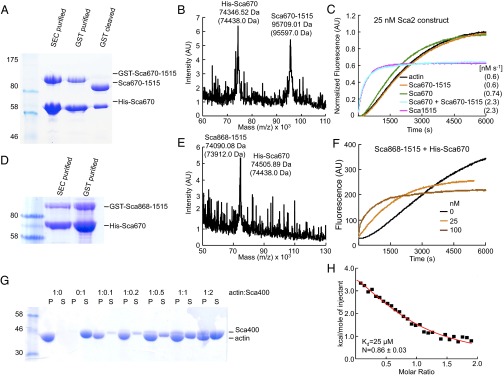
The N- and C-terminal domains of Sca2 interact and cooperate for actin filament assembly. (A) SDS/PAGE analysis of the coexpressed complex of His-Sca670 and GST-Sca670-1515 after successive copurification steps through a Ni-NTA affinity column, SEC, a GST-affinity column, and repurified through a GST affinity column after cleavage of the GST tag. (B) The identity of the two proteins was confirmed by mass spectrometry (theoretical masses are shown in parenthesis). (C) Time course of polymerization of 2 μM Mg-ATP-actin (6% pyrene-labeled) alone (black) or with addition of 25 nM of the complex of Sca670 and Sca670-1515 or the indicated Sca2 fragments. Polymerization rates (calculated as in Fig. 1B) are shown for comparison. (D–F) Same as A–C, but for coexpressed constructs His-Sca670 and GST-Sca868-1515. (G) SDS/PAGE analysis of supernatant (S) and pellet (P) fractions after cosedimentation at 278,000 × g of F-actin (15 μM) with increasing ratios of Sca400. Controls, F-actin, and Sca400 alone, are also shown. (H) Analysis by ITC of the binding of G-actin to Sca400 at 20 °C. The figure shows the binding isotherm derived from the integrated heats of binding plotted against the molar ratio of ligand (Sca400, at 340 μM) added to LatB-actin in the cell (at 17 μM) after subtracting the heat of dilution. The best-fit parameters (solid red line) correspond to a one-site binding model with dissociation constant Kd of 25 μM.
Structure-Function of the N- and C-Terminal Domains of Sca2.
Sca400 was the smallest fragment studied here capable of activating polymerization, albeit weakly (Fig. 1 B and C), and it also contributed to processivity (Fig. 2 B–D). This fragment was found serendipitously as a degradation product in crystallization trials with Sca670, and was subsequently found to account for all of the activity of Sca670, proposed to mimic the FH2 domain. We thus expected Sca400 to bind F-actin, which was confirmed in a high-speed cosedimentation assay (Fig. 3G). The ability to nucleate filaments would be consistent with recruitment of more than one actin monomer by Sca400 to form a nucleus. However, by isothermal titration calorimetry (ITC) we could only detect weak-affinity binding (Kd = 25 μM) to one LatB-actin monomer (Fig. 3H). If Sca400 binds a second monomer, the affinity must be too weak (at least with bound LatB) to be detected by this method, which correlates with the weak nucleation activity of this construct.
The structure of Sca400 was determined using the single-wavelength anomalous dispersion method and X-ray data collected from a selenomethionine-derivatized crystal (Materials and Methods and Table 1). The structure was refined to 2.18 Å resolution and revealed a unique all-helical fold (Fig. 4A and Movie S4), as confirmed by a search for structural homologs with the program Dali (23). Residues 363–400 were disordered and not observed in the structure. The rest of the structure was well-defined in the electron density map, and consisted mainly of a series of helix–loop–helix repeats stacked upon each other, taking on an overall crescent shape. Seven such repeats were identified, including three incomplete repeats (Fig. 4B). The first helix of the repeat was longer, up to seven helical turns, whereas the second helix was approximately two helical turns long. The repeats superimposed well for the last three turns of the first helix and up to the end of the second helix, but large variations were observed at the N termini of the first helix. The repeats were poorly conserved at the sequence level (Fig. 4C), explaining why they had remained undetected until the structure was solved. Sequence conservation was higher around the loop region, where three residues (Tyr, Pro, and Glu) were observed in most of the repeats. Even the few nonrepeat regions of the structure (yellow in Fig. 4A) still loosely resembled the repeat regions.
Table 1.
Crystallographic data and refinement statistics
| Native | Se-Met (peak) | |
| Protein Data Bank ID | 4J7O | |
| Data collection | ||
| Source | CHESS A1 | CHESS A1 |
| Wavelength (Å) | 0.9769 | 0.9769 |
| Space group | P21212 | P21212 |
| Unit cell | ||
| a, b, c (Å) | 82.02 102.87 55.05 | 82.22 102.93 55.17 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 2.18–64.0 (2.18–2.25) | 2.5–43.6 (2.5–2.59) |
| Rmerge (%)* | 4.50 (49.0) | 3.1 (32.2) |
| I/σ I | 13.5 (2.5) | 20.50 (3.2) |
| Completeness (%) | 97.4 (74.0) | 99.70 (99.9) |
| Redundancy | 6.83 (6.03) | 27.4 (26.9) |
| Refinement | ||
| Resolution (Å) | 2.18–64.0 (2.18–2.25) | |
| No. reflections | 24,526 (1,836) | |
| Rwork (%)† | 17.9 (28.6) | |
| Rfree (%)‡ | 22.7 (33.1) | |
| No. atoms | 2,832 | |
| Protein | 2,685 | |
| Ligands | 30 | |
| Solvent | 109 | |
| B-factors (Å2) | 64.20 | |
| Protein | 64.0 | |
| Solvent | 62.50 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.005 | |
| Bond angles (°) | 0.720 | |
| Ramachandran (%) | ||
| Favored | 99.0 | |
| Outliers | 0.3 |
Numbers in parenthesis correspond to last resolution shell.
Rmerge = Σhkl |I − 〈I〉|/Σhkl I, where I and 〈I〉 are the observed and mean intensities of all of the observations of reflection hkl, including its symmetry-related equivalents.
Rwork = Σhkl ||Fobs| − |Fcalc||/Σhkl |Fobs|, where Fobs and Fcalc are the observed and calculated structure factors of reflection hkl.
Rfree, same as Rwork, but calculated for a randomly selected subset of reflections (5%) that were not used in refinement.
Fig. 4.
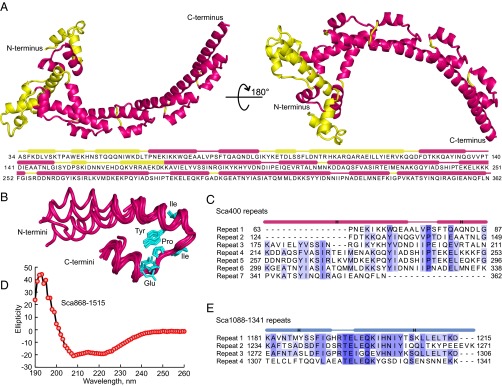
The fold of NRD and CRD consists of helix–loop–helix repeats. (A) Two views of the 2.18 Å crystal structure of NRD rotated by 180°. The structure represents a previously unknown fold, consisting of a series of helix–loop–helix repeats staked upon each other (colored red), giving rise to an overall crescent shaped structure. Regions outside the repeats are colored yellow. The secondary structure assignment based on the crystal structure is shown above the sequence. (B and C) Structural and sequence alignment of the repeats of NRD. The side chains of the most highly conserved residues of the repeat are shown (cyan) and highlighted blue in the sequence alignment. (D) The far-UV circular dichroism spectrum of Sca868-1515 displays minima at 208 and 222 nm, characteristic of all-helical structures. (E) The fragment Sca1088-1341, within this region, also contains a series of predicted helix–loop–helix repeats (four were identifiable from sequence, but more may exist). Like for NRD, sequence conservation is higher in the loop region of the repeat (highlighted in blue).
In contrast to other autotransporters, much of which are predicted to form parallel β-helical structures (24, 25), the entire structure of Sca2 is predicted to consist of α-helices connected by loops or less-ordered regions (Fig. S6). This prediction was experimentally confirmed here for construct Sca868-1515, the far-UV circular dichroism spectrum of which showed a typical α-helical profile with minima at 208 and 222 nm (Fig. 4D). Remarkably, like Sca400 the C-terminal domain also contained predicted helix–loop–helix repeats (Fig. 4E), which is why we named these two regions the N- and C-terminal repeat domains (NRD and CRD). As discussed below, folding conservation among passenger domains results from translocation and folding constraints (24, 26), and the new fold observed in NRD, dominated by local contacts (interacting residues are near in sequence), obeys basic principles of vectorial folding and is most likely preserved throughout most of the Sca2 polypeptide.
The shape of NRD is overall similar to that of one monomer of the FH2 domain, suggesting that the binding site for actin is on the inner concave side of the crescent-shaped structure, making contacts with both the N- and C-terminal ends of NRD (Figs. S7 and S8A). To test this possibility, we generated three deletion mutants: Sca78-400, Sca325, and Sca78-325 (Fig. 1A and Fig. S8A). All three mutants lost the ability to induce actin polymerization (Fig. S8C), confirming that both ends of NRD contribute toward actin assembly. At least one of the constructs, Sca325, still weakly cosedimented with F-actin (Fig. S8D) (Sca78-400 could not be tested because it overlaps with actin in the gel). These findings support the similarity in the overall shapes of Sca400 and the formin FH2 domain, and are consistent with the fact that Sca2 from R. canadensis and R. peacockii, species that do not form actin tails (13), contain deletions near the C terminus of NRD (Figs. S1 and S8B).
Discussion
Differences and Similarities of Sca2 with Other Autotransporter Proteins.
Autotransporters are virulence factors secreted by Gram-negative bacteria. Their C-terminal translocator domain forms a pore at the outer membrane for secretion of the large N-terminal passenger domain, which can be either cleaved or held tethered on the surface of the bacterium after secretion. Translocator domains are either ∼300- or ∼75-aa long, giving rise to two types of autotransporters, monomeric and trimeric, whereby the 12 strands of the β-barrel that forms the translocation pore are all contributed by a single chain or by three chains (14) (Fig. S9). The size of the pore formed by the translocator domain is only ∼10–18 Å in diameter, implying that the passenger domain must be at least partially unfolded during translocation. The small size of the pore imposes major constraints on the folding of passenger domains, 97% of which are predicted to form parallel β-helical structures (also known as β-solenoids) despite wide sequence and functional diversity (24, 25). Existing crystal structures of passenger domains support this prediction (14, 27–31) (Fig. S9). The β-solenoid fold is both repetitive and local in nature, meaning that 3D interactions involve amino acids that are close in sequence. This principle holds true for the single non–β-helical structure of a passenger domain determined thus far, that of the esterase EstA, which displays a fold comprising both α-helices and β-strands (26). Sca2 is unique among autotransporters (Fig. S9); with a 281-aa translocator domain it clearly belongs to the monomeric subgroup of autotransporters, and appears to stay tethered on the surface of the bacterium (13), but its entire passenger domain is predicted to be α-helical (Fig. S6), as established here experimentally for two large constructs, NRD and Sca868-1515 (Fig. 4). Although repeats could not be detected for residues 401–670, this region is similarly predicted to be mostly α-helical (Fig. S6) and contributes toward the nucleation activity by cooperation with the C-terminal region of Sca2. Indeed, separately Sca421-670 and Sca670-1515 had no nucleation activity, but construct Sca401-1515 in which these two fragments were combined had strong polymerization activity (Fig. 1). Sca421-670 was also required for barbed-end binding and elongation (Fig. 2). For all these reasons, we anticipate that residues 401–670 form part of the barbed-end tracking unit. At the C terminus, however, residues 1342–1515 do not appear to conform to the same folding pattern as the rest of the Sca2 passenger domain. Except for a few short predicted α-helices and β-strands, this region seems to be mostly disordered (Fig. S6) and does not contribute to the overall polymerization activity (Fig. 1E). Therefore, it is unlikely that residues 1342–1515 form part of the nucleation-elongation region of Sca2, but may instead function as an autochaperone domain, which are found in this location in most if not all autotransporters (32).
Despite the differences between Sca2 and other passenger domains, the helix–loop–helix repeat observed here for NRD, which we propose is also present in CRD, resembles that of other autotransporters in that it is repetitive and local, suggesting that, like β-helical passengers domains (24, 33), Sca2 also folds in a sequential (or vectorial) manner. In the absence of an external energy source, such as ATP or a proton gradient, it has been proposed that this folding mechanism may additionally provide the energy required for secretion (24, 33), which might also apply to Sca2. Is the new fold of NRD unique to Sca2? Other than Sca2 from other Rickettsia species, we could not find any protein sharing significant sequence identity with Sca2. However, the β-solenoid fold is observed in unrelated proteins, which have converged toward a common solution to the constraints of autotransporter secretion in Gram-negative bacteria. By analogy, we could expect that other proteins may have adopted NRD’s fold, but their number will be necessarily small, probably a small subset of the 3% of non–β-solenoid passenger domains.
Differences and Similarities of Sca2 with Eukaryotic Formins and Proposed Mechanism of Actin Assembly.
Sca2 shares distinctive functional properties with eukaryotic formins, including the ability to promote both actin filament nucleation and profilin-dependent barbed-end elongation, allowing it to play a key role in actin comet tail formation and virulence of various Rickettsia species (13). Based on these similarities, it was proposed that the N-terminal domain of Sca2 might resemble the FH2 domain of formins. Although we have found that this is not the case, and show that Sca2 has adopted formin-like activity through a different fold and mechanism, we also observe interesting parallels between Sca2 and eukaryotic formins. Distinct from formins that function as dimers, Sca2 is monomeric (Fig. S5). Unlike formins, for which the nucleation and elongation activities can be mapped respectively to the FH2-tail and FH1-FH2 domains (20, 34–38), all of the domains of Sca2 participate in these activities (Figs. 1 and 2). Another key difference is that full-length formins, like many cytoskeletal proteins, are internally inhibited and require activation by converging inputs (39), whereas full-length Sca2 is active, reflecting the simplicity of bacterial proteins. The basic fold of Sca2’s NRD is also entirely different from that of the formin FH2 domain (Fig. S7). Curiously, however, the overall crescent shape of the structure of NRD is surprisingly similar to that of a monomer in the FH2 fold, and even the electrostatic surface-charge distribution is similar for both folds. Moreover, our deletion studies suggest that NRD binds actin through its concave surface (Fig. S8), analogous to the interaction of the FH2 domain with actin (Fig. S7). We have shown here that the CRD of Sca2 also has a fold dominated by helices and, like NRD, presents a series of helix–loop–helix repeats that we anticipate give rise to a structure resembling that of NRD (Fig. 4). Indeed, the NRD fold was uniquely observed here, and it obeys basic principles of autotransporter folding (Fig. S9). Because, despite sequence diversity, ∼97% of passenger domains form β-solenoids (24, 25), it appears unlikely that the helix–loop–helix repeat-containing CRD will adopt yet another fold, different from that of NRD. If this prediction is confirmed, the N- and C-terminal domains of Sca2, which as shown here interact with one another (Fig. 3), might form an FH2-like doughnut at the barbed end of the elongating filament (Fig. 5). This model resolves another intriguing difference between Sca2 and formins; the PRDs in Sca2 are found 370-aa apart, positioned C- and N-terminally to our two proposed FH2-like domains. As shown here, the two PRDs contribute synergistically to the profilin-dependent elongation activity of Sca2 (Fig. 2). The interaction between the two ends of Sca2 would bring the PRDs in close proximity to each other, and near the barbed end of the elongating filament, as proposed for the FH1-FH2 of formins (40). We cannot rule out Sca2 dimerization on the surface of the bacterium, but this appears unlikely because, unlike formins (39), the Sca2 monomer is active and intramolecular interactions, as a result of their proximity, are likely to prevail over intermolecular interactions. Moreover, GST-tag induced dimerization had no effect on activity (13).
Fig. 5.
Model of nucleation and elongation of Sca2. The N- and C-terminal domains of Sca2 interact and might give rise to an FH2-like processive cap at the barbed end of the actin filament for elongation. Prorich regions adjacent to these domains mediate the incorporation of profilin-actin complexes. The WH2 domains recruit actin monomers for efficient nucleation. A detailed account of the experimental evidence leading to this model is shown in Fig. S10.
What is the role of the middle WH2 domain repeat of Sca2? A series of studies have recently shown that the FH2 domain of most formins is insufficient to promote efficient nucleation. Some formins, including formin-like 3 (FMNL3) (35), inverted formin 2 (INF2) (34), and mammalian diaphanous-related formin 1 (mDia1) (38), recruit actin monomers for nucleation through sequences C-terminal to the FH2 domain, which in some cases are related to the WH2 domain (34, 35, 41). Other formins use their C-terminal tails to recruit accessory proteins that contribute actin monomers for nucleation (42). For example, the Drosophila formin Cappuccino and its mammalian counterparts Fmn1 and Fmn2 recruit Spire, a protein that has a repeat of four WH2 domains, and cooperate for efficient filament assembly (43). Similarly, mDia1 and the yeast formin Bni1 recruit the actin monomer-binding proteins APC (37) and Bud6 (36), respectively, which contribute actin monomers for efficient nucleation. In Sca2, the WH2 domains sequester actin monomers when analyzed in isolation, but when combined with the N- and C-terminal domains, they contribute strongly to the nucleation activity (Fig. 1).
In summary, the ensemble of our data suggests that Sca2 is a unique type of actin filament nucleation and elongation protein (Fig. 5 and Fig. S10). Unlike formins, Sca2 is monomeric but its N- and C-terminal domains interact to give rise to a barbed-end tracking unit, structurally different from formins but with an overall shape that resembles that of the FH2 domain, its two PRDs converge to function together in the recruitment of profilin-actin, analogous to the FH1 domain of formins, and its middle WH2 domain repeat contributes actin monomers for nucleation, analogous to the tail domains of certain formins.
Materials and Methods
Protein Expression and Purification.
Various protein constructs were amplified by PCR from the R. conorii sca2 gene (GenBank: AAL02648) (Fig. 1A). Most of the constructs were cloned between the SapI and PstI sites of vector pTYB11 (New England BioLabs), containing a chitin affinity tag and an intein domain, and expressed in E. coli BL21 (DE3) cells grown in Terrific Broth media. The longest constructs (Sca1515, Sca1023, Sca1087, Sca702, Sca401-1515) were cloned between the BamHI and XhoI sites of vector pGEX-6P-1 (GE Healthcare), containing an N-terminal GST affinity tag, and expressed in E. coli BL21 (DE3) RPIL cells. Selenomethionine-substituted construct Sca400 was obtained by growing cells in SelenoMet medium (Se-Met; Athena Enzyme Systems), supplemented with 70 mg⋅mL−1 selenomethionine (Acros Organics). Cells were resuspended in either chitin (50 mM Tris⋅HCl pH 8.0, 500 mM NaCl, 4 mM benzamidine HCl, 1 mM EDTA, 1 mM DTT, and 1 mM PMSF) or GST [50 mM Tris⋅HCl, pH 8.0, 200 mM NaCl, 4 mM benzamidine HCl, 5% (vol/vol) Glycerol, 1 mM DTT, 1 mM EDTA, 1 mM PMSF] buffers and lysed using a microfluidizer (MicroFluidics Corporation). After purification on chitin or GST affinity columns, the chitin and GST affinity tags were removed by either self-cleavage of the intein, induced by addition of 50 mM DTT for 16 h at 20 °C, or incubation with PreScission Protease (GE Healthcare) for 4 h at 20 °C. Self-cleavage of the intein leaves no extra residues on the Sca2 constructs, whereas five N-terminal amino acids (GPLGS) remain after cleavage of the GST tag. All of the proteins were additionally purified through a Superdex-200 gel filtration column (GE Healthcare) in 50 mM Tris⋅HCl, pH 7.5, 50 mM NaCl, 4 mM benzamidine HCl and 1 mM DTT. Point mutations into these constructs, including in the proline-rich regions, WH2 domains (Fig. 1A and Fig. S2A), and mutations R400G and K420Q that reduced proteolytic degradation, were introduced using the QuikChange II XL site-directed mutagenesis kit (Strategene). See Fig. S2B for additional information and an SDS/PAGE analysis of the purified proteins.
Coexpression and Copurification of Sca670 and Sca670-1515.
Sca670 and Sca670-1515 (as well as Sca670 and Sca868-1515) were cloned into the compatible vectors pRSFDUET-1 and pGEX-6P-1, respectively, and coexpressed in BL21 (DE3) RPIL cells. Cells were lysed as above, and proteins were purified on a Ni-NTA affinity column. His-Sca670 and GST-Sca670-1515 emerge as a complex, which was additionally purified through an SD200 gel-filtration column. The complex was then purified on a GST affinity column, followed by cleavage of the GST tag and repurification through the GST affinity column to remove any uncleaved protein. The molecular weights of His-Sca670 and Sca670-1515 were confirmed by mass spectrometry analysis of their complex.
Protein Labeling with Qdots.
The AviTag sequence (GGGLNDIFEAQKIEWHE) was added N-terminally to Sca1023 and C-terminally to Sca1515 and Sca401-1515, separated by a 5-aa linker (GAPGS). This sequence is biotinylated at Lys12 by E. coli biotin ligase during protein expression (44). Constructs were coexpressed with E. coli biotin ligase by cotransfection with plasmid pBirAcm (Avidity) (45). Protein expression and purification were conducted as above, in medium supplemented with 100 μM biotin. Conjugation with streptavidin-coupled quantum dot 625 (QD-625; Invitrogen) was carried out according to the manufacturer’s protocol and confirmed by SDS gel electrophoresis.
Actin Polymerization Assay.
Actin polymerization was measured as the fluorescence increase resulting from the incorporation of pyrene-labeled actin into filaments, using a Cary Eclipse Fluorescence Spectrophotometer (Varian). Before data acquisition, 2 μM Mg-ATP-actin (6% pyrene-labeled) was mixed with different concentrations (as indicated in the figures) of Sca2 constructs in F-buffer (10 mM Tris, pH 7.5, 1 mM MgCl2, 50 mM KCl, 1 mM EGTA, 0.1 mM NaN3, 0.02 mg⋅mL−1 BSA, 0.2 mM ATP). Data acquisition started 10 s after mixing. All of the measurements were done at 25 °C. Control experiments were carried out with addition of buffer alone. Polymerization rates were calculated as the slope of the polymerization curve between 0.1 and 0.3 of the fluorescence normalized to the actin control, and converted to nM⋅s−1 (nanomolar monomers adding to filaments per second), assuming a total concentration of polymerizable actin of 1.9 μM, as previously described (18).
Barbed-End Elongation Assay.
Filament barbed-end elongation was monitored by pyrene-actin polymerization as described above, using 1.5 μM phalloidin-stabilized F-actin seeds and 0.5 μM actin monomers (6% pyrene labeled); that is, a monomer concentration that falls in between the critical concentrations for filament elongation at the pointed (0.6 μM) and barbed (0.1 μM) ends (46). The actin seeds were generated by polymerization of 10 μM actin in F-buffer for 2 h at 23 °C, followed by the addition of 20 μM phalloidin and centrifugation at 435,000 × g for 20 min. The pellet was resuspended in 3× F-buffer to a concentration of 6 μM, and passed through a 27½-gauge syringe needle five times to shear the actin filaments. The resuspended F-actin seeds were allowed to reanneal overnight at 23 °C. For each Sca2 construct concentration, initial slopes were taken at 10% completion of the elongation reactions (47).
Sedimentation Assay with F-actin.
Actin (20 μM) in G-buffer (2 mM Tris, pH 7.4, 0.2 mM CaCl2, 0.2 mM ATP, 0.5 mM DTT, 1 mM NaN3) was polymerized with addition of 50 mM KCl, 2 mM MgCl2 and 1 mM EGTA for 30 min at room temperature. Sca2 constructs were centrifuged at 278,000 × g for 30 min to remove potential aggregates. F-actin at 15 μM was incubated with 15 μM Sca2 constructs for 1 h at room temperature. Samples were centrifuged at 278,000 × g for 30 min. Equal volumes of supernatant and pellets were analyzed by SDS/PAGE.
TIRF Microscopy.
TIRF microscopy images were collected at 10-s intervals with an iXon EMCCD camera (Andor Technology) using an Olympus IX-71 microscope fit with through-the-objective TIRF illumination. Mg-ATP-actin (1.5 μM, 33% Oregon Green-labeled) was mixed with 2× TIRF buffer [1×: 10 mM Imidazole pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 50 μM CaCl2, 15 mM glucose, 20 μg⋅mL−1 catalase, 100 μg⋅mL−1 glucose oxidase, and 0.5% (wt/vol) methylcellulose 400 centipoise] and Sca2 or Sca2-Qdot constructs at 0.1 or 0.05 nM concentration (with or without 4 μM profilin), and transferred to a flow cell for imaging at 23 °C. The coverslip was coated with NEM-myosin II to capture the actin filaments.
Isothermal Titration Calorimetry.
Sca400 at 340 μM was titrated in 10 μL injections into 17 μM monomeric actin with bound latrunculin B (LatB) using a VP-ITC calorimeter (MicroCal). The experiments were performed at 20 °C in 50 mM Tris⋅HCl, pH 7.5, 50 mM NaCl, 4 mM benzamidine HCl, 1 mM DTT, and 17 μM LatB. The duration of each injection was 20 s, with an interval of 4 min between injections. The heat of binding was corrected for the small exothermic heat of injection resulting from injecting Sca400 into buffer. Data were analyzed using MicroCal’s scrips in the Origin program (OriginLab).
Crystallization, Data Collection, and Structure Determination.
Sca400 was concentrated to 10 mg⋅mL−1 in 50 mM Tris⋅HCl, pH 7.5, 50 mM NaCl, 4 mM benzamidine HCl, 1 mM DTT. Crystals of unmodified and selenomethionine-substituted Sca400 were obtained by hanging-drop vapor diffusion at 4 °C. A typical 2-μL drop consisted of a 1:1 (vol/vol) mixture of protein solution and a well solution containing 0.2 M sodium citrate, 18–20% (wt/vol) PEG 3350. The crystallization of selenomethionine-substituted Sca400 required in addition 1 mM l-glutathione reduced and 1 mM l-glutathione oxidized (Hampton Research). Crystals were flash-frozen in liquid nitrogen after a short passage through a solution containing 30% (vol/vol) glycerol added to the crystallization buffer. X-ray datasets were collected at the Cornell High Energy Synchrotron Source (CHESS) beamline A1. Data indexation and scaling were done with the XDS package (48). The structure was determined using the single-wavelength anomalous diffraction method. Selenium sites were found and phases were calculated with SHELXD (49). Model building and refinement were done with Coot (50) and Phenix (51) (Table 1). Illustrations of the structure were prepared with PyMOL (Schrödinger).
Circular Dichroism.
The far UV spectrum of Sca868-1515 was obtained at a protein concentration of 5 μM in 50 mM phosphate buffer, pH 7.5, 50 mM NaF, 4 mM benzamidine HCl, and 1 mM DTT, using a Aviv Model 410 Circular Dichroism Spectrometer (Aviv Biomedical, Inc.). Measurements were taken at 22 °C.
Supplementary Material
Acknowledgments
We thank Grzegorz Rebowski for providing the actin for this study; Nageswara Rao Jampani for help with X-ray data collection; Juan J. Martinez for providing the Sca2 cDNA; and Marissa M. Cardwell for help with initial total internal reflection fluorescence microscopy experiments. This work was supported by National Institutes of Health Grants R01 GM073791 (to R.D.) and R01 GM079265 (to D.R.K.); and American Cancer Society Grant PF-13-033-01-DMC (to D.J.K.). Crystal data collection at Cornell High Energy Synchrotron Source was supported by National Science Foundation Grant DMR-0936384 and National Institutes of Health Grant GM103485.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4J7O).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307235110/-/DCSupplemental.
References
- 1.Carabeo R. Bacterial subversion of host actin dynamics at the plasma membrane. Cell Microbiol. 2011;13(10):1460–1469. doi: 10.1111/j.1462-5822.2011.01651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haglund CM, Welch MD. Pathogens and polymers: Microbe-host interactions illuminate the cytoskeleton. J Cell Biol. 2011;195(1):7–17. doi: 10.1083/jcb.201103148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cossart P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci USA. 2011;108(49):19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker DH, Ismail N. Emerging and re-emerging rickettsioses: Endothelial cell infection and early disease events. Nat Rev Microbiol. 2008;6(5):375–386. doi: 10.1038/nrmicro1866. [DOI] [PubMed] [Google Scholar]
- 5.Walker DH. Rickettsiae and Rickettsial infections: The current state of knowledge. Clin Infect Dis. 2007;45(Suppl 1):S39–S44. doi: 10.1086/518145. [DOI] [PubMed] [Google Scholar]
- 6.Gouin E, et al. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J Cell Sci. 1999;112(Pt 11):1697–1708. doi: 10.1242/jcs.112.11.1697. [DOI] [PubMed] [Google Scholar]
- 7.Van Kirk LS, Hayes SF, Heinzen RA. Ultrastructure of Rickettsia rickettsii actin tails and localization of cytoskeletal proteins. Infect Immun. 2000;68(8):4706–4713. doi: 10.1128/iai.68.8.4706-4713.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reed SC, Serio AW, Welch MD. Rickettsia parkeri invasion of diverse host cells involves an Arp2/3 complex, WAVE complex and Rho-family GTPase-dependent pathway. Cell Microbiol. 2012;14(4):529–545. doi: 10.1111/j.1462-5822.2011.01739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouin E, et al. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004;427(6973):457–461. doi: 10.1038/nature02318. [DOI] [PubMed] [Google Scholar]
- 10.Jeng RL, et al. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 2004;6(8):761–769. doi: 10.1111/j.1462-5822.2004.00402.x. [DOI] [PubMed] [Google Scholar]
- 11.Martinez JJ, Cossart P. Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. J Cell Sci. 2004;117(Pt 21):5097–5106. doi: 10.1242/jcs.01382. [DOI] [PubMed] [Google Scholar]
- 12.Kleba B, Clark TR, Lutter EI, Ellison DW, Hackstadt T. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect Immun. 2010;78(5):2240–2247. doi: 10.1128/IAI.00100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haglund CM, Choe JE, Skau CT, Kovar DR, Welch MD. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat Cell Biol. 2010;12(11):1057–1063. doi: 10.1038/ncb2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dautin N, Bernstein HD. Protein secretion in Gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- 15.Cardwell MM, Martinez JJ. The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infect Immun. 2009;77(12):5272–5280. doi: 10.1128/IAI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardwell MM, Martinez JJ. Identification and characterization of the mammalian association and actin-nucleating domains in the Rickettsia conorii autotransporter protein, Sca2. Cell Microbiol. 2012;14(9):1485–1495. doi: 10.1111/j.1462-5822.2012.01815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Namgoong S, et al. Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat Struct Mol Biol. 2011;18(9):1060–1067. doi: 10.1038/nsmb.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris ES, Higgs HN. Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 2006;406:190–214. doi: 10.1016/S0076-6879(06)06015-0. [DOI] [PubMed] [Google Scholar]
- 19.Chereau D, et al. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci USA. 2005;102(46):16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124(2):423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, et al. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116(5):711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 22.Otomo T, et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433(7025):488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Junker M, et al. Pertactin beta-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc Natl Acad Sci USA. 2006;103(13):4918–4923. doi: 10.1073/pnas.0507923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kajava AV, Steven AC. The turn of the screw: Variations of the abundant beta-solenoid motif in passenger domains of Type V secretory proteins. J Struct Biol. 2006;155(2):306–315. doi: 10.1016/j.jsb.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 26.van den Berg B. Crystal structure of a full-length autotransporter. J Mol Biol. 2010;396(3):627–633. doi: 10.1016/j.jmb.2009.12.061. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Charles IG, Fairweather NF, Isaacs NW. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature. 1996;381(6577):90–92. doi: 10.1038/381090a0. [DOI] [PubMed] [Google Scholar]
- 28.Clantin B, et al. The crystal structure of filamentous hemagglutinin secretion domain and its implications for the two-partner secretion pathway. Proc Natl Acad Sci USA. 2004;101(16):6194–6199. doi: 10.1073/pnas.0400291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otto BR, et al. Crystal structure of hemoglobin protease, a heme binding autotransporter protein from pathogenic Escherichia coli. J Biol Chem. 2005;280(17):17339–17345. doi: 10.1074/jbc.M412885200. [DOI] [PubMed] [Google Scholar]
- 30.Johnson TA, Qiu J, Plaut AG, Holyoak T. Active-site gating regulates substrate selectivity in a chymotrypsin-like serine protease the structure of haemophilus influenzae immunoglobulin A1 protease. J Mol Biol. 2009;389(3):559–574. doi: 10.1016/j.jmb.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nummelin H, et al. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. EMBO J. 2004;23(4):701–711. doi: 10.1038/sj.emboj.7600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benz I, Schmidt MA. Structures and functions of autotransporter proteins in microbial pathogens. Int J Med Microbiol. 2011;301(6):461–468. doi: 10.1016/j.ijmm.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Junker M, Besingi RN, Clark PL. Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol Microbiol. 2009;71(5):1323–1332. doi: 10.1111/j.1365-2958.2009.06607.x. [DOI] [PubMed] [Google Scholar]
- 34.Chhabra ES, Higgs HN. INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem. 2006;281(36):26754–26767. doi: 10.1074/jbc.M604666200. [DOI] [PubMed] [Google Scholar]
- 35.Heimsath EG, Jr, Higgs HN. The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. J Biol Chem. 2012;287(5):3087–3098. doi: 10.1074/jbc.M111.312207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tu D, et al. Structure of the formin-interaction domain of the actin nucleation-promoting factor Bud6. Proc Natl Acad Sci USA. 2012;109(50):E3424–E3433. doi: 10.1073/pnas.1203035109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breitsprecher D, et al. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012;336(6085):1164–1168. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gould CJ, et al. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr Biol. 2011;21(5):384–390. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11(1):62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 40.Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18(1):9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaillant DC, et al. Interaction of the N- and C-terminal autoregulatory domains of FRL2 does not inhibit FRL2 activity. J Biol Chem. 2008;283(48):33750–33762. doi: 10.1074/jbc.M803156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanchoin L, Michelot A. Actin cytoskeleton: A team effort during actin assembly. Curr Biol. 2012;22(16):R643–R645. doi: 10.1016/j.cub.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 43.Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179(1):117–128. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8(4):921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith PA, et al. A plasmid expression system for quantitative in vivo biotinylation of thioredoxin fusion proteins in Escherichia coli. Nucleic Acids Res. 1998;26(6):1414–1420. doi: 10.1093/nar/26.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 47.Ramabhadran V, Gurel PS, Higgs HN. Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing. J Biol Chem. 2012;287(41):34234–34245. doi: 10.1074/jbc.M112.365122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55(1):94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.