Significance
Transposons are powerful tools widely used for insertional mutagenesis screens because of the straightforward identification of transposon-induced mutations. We developed a conditionally regulated long interspersed element 1 [tetracycline (tet)-ORFeus] retrotransposon mouse model that may be used for random mutagenesis and mechanistic studies. High-level expression of the retrotransposon during development results in embryonic lethality due to a significant burden of insertions. At lower levels, transgene induction results in a somatic spotting phenotype due to mutations that likely disrupt melanocyte development, suggesting high activity of tet-ORFeus in melanocyte precursors. This model may be used to identify genes and pathways involved in diverse biological processes.
Keywords: L1 retrotransposon, tet-promoter, white-spotted phenotype, mus musculus
Abstract
A codon-optimized mouse LINE-1 element, ORFeus, exhibits dramatically higher retrotransposition frequencies compared with its native long interspersed element 1 counterpart. To establish a retrotransposon-mediated mouse model with regulatable and potent mutagenic capabilities, we generated a tetracycline (tet)-regulated ORFeus element harboring a gene-trap cassette. Here, we show that mice expressing tet-ORFeus broadly exhibit robust retrotransposition in somatic tissues when treated with doxycycline. Consistent with a significant mutagenic burden, we observed a reduced number of double transgenic animals when treated with high-level doxycycline during embryogenesis. Transgene induction in skin resulted in a white spotting phenotype due to somatic ORFeus-mediated mutations that likely disrupt melanocyte development. The data suggest a high level of transposition in melanocyte precursors and consequent mutation of genes important for melanoblast proliferation, differentiation, or migration. These findings reveal the utility of a retrotransposon-based mutagenesis system as an alternative to existing DNA transposon systems. Moreover, breeding these mice to different tet-transactivator/reversible tet-transactivator lines supports broad functionality of tet-ORFeus because of the potential for dose-dependent, tissue-specific, and temporal-specific mutagenesis.
Long interspersed element 1 (LINE-1 or L1) retrotransposons are an abundant class of mobile genetic sequences that constitute ∼17% of the human and mouse genomes (1, 2). In contrast to DNA transposons, L1 elements retrotranspose using a copy-and-paste mechanism in which they are first transcribed into an RNA intermediate before insertion into a new genomic location (3–5). Because L1 retrotransposons are not excised from genomic DNA (gDNA), the donor elements are stable. Furthermore, studies have revealed that L1 elements exhibit relatively unbiased insertion-site selection (6). These findings suggest that retrotransposons may be efficient for utilization in genome-wide insertional mutagenesis screens.
A synthetic mouse L1 element was recently constructed by altering the nucleic acid sequence without changing the amino acid sequence of L1-encoded proteins. These optimized elements abolished transcription-inhibitory sequences and resulted in a >200-fold increase in retrotransposition frequencies when tested in cell culture (7). Subsequently, we generated a mouse model expressing this element, ORFeus, which demonstrated in vivo retrotransposition activity when driven by a constitutive promoter (8). This approach was further modified through the activation of ORFeus using Cre-Lox recombination technology (9). To generate a chemically regulated L1 mouse model with potent mutagenic capabilities, we generated a tetracycline (tet)-regulated ORFeus element harboring a gene-trap cassette designed to truncate target transcripts or activate downstream transcription, depending on the orientation of the gene trap.
We demonstrate that, when mice harboring a tet-ORFeus gene-trap transgene are bred with a reversible tet-transactivator (rtTA) line, double-transgenic progeny express ORFeus only when treated with doxycycline. We observed high levels of retrotransposition in tissues from double-transgenic mice but not in control littermates, and the amount of retrotransposition increases with increased doxycycline dose. Induction of the tet-ORFeus element with high doses of doxycycline during embryogenesis led to a reduced number of double-transgenic mice, likely due to a significant burden of mutations and embryonic lethality in these animals. Unexpectedly, a significant percentage of double-transgenic agouti mice developed white spots, suggesting that somatic mutations occurred at different times in development. Consistent with this, this phenotype is not heritable, and we therefore infer that it occurred somatically in melanocytes or their precursors. We show that the white spots lack melanocytes, suggesting that the ORFeus element has somatically altered a gene(s) involved in melanocyte development, proliferation, or migration. With the development and characterization of the tet-ORFeus model complete, we are poised to use this retrotransposon-based system as a tool for cancer gene discovery and other forward genetic screens. Moreover, this system may be useful as a general tool for mutagenesis in mice.
Results
Generation of a Conditional L1 Retrotransposon Gene-Trap Element.
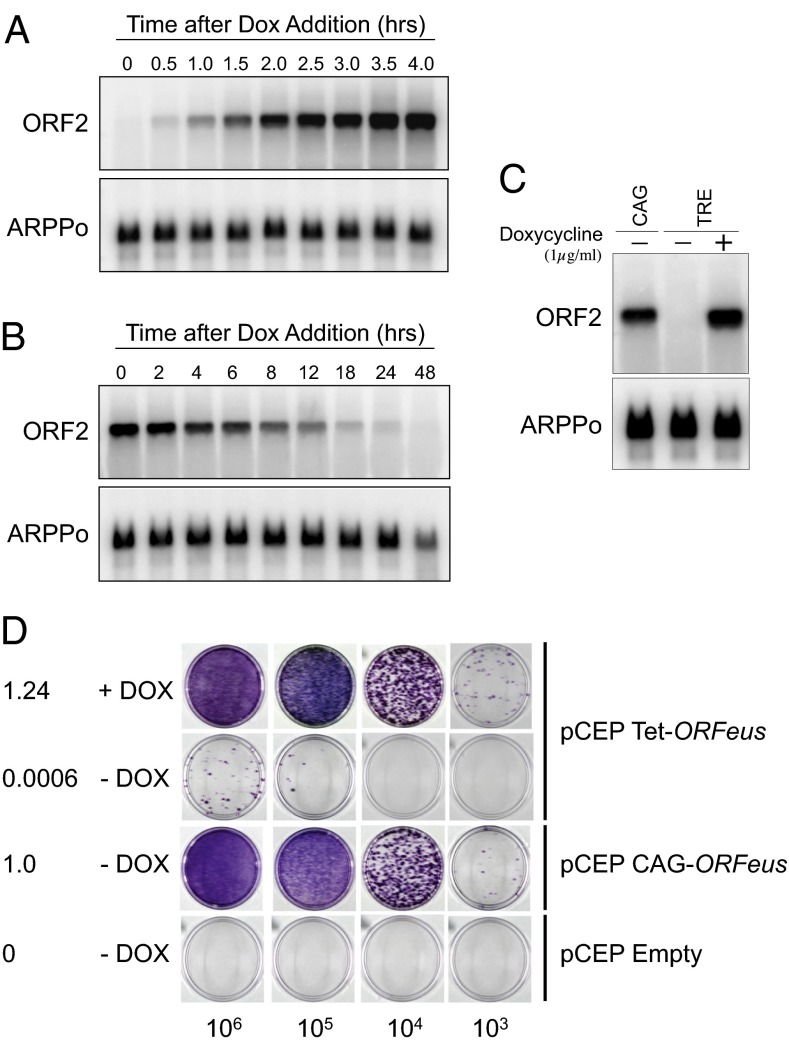
We generated a conditional synthetic L1 retrotransposon by placing the ORFeus element under the control of the tetracycline-responsive promoter (TRE) (10, 11). Confirmation of tight tet-regulated control of ORFeus expression was obtained by RNA blot analysis in Tet-ON and Tet-OFF HeLa cells (Fig. 1 A and B). We also compared the expression of ORFeus when driven by the TRE promoter versus the constitutive cytomegalovirus early enhancer/chicken beta-actin (CAG) promoter and found that mRNA levels were similar (Fig. 1C). We further assessed the retrotransposition frequency of the tet-ORFeus element in a standard cell culture retrotransposition assay. In this system, a functional L1 element is marked with a retrotransposition indicator reporter (5). In this case, we used an indicator gene conferring resistance to blasticidin. Intron removal during splicing of the L1 transcript restores function to a blasticidin resistance gene encoded on the opposite strand. Quantification of blasticidin-resistant colonies demonstrated that the tet-ORFeus element retrotransposed in a doxycycline-dependent manner and at slightly higher frequency than the CAG-ORFeus element (Fig. 1D).
Fig. 1.
Generation of a conditional LINE-1 retrotransposon. (A) Total RNA blot analysis demonstrating doxycycline (Dox)-regulated expression of tet-ORFeus in Tet-ON HeLa cells. ORF2 probe is derived from ORFeus template. ARPPo serves as a loading control. (B) Total RNA blot analysis in Tet-OFF HeLa cells. (C) RNA blot analysis of ORF2 expression in constructs from which the ORFeus transgene is driven by either the CAG promoter or the TRE promoter. (D) Cell culture-based retrotransposition assay in Tet-ON HeLa cells using plasmid pCEP4-Puro (48) carrying the indicated donor ORFeus element in the presence or absence of doxycycline (Dox). The numbers in the first column represent the number of transposition events (i.e., the number of colonies per microgram input of DNA) normalized to the number obtained with pCAG-ORFeus. The numbers on the bottom indicate the number of puromycin-resistant cells plated.
To generate a conditional ORFeus element that also serves as a mutagen, we engineered the tet-ORFeus transgene to contain a gene-trap cassette in its 3′ untranslated region. The gene trap was designed to disrupt gene function in several ways (Fig. S1A). The gene trap contains a splice acceptor followed by a polyadenylation (polyA) signal. When the element transposes into an intron, the host gene may splice to the gene trap. Resulting use of the gene-trap polyA signal will truncate the mRNA, likely resulting in a loss-of-function mutation (Fig. S1B). The gene-trap cassette also contains a long terminal repeat promoter/enhancer element that is designed to activate gene expression when transposed upstream or within a gene, resulting in the activation of either a full-length or a truncated protein, respectively (Fig. S1C). In addition, the gene trap harbors a small chimeric intron, which provides a simple assay for splicing and, by extension, for retrotransposition of the element. Removal of this intron is a signal that retrotransposition has occurred because these elements proceed through an RNA intermediate; thus, a simple PCR-based assay for intron removal gives a quick readout of whether a given cell or mouse line is active for retrotransposition. RT-PCR studies documented high-level expression and efficient splicing of the chimeric intron upon induction of the tet-ORFeus gene-trap element in tet-OFF HeLa cells (Fig. S1D).
We next generated transgenic mice harboring the tet-ORFeus gene-trap element by pronuclear injection into B6.SJL F1 embryos. We screened a total of 162 mice by genotyping PCR and identified five founders. To confirm germ-line transmission of the integrated transgenes, founders were backcrossed to wild-type C57BL/6J mice to generate independent transgenic lines. Southern blotting was used to determine the copy number of the tet-ORFeus transgene in each line. Three independent tet-ORFeus transgenic lines, each with different transgene copy numbers and sites of integration, were established (Table S1).
Confirmation of Somatic Retrotransposition in Vivo.
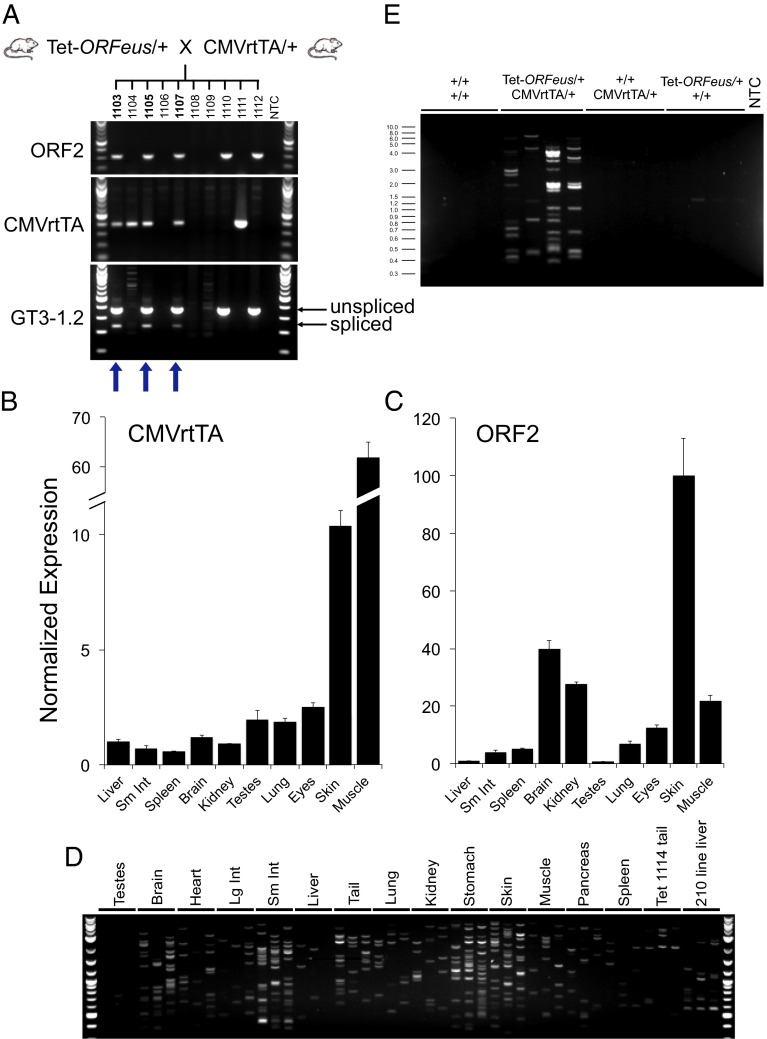
The tet-ORFeus retrotransposon remains inactive until it is combined with a tet transactivator (tTA) or a rtTA allele. To activate the transgene, we bred the L1 mice to a cytomegalovirus rtTA (CMVrtTA) transgenic line, which is broadly expressed in mouse tissues (Fig. 2B) (12, 13). When mice were administered low-dose (0.1 mg/mL) doxycycline in utero via the mother’s drinking water, only double-transgenic progeny exhibited in vivo retrotransposition, as evidenced by intron splicing (Fig. 2A). Quantitative RT-PCR analysis further confirmed that the L1 transgene was expressed in multiple tissues in these animals such as skin, muscle, and brain (Fig. 2C). To profile individual retrotransposition events, we used a modified inverse PCR (iPCR) assay (14). In this assay, one primer spans the gene-trap intron junction, thereby annealing to genomic DNA after splicing and retrotransposition has occurred. This allows for efficient amplification of bona fide somatic insertions without amplification of the donor transgene. We observed high levels of retrotransposition in multiple tissues of double-transgenic mice when treated with doxycycline but not in littermates expressing either transgene alone (Fig. 2 D and E). Furthermore, the unique banding pattern observed when setting up independent PCR reactions from the same ligation reaction indicates a complex population of somatic insertions in these tissues. To map the genomic locations of somatic insertions, we cloned and sequenced a number of the iPCR amplicons from tissues of double-transgenic mice (Table S2). Consistent with earlier studies using a CAG-ORFeus element (8), these insertions are randomly distributed throughout the genome.
Fig. 2.
Confirmation of tet-ORFeus expression and somatic retrotransposition in vivo. (A) Genotyping PCR (ORF2 from tet-ORFeus and CMVrtTA) and gene-trap splicing assay (GT3-1.2) with genomic DNA isolated from tail tissue in progeny of heterozygous tet-ORFeus and CMVrtTA animals. Line 058 mice were treated with 0.1 mg/mL doxycycline during embryogenesis. Blue arrows below the gel images indicate double-transgenic progeny. NTC, no template control. (B and C) Quantitative real-time RT-PCR analysis of CMVrtTA (B) and ORF2 (C) expression in a panel of tissues from the double-transgenic line 058 animal treated with 0.1 mg/mL doxycycline. Bar graphs represent mean ORF2 and CMVrtTA expression relative to actin and normalized to the liver sample for each amplicon, respectively. Error bars represent SDs from three independent measurements. (D) Inverse PCR retrotransposition assay with genomic DNA isolated from a double-transgenic animal (line 058) treated in utero with 0.1 mg/mL doxycycline. For each sample, three independent iPCR reactions were performed using the same ligation mixture. Genomic DNA isolated from liver of the CAG-ORFeus transgenic mouse line (line 210 liver) serves as a reference. (E) Inverse PCR assay with tail gDNA isolated from four littermates (line 058) demonstrating that retrotransposition activity is specific to double-transgenic animals treated with doxycycline. Genotypes are labeled above gel image. For each tail gDNA sample, four independent reactions were performed with the same ligation mix.
We also analyzed several litters of mice that were never exposed to doxycycline. Despite a small degree of leaky retrotransposition in HeLa cells (Fig. 1D), we found no evidence for leaky retrotransposition of the tet-ORFeus transgene in double-transgenic mice in the absence of doxycycline (Fig. S2). This likely results from differences in the copy number of transiently transfected tet-ORFeus in HeLa cells versus single or low copy number in transgenic animals.
Although 100% of double-transgenic mice acquire somatic insertions, we did not observe germ-line retrotransposition in these animals. Accordingly, ORFeus-derived ORF2 expression is extremely low in testes when driven by CMVrtTA (Fig. 2B). Interestingly, CMVrtTA is expressed in testes at levels comparable to tissues where ORFeus expression is robust and retrotransposition is efficient (e.g., brain, kidney, and lung; Fig. 2B). A possible explanation for this discrepancy is that perhaps doxycycline does not efficiently cross the blood–testes barrier.
We further characterized this model by examining the expression and retrotransposition of tet-ORFeus at different times in development and with multiple doses of doxycycline. We confirmed that double-transgenic progeny exhibited high levels of somatic retrotransposition when treated with a low dose (0.1 mg/mL) of doxycycline during embryogenesis (Fig. 2D and Fig. S3A). However, when low-dose doxycycline treatment commenced at birth and was maintained into adulthood, we did not observe somatic retrotransposition by the intron-splicing assay (Fig. S3B). Consistent with these observations, quantitative real-time PCR analysis revealed a significant reduction in ORF2 mRNA expression when the transgene was induced at birth relative to conception (Fig. S3C). However, retrotransposition was detected when animals were treated beginning at birth with a high dose of doxycycline (4 mg/mL, Fig. S3D). These findings demonstrate that doxycycline administration during embryogenesis results in higher expression of the retrotransposon transgene and therefore a greater efficiency of somatic retrotransposition.
Dose-Dependent Retrotransposition and Embryonic Lethality in Mice with a High Insertional Burden.
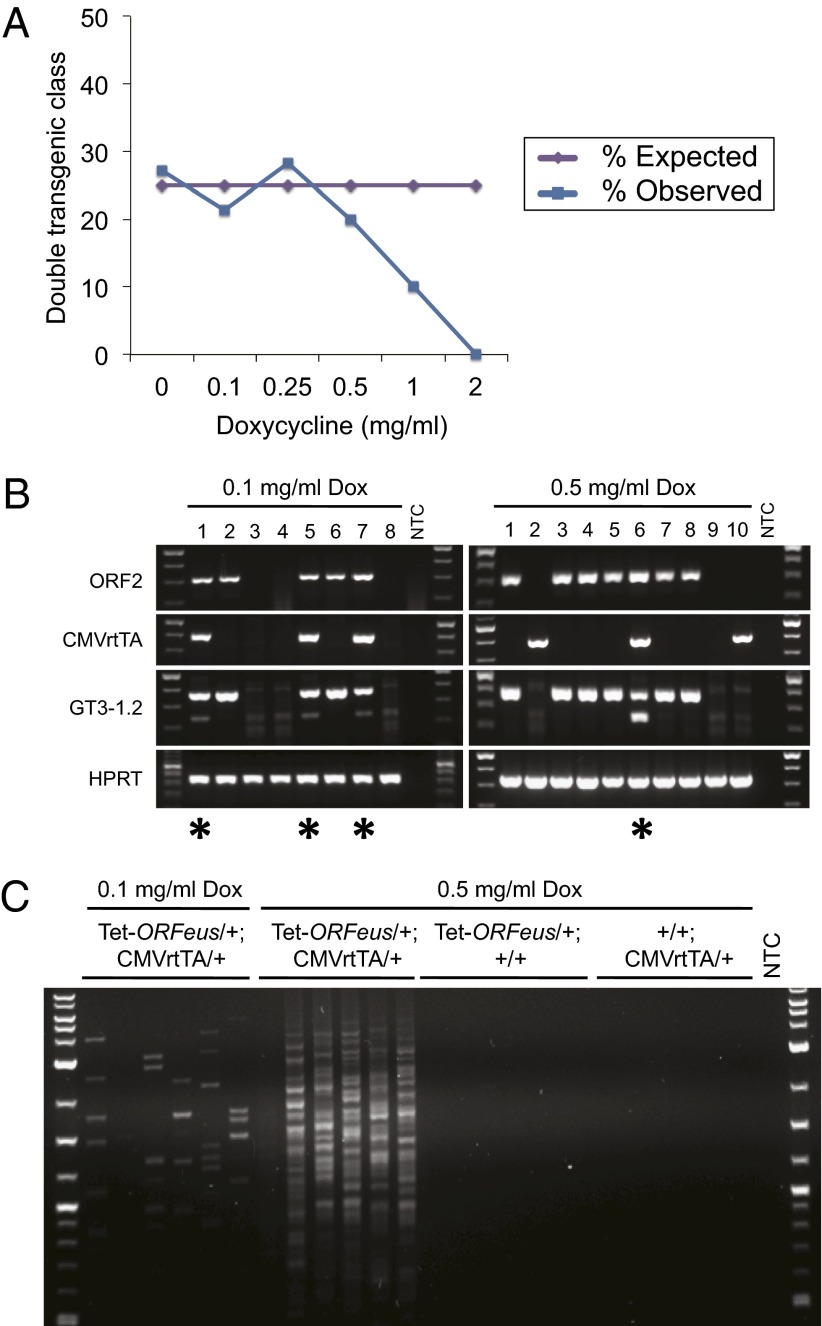
High doses of doxycycline are not reported to cause toxicity in mice (13, 15–17). However, double-transgenic animals did not survive to birth when treated with 4 mg/mL doxycycline during embryogenesis. Moreover, analysis of a range of doxycycline doses between 0.1 and 2.0 mg/mL revealed a clear correlation with greater embryonic lethality of double-transgenic animals with increasing doses of doxycycline (Fig. 3A). We next dissected embryos at embryonic day 14.5 (E14.5) and measured intron splicing and retrotransposition in genomic DNA from embryos treated with either 0.1 or 0.5 mg/mL doxycycline. Using the splicing assay, we observed a greater abundance of spliced product in double-transgenic embryos treated with 0.5 mg/mL doxycycline (Fig. 3B). Consistent with this, ∼5- to 10-fold more insertions were observed at the higher dose by iPCR (Fig. 3C). Taken together, these data demonstrate dose-dependent retrotransposition in embryos expressing the tet-ORFeus element and suggest that a high insertional load of tet-ORFeus carrying a gene trap results in embryonic lethality.
Fig. 3.
Embryonic lethality and dose-dependent retrotransposition in mice with a high mutagenic burden of L1 insertions. (A) The percentage of each genotypic class was calculated with increasing doses of doxycycline. Four possible genotypes are expected with equal representation if there is no effect on viability (25% tet-ORFeus/+; CMVrtTA/+; 25% tet-ORFeus/+; +/+; 25% +/+; CMVrtTA/+; 25% +/+; +/+). Only the double-transgenic class (tet-ORFeus/+; CMVrtTA/+) was under-represented at high doses of doxycycline. The number of animals analyzed at each dose are as follows: untreated–n = 48; 0.1 mg/mL–n = 103; 0.25 mg/mL–n = 92; 0.5 mg/mL–n = 135; 1.0 mg/mL–n = 20; 2.0 mg/mL–n = 14. (B) Genotyping PCR (ORF2 and CMVrtTA) and gene-trap splicing assay (GT3-1.2) with genomic DNA isolated from E14.5 embryos. Animals were treated with either 0.1 or 0.5 mg/mL doxycycline beginning at conception. Numbers represent individual embryos. Asterisks below the gel image indicate double-transgenic embryos. HPRT serves as a control for equal loading of template gDNA. (C) iPCR assay with genomic DNA isolated from four different embryos. The genotype and doxycycline dose for each embryo are labeled above the gel image. For each embryo, six independent iPCR reactions were performed.
Quantitation of L1 Insertion Frequencies in Mouse Embryonic Fibroblasts.
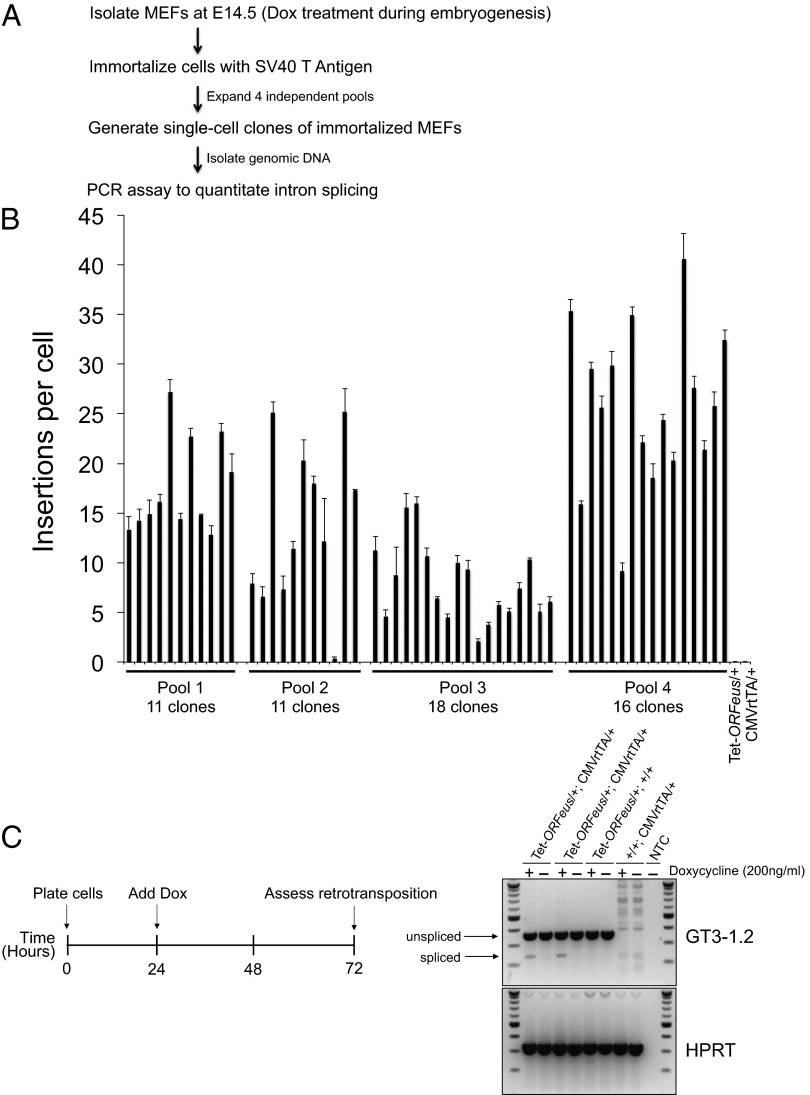
To accurately quantify the frequency of tet-ORFeus insertions in vivo, we harvested mouse embryonic fibroblasts (MEFs) treated with doxycycline throughout the duration of embryogenesis up until the time of MEF isolation at E14.5. To measure the insertion frequency of individual cells, single-cell clones were isolated from immortalized MEFs derived from the single-copy 095 line (Fig. 4A). Only a small number of clones exhibited evidence of intron splicing (2 of 73) when treated with low-dose doxycycline, indicating a retrotransposition efficiency significantly below 1 insertion per cell at this dose. In contrast, 100% of MEF clones isolated from embryos treated with 0.5 mg/mL doxycycline (56 of 56) displayed intron splicing. To quantify insertions in each MEF clone, we designed a quantitative real-time PCR assay incorporating a primer that crosses the gene-trap intron junction. Using this assay, we calculated an average frequency of 16 insertions per cell in fibroblasts treated with a 0.5-mg/mL dose during embryogenesis (Fig. 4B). Although it is expected that the insertion frequency will vary in different tissues depending on the efficiency of the tet-transactivator (tTA or rtTA) line in each tissue, these data provide an estimate of the frequency of tet-ORFeus insertions in fibroblasts from mice with high-level retrotransposition during embryogenesis.
Fig. 4.
Quantitation of L1 insertion frequencies in MEFs. (A) Overview of MEF isolation, immortalization, and generation of single-cell clones for determination of tet-ORFeus insertion frequencies. (B) Bar graph summarizing the frequency of tet-ORFeus insertions per single-cell clone in immortalized MEFs treated with 0.5 mg/mL doxycycline. Animals were exposed to doxycycline during embryogenesis until the time of MEF isolation. (C) Proof-of-principle experiment demonstrating retrotransposition of the tet-ORFeus transgene in MEFs growing in culture. (Right) Gene-trap splicing assay (GT3-1.2) with gDNA isolated from primary MEF cells in the presence (+) or absence of (−) of doxycycline. The genotypes of each cell line are labeled above the image.
We also harvested MEFs from animals not exposed to doxycycline during embryogenesis and then treated the primary cells growing in culture with doxycycline. Retrotransposition was observed 48 h after induction of the transgene in double-transgenic cell lines but not in single-transgenic control cells (Fig. 4C). Currently, most cell culture-based assays for L1 retrotransposition rely on the use of an episomal plasmid and a drug-resistant or fluorescent marker for quantitation of insertions. In contrast, these cells harbor an L1 transgene integrated in a native chromosome in which retrotransposition may be directly quantified. As such, this system complements a recently developed L1 reporter cell line and may be exploited for a wide range of applications including the analysis of retrotransposition kinetics or downstream effects on cell signaling and DNA damage-response pathways in response to L1 transgene induction (18).
A Somatic White-Spotting Phenotype in Tet-ORFeus Mice.
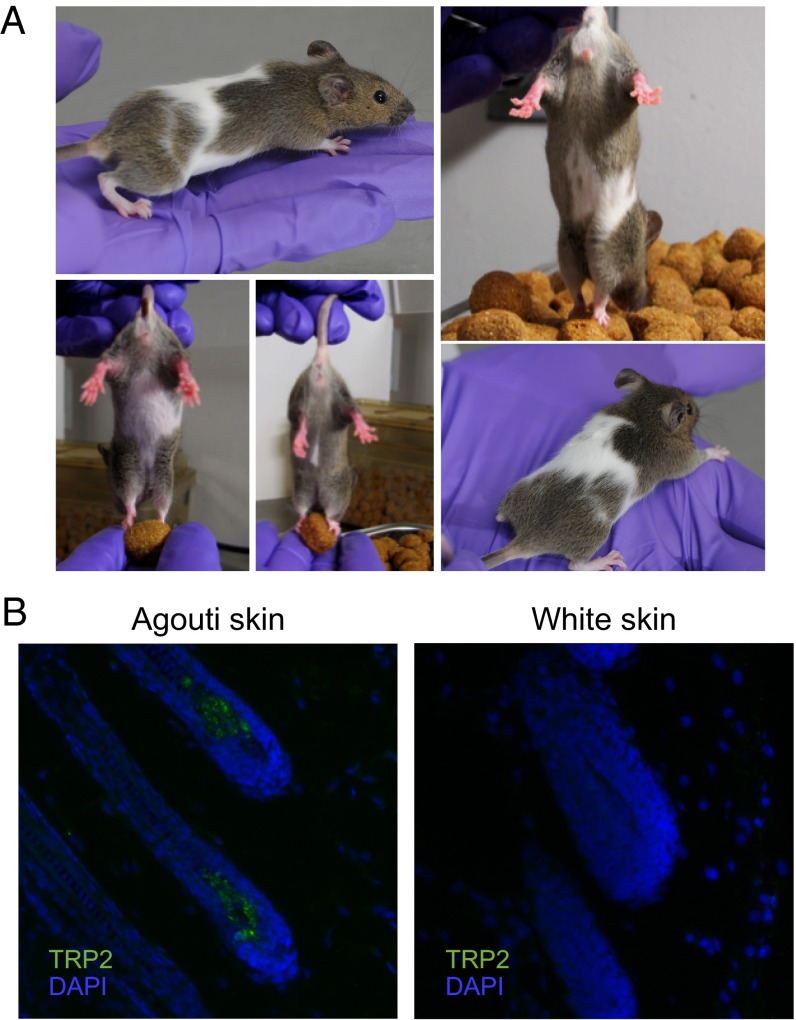
When animals were treated with intermediate doses of doxycycline during embryogenesis, we observed that a significant percentage (∼75%) of double-transgenic animals developed white spots on agouti skin (Fig. 5A). Interestingly, the phenotype is dose-responsive because 58% of double-transgenic mice treated with 0.25 mg/mL doxycycline were spotted, whereas 92% of double-transgenic mice treated with 0.5 mg/mL doxycycline exhibited spotting. These spots were visible within 1 wk after birth and varied in size between animals. White spots were observed in progeny from two independent tet-ORFeus transgenic lines when bred to CMVrtTA mice, demonstrating that the spotting phenotype did not depend on a position effect of the tet-ORFeus transgene integration. Furthermore, white spotting was observed only in double-transgenic animals with active retrotransposition and never in single-transgenic or nontransgenic littermates, nor was it seen in double-transgenic animals treated with low-dose doxycycline, nor in untreated animals. When spotted animals were backcrossed to wild-type C57BL/6J mice, the phenotype was not inherited through the germ line. Taken together, these observations suggest that the tet-ORFeus element is active in somatic cells and that the spotted mice are likely mosaic for the causative retrotransposon insertions.
Fig. 5.
White-spotting phenotype in Tet-ORFeus mice. (A) Representative pictures of double-transgenic animals with white spotting. Animals (line 095) were administered doxycycline water (at doses of 0.25, 0.50, or 1.0 mg/mL) beginning at conception and maintained at the same dose into adulthood. Fifty-eight percent of double-transgenic mice treated with 0.25 mg/mL doxycycline were spotted, whereas 92% of double-transgenic mice treated with 0.5 mg/mL doxycycline exhibited spotting. (B) Representative TRP2 and DAPI-stained hair follicle sections of agouti skin or white skin from a double-transgenic line 095 animal treated with 0.25 mg/mL doxycycline. TRP2 is a melanocyte-specific marker.
Melanocytes are specialized cells derived from the neural crest that produce pigment and specify color of the eye, skin, and hair in mammals (19–21). A reduction in melanocyte number in hair follicles and skin is reported to result in areas of hypopigmentation. A wide variety of genetic defects in the migration, proliferation, survival, or differentiation of melanocyte precursors, known as melanoblasts, cause white-spotting phenotypes in mice (19, 22). We therefore hypothesize that, when induced at high levels, the tet-ORFeus element is highly active in melanoblasts and that insertions of tet-ORFeus into a potentially large number of genes important for melanoblast migration, proliferation, survival, or differentiation result in mosaic loss of these cells, thereby causing the observed mosaic white patches of varying sizes. A prediction of this hypothesis is that melanocytes should be absent from the white patches of skin, rather than present, but defective in melanin production. Accordingly, immunofluorescent staining for tyrosinase-related protein 2 (TRP2), a well-characterized melanocyte marker (20–24), revealed that melanocytes are absent from white skin but are present in normal numbers in the agouti skin of these animals (Fig. 5B). Notably, we detected retrotransposition in double-transgenic embryos as early as E9.5–E10.5 when melanoblast migration is known to occur (Fig. S4 A and B) (25–27). Taken together, our data suggest that L1-mediated mutations alter expression of genes involved in melanocyte development, thus leading to the spotted phenotype.
We aged a cohort of animals on doxycycline for over 1 y and did not observe evidence of lymphoma development in these mice. In contrast, DNA transposon transgenic lines such as Sleeping Beauty (SB) and PiggyBac (PB) succumbed to lymphomas (28–30). Analysis of L1 insertions in hematopoietic tissues by iPCR confirmed that there are few insertions in blood and spleen relative to other tissues such as intestines, skin, and eyes (Fig. S5 A and B). It is therefore likely that the CMVrtTA transgene is not sufficiently active in the hematopoietic system to produce high-level expression of ORFeus to elicit lymphoma development in these mice.
Discussion
The ability to regulate the expression of an insertional mutagen in a spatially and temporally controlled manner provides a significant advantage over a constitutively expressed element. Here we describe the generation of a regulated retrotransposon mouse model and demonstrate that the expression level, retrotransposition activity, and mutagenic load can be controlled with the graded administration of doxycycline. Thus, the model described here expands the repertoire of genetically engineered mice available for controlled in vivo mutagenesis.
Two DNA transposon systems with efficient insertional frequencies in somatic cells have been developed (29, 31, 32). The Sleeping Beauty and PiggyBac transposons are effective tools for cancer gene discovery when used in forward genetic screens in mice (29–31, 33–35). These systems provide powerful reagents for forward genetics, although there are several characteristics of DNA transposons that may limit their full potential as mutagens. For example, these elements use a cut-and-paste mechanism whereby the transposon is excised from a donor site and subsequently inserted into a new genomic location. Several reports demonstrate that empty donor sites are unstable and that excision events may lead to the development of complex rearrangements including deletions and inversions (36–38). The presence of such genomic abnormalities at the transposon excision site may confound the interpretation of mutant phenotypes. Another characteristic, referred to as “local hopping,” describes the tendency of a considerable fraction of insertions of DNA transposons such as SB to lie close (within 1 Mb) to the donor site instead of randomly throughout the genome (39, 40).
L1 retrotransposons may provide several potential advantages compared with DNA transposon mutagenesis systems such as SB and PB. Unlike DNA transposons, L1 elements transpose by reverse transcription of an RNA intermediate. Thus, they can increase in copy number very rapidly due to their mode of reproduction (41). Importantly, it is not possible to remove an L1 insertion after it has jumped, which can easily occur with DNA transposons if the transposase continues to be expressed. Therefore, the mapping of bona fide L1 insertions should prove to be more straightforward in mutagenesis screens because both the donor L1 transgene and the retrotransposon insertions are stable. Finally, because the mechanism of L1 retrotransposition is distinct from that of DNA transposons, it is possible that a different spectrum of insertions may be identified with each system. To assess the relative efficiency of each system, it will be useful to perform genetic screens with the two systems in parallel.
When the tet-ORFeus element was induced at high levels during embryogenesis, the embryonic lethality of double-transgenic animals ensued, most likely due to a very significant burden of insertions. These findings demonstrate the sensitivity of these animals to high-level retrotransposition during development. Additionally, it is clear that doxycycline administration in early postnatal animals results in lower retrotransposition efficiency compared with its administration during embryogenesis. Although this phenomenon remains incompletely understood, our findings are consistent with other tet-regulated transgenic systems. In several reported studies, transgene expression was blunted if induced after embryonic development (42, 43). These observations suggest that low retrotransposition induced with postnatal doxycycline administration is a reflection of the limitations of doxycycline-regulated gene expression systems rather than a general feature of retrotransposition activity.
We discovered a surprising somatic spotting phenotype in animals expressing the L1 transgene at high levels in skin, which represents a defect in melanoblast development and/or migration. Melanocytes were absent from white spots, precluding the identification of causative retrotransposon insertions. The manifestation of a melanoblast phenotype in CMVrtTA;tet-ORFeus mice and the extremely high insertion rate in this population indicates that this line may likely be useful for forward genetic analysis of tumor types such as melanoma arising from this cell population. Indeed, there is an extensive body of literature describing the analysis of spotted mutant animals, which has resulted in the identification of several key genes that are required for melanoblast development and that participate in melanomagenesis, including the c-kit protooncogene (KIT), microphthalmia-associated transcription factor (MITF), and dopachrome tautomerase (DCT) (21, 44). We believe that it is likely that the melanocytes that do complete development in tet-ORFeus;CMVrtTA animals have accumulated mutations that are insufficient to lead to cellular loss, yet may predispose them to oncogenic transformation. Uncovering such cancer-enhancing mutations will likely require introducing an activated oncogene such as BRAF or RAS.
Tet-ORFeus;CMVrtTA mice provide a clear demonstration of a somatic phenotype arising in a mouse model expressing a mutagenic retrotransposon. In this regard, it is interesting to note that various lines of evidence support somatic retrotransposition of L1 elements in neuronal tissues and differentiating neural stem cells (45–47). It is tempting to speculate that neural crest-derived lineages such as melanocytes provide a similarly favorable cellular mileu for retrotransposition, resulting in the spotting phenotype described here.
In summary, the insertional mutagenesis system described here provides an opportunity to regulate the spatial and temporal expression of the retrotransposon in a dose-dependent manner. This suggests that the model may be used for a broad range of applications, including mutagenesis screens for cancer gene identification as well as for the dissection of developmental phenotypes. Thus, the tet-ORFeus system should provide a powerful means to identify genes and pathways involved in many biological processes.
Methods
Transgene and Retrotransposition Plasmid Generation.
The TRE promoter was amplified from pTRE-Tight (Clontech) with the following primers: forward (5′-TAAGCGGCCGCGTACGTCTTCACTCGAGTTTACTCC-3′) containing the NotI and BsiWI restriction sites and reverse (5′-TAAGGCGCGCCAGGCGATCTGACGGTTCA-3′) containing an AscI site. Following amplification and digestion with BsiWI and AscI (New England Biolabs), the TRE promoter sequence was ligated to a fragment containing the mouse ORFeus sequence in pBluescript digested with BsiWI and AscI. The gene-trap cassette (Fig. S1A) consists of the following sequence elements (5′–3′): a chimeric intron from plasmid pCI (Promega), the murine stem-cell virus long-terminal-repeat promoter, the splice donor from the mouse forkhead Foxf2 gene exon 1 (accession Y12293), the SV40 late poly(A) (antisense orientation), and the splice acceptor from human adenovirus 2 (GenBank accession AC000007; antisense orientation). The cassette was de novo synthesized by Codon Devices and subcloned into the 3′ UTR of the transgene plasmid via SfiI-mediated cassette exchange (14). The transgene plasmid was then digested with NotI to remove all bacterial sequences before pronuclear injections were performed. To generate a retrotransposition construct containing tet-ORFeus, the TRE promoter was amplified with the same primers as described above, digested with NotI and AscI (New England Biolabs), and then ligated to a fragment containing the mouse ORFeus sequence in pCEPpuro with an intron-disrupted blasticidin cassette (in the 3′ UTR of ORFeus) digested with NotI and AscI.
Generation and Maintenance of Transgenic Animals.
Tet-ORFeus mice were generated using pronuclear injection and genotyped by PCR. These mice were subsequently maintained on a C57BL/6J background and bred to CMVrtTA mice (FVB/N background). Doxycycline was administered to animals via drinking water at the following concentrations: 0.1, 0.25, 0.5, 1, 2, or 4 mg/mL beginning at conception or at birth. Doxycycline doses of 2 and 4 mg/mL were prepared in a 1% sucrose solution. The Johns Hopkins and UT Southwestern Animal Care and Use Committee approved all procedures described in this work.
Genotyping PCR.
Genomic DNA was isolated from tail clippings using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s instructions. PCR genotyping was performed using 10–100 ng of genomic DNA as template. Primers used for ORF2 were forward (5′-AAGGAGGAAGTGAAGATCAGCCTGT-3′) and reverse (5′-TCCTTGATCTCCTTCTTCAGGCTCT-3′) (amplicon 307 bp); CMVrtTA forward (5′-GTGAAGTGGGTCCGCGTACAG-3′) and reverse (5′-GTACTCGTC AATTCCAAGGGCATCG-3′) (amplicon 400 bp); hypoxanthine phosphoribosyltransferase 1 (Hprt) forward (5′-CTTTCTTGTCACTCCCACTTTTCC-3′) and reverse (5′-CACATCCTTCATTCAGGTGTCACT-3′) (amplicon 340 bp); GT3-1.2 forward (5′- CTAGTCCTGCAGGCCAAAAT-3′) and reverse (5′-TCTGGGGACCATCTGTTCTT-3′) (amplicons 369 bp unspliced and 232 bp spliced). PCR primers, reaction and cycling conditions using ExTaq (Clontech) are shown in Tables S3–S5. PCR products were separated on a 1.2% (wt/vol) agarose gel with ORF2, CMVrtTA, and Hprt amplicons or on a 2.0% (wt/vol) gel with the GT3-1.2 splicing assay. A 2.0% gel is recommended for visualization of the spliced and unspliced products.
Cell Culture.
Tet-ON (M2) and tet-OFF HeLa cells (Clontech) were cultured in high glucose (4.5 g/L) DMEM supplemented with 10% FBS, penicillin, and streptomycin.
Northern Analysis.
RNA was isolated from cell pellets using the RNeasy Kit (Qiagen). Ten micrograms of RNA was loaded on a 1.2% agarose/formaldehyde gel, blotted to a Genescreen plus nylon membrane (Perkin-Elmer) in 10× SSC, cross-linked, and baked for 1 h at 80 °C in a vacuum oven. Hybridizations were performed with ULTRAhyb (Life Technologies) at 42 °C. Washes were performed in 2× SSC, 0.1% SDS and in 0.1× SSC, 0.1% SDS at 42 °C. Radioactive signals were detected with a Typhoon phosphoimager (GE Healthcare) and quantified using ImageQuant software. Northern probes were PCR-amplified, gel-purified, and [α-32P]ATP-labeled using the Random Prime-It II Kit (Agilent). Primers used for ORF2 were forward (5′-TGATCAGCGACAAGATCGAC-3′) and reverse (5′-CCTCGATCTTGTGGAAAAGC-3′); acidic ribosomal phosphoprotein P0 (ARPPo) forward (5′-ACTGTGCCAGCCCAGAACAC-3′) and reverse (5′-GCAGATGGATCAGCCAAGAAG-3′).
Inverse PCR.
Mouse genomic DNA (500 ng) was digested with MspI (New England Biolabs), inactivated, and ligated overnight with T4 DNA ligase (New England Biolabs) at 16 °C in a total volume of 500 µL. Following ligase inactivation, the ligation pool was then concentrated with either Microcon YM-100 or Amicon Ultra 10K columns (Millipore), and the volume was adjusted to 50 µL with water if necessary. One microliter was used for PCR with the following primers: forward (5′-CTAGGTCGGATCCTTTTCCCTCTG-3′) and reverse (5′-ATGGGCCCACCTGCAATTGAAG-3′) that spans the gene-trap intron junction. PCR cycling and reaction conditions are shown in Tables S6–S7. For identification of tet-ORFeus insertion sites, PCR products were purified as a pool, subcloned into a TA-cloning vector (Life Technologies), and sequenced.
Quantitative RT-PCR.
Total RNA was isolated from mouse tissue using TRIzol (Life Technologies) and treated with DNase. Two micrograms of RNA was reverse-transcribed using the SuperScript III First Strand kit (Life Technologies). Briefly, 2 μg total RNA, 50 ng/μL random hexamers, 10 mM dNTP mix, and water were incubated at 65 °C for 5 min in a total volume of 10 μL. For each reaction, 10 μL cDNA synthesis mix containing 10× reverse transcriptase (RT) buffer, 25 mM MgCl2, 0.1 M DTT, 40 U/μL RNaseOUT, and 200 U/μL SuperScript III RT were added to each RNA/primer mixture. Following a 10-min incubation at 25 °C, reverse transcription was performed at 50 °C for 50 min. Reactions were terminated at 85 °C for 5 min, followed by an additional incubation with 1 μL RNaseH for 20 min at 37 °C. All quantitative real-time PCR analysis was performed using the Step One Plus Real Time PCR system (Life Technologies). Analysis of ORF2 and CMVrtTA expression in tissues was performed by SYBR green analysis of cDNA samples. Primers used for ORF2 were forward (5′-TCGGCAAGGAGGAAGTGAAGATCAG-3′) and reverse (5′-GCTCTTGTTGCTGTTGATCTTGTAG-3′); CMVrtTA forward (5′-GCTTAATGAGGTCGGAATCG-3′) and reverse (5′-AGCAAAGCCCGCTTATTTTT-3′); Actin forward (5′-CGGTTCCGATGCCCTGAGGCTCTT-3′) and reverse (5′-CGTCACACTTCATGATGGAATTGA-3′). ORF2 and CMVrtTA expression was normalized to Actin and represented relative to the liver sample for each amplicon, respectively. All SYBR green PCR assays were performed in triplicate.
MEF Isolation.
MEFs were isolated at E14.5 and maintained in DMEM supplemented with 10% FBS, penicillin, streptomycin, and nonessential amino acids. For isolation of single-cell clones, MEFs were first immortalized with the SV40 T antigen and split into four independent pools. After expansion of independent pools, single-cell clones were isolated with sterile glass cloning cylinders (Sigma), and genomic DNA isolation was performed by phenol-choloroform extraction.
Quantitation of Insertional Frequencies.
The frequency of tet-ORFeus insertions was performed by SYBR green analysis of genomic DNA isolated from individual MEF clones relative to a standard curve with a known number of spliced L1 insertions. Importantly, the reverse primer used crosses the gene-trap splice junction and can anneal only to genomic DNA containing a bona fide L1 insertion after splicing and removal of the intron. To generate a standard curve, genomic DNA containing a (spliced) ORFeus insertion was first cloned into the TA cloning vector pCR 2.1 (Life Technologies), and then known copy numbers of the insertion were spiked into 5 ng of nontransgenic C57BL/6J strain mouse genomic DNA. Primers used were forward (5′-CCGAAGCCCCGCTACTAGTC-3′) and reverse (5′-GGCCCACCTGCAATTGAA-3′). All SYBR green PCR assays were performed in triplicate.
Cryosections.
Skin tissues were shaved, dissected, and fixed in 2% (wt/vol) paraformaldehyde/PBS for 30 min and then washed first with PBS (3× 10 min) and then overnight with 10% sucrose/PBS on a rotating platform. Tissue slices were embedded in Tissue-Tek optimum cutting temperature (OCT) compound (VWR) and then frozen with an ethanol/dry-ice bath and cryosectioned at 10 μm for visualization of melanocytes.
Immunofluorescence.
Antibodies for immunofluorescence were as follows: anti-TRP2 goat polyclonal (sc-10451; Santa Cruz Biotechnology) and donkey anti-goat Alexa 488 secondary (A-11055; Life Technologies). After washing, sections were mounted with Prolong Gold with DAPI (Life Technologies).
Supplementary Material
Acknowledgments
We thank C. Hawkins and The Johns Hopkins Transgenic Core Facility for assistance with pronuclear injections and generation of Tet-ORFeus mice; H. Varmus for CMVrtTA mice; J. Shelton and J. Lessard for help with skin cryosection preparation; J. Mendell for help with microscopy; and D. Maag and G. Schumann for sharing reagents. We also thank J. Mendell, L. Dai, and D. Largaespada for critical reading of the manuscript and B. Pavan for helpful discussions on white-spotted mutants. This work was supported by National Institutes of Health Grant CA16519 (to J.D.B.). K.A.O. was a Damon Runyon fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1918-06) and is currently a Cancer Prevention Research Institute of Texas Scholar in Cancer Research.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302504110/-/DCSupplemental.
References
- 1.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Waterston RH, et al. Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 4.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87(5):905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 5.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87(5):917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 6.Ovchinnikov I, Troxel AB, Swergold GD. Genomic characterization of recent human LINE-1 insertions: Evidence supporting random insertion. Genome Res. 2001;11(12):2050–2058. doi: 10.1101/gr.194701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han JS, Boeke JD. A highly active synthetic mammalian retrotransposon. Nature. 2004;429(6989):314–318. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- 8.An W, et al. Active retrotransposition by a synthetic L1 element in mice. Proc Natl Acad Sci USA. 2006;103(49):18662–18667. doi: 10.1073/pnas.0605300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.An W, et al. Conditional activation of a single-copy L1 transgene in mice by Cre. Genesis. 2008;46(7):373–383. doi: 10.1002/dvg.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gossen M, et al. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268(5218):1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 12.Sotillo R, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11(1):9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickins RA, et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat Genet. 2007;39(7):914–921. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.An W, et al. Plug and play modular strategies for synthetic retrotransposons. Methods. 2009;49(3):227–235. doi: 10.1016/j.ymeth.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44(1):23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 16.Gunther EJ, et al. Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev. 2003;17(4):488–501. doi: 10.1101/gad.1051603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vidigal JA, et al. An inducible RNA interference system for the functional dissection of mouse embryogenesis. Nucleic Acids Res. 2010;38(11):e122. doi: 10.1093/nar/gkq199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Y, et al. Cell division promotes efficient retrotransposition in a stable L1 reporter cell line. Mob DNA. 2013;4(1):10. doi: 10.1186/1759-8753-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baxter LL, Hou L, Loftus SK, Pavan WJ. Spotlight on spotted mice: A review of white spotting mouse mutants and associated human pigmentation disorders. Pigment Cell Res. 2004;17(3):215–224. doi: 10.1111/j.1600-0749.2004.00147.x. [DOI] [PubMed] [Google Scholar]
- 20.Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445(7130):843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- 21.White RM, Zon LI. Melanocytes in development, regeneration, and cancer. Cell Stem Cell. 2008;3(3):242–252. doi: 10.1016/j.stem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Wilkie AL, Jordan SA, Jackson IJ. Neural crest progenitors of the melanocyte lineage: Coat colour patterns revisited. Development. 2002;129(14):3349–3357. doi: 10.1242/dev.129.14.3349. [DOI] [PubMed] [Google Scholar]
- 23.Pavan WJ, Tilghman SM. Piebald lethal (sl) acts early to disrupt the development of neural crest-derived melanocytes. Proc Natl Acad Sci USA. 1994;91(15):7159–7163. doi: 10.1073/pnas.91.15.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukamoto K, Jackson IJ, Urabe K, Montague PM, Hearing VJ. A second tyrosinase-related protein, TRP-2, is a melanogenic enzyme termed DOPAchrome tautomerase. EMBO J. 1992;11(2):519–526. doi: 10.1002/j.1460-2075.1992.tb05082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawles ME. Origin of pigment cells from the neural crest in the mouse embryo. Physiol Zool. 1947;20(3):248–266. doi: 10.1086/physzool.20.3.30151958. [DOI] [PubMed] [Google Scholar]
- 26.Serbedzija GN, Bronner-Fraser M, Fraser SE. Developmental potential of trunk neural crest cells in the mouse. Development. 1994;120(7):1709–1718. doi: 10.1242/dev.120.7.1709. [DOI] [PubMed] [Google Scholar]
- 27.Serbedzija GN, Fraser SE, Bronner-Fraser M. Pathways of trunk neural crest cell migration in the mouse embryo as revealed by vital dye labelling. Development. 1990;108(4):605–612. doi: 10.1242/dev.108.4.605. [DOI] [PubMed] [Google Scholar]
- 28.Collier LS, et al. Whole-body sleeping beauty mutagenesis can cause penetrant leukemia/lymphoma and rare high-grade glioma without associated embryonic lethality. Cancer Res. 2009;69(21):8429–8437. doi: 10.1158/0008-5472.CAN-09-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436(7048):221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 30.Rad R, et al. PiggyBac transposon mutagenesis: A tool for cancer gene discovery in mice. Science. 2010;330(6007):1104–1107. doi: 10.1126/science.1193004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436(7048):272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 32.Ding S, et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122(3):473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Keng VW, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27(3):264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starr TK, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323(5922):1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Donnell KA, et al. A Sleeping Beauty mutagenesis screen reveals a tumor suppressor role for Ncoa2/Src-2 in liver cancer. Proc Natl Acad Sci USA. 2012;109(21):E1377–E1386. doi: 10.1073/pnas.1115433109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chia W, McGill S, Karp R, Gubb D, Ashburner M. Spontaneous excision of a large composite transposable element of Drosophila melanogaster. Nature. 1985;316(6023):81–83. doi: 10.1038/316081a0. [DOI] [PubMed] [Google Scholar]
- 37.Lim JK. Intrachromosomal rearrangements mediated by hobo transposons in Drosophila melanogaster. Proc Natl Acad Sci USA. 1988;85(23):9153–9157. doi: 10.1073/pnas.85.23.9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geurts AM, et al. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. PLoS Genet. 2006;2(9):e156. doi: 10.1371/journal.pgen.0020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlson CM, et al. Transposon mutagenesis of the mouse germline. Genetics. 2003;165(1):243–256. doi: 10.1093/genetics/165.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keng VW, et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat Methods. 2005;2(10):763–769. doi: 10.1038/nmeth795. [DOI] [PubMed] [Google Scholar]
- 41.Bestor TH. Transposons reanimated in mice. Cell. 2005;122(3):322–325. doi: 10.1016/j.cell.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 42.Badea TC, et al. New mouse lines for the analysis of neuronal morphology using CreER(T)/loxP-directed sparse labeling. PLoS ONE. 2009;4(11):e7859. doi: 10.1371/journal.pone.0007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bejar R, Yasuda R, Krugers H, Hood K, Mayford M. Transgenic calmodulin-dependent protein kinase II activation: Dose-dependent effects on synaptic plasticity, learning, and memory. J Neurosci. 2002;22(13):5719–5726. doi: 10.1523/JNEUROSCI.22-13-05719.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 45.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479(7374):534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460(7259):1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435(7044):903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 48.Symer DE, et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110(3):327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.