Abstract
Here we report the development of a baculovirus-based delivery system that enables the efficient incorporation of unnatural amino acids into proteins in mammalian cells. We have exploited the large cargo-capacity (>30 kb) and stability of the double-stranded DNA genome of baculovirus to deliver to a variety of cell types all of the components required to genetically incorporate novel amino acids. These include the engineered tRNA/aminoacyl-tRNA synthetase pair and the nonsense mutant of the target gene. Mammalian cell transduction efficiency of baculovirus was significantly improved by incorporating genetic elements from mammalian viruses. Two polyspecific tRNA/aminoacyl-tRNA synthetase pairs were inserted into this expression system, enabling the site-specific incorporation of a variety of unnatural amino acids with novel chemical and biological properties into proteins.
Keywords: genetic code, viral vector, nonstandard amino acid, orthogonal
The genetic incorporation of unnatural amino acids (UAAs) into proteins allows the introduction of biochemical or biophysical probes at defined sites in the proteomes of living cells (1). The UAA of interest is genetically encoded by a reassigned nonsense (typically TAG) or frameshift codon, and is cotranslationally incorporated into proteins using an orthogonal UAA-specific tRNA/aminoacyl-tRNA synthetase (tRNA/aaRS) pair. Three such tRNA/aaRS pairs have been adapted for the site-specific incorporation of UAAs into proteins in eukaryotic cells: tyrosyl (Tyr) and leucyl (Leu) pairs from Escherichia coli and the pyrrolysyl (Pyl) pair from archaea (1). Using these tRNA/aaRS pairs, many UAAs with useful properties have been genetically encoded in eukaryotic cells, including amino acids for bioorthogonal conjugation reactions (e.g., azido, alkynyl and keto moieties), fluorescent amino acids, posttranslationally modified amino acids, photo-caged amino acids, and photoaffinity probes (1).
To apply this methodology to mammalian cells, an orthogonal tRNA/aaRS pair with the desired specificity is evolved in Saccharomyces cerevisiae (E. coli Tyr and Leu) or E. coli (Pyl) and then transferred to the target cells, as technical limitations complicate their directed evolution in mammalian cells (1–3). These genetic components are typically introduced into mammalian cells by transient transfection. However, the low efficiency of transient transfection and its limited applicability to a variety of important mammalian cells significantly restrict the utility of this approach. The development of efficient viral vectors for the delivery of the requisite tRNA, aaRS, and target gene would significantly facilitate the incorporation of UAAs into proteins in mammalian cells. An ideal viral vector should have a large cargo capacity, allowing the accommodation of the orthogonal tRNA/aaRS pair and the mutant gene, and a stable genome tolerant to multiple expression cassettes of the suppressor tRNA, which is required for optimal suppression efficiency. Here we describe a hybrid baculovirus vector, which fulfills these requirements. Two polyspecific tRNA/aaRS pairs, derived from E. coli tyrosyl and archaeal pyrrolysyl pairs, were encoded in this vector, allowing the incorporation of a large number of UAAs into target proteins in a variety of mammalian cells, including primary cells, stem cells and neurons.
Results and Discussion
Development of a Viral Vector for UAA Mutagenesis in Mammalian Cells.
To encode an UAA of interest, the UAA-specific orthogonal tRNA/aaRS pair and the desired nonsense or frameshift mutant of the target gene must be coexpressed in the host cell. The expression level of the orthogonal suppressor tRNA is a limiting factor for amber suppression in mammalian cells, therefore multiple copies of the tRNA must be supplied to achieve efficient UAA incorporation. Consequently, a robust viral vector system for UAA mutagenesis should have a large cargo-capacity and a stable genome that does not readily eliminate multiple copies of the tRNA cassette by recombination. Several viruses have been engineered to efficiently deliver genetic cargos into mammalian cells (4). Retro- and lentiviruses are not ideal due to their highly recombinogenic single-stranded RNA genome (5). In fact, a recent attempt to develop a lentiviral vector for UAA mutagenesis in mammalian cells was limited to a single tRNA expression cassette, and required multiple vectors to deliver all of the required genetic elements, significantly compromising its efficiency and utility (6). Another attractive candidate, adenovirus, is also replicated through a recombinogenic single-stranded DNA intermediate, and likely would encounter similar problems (7). The limited cargo capacity of adeno-associated virus renders it unsuitable for this application as well (4). Baculoviruses comprise a large group of arthropod-viruses, and recombinant versions of a well studied member of this family, Autographa californica nuclear polyhedrosis virus (AcNPV), are widely used to express proteins in insect cells (4, 8). AcNPV is also able to infect some mammalian cells, where its genetic elements remain silent rendering it replication incompetent. Thus, it can be safely used to deliver genetic cargo to a variety of different mammalian cell types both in vitro and in vivo (9–15). Several properties of baculovirus make it attractive as a potential delivery vector for the UAA incorporation machinery, including its very large cargo capacity (>30 kb), stable double-stranded DNA genome, broad host-tropism, ease of production, long shelf-life of the purified virus, intrinsically safe nature, and minimal cytotoxicity to mammalian cells, even when high multiplicity of infection (MOI) is used (9–15).
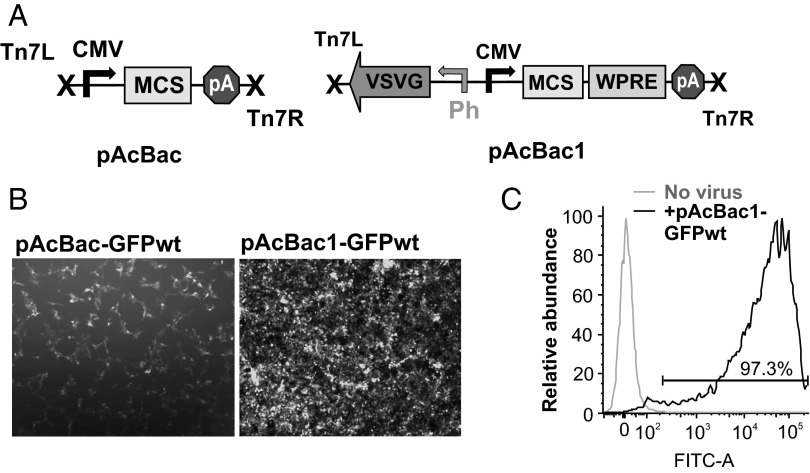
To evaluate the efficiency of baculovirus mediated transduction of mammalian cells we introduced an enhanced green fluorescent protein (eGFP) expression cassette, driven by the strong CMV-IE promoter, into the AcNPV genome using the shuttle-vector pAcBac (Fig. 1A). The recombinant AcNPV genomic DNA was used to produce the corresponding baculovirus in spodoptera frugiperda clonal isolate 9 (Sf9) insect cells. Virus was concentrated and its ability to efficiently deliver and express the eGFP gene in human embryonic kidney 293 (HEK293) cells was evaluated by fluorescence microscopy. Although viral transduction was successful, eGFP expression level was low (Fig. 1B). Addition of histone deacetylase inhibitors (HDACi) sodium butyrate or trichostatin-A resulted in some improvement in expression levels but had associated toxicity.
Fig. 1.
Construction of an enhanced baculovirus vector for transduction of mammalian cells. (A) Baculovirus shuttle plasmids encoding a CMV-promoter driven gene expression cassette for mammalian cells were constructed from the pFastBac (Invitrogen) vector backbone. Additionally, pAcBac1 encodes a VSVG expression cassette for virus-pseudotyping and a WPRE element at the 3′-UTR of the transgene. (B) Expression of eGFP(wt) using pAcBac (Left) and pAcBac1 (Right) derived baculovirus in HEK293 cells. Cells were treated with the indicated virus at an MOI of 500, and the expression profile was analyzed 48 h postinfection. (C) FACS analysis of HEK293 cells infected with pAcBac1-GFPwt 48 h postinfection.
To generate an improved baculovirus vector for mammalian cells, we next investigated pseudotyping of the viral surface with foreign viral envelope proteins, an established method to enhance transduction efficiency and expand host tropism. This strategy has been successfully applied to baculovirus using vesicular stomatitis virus G glycoprotein, VSVG (11–14). Therefore, we constructed the shuttle vector pAcBac1 (Fig. 1A) that includes a VSVG expression cassette under the polyhedrin promoter, which is highly active in insect cells during the late phase of virus production but remains silent in mammalian cells. In addition, a Woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) was included at the 3′-UTR of the transgene, driven by the CMV-IE promoter, as this element was shown to significantly enhance expression levels and duration in mammalian cells (15). We then incorporated wild-type eGFP into this vector to generate pAcBac1-GFPwt, and generated the corresponding recombinant virus in Sf9 cells. Expression of VSVG in Sf9 cells was associated with syncytium formation toward the later stages of virus production (Fig. S1). When the pAcBac1-GFPwt virus was tested for eGFP expression in HEK293 cells, a significant enhancement in eGFP expression levels was observed over pAcBac-GFPwt at the same MOI (Fig. 1B). FACS analysis of the cells infected with pAcBac1-GFPwt virus revealed >97% of the population expressing GFP at high levels (Fig. 1C).
Identification of a Polyspecific EcTyrRS.
Recently, some aaRSs evolved to charge UAAs in bacteria were found to be polyspecific, i.e., they are able to accept a variety of UAAs as substrates, while excluding the canonical 20 amino acids (16). The availability of such a polyspecific aaRS for use in mammalian cells would be attractive, allowing a single construct to be used for the incorporation of a number of distinct UAAs. The wild-type Methanosarcina barkeri pyrrolysyl-tRNA synthetase (MbPylRS) already exhibits significant polyspecificity, incorporating UAAs with bioorthogonal functional groups for conjugation reactions, photoaffinity probes, posttranslational modifications, and others (1, 17). To identify additional polyspecific tRNA/aaRS pairs, we screened eight available E. coli TyrRS (EcTyrRS) variants, previously evolved in yeast for the incorporation of different UAAs (1). A plasmid expressing one TyrRS variant from the CMV-IE promoter and four copies of the suppressor tRNACUATyr was cotransfected into HEK293 cells with another plasmid encoding an eGFP gene with an amber mutation at a permissive site (Tyr39TAG). The ability of the corresponding TyrRS variants to charge a UAA of interest, and produce full-length eGFP in these cells, was measured as an increase in eGFP-fluorescence in the presence of the corresponding UAA (relative to an identical experiment in the absence of the UAA). The specificities of eight different EcTyrRS variants toward 23 different UAAs were evaluated (Fig. S2) and two variants, previously evolved to charge p-O-methyltyrosine (pOMeY; OMeYRS) and p-iodophenylalanine (p-IF; pIF2RS) were found to efficiently accept several other UAAs, including those with bioorthogonal chemical reactivity, photoaffinity probes, IR probes, immunogenic groups and others. (Fig. S2). Successful incorporation of these UAAs into eGFP was verified by isolating the full-length protein using a C-terminal hexahistidine tag followed by electrospray ionization mass spectrometry (ESI-MS) analysis (Table S1). On the basis of these results, OMeYRS was selected for inclusion into the viral vector, as it is able to efficiently charge 10 different UAAs and exhibits minimal background suppression activity in the absence of any added UAA. For the pyrrolysine system, the previously reported polyspecific MbPylRS(wt) was incorporated into the viral vector.
Delivery of the Orthogonal tRNA/aaRS Pair Using Baculovirus.
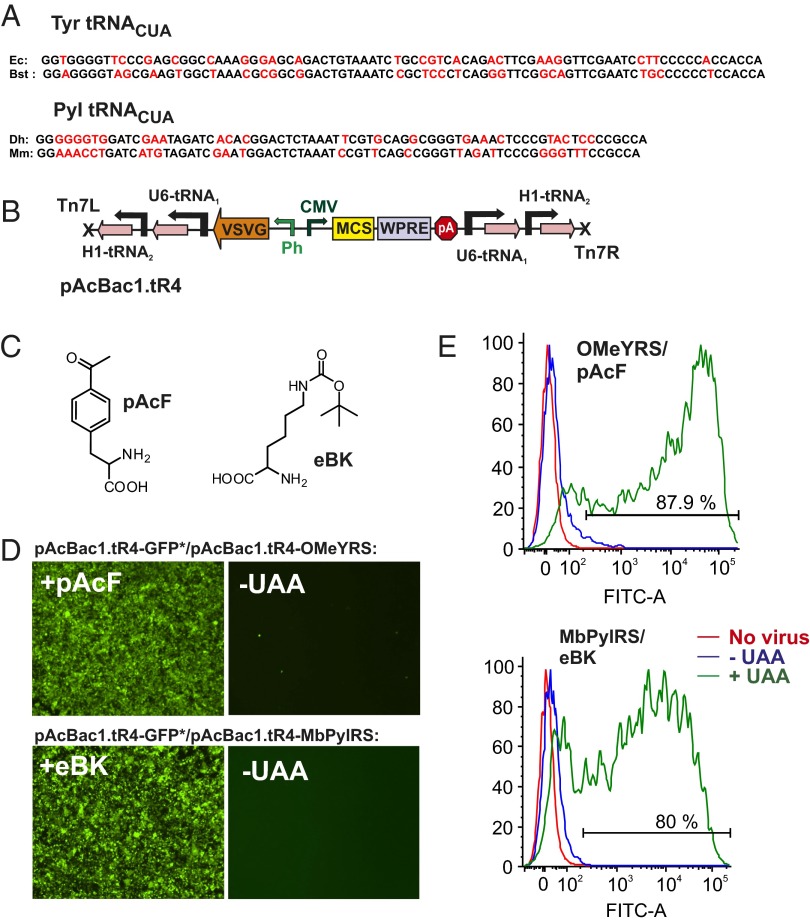
Although baculovirus has a stable dsDNA genome, a multicopy tRNA cassette could still be lost via homologous recombination. To minimize this possibility, we identified two bacterial tRNACUATyrs (from Bacillus stearothermophilus and E. coli) that are orthogonal in mammalian cells, have different primary sequences, and are functional in conjunction with EcTyrRS (Fig. 2A). We expressed these tRNAs using two different polymerase III promoters (H1 and U6) to generate a two-copy tRNA expression cassette, which should have a low propensity for recombination. This two-copy tRNA cassette was then inserted into pAcBac1 in two different orientations to construct pAcBac1.tR4 (Fig. 2B). The corresponding plasmid for the pyrrolysine system was constructed with two tRNACUAPyls from Methanosarcina mazei and Desulfitobacterium hafniense (Fig. 2A).
Fig. 2.
Baculovirus vectors encoding orthogonal tRNA/aaRS pairs. (A) Two different suppressor tRNAs (both for the tyrosine and the pyrrolysine system) were expressed from two different promoters (U6 and H1). (B) Structure of pAcBac1.tR4, which encodes the two-copy tRNA cassette in two different orientations. Either the aaRS or the reporter gene eGFP (Tyr39TAG) was inserted into the multiple cloning site (MCS), downstream of the CMV promoter to generate the suppressor or the reporter virus, respectively. (C) Structures of pAcF and eBK. (D) Expression of eGFP (Tyr39TAG) in HEK293 cells by amber suppression using pAcBac1.tR4-OMeYRS (Upper) or pAcBac1.tR4-MbPylRS (Lower), in the presence or absence of 1 mM pAcF or eBK, respectively. (E) FACS analysis of cells expressing eGFP (Tyr39TAG) as described in D.
Expression cassettes for the two polyspecific synthetases, OMeYRS and MbPylRS(wt), were then incorporated into the pAcBac1.tR4 vector (harboring the Tyr or the Pyl amber suppressor tRNACUAs, respectively) to generate the Tyr- and Pyl-specific suppressor constructs. Separately, an eGFP (Tyr39TAG) expression cassette was incorporated into pAcBac1.tR4, to generate Tyr- or Pyl-specific reporter constructs for the evaluation of UAA incorporation efficiency. High-titer (∼108 mL−1) suppressor and reporter viruses, for both the Tyr and the Pyl systems, were generated using Sf9 cells. HEK293 cells were then coinfected with the suppressor and reporter viruses, in the presence or absence of 1 mM UAA. For the pAcBac1.tR4-OMeYRS virus, p-acetylphenylalanine (pAcF) was used, and for pAcBac1.tR4-MbPylRS virus, ε-tBoc-lysine (eBK) was used (Fig. 2C). For both systems, robust eGFP expression levels were observed only in the presence of the UAA (Fig. 2 D and E). We next optimized the ratio of the suppressor and reporter viruses to achieve the highest expression levels of the reporter gene. Because both the suppressor and the reporter virus encode identical tRNA expression cassettes, changing their ratio for a fixed MOI only changes the relative amounts of the aaRS and the reporter gene (eGFP) delivered to the cell. A suppressor to reporter virus ratio of 1:6 was found to provide the highest expression levels of the reporter (Fig. S3). Under these optimal conditions, >80% cells were found to express eGFP in the presence of the UAA, whereas <5% were fluorescent in its absence (Fig. 2E). These baculovirus stocks can be readily reamplified in Sf9 cells with no apparent loss in quality; and, when stored at 4 °C and protected from light, remain stable for up to 3 mo.
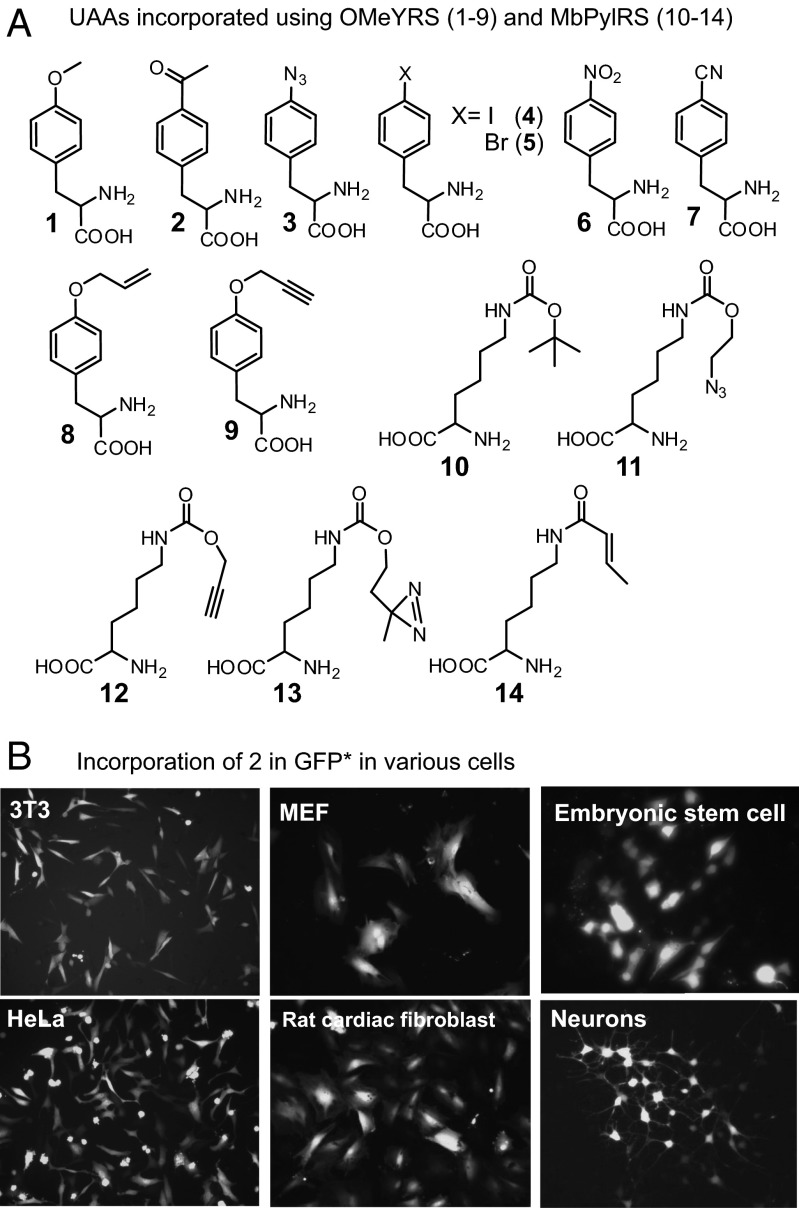
To quantify the levels of protein expression using this system, 106 HEK293 cells were infected with pAcBac1.tR4-OMeY and eGFP(Tyr39TAG) viruses (total MOI of 800, at an optimal ratio of 1:6 for suppressor to reporter virus) in the presence of 1 mM pOMeY (1) (Fig. 3A). The expressed eGFP with a C-terminal hexahistidine affinity tag was isolated 48 h postinfection by nickel-nitrilotriacetic acid (Ni-NTA) chromatography. The purified yield of the reporter protein expressed using this system was significantly higher (∼36 µg per 106 cells; Fig. S4) than the previously reported pSWAN system2 (<1 µg per 107 cells). The mutant protein was characterized by ESI-MS analysis, which revealed the correct mass (Fig. S4). We also successfully used this system with other commonly used cell-lines (CHO, 3T3, HeLa), primary cells (mouse embryonic fibroblast, rat cardiac fibroblast), cultured neurons, and mouse embryonic stem (ES) cells (Fig. 3B). High levels of mutant eGFP expression were observed in >50% of the population in most cases (∼15% for ES cells). Control experiments in the absence of the UAA showed little GFP expression. Using the polyspecific pAcBac1.tR4-OMeYRS or -MbPylRS suppressor viruses, 14 different UAAs were incorporated into eGFP (Tyr39TAG) in HEK293 cells with isolated yields of mutant eGFP ranging from 5 to 35 µg/mL (Fig. 3A and Fig. S5). Additional EcTyrRS or MbPylRS derived variants, evolved to charge other UAAs (1), can be easily incorporated into pAcBac1.tR4 (Tyr or Pyl) using the multiple cloning site downstream of the CMV-IE promoter.
Fig. 3.
Incorporation of different UAAs in various mammalian cells. (A) UAAs incorporated into eGFP (Tyr39TAG) using pAcBac1.tR4 baculovirus encoding polyspecific OMeYRS (1–9) or MbPylRS (10–14) in HEK293T cells. (B) Incorporation of pAcF 2 in eGFP (Tyr39TAG) using pAcBac1.tR4-OMeYRS baculovirus in various cells. Cells were infected with the suppressor and the reporter virus at an optimal 1:6 ratio (MOI of 800) and were analyzed 48 h postinfection.
Generation of a Single Baculovirus Vector Encoding All of the Elements for UAA Mutagenesis.
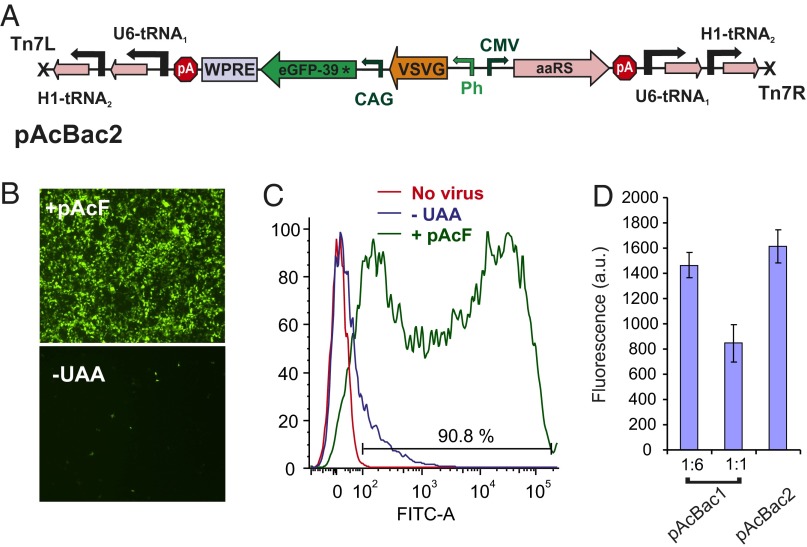
The large cargo capacity of baculovirus should allow the incorporation of the tRNA/aaRS pair as well as the target gene into the same viral vector. Such a vector would be particularly useful for in vivo applications, where it is difficult to efficiently codeliver two different viruses to the same cell in a controlled ratio. To this end, we inserted the expression cassette for the reporter gene eGFP (Tyr39TAG) into pAcBac1.tR4-OMeYRS. Because an excess of the reporter gene over the aaRS was found to be optimal, we used a stronger CAG promoter to drive the expression of the reporter eGFP (Tyr39TAG). The WPRE element was deleted from the 3′-UTR of the OMeYRS expression cassette, but was retained for the reporter expression cassette. The resulting shuttle vector, pAcBac2.tR4-OMeY/GFP*, encodes all of the genetic elements required for UAA mutagenesis in mammalian cells (Fig. 4A). Despite the large size of the genetic cargo incorporated into the AcNPV genome (>10 kb), high-titer virus (∼108 mL−1) was produced in Sf9 cells. The efficiency for incorporation of UAAs into eGFP (Tyr39TAG) was evaluated in HEK293 cells in the presence and absence of 1 mM pAcF. Robust expression of eGFP (Tyr39TAG) was observed in the presence of 1 mM pAcF (Fig. 4B); FACS analysis revealed >90% cells expressing GFP in the presence of pAcF, whereas only 20% cells were fluorescent in the absence of UAA (Fig. 4C). When used at the same MOI, the expression level from pAcBac2.tR4-OMeY/GFP* was comparable to that obtained using pAcBac1.tR4-OMeY and pAcBac1.tR4-GFP* at the optimal 1:6 ratio (Fig. 4D).
Fig. 4.
A single baculovirus vector encoding all of the genetic elements necessary to efficiently incorporate UAA into target proteins in mammalian cells. (A) pAcBac2 encodes a CAG promoter-driven reporter gene expression cassette, which also harbors a WPRE element at the 3′-UTR. Expression of aaRS is driven by a CMV promoter and lacks the WPRE element. (B) HEK293 cells infected with pAcBac2.tR4-OMeYRS/GFP* (MOI of 500) exhibits robust eGFP expression only in the presence of the UAA (1 mM). (C) FACS analysis of cells expressing eGFP in B. (D) Efficiency of GFP (Tyr39TAG) expression using pAcBac1.tR4-OMeYRS or pAcBac2.tR4-OMeYRS/GFP* are similar, when used at similar MOI (500). For expression using pAcBac.tR4-OMeYRS and-GFP*, the suppressor and the reporter viruses were used at the optimal 1:6 ratio or a 1:1 ratio (for a fixed total MOI of 500). Crude cell extracts were prepared 48 h after infection and GFP fluorescence was measured.
Conclusion
The ability to site-specifically incorporate UAAs into proteins in live mammalian cells provides powerful tools to probe and manipulate protein function therein. Here we have developed user-friendly baculovirus-based vectors for the efficient expression of mutant proteins containing UAAs in various mammalian cells. These vectors allow facile integration of any engineered aaRS specific for a desired UAA, as well as the nonsense mutant of any target gene. The use of polyspecific EcTyrRS and MbPylRS facilitates the incorporation of several useful UAAs using a single suppressor virus, including UAAs with bioorthogonal handles for chemical conjugation, photoaffinity labels, and others. Baculovirus, especially the pseudotyped version, has been shown to efficiently transduce a wide range of mammalian cells both in vivo and in vitro, all of which now should be suitable hosts for UAA mutagenesis of target proteins (9–15). The unique baculovirus vectors developed in this study should also be useful for other applications, where multiple genetic elements need to be simultaneously delivered into mammalian cells. Finally, the development of pAcBac2.tR4, a single baculovirus vector encoding all of the genetic elements necessary to express a target protein incorporating a desired UAA, should facilitate the extension of this technology to in vivo applications.
Materials and Methods
For cloning and propagation of plasmid DNA, E. coli DH10B was used. Qiagen spin miniprep kit and Macherey-Nagel Nucleospin columns were used for plasmid DNA isolation and desalting of DNA (from PCR or restriction digest reaction), respectively. DNA oligomers used in this study were obtained from IDT, a list of which can be found in SI Text. Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs. Mammalian cell-culture media were obtained from Corning-Mediatech. Sf9 insect cells were cultured in Sf-900 III SFM serum-free media (Gibco) at 27 °C. Fugene HD (Promega) was used to transfect mammalian or insect cells.
Construction of Viral Vectors.
To construct pAcBac-GFP, pFastBac-Dual (Invitrogen) was digested with AvrII and BstAPI, and an insert amplified from pCDNA3.1 (using oligomers ac1 and ac2), encoding the CMV-IE promoter, a multiple cloning site, and the BGH poly-A sequence, was incorporated. The eGFP gene was subsequently amplified (using oligomers ac3 and ac4) and inserted downstream of the CMV promoter between the EcoRI and NheI restriction sites in pAcBac.
The WPRE element from the lentiviral vector SHC201 (Invitrogen) was amplified (using oligomers ac5 and ac6) and inserted between the EcoRI and the XhoI sites of pAcBac, to generate pAcBac1a. A polyhedrin-promoter driven VSVG expression cassette was then amplified (using oligomers ac7 and ac8) and inserted between the NotI and AvrII restriction sites of pAcBac1a to generate pAcBac1. The two-copy tRNA cassette driven by two different promoters was synthesized by IDT. The first copy of this cassette was amplified (using oligomers ac9 and ac10 for the Tyr tRNAs, or ac9 and ac11 for the Pyl tRNAs), digested with AvrII/NheI, and inserted into the AvrII site in pAcBac1 to generate pAcBac1.tR2. A clone of pAcBac1.tR2 with the appropriate orientation was identified by DNA sequencing analysis, and another unit of the tRNA cassette was amplified (using oligomers ac12 and ac13 for the Tyr tRNAs, or ac12 and ac14 for the Pyl tRNAs), digested with EcoRV and PvuII, and inserted into the PvuII site of pAcBac1.tR2 to generate pAcBac1.tR4. The desired aaRS or eGFP (Tyr39TAG) was amplified (using following oligomers: ac15 and ac16 for EcTyrRS; ac17 and ac18 for MbPylRS; ac3 and ac4 for eGFP-TAG) and inserted into the appropriate pAcBac1.tR4 between the NheI and the EcoRI sites to generate the suppressor and reporter plasmids, respectively. To generate pAcBac2.tR4-OMeYRS/GFP*, the WPRE element from pAcBac1.tR4-OMeYRS was removed by SalI digestion followed by religation. A CAG-promoter driven GFP (Tyr39TAG) expression cassette was then inserted into the AvrII site of the resulting plasmid.
Determination of aaRS Polyspecificity.
HEK293T cells in 24-well tissue-culture plates were transfected with 0.5 μg each of pEGFP-Tyr39TAG and suppression plasmid at 30% confluency. Cells were cultured in 1 mL of DMEM (Dulbecco's Modified Eagle Medium) supplemented with 10% (vol/vol) heat inactivated FBS in the presence or absence of UAA. Culture media was removed 48 h after transfection, and 150 μL of RIPA buffer (Sigma) was added into each well to lyse cells. Clarified crude cell extracts were transferred to a clear bottom 96-well plate, and GFP fluorescence was measured using a plate reader (485-nm excitation and 515-nm emission). For mass-spectrometry characterization, protein was isolated by Ni-NTA affinity chromatography using a C-terminal (His)6 tag from ∼107 cells and analyzed on an Agilent 1100 Series liquid chromatography mass spectrometry (LC/MS) instrument.
Generation of Recombinant Baculovirus.
Recombinant baculovirus carrying the desired insert was generated from shuttle vectors according to the instruction manual for Bac-to-Bac baculovirus expression systems (Invitrogen). In brief, the shuttle vector was transformed into E. coli DH10Bac (Invitrogen) carrying the AcNPV genome as a bacmid. Successful incorporation of the cargo from the shuttle vector to the AcNPV genome by Tn7 transposition results in a disruption of an expression cassette of the reporter protein LacZ, leading to a white phenotype in a blue-white screen in the presence of X-Gal. Recombinant DNA was isolated from white colonies and was transfected into Sf9 cells cultured in Sf-900 III SFM serum-free media. Primary baculovirus stocks were harvested from the supernatant 96 h posttransfection, and amplified by reinfecting Sf9 cells at low MOI (<1). The virus stocks were stored at 4 °C, protected from light, and were typically stable for up to 3 mo. Titers of virus stocks were measured using BacPAK baculovirus rapid titer kit (Clontech). When necessary, virus stock was concentrated using Amicon Ultra 50-kDa molecular weight cutoff centrifugal filters (Millipore). Alternatively, the virus stocks were centrifuged at 45,000 × g for 45 min at 4 °C, and the virus pellet was then resuspended in appropriate volume of Dulbecco’s PBS to achieve desired concentration.
Mammalian Cell Culture.
Cells were incubated at 37 °C in a humidified chamber with 5% CO2. HEK293, HeLa, 3T3, mouse embryonic fibroblast (MEF), and rat cardiac fibroblast (RCF) cells were cultured in DMEM supplemented with 10% heat inactivated FBS. CHO cells were maintained in Ham’s F-12/DMEM supplemented with 10% heat-inactivated FBS. RCFs (Cell Applications) and MEFs (ATCC) were maintained in DMEM (Cellgro) supplemented with 10% FBS (Gibco) and Antibiotic-Antimycotic (Gibco). For infection experiments, cells were seeded at 105 cells per well in six-well dishes and infected with the virus 24 h after plating. The isolation and culture of rat hippocampal neural progenitor cells has been described (18, 19). The cells were maintained and expanded in an undifferentiated state by culturing on polyornithine (10 μg/mL in water; Sigma) and laminin (5 μg/mL in PBS; Invitrogen) coated plates. Propagation and maintenance medium was DMEM/F12 (Cellgro) with N2 (Invitrogen) and basic fibroblast growth factor (20 ng/mL, R&D Systems). For differentiation experiments, cells were seeded at 105 per well in six-well dishes and switched to differentiation medium-DMEM/F12 supplemented with N2, B27 (Invitrogen), and 2 µM all-transretinoic acid (Sigma). Cells were treated with differentiation medium for 5 d before treatment with the virus. Mouse embryonic stem cells (R1) were cultured on gelatin coated dishes, in media containing knockout DMEM, 15% knockout serum replacer, nonessential amino acids, glutamine, 110 µM β-mercaptoethanol, and 103 unit/mL leukemia inhibitory factor (LIF).
Baculovirus-Mediated Expression of Proteins Incorporating UAAs.
To an adherent culture of target cells, baculovirus was added at a desired MOI (between 50 and 1,000) in the presence or absence of the UAA of interest (1 mM). Expression of eGFP was monitored by fluorescence microscopy. Optimal expression levels were observed 48 h postinfection. For FACS analysis, cells were digested with trypsin 48 h postinfection, washed and resuspended in PBS, and analyzed using a BD Biosciences FACSCanto flow cytometer. For isolation of proteins, HEK293 cells were seeded in a six-well plate at 4 × 105 cells per well in DMEM media, supplemented with 10% FBS and Antibiotic-Antimycotic (Invitrogen) at a 1:200 dilution. After overnight incubation, pAcBac1.tR4-OMeYRS and pAcBac1.tR4-GFP* baculovirus were added at approximate MOIs of 150 and 750, respectively. Cells were harvested 48 h postinfection and lysed using 250 µL of RIPA buffer supplemented with 10 unit/mL benzonase, and the crude cell extract was clarified by centrifugation (12,000 × g, 3 min). Mutant eGFP was purified using a C-terminal (His)6 tag by Ni-NTA affinity chromatography. Qiagen Ni-NTA Superflow resin was used according to manufacturer’s protocol. The isolated protein was characterized by SDS/PAGE analysis followed by Coomassie staining and by ESI-MS analysis using an Agilent 1100 series LC/MS instrument.
Supplementary Material
Acknowledgments
We thank Virginia Seely for assistance with manuscript preparation, Dr. Shoutian Zhu for assistance with mouse ES cell culture, and the laboratory of Prof. Ian Wilson for assistance with baculovirus production. This work was funded by National Institutes of Health Grant R01GM062159 (to P.G.S.). This is paper 24006 from The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309584110/-/DCSupplemental.
References
- 1.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 2.Liu W, Brock A, Chen S, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4(3):239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 3.Chen PR, et al. A facile system for encoding unnatural amino acids in mammalian cells. Angew Chem Int Ed Engl. 2009;48(22):4052–4055. doi: 10.1002/anie.200900683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warnock JN, Daigre C, Al-Rubeai M. Introduction to viral vectors. Methods Mol Biol. 2011;737:1–25. doi: 10.1007/978-1-61779-095-9_1. [DOI] [PubMed] [Google Scholar]
- 5.ter Brake O, et al. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol Ther. 2008;16(3):557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- 6.Shen B, et al. Genetically encoding unnatural amino acids in neural stem cells and optically reporting voltage-sensitive domain changes in differentiated neurons. Stem Cells. 2011;29(8):1231–1240. doi: 10.1002/stem.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flint SJ, Berget SM, Sharp PA. Characterization of single-stranded viral DNA sequences present during replication of adenovirus types 2 and 5. Cell. 1976;9(4 Pt 1):559–571. doi: 10.1016/0092-8674(76)90038-6. [DOI] [PubMed] [Google Scholar]
- 8.Jarvis DL. Baculovirus-insect cell expression systems. Methods Enzymol. 2009;463:191–222. doi: 10.1016/S0076-6879(09)63014-7. [DOI] [PubMed] [Google Scholar]
- 9.Kost TA, Condreay JP, Jarvis DL. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat Biotechnol. 2005;23(5):567–575. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fornwald JA, Lu Q, Wang D, Ames RS. Gene expression in mammalian cells using BacMam, a modified baculovirus system. Methods Mol Biol. 2007;388:95–114. doi: 10.1007/978-1-59745-457-5_5. [DOI] [PubMed] [Google Scholar]
- 11.Liu CY, Chen HZ, Chao YC. Maximizing baculovirus-mediated foreign proteins expression in mammalian cells. Curr Gene Ther. 2010;10(3):232–241. doi: 10.2174/156652310791321215. [DOI] [PubMed] [Google Scholar]
- 12.Barsoum J, Brown R, McKee M, Boyce FM. Efficient transduction of mammalian cells by a recombinant baculovirus having the vesicular stomatitis virus G glycoprotein. Hum Gene Ther. 1997;8(17):2011–2018. doi: 10.1089/hum.1997.8.17-2011. [DOI] [PubMed] [Google Scholar]
- 13.Kaikkonen MU, et al. Truncated vesicular stomatitis virus G protein improves baculovirus transduction efficiency in vitro and in vivo. Gene Ther. 2006;13(4):304–312. doi: 10.1038/sj.gt.3302657. [DOI] [PubMed] [Google Scholar]
- 14.Tani H, et al. In vitro and in vivo gene delivery by recombinant baculoviruses. J Virol. 2003;77(18):9799–9808. doi: 10.1128/JVI.77.18.9799-9808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng J, et al. High-efficiency transient transduction of human embryonic stem cell-derived neurons with baculoviral vectors. Mol Ther. 2009;17(9):1585–1593. doi: 10.1038/mt.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young DD, et al. An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry. 2011;50(11):1894–1900. doi: 10.1021/bi101929e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee A, Sun SB, Furman JL, Xiao H, Schultz PG. A versatile platform for single and multiple unnatural amino acid mutagenesis in Escherichia coli. Biochemistry. 2013;52(10):1828–1837. doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warashina M, et al. A synthetic small molecule that induces neuronal differentiation of adult hippocampal neural progenitor cells. Angew Chem Int Ed Engl. 2006;45(4):591–593. doi: 10.1002/anie.200503089. [DOI] [PubMed] [Google Scholar]
- 19.Wurdak H, et al. A small molecule accelerates neuronal differentiation in the adult rat. Proc Natl Acad Sci USA. 2010;107(38):16542–16547. doi: 10.1073/pnas.1010300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.