Abstract
Mammalian cells typically contain thousands of copies of mitochondrial DNA assembled into hundreds of nucleoids. Here we analyzed the dynamic features of nucleoids in terms of mitochondrial membrane dynamics involving balanced fusion and fission. In mitochondrial fission GTPase dynamin-related protein (Drp1)-deficient cells, nucleoids were enlarged by their clustering within hyperfused mitochondria. In normal cells, mitochondrial fission often occurred adjacent to nucleoids, since localization of Mff and Drp1 is dependent on the nucleoids. Thus, mitochondrial fission adjacent to nucleoids should prevent their clustering by maintaining small and fragmented nucleoids. The enhanced clustering of nucleoids resulted in the formation of highly stacked cristae structures in enlarged bulb-like mitochondria (mito-bulbs). Enclosure of proapoptotic factor cytochrome c, but not of Smac/DIABLO, into the highly stacked cristae suppressed its release from mitochondria under apoptotic stimuli. In the absence of nucleoids, Drp1 deficiency failed to form mito-bulbs and to protect against apoptosis. Thus, mitochondrial dynamics by fission and fusion play a critical role in controlling mitochondrial nucleoid structures, contributing to cristae reformation and the proapoptotic status of mitochondria.
Mitochondria are the main source of energy from oxidative phosphorylation and play a role in various cellular processes, including apoptosis and Ca2+ signaling. They are believed to derive from endosymbiosis of bacteria, and contain their own mitochondrial DNA (mtDNA) within the matrix (1, 2). Under fluorescence microscopy, more than 1,000 copies of mtDNAs per cell are observed as hundreds of dot-like structures known as mitochondrial nucleoids (3–5). Nucleoids carry 2–10 copies of mtDNA packaged with proteins, including mitochondrial transcriptional factor A (TFAM), a key component of nucleoids (6–8). Mitochondrial nucleoids appear to be a platform for transcription, translation, and replication of mtDNA; however, the molecular mechanism of their formation and their physiological roles remain poorly understood.
Mitochondria are highly dynamic organelles that change their morphology in response to cellular signaling and differentiation. Mitochondrial morphology is maintained by the balance between fusion and fission (9, 10). In mammals, mitochondrial fusion is regulated by two outer membrane (OM) GTPases, mitofusin (Mfn)1 and Mfn2, whereas the inner membrane (IM) GTPase optic atrophy (OPA)1 regulates fusion of the IM. Deficiencies in Mfn2 or OPA1 result in neurodegenerative conditions, such as Charcot–Marie–Tooth neuropathy type 2a or autosomal dominant optic atrophy type I, respectively. Dynamin-related protein (Drp1), a GTPase, plays a key role in mitochondrial fission, localizes mainly in the cytoplasm and is recruited to mitochondrial fission sites via interaction with OM receptor proteins, such as mitochondrial fission factor (Mff) (11). An autosomal dominant mutation of human Drp1 leads to neonatal lethality (12), and Drp1 KO mice are defective in embryonic development and neural functions (13–15), suggesting the importance of mitochondrial dynamics for development and differentiation in mammals.
Pioneering work on mtDNA dynamics and inheritance using primitive eukaryotes, such as slime mold Physarum polycephalum or red alga Cyanidioschyzon merolae, has shown that highly packed mtDNA division is synchronized with mitochondrial fission and is subsequently passed to two daughter organelles (2, 16). In contrast, live imaging of mitochondrial nucleoids in mammals, higher plants, and yeast has revealed that mitochondrial fission occurs independently of nucleoid fission (2–4, 17). A deficiency in mitochondrial fusion leads to decreased mass and elevated mutation rates of mtDNA and impaired respiratory activity (18, 19). In contrast, mitochondrial fission defects in cultured mammalian cells appear to have less impact on cell viability and on the inheritance of active mitochondria (13–15). How mtDNA is maintained under mitochondrial membrane dynamics remains unclear. To elucidate the dynamic features of mitochondrial nucleoids in the present study, we analyzed their dynamics in conjunction with mitochondrial membrane fission and fusion and examined their relationship with mitochondrial fission, mitochondrial membrane structure, and apoptosis.
Results
Drp1-Mediated Mitochondrial Fission Defects Induce Formation of Enlarged Mitochondrial Nucleoids.
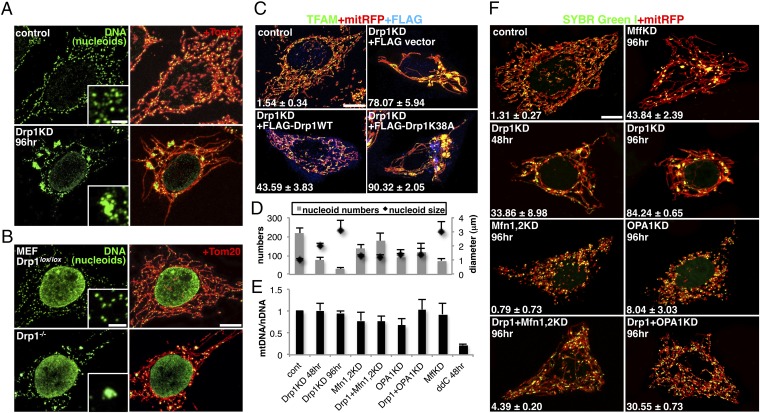
To explore the interplay between mitochondrial fission and nucleoid segregation, we first performed knockdown (KD) of mitochondrial fission GTPase Drp1 in HeLa cells. Immunostaining of Drp1 RNAi cells with an anti-DNA antibody, capable of detecting mitochondrial nucleoids (4, 5), revealed a small number of extremely enlarged nucleoids within highly elongated and connected mitochondrial networks (Fig. 1A and single-channel images in Fig. S1A), as reported previously (20, 21). Drp1 KO mouse embryonic fibroblasts (MEFs) (13) showed the same phenotype, although the effects were weaker (Fig. 1B and single-channel images in Fig. S1B). The phenotype was confirmed using an anti-TFAM antibody, a main protein component of the nucleoid (6, 8) (Fig. 1C), and using the fluorescent stain SYBR Green I (22) (Fig. 1F).
Fig. 1.
Enlarged nucleoids in Drp1-deficient cells. (A) Immunofluorescence staining of HeLa cells treated with Drp1 or control siRNA for 96 h as indicated. (B) Drp1 KO and control MEFs. (C) Rescue experiments. KD HeLa cells stably expressing mitRFP were subsequently transfected with WT or K38A of FLAG-tagged rat Drp1 for 20 h. The cells containing enlarged nucleoids were counted in three independent experiments (n = ∼100). (D–F) HeLa cells stably expressing mitRFP were treated with the indicated siRNAs and stained with SYBR Green I. The average of three independent experiments is shown. The average number and diameter of nucleoids in 30 randomly selected cells (D). mtDNA content was quantified by quantitative PCR (E). The cells containing enlarged nucleoids were counted (n = ∼100) (F). Single-color images of A, B, and C are shown in Fig. S1 A and B and Fig. S2A, respectively. (Scale bar: 10 μm; 2 μm in Insets).
In Drp1 KD cells, we observed a clear inverse correlation between nucleoid number and size in a time-dependent manner (Fig. 1D). Importantly, total mtDNA content did not decrease in mitochondrial fission-deficient cells (Fig. 1E); thus, the reduction of nucleoid numbers in mitochondrial fission-deficient cells is not related to a reduction in the amount of mtDNA. Rescue experiments performed in Drp1 KD cells by transiently expressing WT or Drp1 mutant (K38A) showed that the production of enlarged nucleoids is a reversible process (Fig. 1C and single-channel images in Fig. S2A).
Enlarged Nucleoids in Drp1-Deficient Cells Are Caused by Nucleoid Clustering in Fused Mitochondria.
We next performed KD of other mitochondrial fission or fusion factors. KD of Mff, the OM protein involved in mitochondrial recruitment of Drp1, also induced enlarged nucleoids (Fig. 1 D and F, MffKD). No enlarged nucleoids were observed in cells treated with mitochondrial fusion factors (OPA1KD and Mfn1,2KD). The Drp1 KD-induced enlarged nucleoid phenotype was suppressed by codepletion with mitochondrial fusion factors (Drp1+OPA1KD and Drp1+Mfn1,2KD). Overexpression of Mfn1 also induced enlarged nucleoids (Fig. S2C), suggesting that nucleoid enlargement in mitochondrial fission-deficient cells is related to mitochondrial fusion.
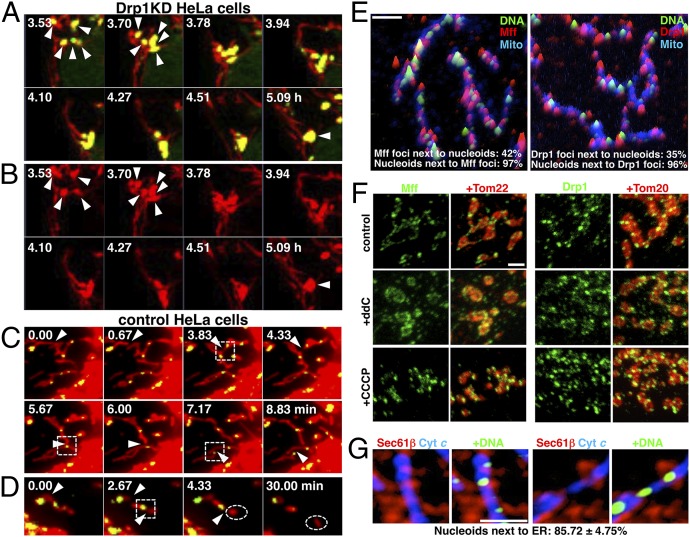
We next performed live-cell imaging and monitored the process of nucleoid enlargement. Nucleoids in HeLa cells expressing mitochondrial red fluorescent protein (mitRFP) were visualized by SYBR Green I. Time-lapse analysis at 5-min intervals in cells after 24 h of Drp1 KD revealed several nucleoids clustered into a single enlarged nucleoid (Fig. 2 A and B, Fig. S3 A–C, and Movies S1, S2, and S3). These changes were not observed in control or Drp1-expressing cells (Fig. 2C and Movies S4 and S5). A subset of single enlarged nucleoids stained by anti-DNA or anti-TFAM antibodies in Drp1 KD cells sometimes could be resolved into clusters containing multiple nucleoids at high resolution (Fig. S1 C–E). Thus, mitochondrial hyperfusion caused by Drp1 KD induced the local clustering of nucleoids.
Fig. 2.
Imaging of mitochondrial fission and nucleoids. (A and B) HeLa cells stably expressing mitRFP treated with Drp1 siRNA for 24 h were stained with SYBR Green I, and live-cell images were obtained by confocal microscopy. The images were obtained every 5 min for 6–10 h as indicated. Magnified images of mitochondria and nucleoids (A) and mitochondria alone (B) in Movie S1 are shown. Red, mitRFP; green, nucleoids (SYBR Green I). Arrowheads indicate clustering of nucleoids. Also see Fig. S3 A–C and Movies S2 and S3. (C and D) Live-cell images of control HeLa cells obtained by CCD camera in Movies S6 (C) and S7 (D). The images were obtained every 10 s. Red, mitRFP; green, nucleoids (SYBR Green I). Arrowheads indicate sites of mitochondrial fission, and dashed squares indicate mitochondrial fission occurring in the vicinity of nucleoids. Dashed circles indicate generated mitochondria devoid of nucleoids. Also see Fig. S3 D–F and Movies S8, S9, and S10. (E) Nucleoids and endogenous Mff or Drp1 localization, shown by intensity profile landscapes. (F) Distribution of Mff and Drp1 in HeLa cells treated with 10 μM ddC for 5 d or with 10 μM CCCP for 45 min. (G) Distribution of ER and nucleoids in control cells. Single-color images of overall landscapes are shown in Fig. S3G. (Scale bar: 2 μm.)
Mitochondrial Fission Often Occurs in the Vicinity of Nucleoids.
We next investigated the relationship between mitochondrial fission and nucleoid segregation in living healthy cells (Fig. 2 C and D, Fig. S3 D–F, and Movies S6, S7, S8, S9, and S10). Although several mitochondrial fission events per 10–30 min were observed (Fig. 2 C and D and Fig. S3 D–F, arrowheads), nucleoid fission accompanied by mitochondrial fission was not observed. Consequently, mitochondria were often divided without nucleoid fission (Fig. 2D and Fig. S3F, dashed circles). Intriguingly, mitochondrial fission tended to occur in the vicinity of nucleoids (∼70% of mitochondrial fissions; Fig. 2 C and D and Fig. S3 D–F, dashed squares).
We further analyzed the distribution of nucleoids and endogenous Mff or Drp1 by immunostaining (Fig. 2E). When we focused on the mitochondrial Drp1 puncta, we found that most of the nucleoids were located in the vicinity of Drp1 (Fig. 2E). Furthermore, more than 96% of nucleoids were located in the vicinity of the Mff foci. Fewer nucleoids were observed compared with Drp1 or Mff foci, and 42% of Mff foci and 35% of Drp1 foci were associated with nucleoids.
We further analyzed the relationship between mitochondrial fission factors and nucleoids. Treatment of HeLa cells with the mtDNA replication inhibitor 2′-3′-dideoxycytidine (ddC) resulted in the loss of detectable levels of nucleoids (here termed “nucleoid-less” cells). In such cells, the distribution of Mff and Drp1 were clearly altered; Mff was dispersed on the edge of the mitochondria, and mitochondrial Drp1 was decreased and redistributed to cytoplasm (Fig. 2F). The mitochondrial morphology was more fragmentary in ddC-treated cells than in normal HeLa cells; however, no changes in distribution of Mff or Drp1 were observed in carbonyl cyanide m-chlorophenylhydrazone (CCCP)-treated cells (Fig. 2F). These data suggest that the distribution of Drp1 and Mff is responsible for the distribution of nucleoids. A recent study found that mitochondrial fission sites are marked by endoplasmic reticulum (ER) (23), and, interestingly, costaining with ER revealed that the majority of the nucleoids (85%) existed adjacent to ER, suggesting a relationship between nucleoids and ER (Fig. 2G; full landscape shown in Fig. S3G). Thus, mitochondrial fission adjacent to nucleoids should be necessary to maintain the small, dispersed nucleoids by preventing nucleoid clustering.
Enlarged Nucleoids in Drp1-Deficient Cells Stabilize IM Cristae Structures.
We next attempted to clarify the physiological significance of the observed nucleoid enlargement. We first analyzed the stability of mtDNA by evaluating the effects of the mtDNA replication inhibitor ddC after Drp1 KD. The rates of reduction in mtDNA mass (Fig. S4A) and TFAM protein level (Fig. S4B) by ddC exposure in Drp1 KD cells were moderate compared with those of control cells, suggesting that mtDNA in enlarged nucleoids should be more stable. We then measured respiration. The maximum oxygen consumption rate was not increased by Drp1 KD, but rather was decreased to a certain extent (Fig. S4C). In addition, Drp1 KD did not affect levels of oxidative phosphorylation (OXPHOS) proteins or mtRNAs (Fig. 3 G–I), suggesting that nucleoid clustering does not increase respiration.
Fig. 3.
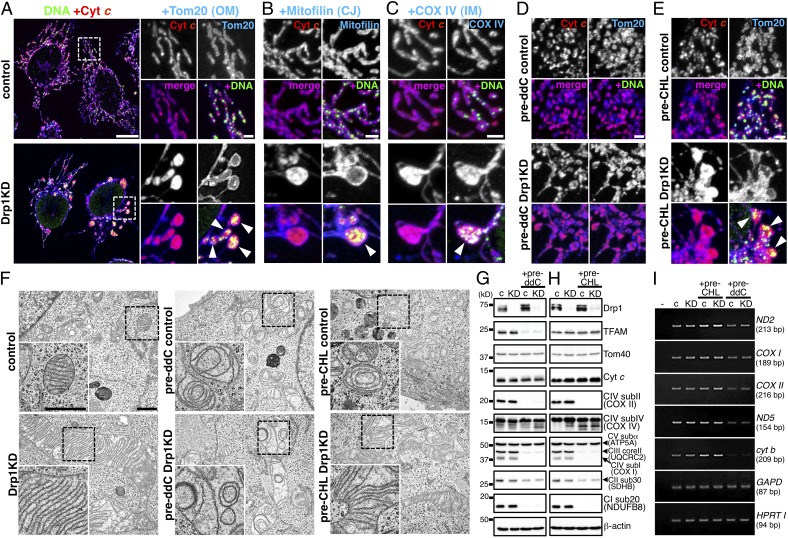
Mitochondrial cristae were highly stacked in the bulb-like mitochondria (mito-bulbs) with clustered nucleoids. HeLa cells were treated with 10 μM ddC (pre-ddC; D, G, and I) or 100 μM chloramphenicol (pre-CHL; E, H, and I) for 48 h, then further treated with siRNA for Drp1 or control for 96 h in the presence of the same reagents. (A–E) Immunofluorescence staining of nucleoids and mitochondrial proteins: red, cyt c; green, mtDNA; blue, Tom20 (A, D, and E); mitofilin (B); and COX IV (C). Arrowheads indicate mito-bulbs. (Scale bars: 10 μm in A, Left; 2 μm in magnified images in A–E). CJ, cristae junction. Overall landscapes of D and E are shown in Fig. S4 D and E. (F) Images obtained by EM. (Lower, Left) Magnified images of the boxed area in each panel. (Scale bar: 1 μm.) Overall landscapes are shown in Fig. S4 F and G. (G and H) Mitochondrial protein levels determined by immunoblot analysis using the indicated antibodies in ddC-treated (G) and CHL-treated (H) cells. Several subunits of OXPHOS complexes, such as the mitochondria-encoded COX I and II (complex IV), as well as nuclear genome-encoded respiratory proteins, such as NDUFB8 (complex I) and UQCRC2 (complex III), were repressed. Nuclear genome-encoded SDHB (complex II), COX IV (complex IV), and ATP5A (complex V) were less affected. (I) RNAs by RT-PCR. Mitochondrial transcripts (ND2, ND5, COX I, COX II, and cyt b) and nuclear genome-encoded transcripts (GAPD and HPRT I) were analyzed. c, control; KD, Drp1 KD.
We next focused on mitochondrial membrane structures. Drp1 KD was previously reported to induce many bulb-like mitochondrial structures (11, 24), although these structures were poorly characterized. We found that most of these bulb-like structures (termed “mito-bulbs” herein) contained enlarged clustered nucleoids (Fig. 1A and Fig. S1). Drp1 KD-induced mito-bulbs were suppressed by reintroduction of Drp1 (Fig. 1C) or codepletion with mitochondrial fusion factors (Fig. 1F). In live imaging, formation of fused mito-bulb structures occurred in parallel with nucleoid clustering (Fig. 2 A and B). Thus, nucleoid clustering in Drp1 KD is clearly associated with mito-bulb formation.
We further characterized the mito-bulbs using immunofluorescence microscopy. Both the OM protein Tom20 (Fig. 3A) and the IM cristae junction protein mitofilin (Fig. 3B) decorated the edges of mito-bulbs in Drp1 KD cells, but were not costained with nucleoids. In contrast, the IM respiratory protein cytochrome c oxidase subunit IV (COX IV) was costained with nucleoids inside the mito-bulb (Fig. 3C). Cytochrome c (cyt c) was costained with nucleoids and COX IV inside the mito-bulbs in Drp1 KD cells (Fig. 3C), but not with Tom20 (Fig. 3A) or mitofilin (Fig. 3B). EM of Drp1 KD cells revealed that mitochondria larger than 2 μm in diameter, corresponding to the mito-bulbs, had tightly stacked cristae (Fig. 3F and full landscapes in Fig. S4E). Although large electron-translucent structures were sometimes observed between cristae stacks, nucleoid distribution was not clearly identified on EM (Fig. S4E, arrows). On fluorescent microscopy, small clustered nucleoids were embedded in the IM structures of mito-bulbs (Fig. 3C and Fig. S1 C–E).
To test whether mito-bulb formation is caused by nucleoid clustering, we further treated nucleoid-less cells with Drp1 KD. We found that ddC treatment before Drp1 KD severely impaired mito-bulb formation (Fig. 3D and full landscapes in Fig. S4D), although Drp1 protein was efficiently depleted (Fig. 3G). Mitofilin, COX IV, and cyt c staining overlapped with Tom20 staining on the outline of elongated mitochondria. EM revealed deformed cristae in the nucleoid-less cells with control RNAi (Fig. 3F and full landscapes in Fig. S4F). The nucleoid-less cells with Drp1 KD had a sparse distribution of deformed cristae in elongated mitochondrial tubules, suggesting that nucleoid clustering results in mito-bulb formation with enriched IM cristae.
MtDNA encodes essential subunits of OXPHOS complexes. We examined whether OXPHOS complexes are required for mito-bulb formation by treatment with chloramphenicol (CHL), which specifically inhibits mitochondrial translation in eukaryotes. In CHL-pretreated Drp1 KD cells, expression of Drp1 and OXPHOS subunits was blocked, as was seen in ddC-pretreated cells (Fig. 3 G and H). Transcription of mitochondrial genome-encoded genes was comparable to that in control cells, although these genes were depressed in ddC-pretreated cells (Fig. 3I). Under these conditions, clustered nucleoids and mito-bulbs were formed by Drp1 KD after CHL treatment (Fig. 3 E and F and full landscapes in Fig. S4 D and F). These results suggest that mtDNA itself, but not its gene products, is required for mito-bulb formation with tightly stacked cristae in mitochondrial fission-deficient cells. Thus, nucleoids should be highly clustered with IM cristae for the formation of cristae-enriched mito-bulb structures.
Nucleoid Clustering Results in Delayed cyt c Release and Apoptosis.
Drp1 deficiency is known to delay the release of cyt c and subsequent apoptosis on challenge with apoptotic stimuli in various types of cultured mammalian cells (13, 25). However, the release of other proapoptotic factors, such as Smac/DIABLO, is not blocked by Drp1 KD (13, 26). The detailed mechanism by which Drp1-mediated mitochondrial fission facilitates apoptosis remains to be clarified.
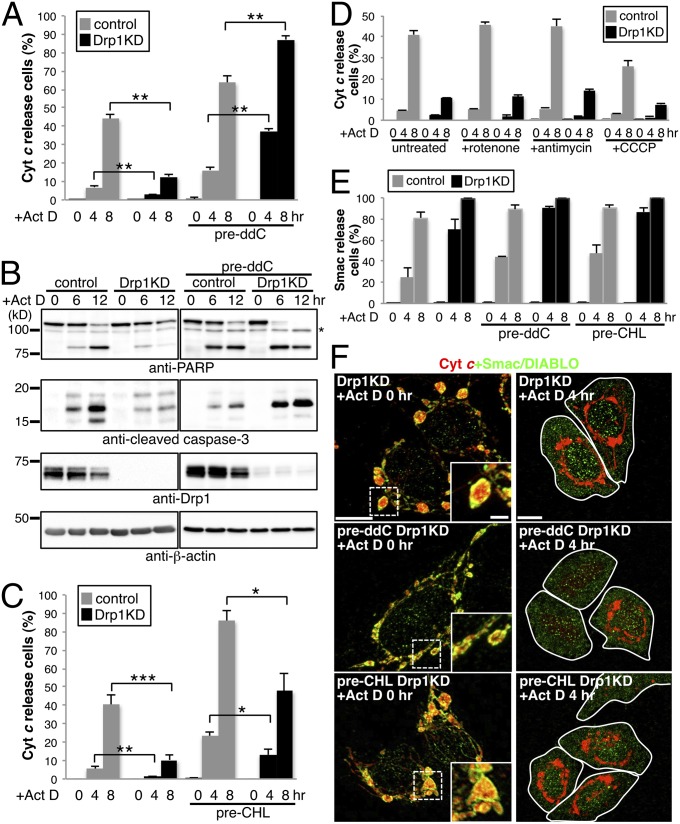
Given our observation of cyt c in the mito-bulbs with clustered nucleoids (Fig. 3), we analyzed the response of these cells to the proapoptotic reagents actinomycin D (Act D) and staurosporine (STS). Drp1 KD efficiently prevented the progression of apoptosis, cyt c release (Act D data in Fig. 4A; STS data in Fig. S5 A and E), caspase-3 activation, and poly(ADP ribose) polymerase (PARP) cleavage (Act D data in Fig. 4B; STS data in Fig. S5B). The nucleoid-less cells had similar susceptibility to apoptotic stimuli as control cells. The cell response was consistent with that reported previously, with apoptosis accompanied by cyt c release seen in ρ0 cells (27). Drp1 KD in nucleoid-less cells, in which cyt c was widely dispersed in the filamentous mitochondria (Fig. 3D), diminished the protective effect against apoptosis and even significantly increased the susceptibility to apoptotic stimuli (Fig. 4 A and B and Fig. S5 A, B, and E). EM analysis after Act D treatment revealed that swollen mitochondria with deformed cristae in nucleoid-less Drp1 KD cells but not in other Drp1 KD cells, suggesting a critical role for nucleoids in apoptotic cristae remodeling in Drp1 KD cells (Fig S5F).
Fig. 4.
Nucleoid clustering was required for the apoptosis delay in Drp1-deficient cells. HeLa cells treated as described in Fig. 3 were further treated with 5 μM Act D, with (A, C, D, and E) or without (B) 50 μM zVAD-fmk for the indicated periods. (A, C, and D) Cyt c and Tom20 were stained, and cells with released cyt c were counted. (E and F) Smac/DIABLO and cyt c were stained, and cells with released Smac/DIABLO were counted. At least 300 cells in three independent experiments were counted. Data are expressed as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001. (B) PARP and cleaved caspase-3 were measured by immunoblot analysis. The asterisk indicates a nonspecific band. (D) Effect of respiratory inhibitors on apoptosis. Drp1 KD cells were treated with 25 μM rotenone (complex I inhibitor), 1 μM antimycin (complex III inhibitor), and 10 μM CCCP (uncoupler) for 1 h. (E and F) Cyt c (red) and Smac/DIABLO (green) were immunostained (F), and the cells with released Smac/DIABLO were counted (E). (Scale bars: 10 μm; 2 μm in Insets).
It is also conceivable that defective respiration leads to annulment of the protective effect of Drp1 KD during apoptosis. We again assessed the rate of cyt c release using cells pretreated with several inhibitors for respiration (Fig. 4D) and mitochondrial translation (Fig. 4C), and found that the apoptosis-protective effect of Drp1 KD was not altered by these manipulations. This finding demonstrates that respiratory activity is not responsible for the delayed apoptosis in Drp1 KD cells. Of note, triple KD cells repressing Drp1, Mfn1, and Mfn2, in which nucleoid clustering and mito-bulb formation were compromised (Fig. 1 D and F), also lost resistance to apoptosis (Fig. S5D), and these proteins were efficiently repressed by RNAi (Fig. S5C). This finding suggests that nucleoid clustering via mitochondrial fusion leads to an antiapoptotic effect in Drp1 KD cells.
Finally, we characterized proapoptotic intermembrane space (IMS) protein Smac/DIABLO in Drp1 KD cells with and without pretreatment with ddC or CHL. Compared with cyt c release, the effect of Drp1 KD cells was similar or more sensitive to Act D in the release of Smac/DIABLO from mitochondria (Fig. 4E). Interestingly, before treatment with Act D, Smac/DIABLO decorated the edges of mito-bulbs in Drp1 KD cells, but these cells showed no costaining with cyt c inside the mito-bulbs (Fig. 4F), likely reflecting distinct states of intramitochondrial localization of two proapoptotic factors: Smac/DIABLO freely within the IMS and cyt c enclosed within cristae. After 4 h of Act D treatment to Drp1 KD cells, we observed mito-bulbs that had lost Smac/DIABLO but still retained cyt c. In contrast, in Drp1 KD cells, pretreatment with ddC, but not with CHL, restored the release of cyt c but not of Smac/DIABLO. Consistent with these results, Bax activation [leading to mitochondrial membrane permeabilization (MOMP)] by proapoptotic stimuli was not blocked by Drp1-KD or ddC (Fig. S5E). Thus, mtDNA nucleoids play important roles in the remodeling of cristae structures and in the regulation of cyt c release, but do not affect the MOMP process.
Discussion
Mitochondrial fission often occurs even in nongrowing cells, but its physiological significance is incompletely understood. Here we focused on the dynamics and distribution of mitochondrial nucleoids and found them to be clustered in mitochondrial fission-deficient cells, leading to the formation of bulb-like mitochondria (i.e, mito-bulbs) with highly stacked cristae that enclose cyt c and resulting in protection against apoptosis.
Recent analysis using superresolution stimulated emission depletion (STED) microscopy revealed that the nucleoids in human fibroblasts are formed by clusters of small (∼100 nm) uniform structures consisting of a single copy of mtDNA (28). Here we observed further clustering of nucleoids in mitochondrial fission-deficient cells via mitochondrial hyperfusion. We also confirmed previous reports that Drp1 neighbors most nucleoids (3, 5), as does Mff. Furthermore, localization of Mff and Drp1 was affected by the nucleoid structures. These data suggest that the mitochondrial fission occurring close to the nucleoids prevents nucleoid clustering. Of note, recent reports have demonstrated a role for the ER in determining mitochondrial fission sites (23). Interestingly, we also found that nucleoids neighbor the ER, suggesting that mitochondrial nucleoids might have some communication with the ER through mitochondrial fission machinery. It will be of interest to determine how mitochondrial fission (or fusion) factors or ER recognize the mitochondrial nucleoids beyond the mitochondrial double membranes.
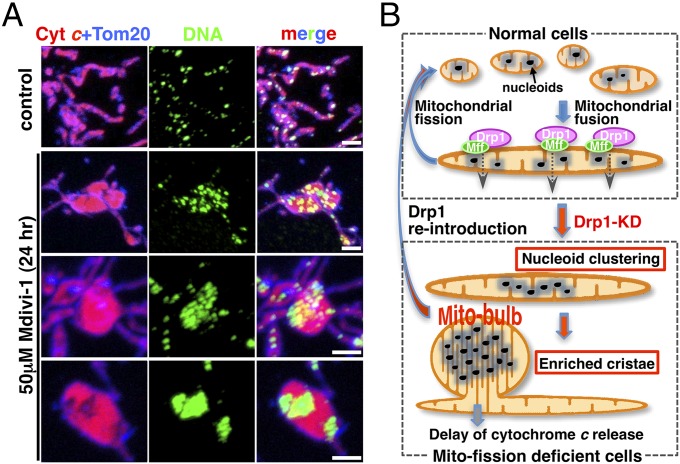
An important question is why nucleoid clustering leads to the formation of mito-bulbs with the development of highly dense IM cristae. We previously observed enlarged mitochondria with tightly stacked cristae in Drp1 KO mice brain (13), suggesting that this phenomenon occurs in vivo. Here we found that mtDNA, but not the respiratory complex/function, is required for mito-bulb formation and cristae reformation. Interestingly, mitochondrial fission inhibitor Mdivi-1 also induced nucleoid clustering and subsequent mito-bulb formation (Fig. 5A). Under these conditions, many intermediate structures containing clustered nucleoids with IM cristae were clearly evident. Because mtDNA is believed to be physically associated with IM (29), nucleoids should be highly clustered with IM cristae in mitochondrial fission-deficient cells, resulting in the formation of cristae-enriched mito-bulb structures (Fig. 5B).
Fig. 5.
Mitochondrial fission regulates nucleoid clustering, mito-bulb formation with IM cristae, and apoptosis. (A) Nucleoids (anti-DNA, green), cyt c (red), and Tom20 (blue) were immunostained in HeLa cells treated with or without Drp1 inhibitor Mdivi-1 for 24 h. (Scale bar: 2 μm.) (B) Schematic representation of our results (Discussion).
Another possibility is that aggregated nucleoids might affect the oligomerization state of mitochondrial ATPase, given that dimerization of the ATPase is reported to correlate with cristae formation (30). It was recently reported that mitochondrial hyperfusion with highly developed cristae occurs during nutrient starvation to protect mitochondria from autophagic degradation and sustained cell viability (31). However, even though mito-bulbs have highly stacked cristae with abundant mtDNA, they do not enhance respiration, and the physiological roles of mito-bulbs in vivo and in healthy cells remain to be conclusively analyzed.
Mitochondrial fission is known to play an important role in the regulation of apoptosis (9, 25). Mitochondria are fragmented in apoptotic cells by Drp1-dependent mitochondrial fission, and Drp1 inactivation affects apoptosis and the release of cyt c from mitochondria (25). How Drp1 regulates release remains unclear, however. Recent in vitro experiments have shown that purified Drp1 stimulates hemifusion of liposomes (32), suggesting that Drp1 directly affects the integrity of OM. However, Drp1 stimulates the release of cyt c after the Bax/Bak-dependent release of Smac/DIABLO, strongly suggesting that Drp1 does not directly regulate MOMP, but does facilitate cyt c release and subsequent apoptosis (13, 26). Here we found that ddC pretreatment overrides the inhibitory effect of Drp1 KD, but not of CHL or respiration inhibitors, in apoptosis, suggesting that nucleoids play unexpected roles in the regulation of cyt c release independent of respiration, although further analysis is needed to exclude the possibility of indirect effects of these reagents. Nucleoid size and mito-bulb formation in MEFs are less affected by Drp1 KO compared with Drp1 KD HeLa cells, which is consistent with previous reports that Drp1 KO MEFs display little or only partial resistance to apoptosis (13, 14). Further analysis is needed to clarify the roles of mitochondrial fission in apoptosis in vivo, considering previous studies suggesting that Drp1 KO neuronal cells are more sensitive to apoptosis, possibly owing to more severe defects in neural functions by Ca2+ signaling (13), and that Drp1 KO Purkinje cells become more sensitive to apoptosis after reactive oxygen species (ROS) generation (15).
Cyt c was previously shown to be enclosed in cristae, which are deformed to release cyt c during apoptosis (33), and Drp1-dependent cristae remodeling is thought to cause this release (34). Using Drp1 KD cells, it is now possible to analyze the detailed submitochondrial distribution in enlarged mito-bulbs under fluorescent microscopy, and we found that two proapoptotic IMS proteins showed distinct distribution patterns, with cyt c inside the cristae, and Smac/DIABLO in the inner boundary area. These distinct topological features reflect the release of proapoptotic factors; that is, the release of Smac/DIABLO by Bax-induced MOMP clearly precedes the release of cyt c facilitated by remodeling of highly stacked cristae. The latter step is regulated by aggregated nucleoids and Drp1-dependent mitochondrial fission.
This report provides fundamental information on the physiological role of mitochondrial fission in controlling mtDNA distribution, which affects IM cristae structures and apoptosis. It will be interesting to determine whether mitochondrial dynamics contribute to the distribution, selection, homeostasis, and inheritance of mtDNA in proliferating cells, differentiating cells, and germ cells in vivo.
Materials and Methods
RNAi and Time-Lapse Microscopy.
For RNAi, siRNAs were transfected by Lipofectamine RNAiMAX (Invitrogen) to cultured HeLa cells according to the manufacturer’s protocols. After 2 d, the cells were transfected again and cultured for a another 2 d. For time-lapse analysis, HeLa cells expressing mitRFP cultured on glass-bottomed dishes were stained with 100,000-fold diluted SYBR Green I (22) for 5 min at 37 °C, washed four times with growth medium, and then changed to fresh growth medium containing 50 mM Hepes buffer (pH 7.4). After 30 min, time-lapse recording was started. Cells were observed under an Olympus IX81 fluorescence microscope or a Zeiss LSM700 confocal microscope, and analyzed using Zeiss ZEN 2010 LSM software or Metamorph software (Molecular Devices). Detained information on materials, immunofluorescence staining, quantitative PCR, and EM is provided in SI Methods.
Supplementary Material
Acknowledgments
This work was supported by Funding Program for Next-Generation World-Leading Researchers (N.I.), the Takeda Science Foundation (N.I.), and the Fukuoka Foundation for Sound Health (R.B.-I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301951110/-/DCSupplemental.
References
- 1.Lang BF, et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387(6632):493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 2.Kuroiwa T. Mechanisms of organelle division and inheritance and their implications regarding the origin of eukaryotic cells. Proc Jpn Acad, Ser B, Phys Biol Sci. 2010;86(5):455–471. doi: 10.2183/pjab.86.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrido N, et al. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell. 2003;14(4):1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legros F, Malka F, Frachon P, Lombès A, Rojo M. Organization and dynamics of human mitochondrial DNA. J Cell Sci. 2004;117(Pt 13):2653–2662. doi: 10.1242/jcs.01134. [DOI] [PubMed] [Google Scholar]
- 5.Iborra FJ, Kimura H, Cook PR. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alam TI, et al. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31(6):1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt IJ, et al. Mammalian mitochondrial nucleoids: Organizing an independently minded genome. Mitochondrion. 2007;7(5):311–321. doi: 10.1016/j.mito.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Kaufman BA, et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18(9):3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McBride HM, Neuspiel M, Wasiak S. Mitochondria: More than just a powerhouse. Curr Biol. 2006;16(14):R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 10.Ishihara N, Otera H, Oka T, Mihara K. 2012. Regulation and physiologic functions of GTPases in mitochondrial fusion and fission in mammals. Antioxid Redox Signal 19(4):389-399.
- 11.Otera H, et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191(6):1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waterham HR, et al. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356(17):1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 13.Ishihara N, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11(8):958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- 14.Wakabayashi J, et al. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186(6):805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kageyama Y, et al. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. J Cell Biol. 2012;197(4):535–551. doi: 10.1083/jcb.201110034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki N, et al. Glom is a novel mitochondrial DNA packaging protein in Physarum polycephalum and causes intense chromatin condensation without suppressing DNA functions. Mol Biol Cell. 2003;14(12):4758–4769. doi: 10.1091/mbc.E03-02-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arimura S, Yamamoto J, Aida GP, Nakazono M, Tsutsumi N. Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc Natl Acad Sci USA. 2004;101(20):7805–7808. doi: 10.1073/pnas.0401077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280(28):26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141(2):280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parone PA, et al. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS ONE. 2008;3(9):e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashley N, Poulton J. Anticancer DNA intercalators cause p53-dependent mitochondrial DNA nucleoid re-modelling. Oncogene. 2009;28(44):3880–3891. doi: 10.1038/onc.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozawa S, Sasaki N. Visualization of mitochondrial nucleoids in living human cells using SYBR Green I. Cytologia (Tokyo) 2009;74:366. [Google Scholar]
- 23.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science. 2011;334(6054):358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Möpert K, et al. Loss of Drp1 function alters OPA1 processing and changes mitochondrial membrane organization. Exp Cell Res. 2009;315(13):2165–2180. doi: 10.1016/j.yexcr.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 25.Frank S, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1(4):515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 26.Parone PA, et al. Inhibiting the mitochondrial fission machinery does not prevent Bax/Bak-dependent apoptosis. Mol Cell Biol. 2006;26(20):7397–7408. doi: 10.1128/MCB.02282-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobson MD, et al. Bcl-2 blocks apoptosis in cells lacking mitochondrial DNA. Nature. 1993;361(6410):365–369. doi: 10.1038/361365a0. [DOI] [PubMed] [Google Scholar]
- 28.Kukat C, et al. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci USA. 2011;108(33):13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopek BG, Shtengel G, Xu CS, Clayton DA, Hess HF. Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc Natl Acad Sci USA. 2012;109(16):6136–6141. doi: 10.1073/pnas.1121558109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paumard P, et al. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21(3):221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13(5):589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montessuit S, et al. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 2010;142(6):889–901. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scorrano L, et al. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev Cell. 2002;2(1):55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- 34.Germain M, et al. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis. EMBO J. 2005;24:1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.