Abstract
Rotaviruses are the single most common cause of fatal and severe childhood diarrheal illness worldwide (>125 million cases annually). Rotavirus shares structural and functional features with many viruses, such as the presence of segmented double-stranded RNA genomes selectively and tightly packed with a conserved number of transcription complexes in icosahedral capsids. Nascent transcripts exit the capsid through 12 channels, but it is unknown whether these channels specialize in specific transcripts or simply act as general exit conduits; a detailed description of this process is needed for understanding viral replication and genomic organization. To this end, we developed a single molecule assay for capturing and identifying transcripts extruded from transcriptionally active viral particles. Our findings support a model in which each channel specializes in extruding transcripts of a specific segment that in turn is linked to a single transcription complex. Our approach can be extended to study other viruses and transcription systems.
Keywords: channel specialization, Reoviridae, single molecule hybridization, single-stranded RNA
Double-stranded RNA (dsRNA) viruses comprise a wide variety of families that vary in genome complexity. These families include Reoviridae with 10–12 genomic segments; Crysoviridae with 4 segments; Cystoviridae with 3 segments; Birnaviridae, Picobirnaviridae, and Partiviridae with 2 segments; and Totiviridae with a single genomic dsRNA. They also vary in their ability to infect diverse hosts from bacteria to humans, yet they share unique features reflecting parallels in their replication; for the Reoviridae, such features include a multicomponent capsid that crosses the host cell membrane and transcription of their dsRNA segments by capsid-attached enzymes. During cell entry, the outer layers of these viruses are lost, while their inner capsids provide a compartment for genome segments (10-12 dsRNAs). Transcript export occurs via channels at the 12 vertices of an icosahedral capsid; although crucial for establishing infection (because the transcripts act as templates for both translation and genomic dsRNA synthesis), the mechanism of transcript export is unclear. In particular, it is unknown whether the nascent transcripts are selectively released through specialized channels, and if so, what the basis of selectivity is.
To address these questions, we studied rotavirus, a major cause of gastroenteritis in infants and children worldwide (1, 2), and a member of the Reoviridae family, which includes many viruses of veterinary and biomedical importance (3). Rotaviruses deliver a 70-nm-diameter double-layered particle (DLP) to host cells following entry; the outer and inner protein layers package transcription complexes (TCs), proteins VP1 (RNA-dependent RNA polymerase) and VP3 (RNA capping enzyme), and 11 dsRNA genomic segments. The outer DLP layer is made of VP6 proteins (4) arranged as pentamers and hexamers forming 132 channels of three classes, including a class of 12 channels, each placed at the fivefold vertices of the icosahedral capsid. Underneath the VP6 layer is the single-layered particle, which comprises VP2 proteins and forms a thin continuous scaffolding layer, except for small pores along the fivefold axis of the icosahedron. These pores are in register with the 12 VP6 channels and serve as RNA exit channels for the newly transcribed positive sense single-stranded RNA (ssRNA). Close to these channels, 12 TCs interact with the viral genomic segments and attach to a hub-like structure formed by VP2 (5, 6). This organization implies that 1 of the 12 TCs could be unoccupied (7–9).
Rotavirus RNA synthesis is thought to be asynchronous and moderately fast [50 nt/s for related orthoreovirus (10)], with some transcripts produced earlier than others (11). It has also been shown that transcripts are released simultaneously through the 12 RNA exit channels (12), but the assignment of specific segments to channels (if any) is unknown, because standard microscopy can only visualize the presence, but not the identity, of the extruded segments per single particle.
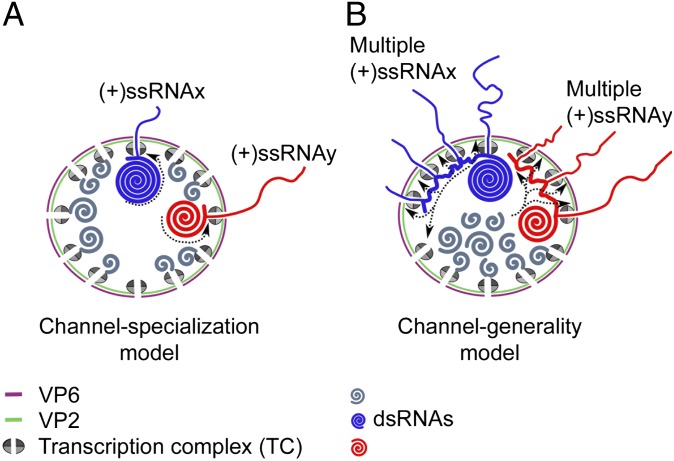
The DLP structural organization is compatible with two mechanistic models for transcript extrusion (Fig. 1). The channel specialization model (Fig. 1A) postulates that each channel is linked to a specific TC and to transcripts of only one specific genomic segment (13); this model predicts that, at any given time, only one transcript of a specific segment can be extruded per channel of a DLP. In contrast, the channel generality model (Fig. 1B) predicts that each channel can extrude transcripts of multiple (perhaps all) genomic segments, but only one at a time. This model also predicts that, at any given time, a genomic segment may interact with several TCs; thus, some particles carry two or more transcripts of a specific segment extruded from two or more channels. Hybrid versions of the two main models are also possible.
Fig. 1.
Models for transcript exit in rotavirus. (A) Channel specialization model: each RNA exit channel is linked to a specific TC and transcripts (thin blue and red lines for segments x and y, respectively) of a single specific genomic segment (thick blue and red lines for segments x and y, respectively). This model predicts that only a single transcript of each viral genomic segment is extruded from each DLP at any given time. (B) Channel generality model: a segment can interact with multiple TCs and their corresponding RNA exit channels during transcription. This model predicts that multiple transcripts from each genomic segment can be extruded from each DLP at any given time.
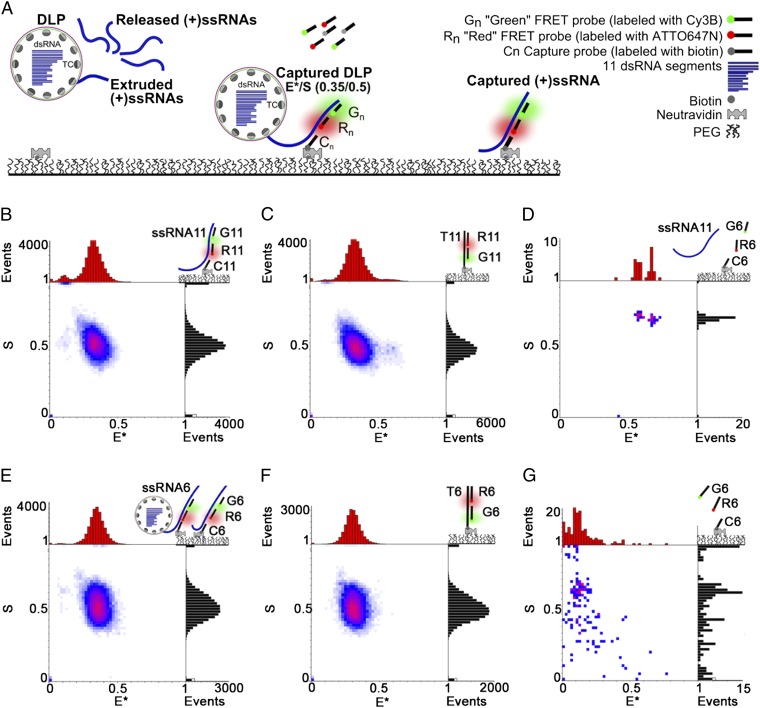
To test these models, we determined the copy number of specific transcripts during their extrusion from transcriptionally active DLPs using a novel single-molecule fluorescence assay for transcript capture and identification (hereafter, CID assay; Fig. 2). CID combines RNA hybridization with single-molecule imaging using alternating-laser excitation (14, 15) on a total internal reflection microscope (TIRF) (16). CID has the advantage of maintaining the transcriptional activity of viral particles, in contrast to approaches that may induce conformational changes in the channels, such as the use of anchor antibodies (17, 18) or labeling methods that target surface amines or cysteines. Our findings support the channel specialization model and exclude alternative models in which an individual segment is transcribed by several complexes and extruded through multiple channels. Our method can be implemented in the sensing of transcripts in other biological model systems.
Fig. 2.
CID detects specific transcripts from transcriptionally active viral particles. (A) Schematic of the CID assay. Transcripts (either attached to DLPs or free) are incubated with complementary capture and FRET probes (donor shown in green; acceptor shown in red), captured to the surface and detected using ALEX-TIRF. Specific transcripts are linked to specific combinations (“codes”) of FRET efficiency E* and relative probe stoichiometry S; in most cases, an E*/S code of 0.35/0.5 is used. (B) The synthetic (+)ssRNA11 using FRET probes (G11 and R11) and capture probe C11 (hybridization strategy, top right) gives a 2D E*/S histogram (n = 1,895 particles) with the expected E*/S code. (C) Positive control using T11-G11-R11, a dsDNA construct made of G11 and R11 hybridized to a complementary biotinylated ssDNA (T11) matching a sequence in (+)ssRNA11. The E*/S histogram (n = 2,043) shows the E*/S code of 0.35/0.5. (D) Negative control does not show a specific signal when incubating (+)ssRNA11 with FRET probes G6, R6 and capture probe C6 targeting (+)ssRNA6. (E) Detection of nascent (+)ssRNA6 transcripts from transcriptionally active DLPs, either while extruded from DLPs or after their release. DLP transcription was allowed to proceed for 2 min in the presence of G6, R6, and C6. E*/S histograms (n = 1,402) clearly show the expected code after 60 min of hybridization. (F) Positive control using construct T6-G6-R6 (designed as in B); the E*/S code for the control matches the signature for transcripts in E. (G) Negative control does not show a specific signal when incubating probes G6, R6, and C6 in the absence of (+)ssRNA6.
Results
RNA Identification Assay Based on Single-Molecule Hybridization.
CID uses three ssDNA probes complementary to sequences close to the 5′ end of a specific transcript: a biotinylated DNA for capturing RNAs on neutravidin-coated surfaces (capture probe C) and two fluorescent DNAs for identifying specific RNAs. The fluorescent DNAs were each labeled using a spectrally distinct fluorophore, with the two fluorophores serving as a donor–acceptor pair for Förster resonance energy transfer (FRET); we refer to these DNA probes as the green (G) and red (R) FRET probes (Fig. 2A). The fluorophore positions were chosen to ensure that target hybridization results in a specific combination of FRET efficiency and stoichiometry, hereafter termed an E*/S code. In most experiments, we used a 0.35/0.5 E*/S code; i.e., a single fully hybridized transcript led to FRET efficiency of 35% (E* ∼ 0.35) and relative probe stoichiometry S of 0.5 (corresponding to a 1:1 donor:acceptor hybrid; see SI Materials and Methods). The probe sequences were varied to target different transcripts (Table S1).
To establish that CID identifies transcripts with high sensitivity and specificity, we targeted a synthetic RNA version of the segment 11 transcript using capture probe C11 and FRET probes G11 and R11 (Fig. 2B; SI Materials and Methods). After hybridization, surface capture, and single-molecule imaging, the E*/S histograms showed a single population of molecules with the designed code of 0.35/0.5 (Fig. 2B). Further experiments to capture synthesized ssRNA11 from RNA transcribed from DLPs also showed the specific E*/S code (Fig. S1).
These results matched exactly those for a positive control, a dsDNA construct (T11-G11-R11) wherein the ssRNA11-specific FRET probes hybridize to a complementary biotinylated DNA (T11) to produce the 0.35/0.5 code (Fig. 2C). As a negative control, we tested for any nonspecific binding of the transcript from segment 11 to a probe set (G6, R6, and C6) complementary to a transcript from segment 6; only two particles were captured, and none with the correct code (Fig. 2D).
We then tested whether CID can detect both extruded and released rotavirus transcripts from transcriptionally active DLPs (11, 13, 19) by monitoring ssRNA6 synthesis after adding nucleotides in mixtures of DLPs carrying probes specific for segment 6 (G6, R6, and C6). After allowing 2 min of transcription, transcripts were hybridized for up to 60 min (Fig. 2E), captured, and imaged. After hybridization for 15 min, ssRNA6 transcripts were clearly visible, matching the positive control (T6-G6-R6; Fig. 2F); the number of detected transcripts increased with hybridization time. All populations had S values of ∼0.5, showing that transcripts do not self-associate to higher-order structures on their release from DLPs; there was also no probe self-association (Fig. 2G). These results showed a high level of transcription from DLPs even for short incubations with nucleotides.
Capture of Extruded Transcripts from DLPs Using CID.
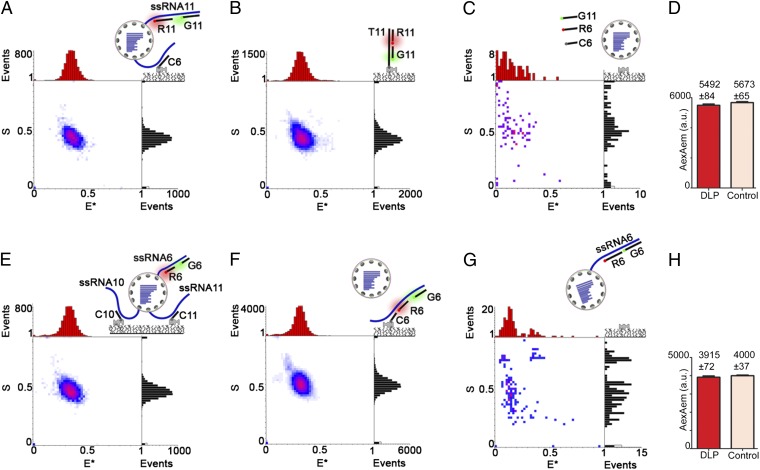
Subsequently, we detected specific transcripts during their extrusion from DLPs (and before their release) by capturing DLPs with transcripts of segments 6 and 11 extruded simultaneously (Fig. 3). To detect such DLPs, we allowed transcription for 1 min (SI Materials and Methods), during which DLPs should produce 10–20 transcripts of each segment [assuming transcription at ∼50 nt/s (9)], with one transcript still attached to a DLP. After fixing the transcribing DLPs (SI Materials and Methods), we hybridized them to an ssRNA6 capture probe and ssRNA11 FRET probes; captured DLPs (311 particles) showed the expected code for a DLP carrying ssRNA11 transcripts (Fig. 3A) and exactly matched the code of the positive control (T11-G11-R11 construct; Fig. 3B). In a negative control (no nucleotides; Fig. 3C), we captured only four molecules, and none with the code. These results showed that DLPs were captured through an extruded ssRNA6 transcript while carrying a yet undetermined number of extruded transcripts of segment 11. However, the S value of ∼0.5 for the main population and the absence of other populations of hybridized particles on the E*/S histogram (Fig. 3A) excluded relative stoichiometries of the two FRET probes other than 1:1 and further suggested the presence of only a single extruded ssRNA11 per DLP. For example, if a significant number of DLPs carried two extruded ssRNA11 transcripts, the bleaching or substoichiometric hybridization for some particles would have resulted in populations with 2:1 or 1:2 G:R stoichiometry and in broadening of the S distribution relative to the 1:1 positive control (Fig. S2). In contrast, the S distribution for the captured DLPs matches exactly that of the 1:1 positive control (cf. black distributions in Fig. 3 A and B).
Fig. 3.
Transcriptionally active DLPs carry a single extruded transcript for segments 6 and 11. (A) CID of actively transcribing DLPs (allowed to transcribe for 2 min) interrogated using probe C6 [which targets extruded (+)ssRNA6] and probes G11 and R11 [which target extruded (+)ssRNA11]. The E*/S histogram (n = 311) shows an E*/S population of 0.35/0.5, matching that of the positive control (B). (B) Positive control. The E*/S histogram (n = 640) for construct T11-G11-R11 (Fig. 2C) shows the expected E*/S code of 0.35/0.5. (C) Negative control. E*/S histogram (n = 4) from DLPs incubated under the same experimental conditions but without nucleotides shows no specific signal. (D) Comparison of AexAem fluorescence intensities (mean ± SEM from experiments in A and B). Hybridized DLPs, red bar; positive control using T11-G11-R11, pink bar. A t-test comparison between the results shows no significant differences in the mean (P > 0.10). (E) CID of actively transcribing DLPs (allowed to transcribe for 2 min) interrogated using probes C10 and C11 [which target extruded (+)ssRNA10 and (+)ssRNA11, respectively] and probes G6 and R6 [which target extruded (+)ssRNA6]. The E*/S histogram (n = 322) shows an E*/S population of 0.35/0.5, matching that of the positive control (F). (F) Positive control. The E*/S histogram (n = 1,832) for captured (+)ssRNA6 (synthesized under the same conditions as in E and detected using C6, G6, and R6) shows the expected E*/S code of 0.35/0.5. (G) Negative control performed with FRET probes but in the absence of a capture probe do not show any specific hybridization (n = 8). (H) Comparison of AexAem fluorescence intensities (mean ± SEM from experiments in E and F). Hybridized DLPs, red bar; positive control using (+)ssRNA6, pink bar. A t-test comparison between the results shows no significant differences in the mean (P > 0.35).
To confirm the presence of a single extruded transcript of ssRNA11 per DLP, we compared the DLP-based acceptor emission intensity on acceptor excitation (AexAem, a FRET-independent measure of the number of acceptor fluorophores per particle; using data from Fig. 3A) with that of the positive control (T11-G11-R11 construct, with an absolute G:R stoichiometry of 1:1; Fig. 3B). For a single extruded transcript per DLP, the mean AexAem intensity for the captured DLPs should match that of the 1:1 control; however, if the DLPs carry two (or more) extruded transcripts for ssRNA11, the mean AexAem for DLPs should be twofold higher (or more) than that of the 1:1 control (see Fig. S2 C and G for the AexAem distribution for standards with G:R stoichiometries of 1:1 and 1:2, respectively; also see Fig. S2J for the mean AexAem of the same standards). Our results showed no differences between the mean AexAem intensity per captured DLP and the control (Fig. 3D; 5,492 ± 84 counts for DLPs; 5,673 ± 65 for the 1:1 control), leading us to conclude that only a single ssRNA11 transcript was present on the surface of each DLP during transcription.
To examine whether the extrusion of a single transcript per segment on transcribing DLPs applied to other segments, we targeted DLPs by swapping the capture and FRET probes used previously (i.e., we used a capture probe for ssRNA11 and FRET probes for ssRNA6). We performed the hybridization under the same conditions as those carried out to capture ssRNA6. To improve DLP capture, we used an additional capture probe (C10) targeting ssRNA10; the results also showed the same ssRNA6-specific signal (Fig. 3E; 322 particles). The presence of a single capture probe (C11) under these hybridization conditions resulted in a specific signal but less efficient capture of transcripts (38 particles; Fig. S3). The signal specificity was supported by comparisons with the positive control (matching the signal for released ssRNA6; Fig. 3F; 1,832 particles) and the negative control (8 particles showing no specific signal without capture probes; Fig. 3G). Finally, as with ssRNA11 (Fig. 3H), the mean AexAem intensity for probe R6 hybridized in DLPs matched that of the positive control (3,915 ± 72 counts for DLPs; 4,000 ± 37 for the 1:1 control), suggesting that the captured DLPs carry only a single extruded ssRNA6 transcript. Additional experiments aimed at the detection of a third rotavirus segment (ssRNA2) also showed the extrusion of a single transcript (Fig. S4). These results establish that, for segments 2, 6, and 11, at most one single transcript of each of these rotavirus genes is being extruded from a DLP at any given time, and argue for a general mechanism in which at most one single transcript of any segment is synthesized and released from a DLP at any given time.
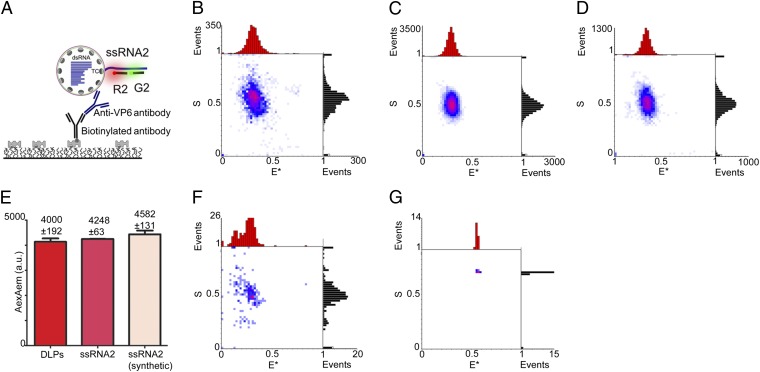
To confirm that the detected ssRNAs colocalize with DLPs, we performed CID experiments using antibodies for DLP capture (Fig. 4) instead of DNA capture probes. Specifically, we captured transcribing DLPs using a sheep antibody targeting VP6 (the protein covering the DLP surface with 780 copies) and a rabbit biotinylated antibody against sheep IgG; the DLPs were probed for the presence of extruded ssRNA2 (Fig. 4A). The E*/S histogram for the captured DLPs (Fig. 4B) displays a single population (n = 120) with the ssRNA2-specific E*/S code of ∼0.35/0.5, thus matching the results for DLPs captured using DNA-based capture probes (Fig. S4). This single population matches the E*/S populations in two positive controls: probed ssRNA2 released from DLPs (Fig. 4C) and probed synthetic ssRNA2 (Fig. 4D), both captured using the C2 probe. Furthermore, the mean AexAem intensity for probe R2 hybridized on antibody-captured DLPs matches those of the positive controls (Fig. 4E), showing that only a single ssRNA2 is being extruded from a DLP at a given time.
Fig. 4.
Antibody-captured transcriptionally active DLPs carry a single extruded transcript for segment 2. (A) Schematic of the antibody-based CID assay. Actively transcribing DLPs (allowed to transcribe for 2 min) were interrogated using FRET probes G2 and R2 [which target (+)ssRNA2]. After incubation with a VP6-specific sheep antibody, the DLPs were added to a coverslip coated with a biotinylated rabbit anti-sheep antibody. For details, see SI Materials and Methods. (B) CID of antibody-captured actively transcribing DLPs interrogated using probes G2 and R2. The E*/S histogram (n = 120) shows a single E*/S population of 0.35/0.5, matching the main populations of the positive controls (C and D). (C) Positive control. The E*/S histogram (n = 879) for (+)ssRNA2 transcripts captured from transcribing DLPs using FRET probes R2, G2 and capture probe C2 shows a single E*/S population of 0.35/0.5. (D) Positive control. The E*/S histogram (n = 414) for synthetic (+)ssRNA2 transcripts captured using the same probes as in C shows a single E*/S population of 0.35/0.5. (E) Pairwise comparison of the AexAem fluorescence intensities (mean ± SEM) from experiments in B (red bar), C (dark pink bar), and D (light pink bar). A one-way ANOVA using a Tukey’s test shows no significant differences between DLPs and the ssRNA control (P > 0.05), supporting that only one ssRNA2 is extruded from the DLP at any given time. (F) Negative control. A CID assay performed as in A and B, but with the primary antibody being substituted by rabbit IgG (which does not bind to VP6). The E*/S histogram shows only few particles (n = 12) with an E*/S population of 0.35/0.5. (G) Negative control. A CID assay performed as in A and B but in the absence of a secondary antibody. Only a single particle is detected on the surface.
We also performed two negative controls. First, we substituted the VP6-specific antibody with rabbit γ-globulins and found only 12 colocalizations (some with the right code, possibly due to nonspecific interactions between DLPs and γ-globulins). Second, we performed immune-capture in the absence of the biotinylated secondary antibody; this pulled down only one particle and it did not have the correct E*/S code. These results confirmed that the vast majority of probed ssRNA molecules are attached to transcriptionally active surface-captured DLPs.
Actively Transcribing DLPs Carry Single Extruded Transcripts of Segments 6 and 11.
To confirm that specific transcripts are extruded through single channels, we modified our CID assay to directly observe colocalization of ssRNA6 and ssRNA11 transcripts at the single DLP level (Fig. S5). This modification was achieved by using an ssRNA6-specific probe pair (capture probe C6 and reporter probe G6) and an ssRNA11-specific probe pair (capture probe C11 and reporter probe R11). If our predictions were correct, the probing of actively transcribing DLPs using these four probes should capture particles with an E*/S value of 0.1/0.5, because independent transcripts extruded through independent channels should be beyond FRET range (>10 nm; the E* of ∼0.1 is due to cross-talk between emission channels; SI Materials and Methods). Indeed, the DLP sample (357 particles) showed E*/S values of ∼0.1/0.5 (Fig. S5A). Due to the nature of the sample (which contains both DLPs with extruded RNAs and released RNAs), the colocalization is in part due to captured DLPs and in part due to random coincidence of released transcripts. The degree of ssRNA6–ssRNA11 colocalization for the DLP sample was higher (P = 0.0004; Fig. S5B) than what was expected on the basis of random coincidence (SI Materials and Methods), supporting the presence of significant specific transcript colocalization on the captured DLPs. These colocalization experiments provided an opportunity to test the hypothesis that rotavirus transcripts can associate independently and guide sequence assortment during encapsidation (7). We thus used our CID assay to test for such an association between ssRNA6 and ssRNA11. We synthesized all transcripts (preparing a “total ssRNA” sample) by DLP transcription, followed by DLP removal (Fig. S5B). The number of colocalizations for total ssRNA was then compared with the colocalizations expected due to random coincidence; the absence of any statistically significant difference further supported that the DLP-dependent colocalisation arises due to specific colocalization of transcripts on DLPs as opposed to colocalization due to capture of higher-order complexes of transcripts; we also conclude that, under our conditions, there is no detectable direct or indirect (within a higher-order self-assembled structure) association of ssRNA6 and ssRNA11.
Discussion
In this study, we introduce a CID assay for studying the synthesis of specific transcripts from rotavirus particles. The assay has a simple design and allows unambiguous identification and quantification of specific ssRNAs. We implemented the CID assay to study the extrusion and release of specific transcripts from rotavirus particles and characterized the specialization of the transcript exit channels for two specific segments.
CID Assay.
The assay is based on a coding strategy that relies on two single-molecule fluorescence ratios (E* and S, forming the E*/S code) to identify RNA targets with high specificity and very low false positives, a task difficult for standard hybridization techniques that are often hampered by nonspecific hybridization due to use of either multiple short probes or very long probes. The CID assay circumvents the need for complex multicolor labeling that may complicate signal interpretation due to cross-talk between overlapping emission spectra. Although the DLP capture is indirect (because it mainly depends on interactions with the transcripts rather than the capsid itself), it has the advantage of maintaining the transcriptional activity of DLPs and the structure of the exit channels. The CID assay can also be applied to other Reoviridae members, RNA viruses, and transcription systems; its applicability will also increase as single-molecule fluorescence microscopes become more robust, compact, and accessible to biology laboratories.
Channel Specialization in Rotavirus.
Our CID studies of three rotavirus transcripts that differ in sequence, type of proteins encoded, and length (S2, 2,691 nt; S6: 1,356 nt; S11: 667 nt, representing long, intermediate length, and short segments, respectively) provide unambiguous evidence that rotavirus channels specialize in extruding specific transcripts from DLPs. Our results provide direct evidence for the channel specialization model, which is also supported by the fact that ssRNA TCs are thought to form before replication within the capsid and that RNA TCs are very tightly packed within the viral particles (9, 20). Furthermore, our results, along with studies showing that RNA segments interact with unique TCs close to an exit channel (21, 22) while tightly packed in a semicrystalline state (23), preclude the possibility that extrusion of a specific transcript occurs through different channels each time.
These RNA–TC interactions, mediated through the VP1 component of TC, may guide packaging of RNA segments in the capsid; the importance of these interactions suggests that our observations on ssRNA2, ssRNA6, and ssRNA11 are likely to reflect the behavior of all rotavirus transcripts. Our results, along with structural studies, also support the suggestion that 1 of the 12 TCs is not engaged with an RNA segment.
Our results also agree with the capsid organization suggested in studies of bluetongue virus (24), another member of the Reoviridae family, which showed that the virus recruits a complete set of 10 RNA segments in the infectious capsid; notably, the absence of even a single ssRNA abrogated recruitment of the segments in the inner layered particle. The same report also suggested a hierarchy in the uncapped ssRNAs that leads to the formation of an RNA scaffold with a conserved set of RNA molecules per capsid. Although the mechanism by which rotavirus packages exactly 11 segments is not understood, it is thought that cis-acting sequences in the transcripts drive this process (7), as shown in influenza virus, which has eight negative-strand ssRNA segments and where it was shown (using FISH) that there is selective packaging driven by cis-acting regions located at the segment termini (25). In rotavirus, in silico modeling revealed the presence of regions with codon conservation that may act as structural motifs of cis-acting sequences (26). RNA sequences within viral segments are predicted to form conserved long-range interactions, stem loops, and codon conservation, but also dynamic ssRNA structures that may form intermolecular interactions or bind proteins to accommodate the segments in the confined space of the capsid. Under our experimental conditions, no evidence of such a scaffold was observed for total ssRNA (although we cannot discard the possibility of smaller oligomeric subunits). One possibility is that the assembly requires different reaction conditions. For example, our experiments were performed in the absence of ancillary proteins present in the DLP that may stabilize or unveil cryptic binding sites in the ssRNAs. Furthermore, the low ssRNA concentration used might have been unable to drive the formation of RNA–RNA interactions that can take place in the viroplasm (where the local RNA concentration may be very high). Further insight on the assortment process should arise from future studies on the RNA sequence, structures, or interactions that drive oligomerization; such studies will no doubt benefit from the CID assay.
Materials and Methods
Standard techniques for DNA labeling, synthesis of RNA transcripts, and DLP purification were used and are described in detail in SI Materials and Methods. The capture of transcripts in solution and extruded on the DLPs is described in SI Materials and Methods. Single molecule experiments are described in SI Materials and Methods. Details and procedures for data analysis are also presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
J.P. was funded by a UK Medical Research Council studentship. J.N.M.P. was funded by a UK Engineering and Physical Sciences Research Council studentship through the Oxford Doctoral Training Center at the Life Sciences Interface. C.C. and P.R. were funded by the Wellcome Trust (WT093007). A.N.K. was supported by the European Commission Seventh Framework Program (Grant FP7/2007-2013 HEALTH-F4-2008-201418), the UK Biotechnology and Biological Research Council (Grant BB/H01795X/1), and the European Research Council (Starter Grant 261227).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220345110/-/DCSupplemental.
References
- 1.Lawton JA, Estes MK, Prasad BV. Mechanism of genome transcription in segmented dsRNA viruses. Adv Virus Res. 2000;55:185–229. doi: 10.1016/S0065-3527(00)55004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tate JE, et al. WHO-coordinated Global Rotavirus Surveillance Network 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: A systematic review and meta-analysis. Lancet Infect Dis. 2012;12(2):136–141. doi: 10.1016/S1473-3099(11)70253-5. [DOI] [PubMed] [Google Scholar]
- 3.Trask SD, McDonald SM, Patton JT. Structural insights into the coupling of virion assembly and rotavirus replication. Nat Rev Microbiol. 2012;10(3):165–177. doi: 10.1038/nrmicro2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathieu M, et al. Atomic structure of the major capsid protein of rotavirus: Implications for the architecture of the virion. EMBO J. 2001;20(7):1485–1497. doi: 10.1093/emboj/20.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen D, Luongo CL, Nibert ML, Patton JT. Rotavirus open cores catalyze 5′-capping and methylation of exogenous RNA: Evidence that VP3 is a methyltransferase. Virology. 1999;265(1):120–130. doi: 10.1006/viro.1999.0029. [DOI] [PubMed] [Google Scholar]
- 6.Prasad BV, et al. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382(6590):471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 7.Trask SD, Ogden KM, Patton JT. Interactions among capsid proteins orchestrate rotavirus particle functions. Curr Opin Virol. 2012;2(4):373–379. doi: 10.1016/j.coviro.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. Atomic model of an infectious rotavirus particle. EMBO J. 2011;30(2):408–416. doi: 10.1038/emboj.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estrozi LF, et al. Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles. J Mol Biol. 2013;425(1):124–132. doi: 10.1016/j.jmb.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayaram H, Estes MK, Prasad BV. Emerging themes in rotavirus cell entry, genome organization, transcription and replication. Virus Res. 2004;101(1):67–81. doi: 10.1016/j.virusres.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Johnson MA, McCrae MA. Molecular biology of rotaviruses. VIII. Quantitative analysis of regulation of gene expression during virus replication. J Virol. 1989;63(5):2048–2055. doi: 10.1128/jvi.63.5.2048-2055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawton JA, Estes MK, Prasad BV. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4(2):118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 13.Guglielmi KM, McDonald SM, Patton JT. Mechanism of intraparticle synthesis of the rotavirus double-stranded RNA genome. J Biol Chem. 2010;285(24):18123–18128. doi: 10.1074/jbc.R110.117671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapanidis AN, et al. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci USA. 2004;101(24):8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapanidis AN, et al. Alternating-laser excitation of single molecules. Acc Chem Res. 2005;38(7):523–533. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- 16.Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2001;2(11):764–774. doi: 10.1034/j.1600-0854.2001.21104.x. [DOI] [PubMed] [Google Scholar]
- 17.Lawton JA, Estes MK, Prasad BV. Comparative structural analysis of transcriptionally competent and incompetent rotavirus-antibody complexes. Proc Natl Acad Sci USA. 1999;96(10):5428–5433. doi: 10.1073/pnas.96.10.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thouvenin E, et al. Antibody inhibition of the transcriptase activity of the rotavirus DLP: A structural view. J Mol Biol. 2001;307(1):161–172. doi: 10.1006/jmbi.2000.4479. [DOI] [PubMed] [Google Scholar]
- 19.Mason BB, Graham DY, Estes MK. In vitro transcription and translation of simian rotavirus SA11 gene products. J Virol. 1980;33(3):1111–1121. doi: 10.1128/jvi.33.3.1111-1121.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pesavento JB, Lawton JA, Estes ME, Venkataram Prasad BV. The reversible condensation and expansion of the rotavirus genome. Proc Natl Acad Sci USA. 2001;98(4):1381–1386. doi: 10.1073/pnas.98.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClain B, Settembre E, Temple BR, Bellamy AR, Harrison SC. X-ray crystal structure of the rotavirus inner capsid particle at 3.8 A resolution. J Mol Biol. 2010;397(2):587–599. doi: 10.1016/j.jmb.2010.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey JD, Bellamy AR, Earnshaw WC, Schutt C. Biophysical studies of reovirus type 3. IV. Low-angle x-ray diffraction studies. Virology. 1981;112(1):240–249. doi: 10.1016/0042-6822(81)90629-2. [DOI] [PubMed] [Google Scholar]
- 23.Gouet P, et al. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell. 1999;97(4):481–490. doi: 10.1016/s0092-8674(00)80758-8. [DOI] [PubMed] [Google Scholar]
- 24.Lourenco S, Roy P. In vitro reconstitution of Bluetongue virus infectious cores. Proc Natl Acad Sci USA. 2011;108(33):13746–13751. doi: 10.1073/pnas.1108667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou YY, et al. One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proc Natl Acad Sci USA. 2012;109(23):9101–9106. doi: 10.1073/pnas.1206069109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, et al. Genomic analysis of codon, sequence and structural conservation with selective biochemical-structure mapping reveals highly conserved and dynamic structures in rotavirus RNAs with potential cis-acting functions. Nucleic Acids Res. 2010;38(21):7718–7735. doi: 10.1093/nar/gkq663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.