Abstract
Liquid–liquid phase transitions in complex mixtures of proteins and other molecules produce crowded compartments supporting in vitro transcription and translation. We developed a method based on picoliter water-in-oil droplets to induce coacervation in Escherichia coli cell lysate and follow gene expression under crowded and noncrowded conditions. Coacervation creates an artificial cell-like environment in which the rate of mRNA production is increased significantly. Fits to the measured transcription rates show a two orders of magnitude larger binding constant between DNA and T7 RNA polymerase, and five to six times larger rate constant for transcription in crowded environments, strikingly similar to in vivo rates. The effect of crowding on interactions and kinetics of the fundamental machinery of gene expression has a direct impact on our understanding of biochemical networks in vivo. Moreover, our results show the intrinsic potential of cellular components to facilitate macromolecular organization into membrane-free compartments by phase separation.
Keywords: microdroplets, macromolecular crowding
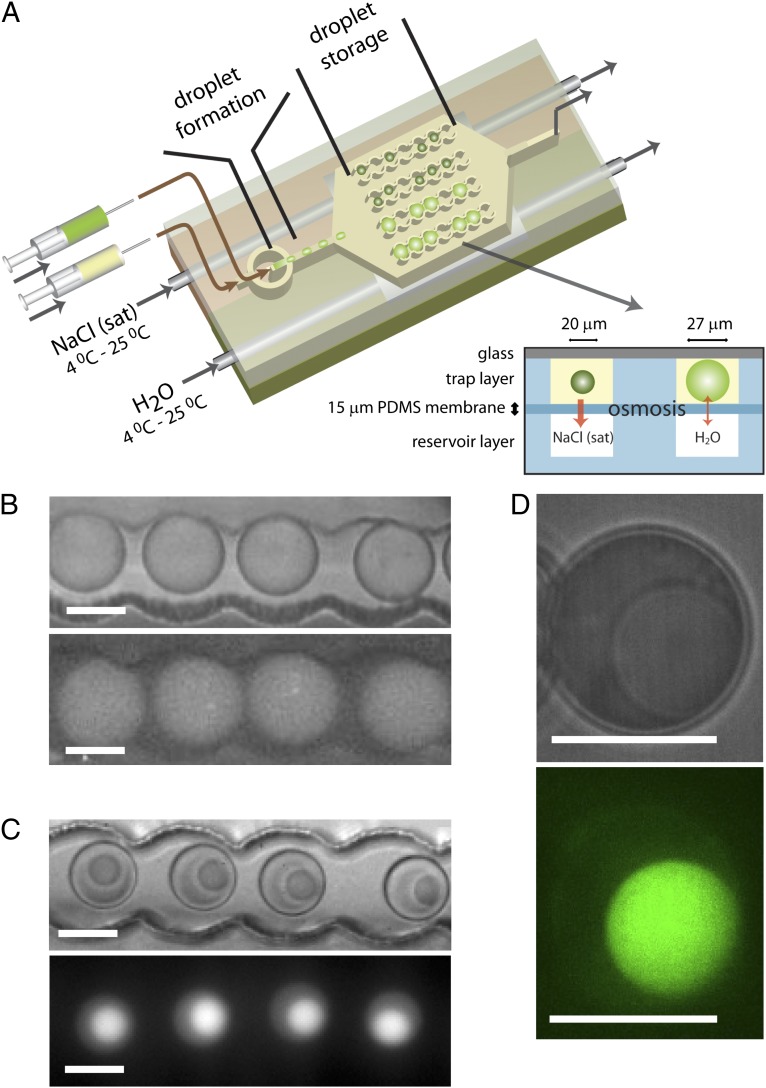
Protocells are minimal compartmentalized systems exhibiting key characteristics of cellular function, including metabolism and replication (1, 2). Lipid vesicles are considered the prototypical protocell as they can form functional microscopic spherical assemblies suited for in vitro gene expression (3, 4). Compartmentalization via lipid bilayers is considered essential for the emergence of cells (4), but there are alternative models based on liquid–liquid phase transitions that lead to the emergence of compartments (5, 6). Compartmentalization is but one characteristic, as protocells ideally also mimic the highly crowded interior of living cells, which have total macromolecule concentrations in excess of 300 g/L (7). Examples in which compartmentalization and high local concentrations are obtained concurrently, include DNA brushes (8), aqueous two-phase systems (9), and liquid coacervates (10). Phase separation or coacervation occurs in a wide range of polymer and protein solutions, often triggered by changes in temperature or salt concentration, or by the addition of coacervating agents (11). The (complex) coacervate droplets that are formed in such systems present macromolecularly crowded, aqueous, physical compartments, 1–100 μm in diameter (12). Recent work has identified similar liquid phase transitions in vivo in the formation of intracellular non–membrane-bound compartments exhibiting liquid-like properties, slowed down diffusion, and strongly interacting macromolecular components (13, 14). Well-studied examples are the intracellular localization of DNA or RNA and proteins in Cajal bodies, P granules, and nucleoli (15–17), which can contain over 100 components. Such complexity has not been achieved in two-phase systems in vitro (18, 19). Although the physics of coacervates is well understood, progress in their development as protocell models has stalled, because of the lack of demonstrations of complex biochemical processes inside coacervates and the small number of different biochemical components involved. Here, we demonstrate the formation of crowded coacervate compartments composed of cell lysate and show how crowding affects the kinetics of key cellular processes inside the coacervates. We took advantage of microfluidics technology to form monodisperse picoliter water-in-oil droplets (20), which allow for reactions to be studied systematically under precisely controlled conditions (SI Text, S1–S5 and Fig. S1). Among the most important complex sets of reactions in the cell is protein synthesis (the result of transcription and translation), a process that functions in vitro despite almost two orders of magnitude lower total protein concentration than found in vivo. We and others have previously shown how droplet-based microfluidic devices can be used to study the kinetics of in vitro expression of a reporter protein using commercial in vitro transcription and translation kits (21–23). The volume of droplets can be controlled via osmotic transport of water to and from reservoir channels filled with concentrated salt solutions (24, 25) and separated from the droplet traps (25) by a thin (15-μm) polydimethylsiloxane (PDMS) membrane (Fig. S1F). Here, we have adapted this approach to raise the salt concentration and concentrate the contents in the droplets, and thus induce coacervation, via controlled withdrawal of water from water-in-oil droplets that are trapped inside microfluidic channels made out of PDMS (Fig. 1A). This approach allows us to study monodisperse droplets (27-μm diameter, 10.3 pL) with identical starting compositions (Escherichia coli cell-free expression kit and plasmid DNA for GFP production; see SI Text, S6–S8 and Figs. S1 and S2 for experimental details) but with different final volumes simultaneously, as droplets in osmotic contact with the saturated NaCl reservoir shrunk from 27 to 20 μm within 20 min. When following the production of GFP using fluorescence microscopy, we find that in the shrunk droplets a second, much smaller (∼13 µm in diameter), highly fluorescent, liquid droplet appeared, before significant fluorescence due to GFP production was observed in the nonshrinking droplets (Fig. 1 B and C).
Fig. 1.
Phase separation in picoliter droplet by controlled osmotic shrinkage. (A) Schematic drawing of the microfluidic device in which two droplet populations with identical contents coexist: nonshrunk, homogeneous droplets, and shrunk, phase-separated droplets. (B and C) Optical microscopy images (Upper) and fluorescence images (Lower) of nonshrunk, homogeneous droplets (B) and shrunk, phase-separated droplets (C) in droplets traps. (D) Zoomed optical microscopy (Upper) and false-color confocal microscopy (Lower) image of a phase-separated droplet, showing that the coacervate is homogeneous on length scales down to the resolution of the microscope. (All scale bars: 20 μm.)
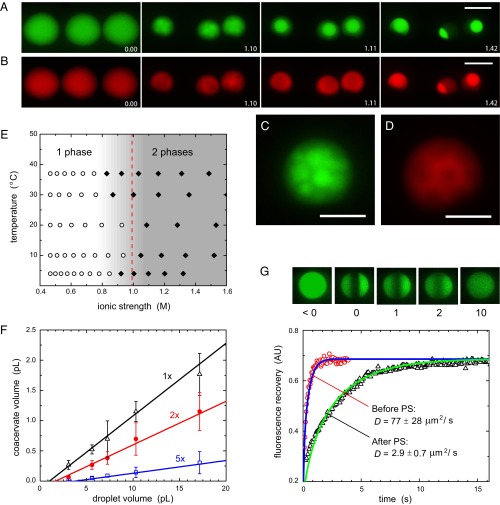
We first investigated the origin and properties of this coacervate droplet. In home-made lysate (SI Text, S9) (26), we did not observe the emergence of two phases, unless small amounts (∼2 wt%) of PEG 8000 were added, similar to the presence of PEG in commercial cell-free expression kits (Fig. S3A). PEG is known to form aqueous two-phase systems in the presence of certain inorganic salts (27). To elucidate the role of PEG in the coacervation process, we covalently labeled all proteins in commercial cell lysate with DyLight 550 and added 0.1 wt% additional fluorescein-labeled PEG 8000 (SI Text, S10). Upon shrinking the droplets, we first observed the formation of small PEG-rich droplets, which subsequently merged to form a single large coacervate (Fig. 2 A–D and Movie S1). The coacervates we observed in stained or nonstained lysates had volumes 8–20 times smaller than the initially generated droplets, depending on the exact composition of the starting mixture (Table S1). The (fluorescently labeled) cell lysate then accumulated in the liquid coacervate, as demonstrated by the rapid increase of DyLight 550 fluorescence intensity in the coacervate. Upon further incubation, more and more macromolecules partitioned into the coacervate and eventually 75% of the protein content of the lysate and 78% of the PEG was compartmentalized in the coacervate, corresponding to a total macromolecule concentration of 375 g/L, of which 290 g/L is PEG 8000 and 85 g/L are lysate proteins (SI Text, S10). In contrast, the total macromolecule concentrations of the dilute phase is 45 g/L, of which 33 g/L is PEG 8000 and 12 g/L are lysate proteins. In a complementary experiment in which we spiked the same, commercial, nonlabeled cell lysate with recombinant GFP (rGFP), we observed that 81% of the rGFP molecules partitioned into the coacervate, which shows that partitioning is not driven by the fluorescent labeling (SI Text, S11 and Fig. S3B). Interestingly, the coacervation of fluorescently labeled PEG 8000 in buffer alone, took place at an internal salt concentration around 1.9 M, leading to coacervate droplets that contain ∼180 g/L PEG (SI Text, S12 and Fig. S3 C and D). The coacervate and “dilute” phase are osmotically balanced, and as the coacervate contains most of the macromolecules (PEG and proteins combined) we expect the dilute phase to be enriched in salts, in agreement with classical examples of liquid–liquid phase separation in PEG–salt mixtures (28). Measurements of the concentrations of various salts using inductively coupled plasma-optical emission spectrometry (SI Text, S13 and Table S2) confirms that salt concentrations in the coacervate are a little lower (∼1.06 M for K+, Na+, and Mg2+ combined, compared with 1.22 M in the dilute phase), which means that 77% of the salts remain in the dilute phase. We also measured DNA distribution over the phase separated droplets by covalently attaching a dye to plasmid DNA (SI Text, S14). We found that a small amount of DNA adsorbed to the oil–water interface (Fig. S4 A and B) and, somewhat unexpectedly, no strong preference of the DNA for either phase [54% vs. 46% in the dilute and coacervate phases, respectively (Fig. S4C)].
Fig. 2.
Coacervates are cell-like compartments. (A and B) Process of coacervation in shrinking droplets showing the concentrations of PEG (A) and DyLight 550-stained cell lysate (B) by false-color fluorescence microscopy. (Scale bars: 20 μm.) The labels indicate time (h:mm). (C and D) Zoomed images of the nucleation the coacervate for PEG (C) and cell lysate (D). (Scale bars: 10 μm.) (E) Phase diagram of cell-free expression kit in droplets as a function of ionic strength and temperature. The open symbols represent single-phase droplets, and the closed symbols represent phase-separated droplets. (F) The volume of the coacervate depends linearly on the volume of the original droplet and scales with the concentration of the cell-free expression kit. Different datasets correspond to different dilutions (1×, 2×, 5×) of the kit before droplet formation. The solid lines are linear fits of the data. (G) Fluorescence recovery of eGFP in single-phase droplets and coacervates after photobleaching. The solid lines are fits of the recovery curves to a 1D diffusion problem (see SI Text, S15 for details). Small images are false-color confocal microscopy images of coacervates, taken at times as indicated by the labels (in seconds).
The characterization of the coacervates reveals that combination of E. coli lysate and PEG has a remarkable effect on the distribution of the biologically active components. This system presents a unique example of a salt-induced phase separation in a complex mixture where most components partition in a single crowded coacervate compartment. Such a process could also drive the formation of nucleoli and other complex non–membrane-bound intracellular compartments, where coacervation is driven by interaction between multivalent proteins, followed by accumulation of other classes of proteins via selective interactions or physical properties (5). We find that the phase separation of droplets from the commercial gene expression kit is triggered at an internal salt concentration in the range of 1.0 M, with little dependence on the temperature at which the controlled evaporation is carried out (Fig. 2E). Coacervate droplets formed from the kit have an 8- to 10-fold smaller volume than the initial droplets (Table S1), corresponding to a total macromolecule concentration of ∼200 g/L, of which ∼155 g/L is PEG 8000 and ∼45 g/L are lysate proteins, assuming the partition coefficients determined in the above-mentioned experiment using fluorescently labeled PEG and proteins (SI Text, S10). The difference between these values and those for coacervates containing fluorescently labeled lysate proteins and PEG is likely due to the presence of dye leading to the formation of smaller coacervates (Table S1). There is a clear correlation between coacervate volume and amount of lysate in the initial droplet (Fig. 2F). Fluorescence recovery after photobleaching experiments probing the diffusion of two variants of GFP [enhanced GFP (eGFP) and rGFP] added to the coacervates (see SI Text, S15 for experimental details) confirm that the coacervate is a liquid phase with a density and viscosity strongly resembling the environment within living cells (Fig. 2G). We find a diffusion coefficient of 2.9 ± 0.7 μm2⋅s−1 for eGFP (1.4 ± 0.5 μm2⋅s−1 for rGFP) compared with 3.6 ± 0.7–7.7 ± 2.5 μm2⋅s−1 (depending on the levels of expression) in E. coli (29). Before phase separation, the diffusion coefficient in the dilute lysate is around 77 ± 28 μm2⋅s−1 (59 ± 24 μm2⋅s−1 for rGFP) compared with 87 μm2⋅s−1 in pure water.
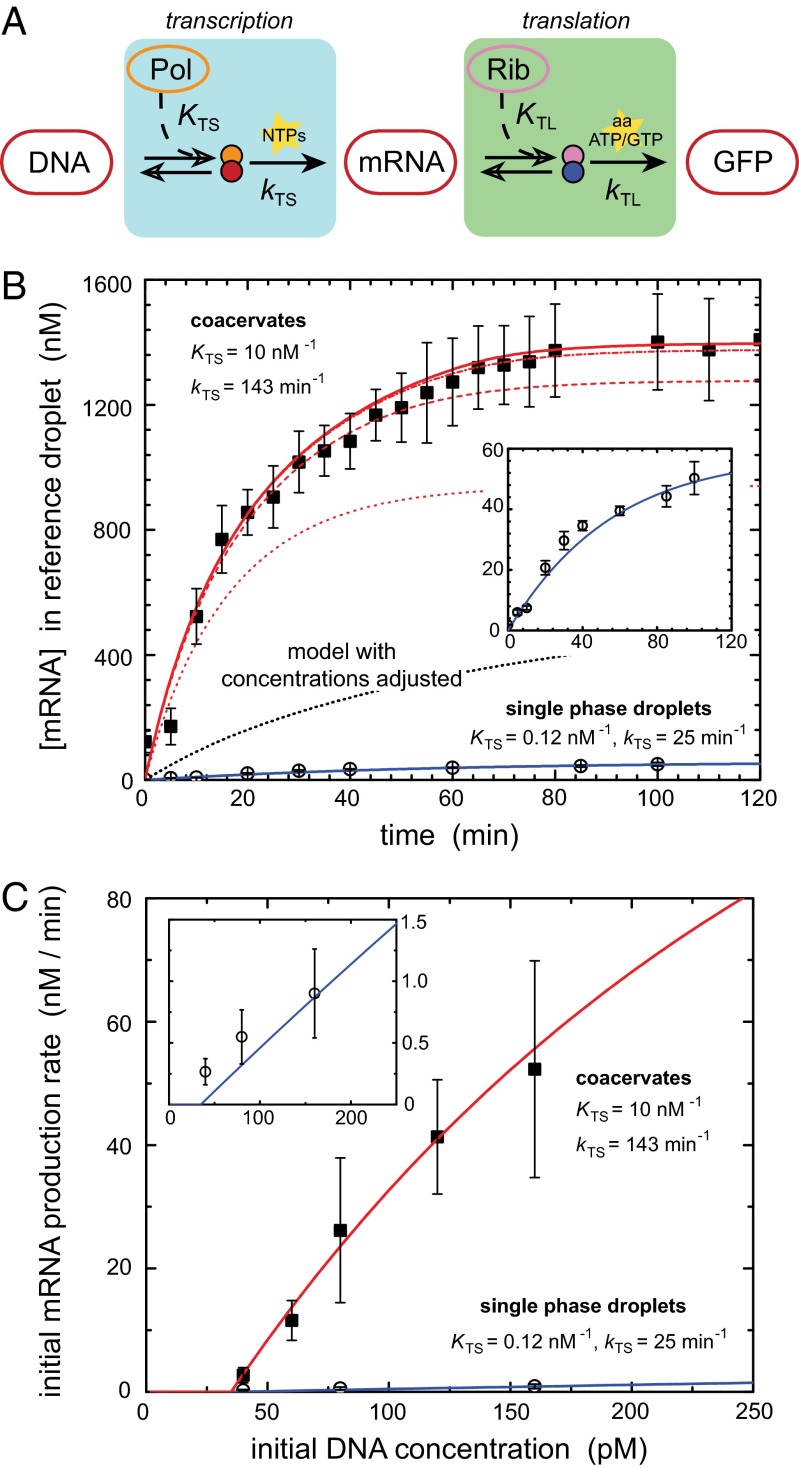
We have shown that the coacervates are crowded compartments, which, despite the large changes in buffer and lysate concentrations during droplet shrinking and phase transition, form a functional transcription and translation compartment as evidenced by the observed production of GFP. To directly compare mRNA and protein synthesis rates in single-phase droplets and in the more crowded coacervates, we proceed as follows: droplets were formed at 4 °C to prevent transcription and translation, and only after phase separation and partitioning of the cell lysate and the DNA (typically after 40 min), the temperature was raised to 25 °C and transcription and translation were initiated. To quantify the effect of compartmentalization and crowding on the production of mRNA and GFP, we constructed a deterministic model, based on the underlying biochemical reactions. Inspired by similar models in recent experiments on cell-free expression kits (30, 31), we write both transcription and translation as a two-step reaction: after a fast-equilibrium complexation of DNA to T7 RNA polymerase (KTS) or mRNA to the ribosome (KTL), transcription and translation proceeds with an overall rate of catalysis kTS and kTL, respectively (SI Text, S16). The key features of this model are shown schematically in Fig. 3A. We further assume that degradation of mRNA and protein can be neglected during our experiments (Fig. S5 A and B).
Fig. 3.
Compartmentalization enhances transcription. (A) Schematic reaction pathway of transcription and translation, as used to model our data (see SI Text, S16 for details). (B) mRNA production in droplets and coacervates. The open symbols represent single-phase droplets, and the closed symbols represent coacervates. Inset shows a zoom-in of the production in single-phase droplets. To allow comparison between different DNA concentrations, all mRNA concentrations in the coacervates have been normalized to a reference droplet of 27 μm and coacervate of 13 μm. The lines are model predictions for equilibrium T7 RNA polymerase binding constants KTS and transcription rate constants kTS as indicated by the labels (see SI Text, S16 for details on other parameters). The single-phase droplet data are modeled with a binding constant KTS = 0.12 nM−1 and a transcription rate constant kTS = 25 min−1 (solid blue); the coacervate data are modeled with KTS = 0.12 (dotted), 1.0 (dashed), 10 (dash-dot), and 100 (solid red) nM−1 and kTS = 143 min−1. The black dotted line shows the predicted mRNA production for the case in which all macromolecular concentrations are increased to their actual value in the coacervates (SI Text, S10), but all binding constants and rate constants are unchanged with respect to single-phase droplets. (C) Data (symbols) and model predictions (lines) of the initial rate of mRNA production in droplets and coacervates as a function of the initial plasmid DNA concentration (see Fig. S6 for corresponding mRNA production curves). We assume that a small amount of DNA (0.095 µm−2, corresponding to 35 pM in a droplet of 27 µm) is adsorbed to the oil–water interface (SI Text, S14 and Fig. S4).
We first focused our studies on the kinetics of mRNA production as a function of DNA concentration using a molecular beacon that is complementary to a part of the GFP mRNA sequence (SI Text, S8). Preliminary experiments showed that increasing the amount of PEG from 0 wt% to between 5 and 8 wt% increases transcription rates, but that mRNA production is strongly inhibited when the concentration of PEG is raised further (>10 wt%), even at high salt concentrations, as shown in SI Text, S17 and Fig. S5 C and D. However, in coacervates (where the PEG concentration exceeds 12–15 wt%), transcription is fully active: Fig. 3B shows the increase in fluorescence intensity for dilute lysate and coacervates as a function of time for a DNA concentration of 160 pM in the starting droplet (see Fig. S6 for mRNA production from 40, 60, 80, 120, and 160 pM DNA). All curves exhibit a delay time of ∼10–20 min and a slowdown in mRNA production after 80–120 min, probably due to depletion of transcription resources and inactivation of some of the components from the lysate (30, 31). Clearly, the rate of mRNA production in the coacervate is increased about 50-fold compared with the nonshrunk droplets, and micromolar concentrations of mRNA are obtained in coacervate droplets from subnanomolar concentrations of DNA in the starting droplet. The fact that the coacervates act as compartments in which most of the relevant macromolecular components of transcription have been accumulated (Fig. 2 A and B), accounts for part of the increase in transcription rate. However, our kinetic model shows that this concentrating effect is not all. Even if we take into account the actual concentrations of DNA, T7 RNA polymerase (and the ribosomes and all other proteins) in the coacervates as determined above, we cannot predict as high a rate of transcription as we find experimentally. Two additional effects are likely to play a role in the coacervates. Crowding generally alters the association constants of complexation equilibria (32) such as the T7 RNA polymerase binding here. Second, the transcription rate constant kTS may be increased in the coacervates. The fits in Fig. 3B show that a combination of these effects, that is, an increase in the DNA association constant by two orders of magnitude, and a fivefold to sixfold increase in kTS, from 25 to 143 min−1, leads to a predicted transcription rate that matches our experiments. The concentration of DNA strongly affects the rate of mRNA production, both in single-phase droplets and in coacervates. Fig. 3C shows that our model correctly predicts the mRNA production rates for all DNA concentrations we investigated. In all cases, the rate of mRNA production in the coacervates is enhanced by nearly two orders of magnitude, compared with the single-phase droplets. At very high DNA concentrations, the rate of mRNA production will level off because every RNA polymerase is already bound to a DNA molecule. The DNA concentration at which leveling off occurs is similar to previous bulk experiments using another E. coli-based cell-free expression kit (31). At very low DNA concentrations, the mRNA production in single-phase droplets is too small to be detected reliably. Remarkably, both the RNA polymerase binding constant and the transcription rate constant in coacervates are of the same order of magnitude as the values typically found for E. coli in vivo (33), whereas two orders of magnitude lower constants are usually found in bulk experiments on cell-free expression kits with the same RNA polymerase in vitro (30, 31). This supports our conclusion that the coacervates are crowded environments that mimic the conditions necessary for in vivo transcription.
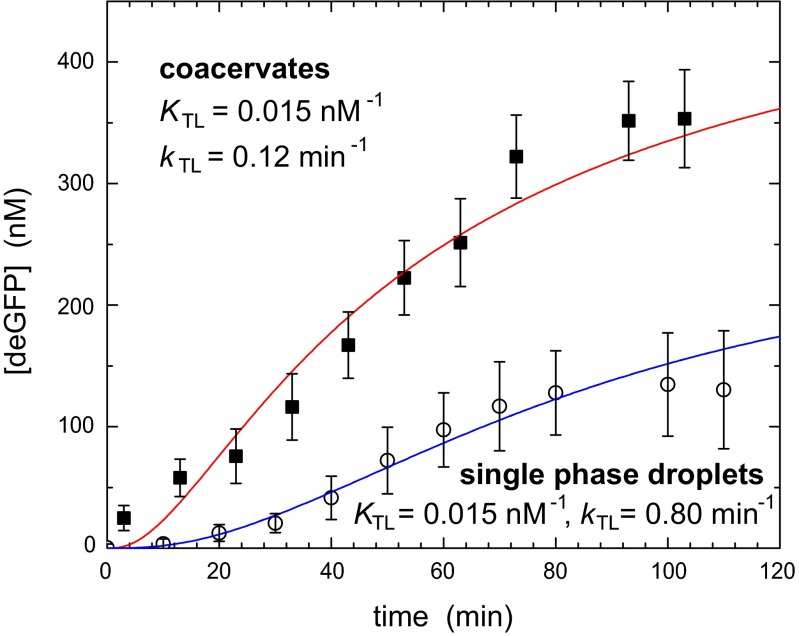
In contrast to the enhanced transcription, protein synthesis rates in coacervates are adversely affected by the high PEG concentrations. Although small amounts of PEG are typically added to optimize cell-free protein synthesis rates, higher concentrations completely prevent protein synthesis (34). Indeed, we observed no appreciable production of GFP in home-made lysate when >10% PEG 8000 was added (SI Text, S18 and Fig. S5E). Nevertheless, we did observe higher concentrations of GFP in the coacervates than in dilute droplets (Fig. 1 B and C), as indicated by the high fluorescence intensity. As GFP is a slow-folding protein, the increased concentrations observed in Fig. 1C could be due to delayed folding of already produced proteins in the coacervate. We therefore extended our studies to include a fast folding eGFP mutant, deGFP, with a maturation time of 8.5 min (26) (see SI Text, S7 and Fig. S2B for calibration details) to decouple translation and fluorophore maturation. Fig. 4 shows that the rate of fluorescence increase resulting from the fast-folding deGFP is significantly larger in the coacervates, although the difference with single-phase droplets is smaller than for transcription. In fact, if we again assume that all protein and RNA components necessary for translation have been accumulated to the same extent in the coacervates, we can account for the increased rate of translation with a translation binding constant KTL that is unchanged and an overall rate constant kTL that is sixfold to sevenfold lower, as expected from the adverse effect of PEG. We emphasize that this assumption is much stricter in case of translation, because tens of proteins and RNA fragments need to assemble before translation can take place. Nevertheless, the coacervates clearly form fully functional compartmentalized systems capable of protein synthesis, which is truly remarkable considering the high concentrations of PEG.
Fig. 4.
Protein expression in coacervates. deGFP production in droplets and coacervates. The open symbols represent single-phase droplets, and the closed symbols represent coacervates. The solid lines are model predictions for equilibrium ribosome binding constants and translation rate constants as indicated by the labels (see SI Text, S16 for details on other parameters). The coacervate data are modeled with the actual concentrations of all macromolecular components and enhanced transcription binding and rate constants KTS = 10 nM−1 and kTS = 143 min−1, respectively (Fig. 3B), compared with the single-phase droplets.
The phase separation of E. coli cell lysate into a dense liquid coacervate containing all macromolecular components has enabled a direct comparison between mRNA synthesis in dilute and crowded environments. We demonstrate that crowding significantly enhances the binding constant of T7 RNA polymerase to DNA and the transcription rate constant, which are direct results of crowding affecting the kinetics of the rate-determining steps of the fundamental machinery of gene expression in the cell. Our results thus impact on our understanding of how biochemical networks function, as rates of enzymatic reactions determined in dilute solution do not necessarily reflect in vivo rates. Our experimental platform enables a systematic study into the effects of crowding on key cellular processes such as transcription and translation in membrane-free protocells. It is striking that both the mRNA (and the molecular beacon) and the GFP remain associated with the coacervate during their production, even in the absence of lipid bilayer membrane around the droplets (4). We hypothesize that a functional coacervate forming spontaneously due to salt-driven liquid–liquid phase separation could be of interest in new theories for the compartmentalization of (proto)cellular components (6, 35, 36).
Supplementary Material
Acknowledgments
We thank Vincent Noireaux and Jonghyeon Shin (University of Minnesota) for donating the pBEST-OR2-OR1-UTR1-eGFP-Del6-229-T500 plasmid and their assistance with lysate preparation. We also thank Kerstin Blank and David Foschepoth (Radboud University Nijmegen) for assistance with GFP vectors. This work was supported by a European Research Council Advanced Grant (246812 Intercom) and a VICI grant of the Netherlands Organisation for Scientific Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222321110/-/DCSupplemental.
References
- 1.Rasmussen S, et al. Evolution. Transitions from nonliving to living matter. Science. 2004;303(5660):963–965. doi: 10.1126/science.1093669. [DOI] [PubMed] [Google Scholar]
- 2.Rasmussen S, Chen LH, Nilsson M, Abe S. Bridging nonliving and living matter. Artif Life. 2003;9(3):269–316. doi: 10.1162/106454603322392479. [DOI] [PubMed] [Google Scholar]
- 3.Noireaux V, Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci USA. 2004;101(51):17669–17674. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature. 2001;409(6818):387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 5.Hyman AA, Simons K. Cell biology. Beyond oil and water—phase transitions in cells. Science. 2012;337(6098):1047–1049. doi: 10.1126/science.1223728. [DOI] [PubMed] [Google Scholar]
- 6.Oparin AI. The Origin of Life. 2nd Ed. New York: Dover Publications; 1953. p. 270. [Google Scholar]
- 7.Fulton AB. How crowded is the cytoplasm? Cell. 1982;30(2):345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- 8.Daube SS, Bracha D, Buxboim A, Bar-Ziv RH. Compartmentalization by directional gene expression. Proc Natl Acad Sci USA. 2010;107(7):2836–2841. doi: 10.1073/pnas.0908919107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strulson CA, Molden RC, Keating CD, Bevilacqua PC. RNA catalysis through compartmentalization. Nat Chem. 2012;4(11):941–946. doi: 10.1038/nchem.1466. [DOI] [PubMed] [Google Scholar]
- 10.Koga S, Williams DS, Perriman AW, Mann S. Peptide-nucleotide microdroplets as a step towards a membrane-free protocell model. Nat Chem. 2011;3(9):720–724. doi: 10.1038/nchem.1110. [DOI] [PubMed] [Google Scholar]
- 11. Bungenberg-de Jong HG, Kruyt HR (1929) Coacervation (partial miscibility in colloid systems). Proc Koninklijke Nederlandse Akademie Wetenschappen 32:849-856.
- 12.Walter H, Brooks DE. Phase separation in cytoplasm, due to macromolecular crowding, is the basis for microcompartmentation. FEBS Lett. 1995;361(2–3):135–139. doi: 10.1016/0014-5793(95)00159-7. [DOI] [PubMed] [Google Scholar]
- 13.Li PL, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483(7389):336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149(6):1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 15.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA. 2011;108(11):4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322(5908):1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- 17.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25(5):635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Albertsson PA. Partition of proteins in liquid polymer-polymer two-phase systems. Nature. 1958;182(4637):709–711. doi: 10.1038/182709a0. [DOI] [PubMed] [Google Scholar]
- 19.Baumgart T, et al. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci USA. 2007;104(9):3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theberge AB, et al. Microdroplets in microfluidics: An evolving platform for discoveries in chemistry and biology. Angew Chem Int Ed Engl. 2010;49(34):5846–5868. doi: 10.1002/anie.200906653. [DOI] [PubMed] [Google Scholar]
- 21.Courtois F, et al. An integrated device for monitoring time-dependent in vitro expression from single genes in picolitre droplets. ChemBioChem. 2008;9(3):439–446. doi: 10.1002/cbic.200700536. [DOI] [PubMed] [Google Scholar]
- 22.Fallah-Araghi A, Baret JC, Ryckelynck M, Griffiths AD. A completely in vitro ultrahigh-throughput droplet-based microfluidic screening system for protein engineering and directed evolution. Lab Chip. 2012;12(5):882–891. doi: 10.1039/c2lc21035e. [DOI] [PubMed] [Google Scholar]
- 23.Agresti JJ, et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci USA. 2010;107(9):4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shim JU, et al. Control and measurement of the phase behavior of aqueous solutions using microfluidics. J Am Chem Soc. 2007;129(28):8825–8835. doi: 10.1021/ja071820f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shim JU, et al. Simultaneous determination of gene expression and enzymatic activity in individual bacterial cells in microdroplet compartments. J Am Chem Soc. 2009;131(42):15251–15256. doi: 10.1021/ja904823z. [DOI] [PubMed] [Google Scholar]
- 26.Shin J, Noireaux V. Efficient cell-free expression with the endogenous E. coli RNA polymerase and sigma factor 70. J Biol Eng. 2010;4:8. doi: 10.1186/1754-1611-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ananthapadmanabhan KP, Goddard ED. Aqueous biphase formation in polyethylene oxide–inorganic salt systems. Langmuir. 1987;3(1):25–31. [Google Scholar]
- 28.Ferreira LA, Teixeira JA. Salt effect on the aqueous two-phase system PEG 8000-sodium sulfate. J Chem Eng Data. 2011;56(1):133–137. [Google Scholar]
- 29.Elowitz MB, Surette MG, Wolf PE, Stock JB, Leibler S. Protein mobility in the cytoplasm of Escherichia coli. J Bacteriol. 1999;181(1):197–203. doi: 10.1128/jb.181.1.197-203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karzbrun E, Shin J, Bar-Ziv RH, Noireaux V. Coarse-grained dynamics of protein synthesis in a cell-free system. Phys Rev Lett. 2011;106(4):048104–048108. doi: 10.1103/PhysRevLett.106.048104. [DOI] [PubMed] [Google Scholar]
- 31.Stögbauer T, Windhager L, Zimmer R, Rädler JO. Experiment and mathematical modeling of gene expression dynamics in a cell-free system. Integr Biol (Camb) 2012;4(5):494–501. doi: 10.1039/c2ib00102k. [DOI] [PubMed] [Google Scholar]
- 32.Minton AP. Macromolecular crowding. Curr Biol. 2006;16(8):R269–R271. doi: 10.1016/j.cub.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 33.Sastry SS, Ross BM. Nuclease activity of T7 RNA polymerase and the heterogeneity of transcription elongation complexes. J Biol Chem. 1997;272(13):8644–8652. doi: 10.1074/jbc.272.13.8644. [DOI] [PubMed] [Google Scholar]
- 34.Ge XM, Luo D, Xu JF. Cell-free protein expression under macromolecular crowding conditions. PLoS One. 2011;6(12):e28707. doi: 10.1371/journal.pone.0028707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fox SW. A theory of macromolecular and cellular origins. Nature. 1965;205(4969):328–340. doi: 10.1038/205328a0. [DOI] [PubMed] [Google Scholar]
- 36.Long MS, Jones CD, Helfrich MR, Mangeney-Slavin LK, Keating CD. Dynamic microcompartmentation in synthetic cells. Proc Natl Acad Sci USA. 2005;102(17):5920–5925. doi: 10.1073/pnas.0409333102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.