Abstract
Mpr1 (sigma1278b gene for proline-analog resistance 1), which was originally isolated as N-acetyltransferase detoxifying the proline analog l-azetidine-2-carboxylate, protects yeast cells from various oxidative stresses. Mpr1 mediates the l-proline and l-arginine metabolism by acetylating l-Δ1-pyrroline-5-carboxylate, leading to the l-arginine–dependent production of nitric oxide, which confers oxidative stress tolerance. Mpr1 belongs to the Gcn5-related N-acetyltransferase (GNAT) superfamily, but exhibits poor sequence homology with the GNAT enzymes and unique substrate specificity. Here, we present the X-ray crystal structure of Mpr1 and its complex with the substrate cis-4-hydroxy-l-proline at 1.9 and 2.3 Å resolution, respectively. Mpr1 is folded into α/β-structure with eight-stranded mixed β-sheets and six α-helices. The substrate binds to Asn135 and the backbone amide of Asn172 and Leu173, and the predicted acetyl-CoA–binding site is located near the backbone amide of Phe138 and the side chain of Asn178. Alanine substitution of Asn178, which can interact with the sulfur of acetyl-CoA, caused a large reduction in the apparent kcat value. The replacement of Asn135 led to a remarkable increase in the apparent Km value. These results indicate that Asn178 and Asn135 play an important role in catalysis and substrate recognition, respectively. Such a catalytic mechanism has not been reported in the GNAT proteins. Importantly, the amino acid substitutions in these residues increased the l-Δ1-pyrroline-5-carboxylate level in yeast cells exposed to heat stress, indicating that these residues are also crucial for its physiological functions. These studies provide some benefits of Mpr1 applications, such as the breeding of industrial yeasts and the development of antifungal drugs.
Keywords: cyclic amine N-acetyltransferase, X-ray crystallography, reaction mechanism, antioxidant enzyme
In yeast Saccharomyces cerevisiae Σ1278b, Takagi et al. discovered genes involved in the detoxification of the proline analog l-azetidine-2-carboxylic acid (AZC) (1, 2). These genes, MPR1 and MPR2 (sigma1278b gene for proline-analog resistance), encode N-acetyltransferase Mpr1, which can convert AZC into N-acetyl-AZC (Fig. S1A). Only one base change occurs between MPR1 and MPR2, and the two genes have similar functions. The genes homologous to MPR1 were found in the genomes of many yeasts and fungi, and AZC acetyltransferase activity has already been detected in a number of yeast strains (3), suggesting that the Mpr1 homologs are widely present in yeasts and fungi. However, it is unlikely that AZC is a physiological substrate for Mpr1 because AZC occurs only in certain plants (4–6).
Mpr1 protects yeast cells by reducing reactive oxygen species (ROS) levels under oxidative stress conditions, such as H2O2, heat shock, freeze–thawing, or ethanol treatment (7–9). In addition to AZC, Mpr1 can acetylate the l-proline catabolism intermediate l-Δ1-pyrroline-5-carboxylate (P5C), the tautomer l-glutamate-γ-semialdehyde (GSA), or the intermediate 5-hydroxy-l-proline in the equilibrium reaction of P5C into N-acetyl-GSA, indicating that Mpr1 mediates the l-proline and l-arginine metabolism (7, 10) (Fig. S1B). Interestingly, we found that Mpr1-dependent conversion of l-proline into l-arginine led to the production of nitric oxide (NO), which confers oxidative stress tolerance on yeast cells (10, 11) (Fig. S1C). Recently, P5C was shown to directly inhibit mitochondrial respiration, leading to ROS generation in yeast (12). We concluded that Mpr1 is an antioxidant enzyme involved in P5C detoxification and NO production.
Mpr1 belongs to the Gcn5 (a histone acetyltransferase)-related N-acetyltransferase (GNAT) superfamily, whereas Mpr1 displays poor sequence homology with other structurally determined GNAT proteins. In addition, both of the acetyl receptors in the reaction of Mpr1 identified so far, AZC and cis-4-hydroxy-l-proline (CHOP) (Fig. S1A) (13), are cyclic secondary amines, whereas most of the acetyl receptors of the proteins in the GNAT superfamily identified so far are primary amines (14). These facts suggest that Mpr1 has a unique structure and/or reaction mechanism for catalytic reaction and substrate recognition. Therefore, the structure determination of Mpr1 can lead to important insights for understanding both the structure–function and the evolutional relationships among various N-acetyltransferases.
Here, we determined the crystal structure of Mpr1 and its complex with the substrate CHOP. Although the overall structure of Mpr1 showed a folding that is typical among proteins in the GNAT superfamily, Mpr1 showed a unique reaction mechanism. The structure and the kinetic analyses using the Mpr1 variants indicate that Asn178 and Asn135 participate in catalysis and in substrate recognition, respectively. Additionally, the amino acid substitutions in these residues increased intracellular P5C levels after exposure to heat shock in yeast. Thus, these residues are definitely involved in the physiological function of Mpr1.
Results
Protein Preparation, Crystallization, and Structure Determination of Mpr1.
Although we previously tried to determine the crystal structure of Mpr1 by the molecular replacement method (15), the structure could not be solved probably due to the low sequence identity between Mpr1 and the search models of other acetyltransferases. To resolve this, we then applied the single-wavelength anomalous dispersion (SAD) method using a selenomethionine (SeMet)-incorporated protein (Se-SAD) here. The wild-type Mpr1 (WT-Mpr1) has two Met residues, which are located at the N terminus (Met1) and near the C terminus (Met228). SeMet residues introduced into these regions may not be effective in phasing because of disordering in crystal. Thus, the mutant Mpr1 in which Leu87 is replaced by Met was constructed, and Leu87SeMet-Mpr1 was prepared according to the procedures described in SI Materials and Methods. We confirmed the incorporation of SeMet by MALDI-TOF mass spectrometry (Table S1), the removal of His-tag by the N-terminal sequencing analysis (Table S2), and sufficient purity for crystallization by SDS/PAGE and Coomassie Brilliant Blue staining (Fig. S2A). Next, we screened the crystallization conditions and obtained crystals in the optimal condition containing 100 mM Bistris-HCl (pH 5.5), 240 mM MgCl2, 20.5% (wt/vol) PEG 3350, and 50 mM AZC. X-ray diffraction data were collected using a Rayonix MX225HE CCD detector installed on the BL41XU beamline at SPring-8. Finally, the 3D structure of Mpr1 was determined at the resolution of 2.1 Å by the Se-SAD method (Table S3). The crystal structure of WT-Mpr1 (15) and its complex with WT-Mpr1-CHOP were also solved by molecular replacement using the structure of the SeMet mutant as a search model at the resolution of 1.9 and 2.3 Å, respectively (Table S3).
Overall Structure and Substrate Binding Site of Mpr1.
The structure of Mpr1 showed an α/β-structure with eight-stranded mixed β-sheet and six α-helices, which is similar to those of other members in the GNAT superfamily reported so far (Fig. S3A), indicating that Mpr1 definitely belongs to the GNAT superfamily, even though Mpr1 exhibits poor sequence homology with other typical GNAT proteins. Mpr1 exists as a dimer form in solution (Fig. S2B), indicating that the assembly state of Mpr1 is also similar to those of other GNAT proteins (14). Our 3D homology search using the Dali server (16) indicated that the structure of Mpr1 is more similar to those of bacterial N-acetyltransferases than to those of eukaryotic N-acetyltransferases (Table S4).
The bound CHOP molecule was found in the predicted active-site crevice outside the edges of two adjacent β-strands, and its conformation was well defined in the electron-density maps (Fig. 1A). The final substrate model was proposed to form hydrogen bonds with the side chain of Asn135 and the backbone amide N-H groups of Asn172 and Leu173 through its carboxyl group and with a water molecule bound to Phe138 through its amino group, predicted by the PISA server (18). Moreover, it was suggested that the van der Waals contact between the Cγ atom of CHOP and the phenolic side chain of Tyr75 (3.6 Å) is involved in the recognition of the cyclic secondary amine (Fig. 1A and Fig. S4). To predict the binding site of acetyl-CoA (AcCoA) in Mpr1, we used the structure of an aminoglycoside 6′-N-acetyltransferase from Enterococcus faecium [Protein Data Bank (PDB) ID code 1B87] (17), which was reported to form a dimer in solution as Mpr1 does (19). This structure is most similar to that of Mpr1 among the proteins, which are structurally determined as an AcCoA-binding form and form a dimer (Table S4). The structure of Mpr1-CHOP including seven β-strands (from second to eighth) and four α-helices (from second to fifth) overlapped nicely with that of 1B87 (Fig. S3B). In particular, a high degree of overlapping was observed near AcCoA (Fig. S3B). Based on the superimposition of Mpr1 with the structure of 1B87, the pyrophosphate moiety of AcCoA was predicted to be located near the region corresponding to the conserved motif A of Mpr1 (Fig. 1 B and C), which is believed to be involved in AcCoA binding through its pyrophosphate (14), suggesting that the predicted AcCoA-binding site is almost correct. In the ternary complex model of Mpr1 with CHOP and AcCoA, the backbone amide group of Phe138 and the side chain of Asn178 and Trp185 were positioned near the carbonyl group of AcCoA (Fig. 1B). In many GNAT proteins, it is reported that the backbone amide of a conserved hydrophobic residue and the phenolic hydroxyl group of a conserved Tyr residue, which correspond to Phe138 and Trp185 in Mpr1 respectively, function as catalytic residues (14), suggesting that Phe138 and Trp185 could be also involved in the catalysis in Mpr1. Additionally, Mpr1 has a unique structural characteristic. Many GNAT proteins, including ribosomal protein acetyltransferase (20), aminoglycoside N-acetyltransferase (21), serotonin N-acetyltransferase (22), glucosamine-6-phosphate N-acetyltransferase (23), histone acetyltransferase (24), and mycothiol synthase (25), have a β-bulge structure on the β-strand containing the catalytic hydrophobic residue, which corresponds to Phe138 in Mpr1, near the active site. In contrast, Mpr1 lacks such a β-bulge structure, which probably gives an advantage to reduce steric interference with the cyclic secondary amine due to its binding (Fig. S3C). Based on the unique structural features, Mpr1 is classified into a group different from other GNAT proteins.
Fig. 1.
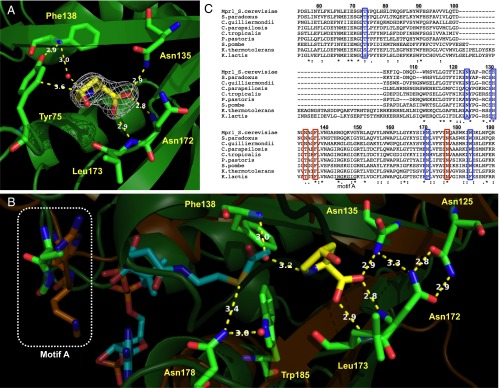
Structure determination and the substrate-binding site of Mpr1. (A) Simulated-annealing |Fo|-|Fc| omit map contoured at 2.5 sigma. CHOP, Tyr75, Asn135, Phe138, Asn172, and Leu173 are shown as a stick model. Judging from the electron density of CHOP, CHOP is likely to interact with these residues. White numbers show distances for hydrogen bonds, and a red ball shows a water molecule. (B) The predicted AcCoA-binding site on Mpr1 constructed by the superimposition of WT-Mpr1-CHOP structure with the structure 1B87 (17). The structure of Mpr1 (green) and 1B87 (orange) are shown as a ribbon model. CHOP, AcCoA, and some residues are shown as a stick model. White numbers show distances for hydrogen bonds. The conserved residues in motif A are indicated by a white dashed box. (C) The multiple alignments of Mpr1 homologs from S. cerevisiae, Saccharomyces paradoxus, Candida guilliermondii, Candida parapsilosis, Candida tropicalis, Pichia pastoris, Schizosaccharomyces pombe (Ppr1), Kluyveromyces thermotolerans, and Kluyveromyces lactis. Asterisk, colon, and dots indicate completely, strongly, and weakly conserved amino acid residues, respectively. Blue boxes indicate the amino acid residues mutated in this study, and a red box shows Asn135, Phe138, and Asn178. Dashes indicate the absence of corresponding amino acid residues at those positions.
Kinetics of Acetylation Catalyzed by Mpr1.
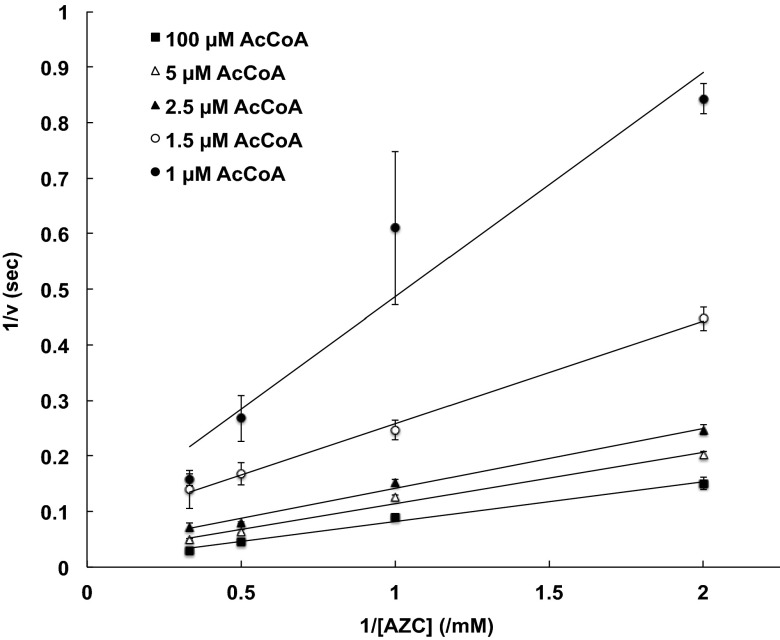
To understand the reaction mechanism, we determined the initial velocity of Mpr1 at various concentrations of the bisubstrates AZC and AcCoA. The Lineweaver–Burk plot of Mpr1, which shows the intersecting lines at one point (Fig. 2), revealed that the Bi-Bi reaction catalyzed by Mpr1 proceeds via a sequential mechanism through the formation of the Mpr1-AcCoA-AZC ternary complex. This indicates that the reaction mechanism of Mpr1 is similar to those of many other proteins in the GNAT superfamily reported thus far (14).
Fig. 2.
Kinetic mechanism of Mpr1. Lienweaver–Burk plot of the initial velocity data at various concentrations of AZC and fixed concentrations of AcCoA at 1 μM (●), 1.5 μM (○), 2.5 μM (▲), 5 μM (△), and 100 μM (■) was determined as described in SI Materials and Methods. The values at each point are the means and SDs from three independent experiments. The intersection of each line at one point indicates a sequential kinetic mechanism.
Analysis of Mpr1 Mutants.
Based on the structure of Mpr1-CHOP and the predicted binding site of AcCoA on Mpr1, we constructed several Mpr1 mutants with amino acid substitution at positions near CHOP or the acetyl group of AcCoA, both of which are highly conserved among Mpr1 homologs (3, 26, 27) (Fig. 1C). These residues are likely to be important for catalysis and substrate recognition. The mutant enzymes were purified to apparent homogeneity from the soluble fractions of Escherichia coli transformant cells. We analyzed these mutants by steady-state kinetic analysis, and the results are summarized in Table 1. The replacement of Asn135 by Ala most severely affected the apparent Km value, leading to an ∼20-fold increase in the value against AZC compared with that of WT-Mpr1. This indicates that the side chain of Asn135 is important for the substrate recognition, in agreement with the observation that the side chain of Asn135 recognizes CHOP in the crystal structure. The Asn135Asp mutant did not show detectable AZC acetyltransferase activity, suggesting that the side chain of Asn135 recognizes AZC through its carboxyl anion group. Asparagine residues at positions 125 and 172 interact each other, and Asn172 is located at the position to interact with Asn135 (Fig. 1B). The replacement of Asn125 and Asn172 by Ala drastically decreased the specific activities (Fig. S2C). Thus, it seems that Asn125 and Asn172 assist the function of Asn135 by maintaining its correct orientation and location. In the Asn178Ala variant, the apparent kcat value was greatly decreased (40-fold). This indicates that the side chain of Asn178 is crucial for catalysis. This is consistent with the prediction that the sulfur atom on AcCoA interacts with the side-chain amide of this residue (Fig. 1B). In many GNAT proteins, it is suggested that the thiolate anion became protonated after degradation of the tetrahedral intermediate by the phenolic hydroxyl group of the conserved Tyr residue, which corresponds to Trp185 in Mpr1. Although this protonation is thought to be essential in the catalytic action, Trp185 is unlikely to be a catalytic residue because it is not conserved among Mpr1 homologs. Considering the pKa values for thiol and carboxyamide groups, it is unlikely that Asn178 directly protonates the thiolate anion. The lack of any measurable activity in the Asn178Asp mutant (Table 1) indicates that the negative charge at position 178 is unfavorable for catalysis. In the complex model of Mpr1 with AcCoA, the side-chain amide of Asn178 interacts with the AcCoA sulfur atom. From these results, we concluded that the stabilization by Asn178 and the protonation by a water molecule bound to Asn178 of thiolate anion are driving forces in catalysis.
Table 1.
Kinetic analysis of WT and mutant Mpr1
| Mpr1 | kcat (s−1) | KmAZC (mM) | KiAZC (mM) | KmAcCoA (μM) |
| WT | 287.2 (26.7) | 20.9 (2.4) | 3.0 (0.3) | 4.3 (0.6) |
| N135A | 38.6 (10.6) | 407.0 (91.2) | — | 2.2 (0.5) |
| N135D | — | — | — | — |
| N178A | 7.2 (1.2) | 135.8 (24.6) | — | 7.3 (0.2) |
| N178D | — | — | — | — |
The apparent kinetic parameters were determined. The kinetic parameters for AZC were determined at the fixed concentration of AcCoA (100 μM), and those for AcCoA were determined at the fixed concentration of AZC (5 mM). The values are the means and SDs shown in parentheses from three independent experiments. “—” indicates that the values were not determined. The Mpr1 variants except for WT did not exhibit the substrate inhibition. The N135D and N178D variants had no detectable activity (less than 0.003 U/mg).
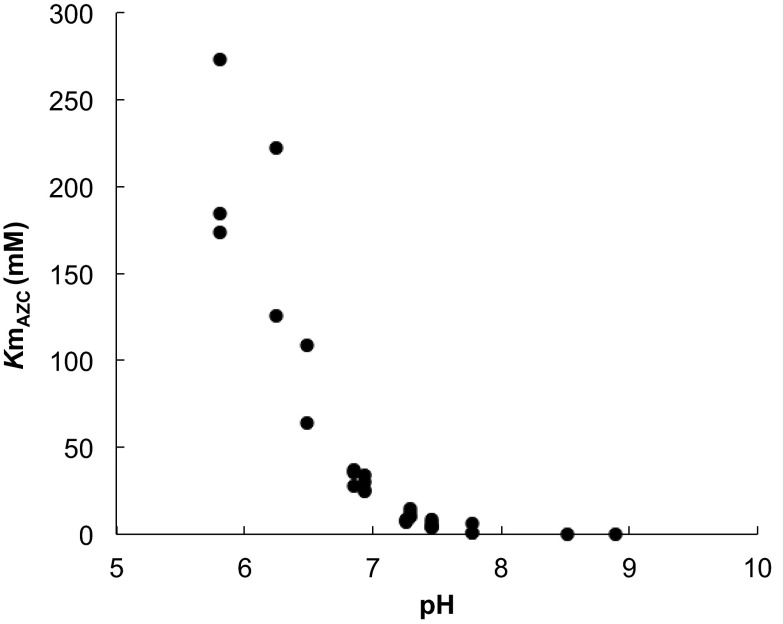
To analyze the pH dependency of the apparent Km value for AZC of Mpr1, we determined the Km value for AZC under various pH conditions (Fig. 3). The Km value increased as pH of the reaction solution decreased. In combination with the observation that CHOP interacts with the side-chain amide of Asn135 through its carboxyl group (Fig. 1B) and the result that Asn135Asp-Mpr1 did not show any measurable activity (Table 1), it was demonstrated that the side-chain amide of Asn135 in Mpr1 recognizes the substrate through its carboxyl anion group.
Fig. 3.
The pH dependency of the Km value for AZC of Mpr1. The apparent Km value for AZC was determined at various pH conditions as described in SI Materials and Methods and plotted.
Physiological Functions of Mpr1 Mutants in Yeast.
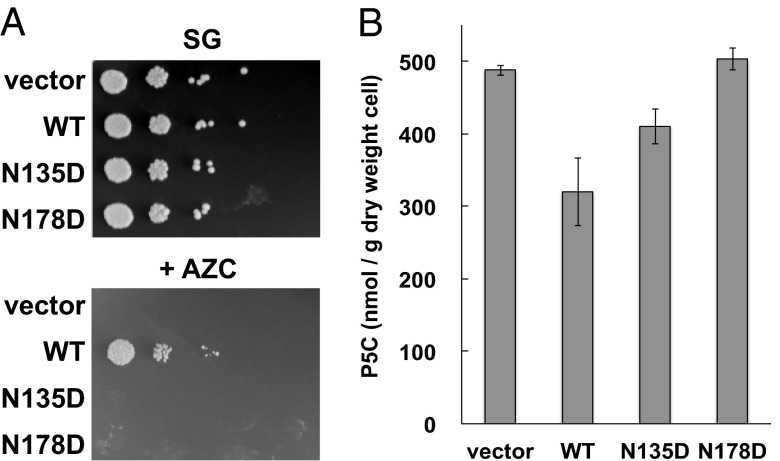
To analyze the function of mutant Mpr1 in vivo, we tested the growth phenotypes of the S. cerevisiae LD1014ura3 strain, in which the endogenous MPR1/2 genes were disrupted, overexpressing mutant Mpr1 (Fig. 4A). Following induction of the Mpr1 expression, yeast cells were spotted onto SG agar plates in the presence or absence of 5 mM AZC and cultured at 30 °C for 4 d. Although WT-Mpr1 conferred resistance to AZC on S. cerevisiae, yeast cells expressing Asn135Asp and Asn178Asp-Mpr1, as well as those carrying only the vector, did not grow on the AZC-containing medium (Fig. 4A). These results indicate that Asn135 and Asn178 in Mpr1 are indispensable for the detoxification of AZC.
Fig. 4.
Functional analyses of WT and mutant Mpr1 in vivo. (A) Growth phenotypes on SG agar medium of S. cerevisiae cells that express WT and mutant Mpr1. Strains LD1014ura3 expressing WT, Asn135Asp, or Asn178Asp or harboring the empty vector were cultured, and the serial dilutions of ∼105 cells to 10 cells (from left to right) were spotted onto SG agar medium in the presence or absence of 5 mM AZC. The plates were incubated at 30 °C for 100 h. The experiments were triplicated, and the picture shows the representative result. (B) The P5C accumulation after heat-shock stress in S. cerevisiae cells that express WT and mutant Mpr1. Strains LD1014ura3 expressing WT, Asn135Asp, or Asn178Asp or harboring the empty vector were cultured and exposed to heat stress (at 39 °C for 5 h), and then the intracellular P5C content was measured. The values are the means and SDs from two independent experiments.
Mpr1 was shown to acetylate P5C/GSA in vitro and in vivo (7, 10, 11). Therefore, we determined P5C levels in yeast cells after exposure to heat stress (at 39 °C for 5 h) (Fig. 4B). In agreement with the previous result (7), yeast cells expressing WT-Mpr1 showed an ∼40% decrease in P5C content compared with the mpr1/mpr2 disruptant, indicating that Mpr1 can catabolize P5C. Contrastingly, yeast cells expressing the Asn135Asp and Asn178Asp variants accumulated higher P5C levels than did those expressing WT-Mpr1. The P5C contents in Asn178Asp variant cells were close to those in cells carrying only the vector. These results indicate that Asn135 and Asn178 in Mpr1 are necessary to exert the physiological function involved in oxidative stress tolerance, in addition to the detoxification of AZC. It was also demonstrated that the enzymatic activity of Mpr1 is definitely important for P5C catabolism.
The Proposed Reaction Mechanism Catalyzed by Mpr1.
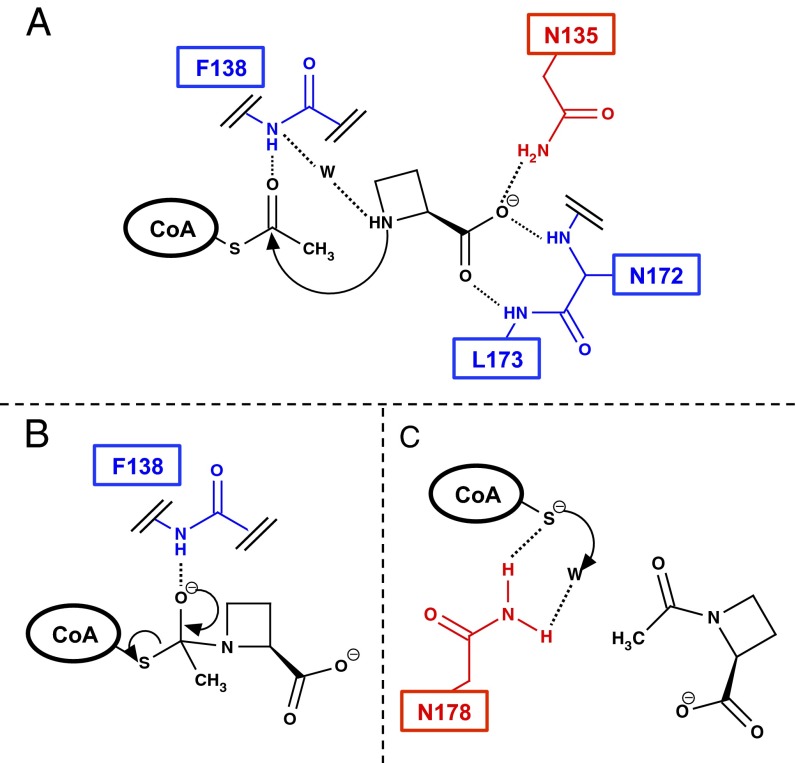
Based on the results shown above, we proposed the reaction mechanism catalyzed by Mpr1 (Fig. 5). First, the side-chain amide of Asn135 and the backbone amide of Asn172 and Leu173 in Mpr1 recognize the substrate AZC through its carboxyl anion group, and AcCoA also binds to Mpr1, randomly or in an ordered manner (Fig. 5A). Next, AZC attacks to the carbonyl carbon of AcCoA nucleophilically, forming the tetrahedral intermediate, which is stabilized by the interaction with the NH group on the backbone of Phe138 (Fig. 5B). The degradation of the intermediate generates a thiolate anion of CoA and N-acetyl AZC. The thiolate anion is stabilized by the side-chain amide group of Asn178 and then protonated by a water molecule bound to Asn178 (Fig. 5C). Finally, the generated CoA and N-acetyl AZC are released.
Fig. 5.
The proposed catalytic reaction mechanism of Mpr1. (A) AZC is recognized by the side-chain amide of Asn135 and the backbone amide of Phe138, Asn172, and Leu173. (B) The tetrahedral intermediate is stabilized by the backbone NH group of Phe138. (C) The thiolate anion generated after the breakdown of the intermediate is stabilized by the side-chain amide of Asn178, and then the thiolate is protonated by a water molecule bound to Asn178. The letter “W,” black dotted lines, and the arrows indicate a water molecule, the possible interactions, and the electron flow, respectively. The red and blue residues interact with the substrates through their side chain and backbone, respectively.
Discussion
In this study, we determined the crystal structure and the kinetics of the enzymatic reaction of the yeast N-acetyltransferase Mpr1 and proposed a reaction mechanism by analyses of the mutant enzymes. The overall structure of Mpr1 resembles other members of the GNAT superfamily, although the sequence homology is poor.
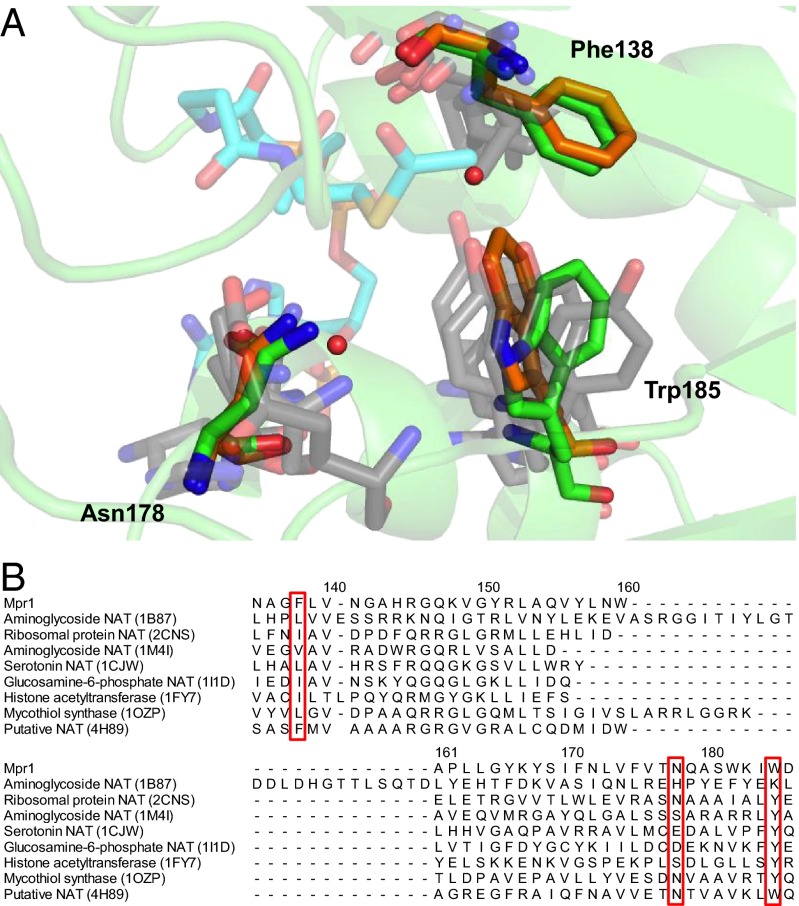
Although in many other GNAT proteins the β-bulge structure forms an oxyanion hole to stabilize the tetrahedral intermediate, the Mpr1 protein does not have a β-bulge structure. Phe138 and its neighboring residues (Ala136, Leu139, and Val140) on the fifth β-strand lacking a β-bulge structure are conserved among the Mpr1 homologs (Fig. 1C), suggesting that the structure lacking a β-bulge is unique and conserved in Mpr1. The side-chain amide group of Asn178 was suggested to act as a catalyst in Mpr1. It was reported that a phenolic hydroxyl group of a Tyr residue in many other GNAT proteins functions as a catalyst to protonate the thiolate anion generated after the tetrahedral intermediate breaks down (14), although in some GNAT proteins, whose active sites do not have tyrosine residues as acidic catalysts to protonate thiolate, there were no possible catalytic acids like those needed to accelerate the reaction. Instead, it was concluded that thiolate is protonated by bulk water (14). However, our kinetic analysis and the complex structure model with AcCoA suggest that Asn178, which is a well-conserved residue among the Mpr1 homologs (Fig. 1C) (3), functions as a catalyst. The residue corresponding to Asn178 in Mpr1 is not conserved in other GNAT proteins (Fig. 6), suggesting that the Asn-involved catalysis is unique to the Mpr1 protein.
Fig. 6.
Multiple alignment of Mpr1 with other members in the GNAT superfamily. (A) The structure of Mpr1 is superimposed with those of other GNAT proteins described below. Mpr1, a putative N-acetyltransferase from K. flavida (PDB ID code 4H89), and the other GNAT proteins are shown by green, orange, and gray color, respectively. Phe138, Asn178, and Trp185 in Mpr1 and AcCoA from the structure 1B87 are shown by a stick model and overlapped with the corresponding residues in the other proteins, which are also shown by a stick model. Red balls indicate water molecules bound to Mpr1. (B) The multiple alignment of Mpr1 with the other proteins. Red boxes indicate Phe138, Asn178, and Trp185. Other GNAT proteins are aminoglycoside 6′-N-acetyltransferase (PDB ID code 1B87) (17), ribosomal S18 N-α-protein acetyltransferase RimI (PDB ID code 2CNS) (20), aminoglycoside 2′-N-acetyltransferase (PDB ID code 1M4I) (21), serotonin N-acetyltransferase (PDB ID code 1CJW) (22), glucosamine-6-phosphate N-acetyltransferase (PDB ID code 1I1D) (23), histone acetyltransferase (PDB ID code 1FY7) (24), and mycothiol synthase (PDB ID code 1OZP) (25).
Two characteristics of Mpr1—participation of an Asn residue in the catalytic activity and the absence of the β-bulge structure—have not been reported in any other GNAT proteins. Such features of Mpr1 could be related to the shapes and/or the nucleophilicities of the substrates, secondary cyclic amines. The structure lacking the β-bulge of Mpr1 has the advantage for the enzymatic activity to reduce steric interference. However, the Asn-dependent catalytic mechanism seems to be inefficient for catalysis because the side-chain amide of Asn cannot protonate the thiolate anion directly. Generally, secondary cyclic amines possess higher nuclephilicities than primary amines. The inefficiency of Asn-involved catalysis of Mpr1 may be necessary to maintain an appropriate reaction rate to modify the substrates with higher nucleophilicities. These unique features indicate that Mpr1 is classified to an unreported group of GNAT superfamily, a cyclic secondary amine N-acetyltransferase. Interestingly, the Km value for AZC (20 mM) is significantly higher than the Ki value for AZC (3 mM). It is also possible that the Km value that is higher than the Ki value is not only due to the fact that AZC is not a physiological substrate for Mpr1 but also because such a balance between Km and Ki is required to control the reaction rate. The Mpr1-mediated proline–arginine metabolism leads to NO synthesis in vivo (Fig. S1C). The excess amount of NO can be converted into highly toxic peroxynitrite by the reaction with ROS (28). The regulation of the reaction rate in Mpr1 may control the NO level to protect the cell from its toxic effect.
Recently, the crystal structure of a putative N-acetyltransferase from the bacteria Kribbella flavida was solved (PDB ID code 4H89) (Fig. 6A). The amino acid sequence (29% of identity) and the 3D structure (rmsd is 1.8 Å) of 4H89 are significantly similar to those of Mpr1. Moreover, the expected crucial residues, which correspond to Asn135, Phe138, and Asn178 in Mpr1 (Fig. 6B), and the small loop for the recognition of the substrate’s carboxyl group (Fig. S3D) are conserved in the structure of 4H89. This putative N-acetyltransferase may catalyze the reaction in the same manner as Mpr1 and have a similar substrate specificity with Mpr1, although its substrates are not identified. This fact suggests that Mpr1 is evolutionarily close to the bacterial N-acetyltransferase and raises the possibility that the Mpr1-mediated stress-tolerant mechanism exists in other microorganisms in addition to yeasts and fungi.
We also found that Asn135 and Asn178 are important residues not only for AZC tolerance but also for P5C metabolism. The P5C level in cells expressing the Asn135Asp mutant was lower than that in cells expressing the Asn178Asp mutant, even though AZC acetyltransferase activity was not detected in either mutant in vitro. This difference suggests that the roles of Asn135 in recognition of AZC and P5C/GSA are different. In the structure of Mpr1-CHOP, CHOP is recognized by Asn135, Asn172, and Leu173 through its carboxyl group and by Phe138 through its amine group. It is possible that the additional interactions without Asn135 contribute to the substrate recognition and/or the catalysis in the case of P5C/GSA.
Previously, we isolated two Mpr1 variants with higher AZC resistance (Lys63Arg, Phe65Leu) (29). Interestingly, the Lys63Arg variant confers higher oxidative stress tolerance than WT-Mpr1 on yeast cells. The Phe65Leu mutation enhanced the thermal stability of Mpr1 (29). In the structure of Mpr1, Phe65 is buried in the hydrophobic region (Fig. S3E), suggesting that the replacement of Phe65 by Leu improves the packing of the side chain in this region, leading to the enhancement of stability. In contrast, we have no clear explanation of why the Lys63Arg mutant showed improved stress tolerance because no functionally crucial region exists around a nonconserved Lys63 (Fig. S3E).
Mpr1 confers oxidative stress tolerance on yeast cells by the conversion of P5C/GSA into N-acetyl GSA (Fig. S1C). The Km value of the recombinant Mpr1 for P5C is 7 mM (7), although the intracellular P5C concentration shown here was estimated at ∼50 μM even under the stress condition, suggesting that Mpr1 itself is not enough to exert its physiological function: unknown factors, such as certain proteins or chemical compounds, should be required to express the full activity. In fact, previous reports revealed that some GNAT enzymes require protein partners or small cofactors to exert their functions. For example, Rtt109, a yeast histone acetyltransferase, interacts with the histone chaperones Aft1 and Vps75 to acetylate the ε-amino group of Lys56 on histone H3 (30, 31), and the aminoalkylphosphonic acid N-acetyltransferase PhnO from Salmonella enterica requires a divalent metal ion for its activity (32). Taking into account our discussion above, we believe that Mpr1 should have some partner molecule(s) such as proteins and/or small-molecular-weight compounds and that the protein complex containing Mpr1 should be required for full display of its physiological function.
An understanding of the reaction mechanism of Mpr1 provides some benefits in its application. Because Mpr1 confers AZC resistance on various organisms, selectable marker systems using the MPR1 gene and AZC have been constructed for yeast and plant transformation (33–35). We recently showed that industrial yeast strains expressing the Mpr1 variant are more tolerant to oxidative stresses, such as ethanol, freezing, and desiccation, than those expressing WT-Mpr1 (29, 36, 37). Engineered Mpr1 might be useful in breeding oxidative stress-tolerant yeast strains with improved fermentation ability. Because the Mpr1 homologs are found only in yeasts and fungi, it appears that the Mpr1-mediated antioxidative mechanism is highly conserved in yeasts and fungi (10, 11). Thus, the inhibition of Mpr1 activity could be a promising target for the development of antifungal drugs.
Materials and Methods
Details for strains, plasmids, and medium used in this study and methods for protein expression and purification can be found in SI Materials and Methods. Procedures detailing crystallization, data collection, structure determination, and refinement can be found in SI Materials and Methods. Protocols for acetyltransferase assay and functional analysis of Mpr1 are also available in SI Materials and Methods.
Supplementary Material
Acknowledgments
The synchrotron radiation experiments were performed at the BL41XU of SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (Proposal 2012A1058). We thank the Photon Factory Program Advisory Committee for the provision of synchrotron data-collection facilities (Proposal 2008G602, 2010G005). This work was supported by a Grant-in-Aid for Scientific Research (B) (22380061) from the Japan Society for the Promotion of Science and by a Grant-in-Aid for Scientific Research on Innovative Area (ROS Signal) (23117711).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3W91, 3W6S, and 3W6X).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300558110/-/DCSupplemental.
References
- 1.Takagi H, Shichiri M, Takemura M, Mohri M, Nakamori S. Saccharomyces cerevisiae sigma 1278b has novel genes of the N-acetyltransferase gene superfamily required for L-proline analogue resistance. J Bacteriol. 2000;182(15):4249–4256. doi: 10.1128/jb.182.15.4249-4256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shichiri M, Hoshikawa C, Nakamori S, Takagi H. A novel acetyltransferase found in Saccharomyces cerevisiae Σ1278b that detoxifies a proline analogue, azetidine-2-carboxylic acid. J Biol Chem. 2001;276(45):41998–42002. doi: 10.1074/jbc.C100487200. [DOI] [PubMed] [Google Scholar]
- 3.Wada M, et al. Distribution of l-azetidine-2-carboxylate N-acetyltransferase in yeast. Biosci Biotechnol Biochem. 2008;72(2):582–586. doi: 10.1271/bbb.70534. [DOI] [PubMed] [Google Scholar]
- 4.Fowden L. Azetidine-2-carboxylic acid: A new cyclic imino acid occurring in plants. Biochem J. 1956;64(2):323–332. doi: 10.1042/bj0640323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fowden L, Bryant M. Nitrogenous compounds and nitrogen metabolism in the Liliaceae. 5. The metabolism of azetidine-2-carboxylic acid. Biochem J. 1959;71(2):210–217. doi: 10.1042/bj0710210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Troxler RF, Brown AS. Metabolism of L-azetidine-2-carboxylic acid in the alga Cyanidium caldarium. Biochim Biophys Acta. 1974;366(3):341–349. doi: 10.1016/0005-2787(74)90294-9. [DOI] [PubMed] [Google Scholar]
- 7.Nomura M, Takagi H. Role of the yeast acetyltransferase Mpr1 in oxidative stress: Regulation of oxygen reactive species caused by a toxic proline catabolism intermediate. Proc Natl Acad Sci USA. 2004;101(34):12616–12621. doi: 10.1073/pnas.0403349101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du X, Takagi H. N-acetyltransferase Mpr1 confers freeze tolerance on Saccharomyces cerevisiae by reducing reactive oxygen species. J Biochem. 2005;138(4):391–397. doi: 10.1093/jb/mvi134. [DOI] [PubMed] [Google Scholar]
- 9.Du X, Takagi H. N-Acetyltransferase Mpr1 confers ethanol tolerance on Saccharomyces cerevisiae by reducing reactive oxygen species. Appl Microbiol Biotechnol. 2007;75(6):1343–1351. doi: 10.1007/s00253-007-0940-x. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura A, Kotani T, Sasano Y, Takagi H. An antioxidative mechanism mediated by the yeast N-acetyltransferase Mpr1: Oxidative stress-induced arginine synthesis and its physiological role. FEMS Yeast Res. 2010;10(6):687–698. doi: 10.1111/j.1567-1364.2010.00650.x. [DOI] [PubMed] [Google Scholar]
- 11.Nishimura A, Kawahara N, Takagi H. The flavoprotein Tah18-dependent NO synthesis confers high-temperature stress tolerance on yeast cells. Biochem Biophys Res Commun. 2013;430(1):137–143. doi: 10.1016/j.bbrc.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura A, Nasuno R, Takagi H. The proline metabolism intermediate Δ1-pyrroline-5-carboxylate directly inhibits the mitochondrial respiration in budding yeast. FEBS Lett. 2012;586(16):2411–2416. doi: 10.1016/j.febslet.2012.05.056. [DOI] [PubMed] [Google Scholar]
- 13.Hoa BT, et al. Production of N-acetyl cis-4-hydroxy-l-proline by the yeast N-acetyltransferase Mpr1. J Biosci Bioeng. 2012;114(2):160–165. doi: 10.1016/j.jbiosc.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Vetting MW, et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433(1):212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Hibi T, Yamamoto H, Nakamura G, Takagi H. Crystallization and preliminary crystallographic analysis of N-acetyltransferase Mpr1 from Saccharomyces cerevisiae. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65(Pt 2):169–172. doi: 10.1107/S1744309109000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm L, Rosenström P. 2010. Dali server: Conservation mapping in 3D. Nucleic Acids Res 38(Web Server issue): W545–W549.
- 17.Wybenga-Groot LE, Draker K, Wright GD, Berghuis AM. Crystal structure of an aminoglycoside 6′-N-acetyltransferase: Defining the GCN5-related N-acetyltransferase superfamily fold. Structure. 1999;7(5):497–507. doi: 10.1016/s0969-2126(99)80066-5. [DOI] [PubMed] [Google Scholar]
- 18.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Burk DL, Ghuman N, Wybenga-Groot LE, Berghuis AM. X-ray structure of the AAC(6′)-Ii antibiotic resistance enzyme at 1.8 A resolution: Examination of oligomeric arrangements in GNAT superfamily members. Protein Sci. 2003;12(3):426–437. doi: 10.1110/ps.0233503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vetting MW, Bareich DC, Yu M, Blanchard JS. Crystal structure of RimI from Salmonella typhimurium LT2, the GNAT responsible for N(alpha)-acetylation of ribosomal protein S18. Protein Sci. 2008;17(10):1781–1790. doi: 10.1110/ps.035899.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vetting MW, Hegde SS, Javid-Majd F, Blanchard JS, Roderick SL. Aminoglycoside 2′-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates. Nat Struct Biol. 2002;9(9):653–658. doi: 10.1038/nsb830. [DOI] [PubMed] [Google Scholar]
- 22.Hickman AB, Namboodiri MA, Klein DC, Dyda F. The structural basis of ordered substrate binding by serotonin N-acetyltransferase: Enzyme complex at 1.8 A resolution with a bisubstrate analog. Cell. 1999;97(3):361–369. doi: 10.1016/s0092-8674(00)80745-x. [DOI] [PubMed] [Google Scholar]
- 23.Peneff C, Mengin-Lecreulx D, Bourne Y. The crystal structures of Apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase. J Biol Chem. 2001;276(19):16328–16334. doi: 10.1074/jbc.M009988200. [DOI] [PubMed] [Google Scholar]
- 24.Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V. Structure of the histone acetyltransferase Hat1: A paradigm for the GCN5-related N-acetyltransferase superfamily. Cell. 1998;94(4):427–438. doi: 10.1016/s0092-8674(00)81584-6. [DOI] [PubMed] [Google Scholar]
- 25.Vetting MW, Roderick SL, Yu M, Blanchard JS. Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases. Protein Sci. 2003;12(9):1954–1959. doi: 10.1110/ps.03153703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura Y, Nakamori S, Takagi H. Polymorphism of the MPR1 gene required for toxic proline analogue resistance in the Saccharomyces cerevisiae complex species. Yeast. 2002;19(16):1437–1445. doi: 10.1002/yea.927. [DOI] [PubMed] [Google Scholar]
- 27.Nomura M, Nakamori S, Takagi H. Characterization of novel acetyltransferases found in budding and fission yeasts that detoxify a proline analogue, azetidine-2-carboxylic acid. J Biochem. 2003;133(1):67–74. doi: 10.1093/jb/mvg003. [DOI] [PubMed] [Google Scholar]
- 28.Ferrer-Sueta G, Radi R. Chemical biology of peroxynitrite: Kinetics, diffusion, and radicals. ACS Chem Biol. 2009;4(3):161–177. doi: 10.1021/cb800279q. [DOI] [PubMed] [Google Scholar]
- 29.Iinoya K, Kotani T, Sasano Y, Takagi H. Engineering of the yeast antioxidant enzyme Mpr1 for enhanced activity and stability. Biotechnol Bioeng. 2009;103(2):341–352. doi: 10.1002/bit.22247. [DOI] [PubMed] [Google Scholar]
- 30.Fillingham J, et al. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol. 2008;28(13):4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berndsen CE, et al. Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75. Nat Struct Mol Biol. 2008;15(9):948–956. doi: 10.1038/nsmb.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Errey JC, Blanchard JS. Functional annotation and kinetic characterization of PhnO from Salmonella enterica. Biochemistry. 2006;45(9):3033–3039. doi: 10.1021/bi052297p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishchuk OP, Abbas CA, Sibirny AA. Heterologous expression of Saccharomyces cerevisiae MPR1 gene confers tolerance to ethanol and L: -Azetidine-2-carboxylic acid in Hansenula polymorpha. J Ind Microbiol Biotechnol. 2010;37(2):213–218. doi: 10.1007/s10295-009-0674-0. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa-Mitsuhashi K, Sagane K, Kuromitsu J, Takagi H, Tsukahara K. MPR1 as a novel selection marker in Saccharomyces cerevisiae. Yeast. 2009;26(11):587–593. doi: 10.1002/yea.1708. [DOI] [PubMed] [Google Scholar]
- 35.Tsai FY, Zhang XH, Ulanov A, Widholm JM. The application of the yeast N-acetyltransferase MPR1 gene and the proline analogue L-azetidine-2-carboxylic acid as a selectable marker system for plant transformation. J Exp Bot. 2010;61(10):2561–2573. doi: 10.1093/jxb/erq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasano Y, Takahashi S, Shima J, Takagi H. Antioxidant N-acetyltransferase Mpr1/2 of industrial baker’s yeast enhances fermentation ability after air-drying stress in bread dough. Int J Food Microbiol. 2010;138(1-2):181–185. doi: 10.1016/j.ijfoodmicro.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Sasano Y, et al. Enhancement of the proline and nitric oxide synthetic pathway improves fermentation ability under multiple baking-associated stress conditions in industrial baker’s yeast. Microb Cell Fact. 2012;11:40. doi: 10.1186/1475-2859-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.