Abstract
Isoprenylcysteine carboxyl methyltransferases (Icmts) are a class of integral membrane protein methyltransferases localized to the endoplasmic reticulum (ER) membrane in eukaryotes. The Icmts from human (hIcmt) and S. cerevisae (Ste14p) catalyze the α-carboxyl methyl esterification step in the post-translational processing of CaaX proteins, including the yeast a-factor mating pheromones and both human and yeast Ras proteins. Herein, we evaluated synthetic analogs of two well-characterized Icmt substrates, N-acetyl-S-farnesyl-L-cysteine (AFC) and the yeast a-factor peptide mating pheromone, that contain photoactive benzophenone moieties in either the lipid or peptide portion of the molecule. The AFC based-compounds were substrates for both hIcmt and Ste14p, whereas the a-factor analogs were only substrates for Ste14p. However, the a-factor analogs were found to be micromolar inhibitors of hIcmt. Together, these data suggest that the Icmt substrate binding site is dependent upon features in both the isoprenyl moiety and upstream amino acid composition and that hIcmt and Ste14p have overlapping, yet distinct, substrate specificities. Photocrosslinking and neutravidin-agarose capture experiments with these analogs revealed that both hIcmt and Ste14p were specifically photolabeled to varying degrees with all of the compounds tested. These data suggest that these analogs will be useful for the future identification of the Icmt substrate binding sites.
Keywords: Icmt, Ste14p, a-factor, photocrosslinking, benzophenone, methyltransferase
INTRODUCTION
Many eukaryotic proteins are initially synthesized with a C-terminal amino acids CaaX motif that signals a series of post-translational modifications including isoprenylation of the cysteine (C) by either a farnesyl or geranylgeranyl moiety, proteolysis of the –aaX residues and α-carboxyl methyl esterification of the newly exposed cysteine residue [1-3]. CaaX proteins include the Ras superfamily of small GTPases [4, 5], Rheb, the nuclear lamins, the Rho family of GTPases and the γ subunits of heterotrimeric G proteins [2, 6-8].
The only enzymes known to methyl esterify the α-carboxylate group of CaaX proteins are the isoprenylcysteine carboxyl methyltransferases (Icmts), a family of integral membrane proteins localized to the endoplasmic reticulum (ER) [2, 6, 9-12]. Ste14p from S. cerevisiae, the founding member of the Icmt family of enzymes, is a 26-kDa integral membrane protein with six putative transmembrane helices [13, 14]. The human enzyme, hIcmt, which shares 41% identity and 63% similarity with Ste14p, is a 33-kDa membrane protein with putative eight transmembrane helices [15-17]. Interestingly, hIcmt functionally complements the mating defect of a Δste14 strain by methylating the a-factor peptide, suggesting that the enzymes have overlapping substrate specificities [15]. In addition to CaaX proteins and peptides, numerous small molecules such as N-acetyl-S-farnesyl-L-cysteine (AFC), N-acetyl-S-geranylgeranyl-L-cysteine (AGGC), and farnesyl thiopropionic acid (FTP) have been shown to be substrates for both human and yeast Icmts [18, 19], while other compounds have shown specificity for the yeast enzyme [17].
Aside from these few examples, little is known about the differences in substrate specificity between the yeast and human enzymes, nor is it known how and where the substrates bind to these Icmts. A recent 3.4 Å crystal structure of the prokaryotic Icmt ortholog Ma-ICMT has been published that revealed important well-conserved structural features of the binding pocket for the co-substrate S-adenosyl-L-methionine (SAM) [20]. The structure also showed a conserved access tunnel for lipidated substrates that is comprised of residues both in the C-terminal SAM-binding domain and those in the N-terminal half of the protein [20]. The N-terminal segment of the protein is thought to confer substrate specificity for lipid substrates. However, poor sequence conservation between Ma-ICMT and the eukaryotic Icmts in the N-terminal half precluded a definitive analysis of residues important for binding the isoprene moieties and thus, different approaches must be taken to identify these key amino acids.
Benzophenone-modified substrate analogs have been used previously to interrogate the protein binding sites for farnesyl and geranylgeranyl moieties. The utility of this approach was first demonstrated in experiments designed to probe the interaction between the CaaX protein Rho and its regulator RhoGDI [21]. In that study, an isoprenoid-containing cysteine analog bearing the benzophenone label in the lipid itself was used to demonstrate that the isoprene group itself specifically interacts with RhoGDI. Most recently, a series of benzophenone-modified peptide analogs based on the yeast a-factor sequence was developed and used to examine the activity of yeast CaaX protease Ras converting enzyme 1 (Rce1p). These experiments demonstrated that yeast Rce1p recognized the modified analogs as substrates and each analog specifically photoaffinity labeled the protein [22] [23]. Similarly, carboxyl methylated, photoactive analogs of the a-factor mating pheromone peptide were shown to functionally interact with the a-factor receptor, Ste3p [24].
In this study, we synthesized and examined several analogs of AFC and farnesylated a-factor peptides that contained both a biotin tag and a photoactive benzophenone moiety, either in the isoprene unit or in the peptide region, for their ability to bind and act as substrates or inhibitors of hIcmt and Ste14p. The benzophenone group allowed for specific photoaffinity labeling of the substrate binding site and the biotin moiety allowed for isolation of the labeled protein from a crude membrane mixture. Using kinetic studies, we determined that the AFC analogs were substrates for both enzymes. The a-factor analogs were only substrates for Ste14p but were competitive inhibitors of hIcmt. Furthermore, using photocrosslinking experiments followed by isolation with neutravidin-agarose beads, we determined that both Ste14p and hIcmt were labeled by all of the analogs tested, albeit to varying degrees, under saturating conditions.
MATERIALS AND METHODS
Chemistry
Materials
All solvents and reagents used for the solid-phase peptide synthesis of the photoactivatable peptides were of analytical grade and purchased from Peptides International (Louisville, KY), NovaBioChem® (Nohenbrunn, Germany), or Sigma-Aldrich (St. Louis, MO). N-acetyl-S-farnesyl-L-cysteine (AFC) was synthesized in the Gibbs laboratory (Purdue University) as previously described [25]. High performance liquid chromatography grade acetonitrile (CH3CN), dichloromethane (DCM), N,N-dimethylformamide (DMF), trifluoroacetic acid (TFA), and H2O were purchased from Fisher Scientific (Springfield, NJ), OmniSolv® (Charlotte, NC), or Sigma-Aldrich. NHS-PEG4-Biotin was obtained from Thermo Scientific (Waltham, MA).
Synthesis and Chemical Characterization of Photoaffinity Analogs
Detailed descriptions for the synthesis and chemical characterization of the photoaffinity analogs Am-bpBFC BPA Analog (2), F-bpBFC AFC Analog (3), Biotin-Peg4-YIIKGVFWDPAC (4), Biotin-Peg4-YIIKGVFWDPAC(C5-meta-Bp) (7a), Biotin-Peg4-YIIKGVFWDPAC(C5-para-Bp) (7b), Biotin-Peg 4-YIIKGVFWDPAC(C10-meta-Bp) (8a) and Biotin-Peg4-YIIKGVFWDPAC(C10-para-Bp) (8b) are found in Supplementary Data.
Biochemical Evaluation
Materials
S-adenosyl-L-[14C-methyl] methionine ([14C]-SAM) was purchased from Perkin Elmer (Waltham, MA). α-myc monoclonal antibody, goat α-mouse IgG and goat α-rabbit IgG were purchased from Invitrogen (Carlsbad, CA). The α-Ste14 polyclonal antibody was a gift from Dr. S. Michaelis (The Johns Hopkins University School of Medicine). The neutravidin coated agarose beads and SuperSignal West Pico enhanced chemiluminescence (ECL) were purchased from Pierce (Rockford, IL).
Yeast strains and crude membrane preparations from yeast cells
His10myc3N-Ste14p (His-Ste14p) and His10myc3N-hIcmt (His-hIcmt) yeast strains were cloned and expressed as previously described [9, 17]. Crude membranes were prepared as described previously with minor modifications [9]. After centrifugation at 100,000 × g, the membrane pellet was resuspended in 10 mM Tris-HCl, pH 7.5, aliquoted, flash frozen in liquid N2 and stored at −80°C.
In Vitro Methyltransferase Vapor Diffusion Assay
Reactions were performed as described previously [26]. All inhibition studies were completed as detailed earlier [17]. Crude membrane preparations derived from a Δste14 strain transformed with an empty vector were used as the negative control for these experiments.
Photocrosslinking and neutravidin-agarose pull-down assays
Photocrosslinking assays were performed as described previously, with minor modifications [22]. Briefly, 100 μg of crude membrane preparation expressing either His-Ste14p or His-hIcmt in 100 mM Tris-HCl, pH 7.5 were incubated in the presence of saturating concentrations of the photoaffinity analogs and incubated at 4°C for 10 min. After incubation, the samples were irradiated with UV light (365 removed nm) in 96-well plates for 40 min on ice. Following photocrosslinking, unreacted analog was by chloroform/methanol extraction [27]. The resulting protein samples were solubilized in 400 μL of radioimmunoprecipitation assay (RIPA) buffer (25mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecylsulfate) /10% SDS and incubated with 50 μL of a 50% neutravidin/RIPA bead slurry for 2 h at room temperature. Following incubation, the beads were centrifuged at 13,000 × g for 1 min and washed three times with RIPA/10% SDS. The crosslinked protein was eluted from the neutravidin beads by the addition of 50 μL of 2X SDS sample buffer (0.5 M Tris-HCl, pH 6.8, 30% sucrose (w/v), 10% sodium dodecylsulfate (w/v), 3.5 M 2-mercaptoethanol and 0.1% bromophenol blue (w/v)). The His-Ste14p samples were heated for 30 min at 65°C and the His-hIcmt samples were incubated at room temperature overnight. The proteins were separated by 10% SDS-PAGE and transferred to nitrocellulose (0.22 μm). The nitrocellulose membrane was blocked at room temperature for 2 h in 20% (w/v) non-fat dry milk in phosphate-buffered saline with Tween-20 (137 mM NaCl, 2.7 mM KCl, 4 mM Na2HPO4, 1.8 mM KH2PO4 and 0.05% (v/v) Tween-20, pH 7.4) (PBST). The blocked membrane was then incubated for 2 h at room temperature with primary antibody (1:1,000 α-Ste14p) or (1:10,000 α-myc) in 5% (w/v) non-fat dry milk in PBST for His-Ste14p or His-hIcmt, respectively. The membrane was washed in PBST three times and then incubated for 1 h at room temperature with secondary antibody (1:10,000 goat α-rabbit IgG-HRP for α-Ste14p or 1:4000 goat α-mouse IgG-HRP for α-myc) in 5% (w/v) dry milk in PBST. The membrane was washed three times with PBST and the proteins were visualized using ECL.
RESULTS AND DISCUSSION
Probe synthesis
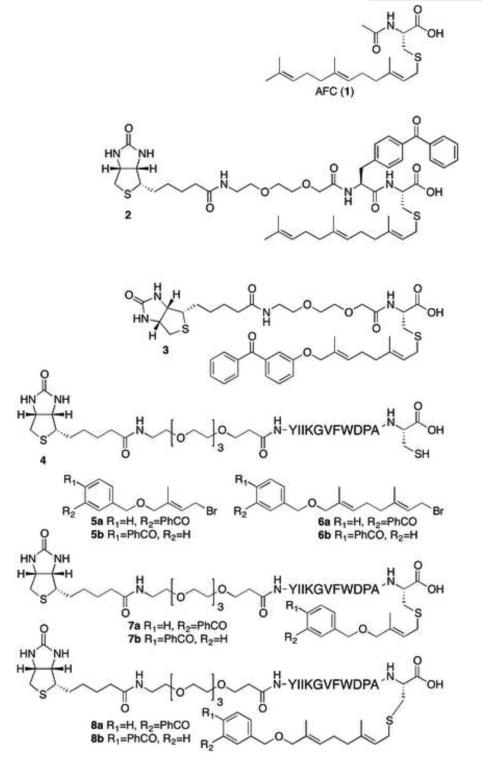
All of the photoactive probes used in the studies described here were prepared via solid phase synthesis (Fig.1). For the preparation of the AFC analogs, the compounds were assembled starting with Fmoc-Cys(S-t-Bu)-OH that had been immobilized on 2-chlorotrityl resin [28]. Side chain deprotection followed by S-alkylation was used to install either a farnesyl group (for 2) or a benzophenone-based photolabel (for 3) [29]. Photolabel 3 was based on a previously described AFC analog [21]. Fmoc deprotection and subsequent chain elongation using standard solid phase synthetic procedures followed by cleavage under mild acidic conditions afforded the desired probes. For the preparation of the a-factor-based analogs, the biotinylated peptide 4 was first prepared via standard solid phase synthetic procedures. That compound was then alkylated with the short chain benzophenone-functionalized isoprenoids 5a or 5b [30, 31]to yield 7a or 7b, respectively; similarly, alkylation of 4 with 6a or 6b [32, 33]yielded 7a or 7b, respectively.
Figure 1. Structures of Photoactive Analogs of AFC and the a-factor peptide from S.
cerevisiae.
All photoaffinity analogs were substrates of His-Ste14p
We first examined the ability of the benzophenone-labeled AFC (1) analogs (2 and 3) and a-factor mimetics (7a, 7b, 8a and 8b) to act as substrates for yeast His10myc3N-Ste14p (His-Ste14p) using an in vitro methyltransferase vapor diffusion assay. We determined the Km and Vmax values for each analog using crude membrane preparations from yeast expressing His-Ste14p (5μg), using AFC as the positive control [17]. The small molecule photoaffinity compounds based on AFC (2 and 3), demonstrated similar Km values to that of AFC (Table 1) [17]. The a-factor peptide photoaffinity analogs with the benzophenone substituted in the meta position (7a and 8a) also demonstrated similar Km values to AFC (Table 1). Interestingly, the Km values for the a-factor analogs containing a para-substituted benzophenone moiety (7b and 8b) were 2 – 3 times lower than the meta-substituted analogs. These data suggest that 7b and 8b interact with the His-Ste14p binding site with higher affinity, possibly as a result of the increased linear and aromatic hydrophobicity of the analogs. AFC and the small molecule AFC-based analogs substrates may demonstrate greater Km values than the a-factor analogs because they lack favorable downstream amino acid contacts with the His-Ste14p substrate binding site. Together, these data suggest that features of both the lipid moiety and the peptide sequence affect binding to His-Ste14p. However, it is important to note that while the isoprene moiety is absolutely essential for recognition by His-Ste14p and other Icmts, the peptide sequence is dispensable [8], and may serve as a more subtle recognition factor for the selection of substrates in vivo.
Table 1.
Kinetic Parameters for AFC and a-Factor Photoaffinity Analogs as Substrates for His-Ste14p.
| Compound | Km(app)a,b | Vmaxa,b |
|---|---|---|
| μM | pmol methyl groups transferred/min/mg |
|
| 1 | 15.9 ± 0.9[17] | 869.6 ± 14.9 |
| 2 | 17.9 ± 0.5 | 2072 ± 112 |
| 3 | 18.3 ± 0.4 | 362 ± 4.2 |
| 7a | 14.6 ± 0.3 | 762 ± 28 |
| 7b | 6.1 ± 0.6 | 320 ± 16 |
| 8a | 15 ± 0.2 | 473 ± 8 |
| 8b | 5.1 ± 0.6 | 240 ± 3.8 |
Data are the average of three experiments performed in duplicate ± standard deviation (S.D.)
Values were determined by fitting data to classical (Michaelis-Menten) enzyme-catalyzed curve data using GraphPad Prism 4.
Although the Km values for 2, 3, 7a and 8a were similar for His-Ste14p, the Vmax values varied greatly (Table 1). Analog 2, which contains a photoactive a p-benzoyl-L-phenylalanine upstream of the cysteine residue, demonstrated the greatest Vmax (2072 pmol/min/mg). This value was more than 2-fold greater than that of AFC (869 pmol/min/mg), the compound defined as the minimal substrate for Icmts that contains peptide character [8]. These data suggest that His-Ste14p can turn over a substrate containing a bulky hydrophobic moiety next to the lipidated cysteine residue more quickly and may point to a mechanism in which product release is rate limiting. Analog 3 manifested the lowest Vmax, 362 pmol/min/mg, with analogs 7a and 8a showing midrange Vmax values of 473 and 762 pmol/min/mg, respectively (Table 1). Analogs 7b and 8b, which had the lowest Km values, showed the lowest Vmax values of all the substrates tested against His-Ste14p. This reduction in substrate turnover may reflect a suboptimal conformation for effective catalysis or a decreased ability to release product following the reaction.
AFC photoaffinity analogs were substrates for His-hIcmt but the a-factor photoaffinity peptide analogs were inhibitors
We next examined the ability of these analogs to act as substrates for His10myc3N-hIcmt (His-hIcmt). We performed kinetic analyses to determine the Km and Vmax values for each of the analogs and to establish if differences in substrate specificity existed between the yeast and human enzymes (Table 2). We found that 2 is a substrate for His-hIcmt with a Km lower than that for AFC and a Vmax that was only slightly decreased. However, 3, which contains the modified isoprenyl group, was not a substrate for His-hIcmt to any measurable extent and thus, the Km value is not particularly relevant (Table 2). Furthermore, even though the native a-factor peptide is a substrate for His-hIcmt [15], none of the modified a-factor analogs displayed any measurable activity (data not shown). However, all of a-factor peptides were inhibitors of His-hIcmt, to varying degrees (Table 2). Analogs 7a and 7b, which have a shorter 5-carbon isoprene spacer, were poor inhibitors of His-hIcmt (IC50 >100 μM) whereas 8a and 8b, which have a 10-carbon isoprene spacer, were relatively potent, displaying IC50 values of 25.7 ± 1.6 μM and 13.7 ± 0.8 μM, respectively (Table 2). The calculated Ki values for 8a and 8b using the Cheng – Prusoff method were 13.0 ± 0.8 μM and 7.0 ± 0.4 μM, respectively [34]. These marked differences between the activities of these photoaffinity analogs and His-Ste14p and His-Icmt highlight previously unappreciated substrate specificity differences between the different Icmts. Furthermore, these data suggest that the hIcmt binding site is less tolerant of changes in the isoprene architecture than is Ste14p.
Table 2.
Kinetic Parameters for AFC and a-Factor Photoaffinity Analogs as Substrates for His-hIcmt.
| Compound | Kma,b | Vmaxa,b | IC50a,c |
|---|---|---|---|
| μM | pmol methyl groups transferred/min/mg |
μM | |
| 1 | 10.3 ± 0.5[17] | 884 ± 49 | N. A. |
| 2 | 5.9 ± 0.5 | 504 ± 27 | N. A. |
| 3 | 3.4 ± 0.3 | 24 ± 2 | N. A. |
| 7a | N. D. | N. D. | > 100 |
| 7b | N. D. | N. D. | > 100 |
| 8a | N. D. | N. D. | 25.7 ± 1.6 |
| 8b | N. D. | N. D. | 13.7 ± 0.8 |
Data are the average of three experiments performed in duplicate ± standard deviation (S.D.)
Values were determined by fitting data to classical (Michaelis-Menten) enzyme-catalyzed curve data using GraphPad Prism 4.
Values were found by fitting data to variable slope sigmoidal dose-response curve using GraphPad Prism 4.
His-Ste14p specifically interacted with each of the benzophenone-modified analogs
To evaluate the ability of His-Ste14p to be labeled with the analogs, we performed photoaffinity labeling studies. Crude membranes expressing His-Ste14p (100 μg) were incubated with saturating concentrations of each substrate, based on kinetic parameters, and photocrosslinked with UV light (365 nm) for 40 min [22]. The crosslinked proteins were isolated by neutravidin-agarose capture exploiting the biotin tag on each analog and resolved by SDS-PAGE. Incubation with AFC served as the negative control and membrane preparations from a Δste14 deletion strain served as the control for non-specific binding. The proteins were visualized by enhanced chemiluminescence (ECL) following immunoblot analysis. The signal corresponded to the amount of analog covalently crosslinked to His-Ste14p (Fig 2A). Although His-Ste14p was labeled with all of the analogs, it is noteworthy that 2 and 7a crosslinked to the protein less efficiently than the other compounds, which labeled His-Ste14p with approximately equal efficiency. Paradoxically, 2 was the best substrate for His-Ste14p enzymatically, suggesting that the benzophenone moiety was not in an ideal position in the binding pocket for efficient labeling.
Figure 2. Immumoblot of Analyses of His-Ste14p (A) or His-hIcmt (B) photocrosslinked with benzophenone-containing analogs.
One hundred μg of crude membrane protein were incubated with each substrate for 40 min on ice under UV irradiation (365nm). The samples were extracted and re-solubilized in RIPA/10% SDS before enrichment using neutravidin-agarose beads. Proteins were eluted, resolved by 10% SDS-PAGE, and visualized as described in Materials and Methods. A: His-Ste14p: lane 1: 200 μM 1, lane 2:100 μM 2, lane 3:100 μM 3, lane 4: 50 μM 7b, lane 5: 100 μM 7a , lane 6: 50 μM probe 8b, lane 7: 100 μM 8a, lane 8: empty vector (Δste14) + 100 μM 2. B: His-hIcmt: lane 1: 200 μM 1, lane 2: 50 μM 2, lane 3: 50 μM 3, lane 4: 200 μM 7b, lane 5: 200 μM 7a, lane 6: 200 μM 8b, lane 7: 200 μM 8a, lane 8: empty vector (Δste14) + 50 μM 2.
Crosslinking experiments of modified substrates exhibit the ability to covalently crosslink to His-hIcmt
To determine if His-hIcmt interacted similarly with the analogs, crude membrane extracts expressing His-hIcmt (100 μg) were incubated with saturating concentrations of each of the photoactive reagents (200 μM). Samples with His-hIcmt expressing membranes incubated with AFC were used as the negative control and the membrane preparation from the Δste14 strain was used as a non-specific binding control. All of the samples were crosslinked, enriched, and resolved under the same conditions as described above for His-Ste14p. The amount of each crosslinked protein was determined by immunoblot analysis followed by visualization by ECL (Figure 2B). These data indicated that all of the analogs labeled the protein but that the degree of crosslinking varied with the structure of these molecules. Analog 2 crosslinked more efficiently than 3, possibly due to the fact that 3 was a poor substrate for His-hIcmt (Table 2). For the peptide analogs, 8a labeled His-hIcmt most efficiently followed by 7b, 8b and 7a. In an effort to determine the nature of inhibition demonstrated by the a-factor analogs, we performed competition experiments with His-Icmt and 8b (200 μM) using increasing concentrations of the substrate AFC. The intensity of photolabeling of His-Icmt with 8b decreased with increasing AFC concentration, suggesting that 8b acted as a competitive inhibitor (data not shown). These data suggest that the a-factor analogs interact and specifically label residues within the His-Icmt binding pocket
It is clear from our data that the labeling efficiencies for the benzophenone-containing substrate analogs did not correlate completely to the kinetic data. For example, the a-factor peptide that photolabeled His-hIcmt most efficiently was one of the poorest inhibitors (Table 2 and Fig. 2B). Thus, care must be taken not to over interpret the crosslinking data. Since variables such as proximity and orientation of the moiety to the residues in the binding pocket affect the ability to efficiently label a protein [35] [36], the observed variances in crosslinking to His-Ste14p and His-hIcmt may be due to the position of the photolabel in the substrate binding site rather than a measure of their affinity for the enzymes. However, since little structural information is available for these integral membrane proteins, these analogs will be invaluable tools in our future efforts to identify and define the substrate binding sites for both yeast and human Icmts.
Supplementary Material
Highlights.
– Novel benzophenone and biotin analogs of AFC and a-factor were synthesized.
– AFC analogs were substrates of Ste14p and hIcmt, to varying degrees.
– The a-factor analogs were substrates for Ste14p but weak inhibitors of hIcmt.
– Analogs photocrosslinked specifically to the substrate binding sites of hIcmt and Ste14p.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Hrycyna CA, Clarke S. Modification of eukaryotic signaling proteins by C-terminal methylation reactions. Pharmacol Ther. 1993;59:281–300. doi: 10.1016/0163-7258(93)90071-k. [DOI] [PubMed] [Google Scholar]
- [2].Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- [3].Clarke S. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu Rev Biochem. 1992;61:355–386. doi: 10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- [4].Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- [5].Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- [6].Glomset JA, Gelb MH, Farnsworth CC. Prenyl proteins in eukaryotic cells: a new type of membrane anchor. Trends Biochem Sci. 1990;15:139–142. doi: 10.1016/0968-0004(90)90213-u. [DOI] [PubMed] [Google Scholar]
- [7].Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- [8].Tan EW, Perez-Sala D, Canada FJ, Rando RR. Identifying the recognition unit for G protein methylation. J Biol Chem. 1991;266:10719–10722. [PubMed] [Google Scholar]
- [9].Anderson JL, Frase H, Michaelis S, Hrycyna CA. Purification, functional reconstitution, and characterization of the Saccharomyces cerevisiae isoprenylcysteine carboxylmethyltransferase Ste14p. J Biol Chem. 2005;280:7336–7345. doi: 10.1074/jbc.M410292200. [DOI] [PubMed] [Google Scholar]
- [10].Hrycyna CA, Wait SJ, Backlund PS, Jr., Michaelis S. Yeast STE14 methyltransferase, expressed as TrpE-STE14 fusion protein in Escherichia coli, for in vitro carboxylmethylation of prenylated polypeptides. Methods Enzymol. 1995;250:251–266. doi: 10.1016/0076-6879(95)50077-4. [DOI] [PubMed] [Google Scholar]
- [11].Court H, Hahne K, Philips Mark R., Hrycyna CA. Biochemical and Biological Fuctions of Isoprenylcysteine Carboxyl Methyltransferase. In: Christine MOB, Hrycyna A, Tamanoi Fuyuhiko, editors. The Enzymes. vol. 30. Elsveir; 2011. pp. 1–329. [Google Scholar]
- [12].Reid TS, Terry KL, Casey PJ, Beese LS. Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity. J Mol Biol. 2004;343:417–433. doi: 10.1016/j.jmb.2004.08.056. [DOI] [PubMed] [Google Scholar]
- [13].Romano JD, Michaelis S. Topological and mutational analysis of Saccharomyces cerevisiae Ste14p, founding member of the isoprenylcysteine carboxyl methyltransferase family. Mol Biol Cell. 2001;12:1957–1971. doi: 10.1091/mbc.12.7.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hrycyna CA, Sapperstein SK, Clarke S, Michaelis S. The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and RAS proteins. EMBO J. 1991;10:1699–1709. doi: 10.1002/j.1460-2075.1991.tb07694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dai Q, Choy E, Chiu V, Romano J, Slivka SR, Steitz SA, Michaelis S, Philips MR. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J Biol Chem. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- [16].Wright LP, Court H, Mor A, Ahearn IM, Casey PJ, Philips MR. Topology of mammalian isoprenylcysteine carboxyl methyltransferase determined in live cells with a fluorescent probe. Mol Cell Biol. 2009;29:1826–1833. doi: 10.1128/MCB.01719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Anderson JL, Henriksen BS, Gibbs RA, Hrycyna CA. The isoprenoid substrate specificity of isoprenylcysteine carboxylmethyltransferase: development of novel inhibitors. J Biol Chem. 2005;280:29454–29461. doi: 10.1074/jbc.M504982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Volker C, Lane P, Kwee C, Johnson M, Stock J. A single activity carboxyl methylates both farnesyl and geranylgeranyl cysteine residues. FEBS Lett. 1991;295:189–194. doi: 10.1016/0014-5793(91)81415-5. [DOI] [PubMed] [Google Scholar]
- [19].Volker C, Miller RA, McCleary WR, Rao A, Poenie M, Backer JM, Stock JB. Effects of farnesylcysteine analogs on protein carboxyl methylation and signal transduction. J Biol Chem. 1991;266:21515–21522. [PubMed] [Google Scholar]
- [20].Yang J, Kulkarni K, Manolaridis I, Zhang Z, Dodd RB, Mas-Droux C, Barford D. Mechanism of isoprenylcysteine carboxyl methylation from the crystal structure of the integral membrane methyltransferase ICMT. Mol Cell. 2011;44:997–1004. doi: 10.1016/j.molcel.2011.10.020. [DOI] [PubMed] [Google Scholar]
- [21].Kale TA, Raab C, Yu N, Dean DC, Distefano MD. A photoactivatable prenylated cysteine designed to study isoprenoid recognition. J Am Chem Soc. 2001;123:4373–4381. doi: 10.1021/ja0012016. [DOI] [PubMed] [Google Scholar]
- [22].Kyro K, Manandhar SP, Mullen D, Schmidt WK, Distefano MD. Photoaffinity labeling of Ras converting enzyme 1 (Rce1p) using a benzophenone-containing peptide substrate. Bioorg Med Chem. 2010;18:5675–5684. doi: 10.1016/j.bmc.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kyro K, Manandhar SP, Mullen D, Schmidt WK, Distefano MD. Photoaffinity labeling of Ras converting enzyme using peptide substrates that incorporate benzoylphenylalanine (Bpa) residues: improved labeling and structural implications. Bioorg Med Chem. 2011;19:7559–7569. doi: 10.1016/j.bmc.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mullen DG, Kyro K, Hauser M, Gustavsson M, Veglia G, Becker JM, Naider F, Distefano MD. Synthesis of a-factor peptide from Saccharomyces cerevisiae and photoactive analogues via Fmoc solid phase methodology. Bioorg Med Chem. 2011;19:490–497. doi: 10.1016/j.bmc.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mark PDM, Brown J, Lever David C., Epstein William W., Poulter Dale. Prenylated Proteins. A Convenient Synthesis of Farnesyl Cysteinyl Thioethers. Journal of the American Chemical Society. 1991;113:3176–3177. [Google Scholar]
- [26].Griggs AM, Hahne K, Hrycyna CA. Functional oligomerization of the Saccharomyce cerevisiae isoprenylcysteine carboxyl methyltransferase, Ste14p. J Biol Chem. 2010;285:13380–13387. doi: 10.1074/jbc.M109.061366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- [28].Donelson JL, Hodges-Loaiza HB, Henriksen BS, Hrycyna CA, Gibbs RA. Solid-phase synthesis of prenylcysteine analogs. J Org Chem. 2009;74:2975–2981. doi: 10.1021/jo8021692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Volkert M, Uwai K, Tebbe A, Popkirova B, Wagner M, Kuhlmann J, Waldmann H. Synthesis and biological activity of photoactivatable N-ras peptides and proteins. J Am Chem Soc. 2003;125:12749–12758. doi: 10.1021/ja036178d. [DOI] [PubMed] [Google Scholar]
- [30].Turek TC, Gaon I, Distefano MD. Analogs of farnesyl pyrophosphate incorporating internal benzoylbenzoate esters: Synthesis, inhibition kinetics and photoinactivation of yeast protein farnesyltransferase. Tetrahedron Letters. 1996;37:4845–4848. doi: 10.1021/jo9602736. [DOI] [PubMed] [Google Scholar]
- [31].Turek TC, Gaon I, Distefano MD, Strickland CL. Synthesis of farnesyl diphosphate analogues containing ether-linked photoactive benzophenones and their application in studies of protein prenyltransferases. Journal of Organic Chemistry. 2001;66:3253–3264. doi: 10.1021/jo991130x. [DOI] [PubMed] [Google Scholar]
- [32].Gaon I, Turek TC, Weller VA, Edelstein RL, Singh SK, Distefano MD. Photoactive analogs of farnesyl pyrophosphate containing benzoylbenzoate esters: Synthesis and application to photoaffinity labeling of yeast protein farnesyltransferase. Journal of Organic Chemistry. 1996;61:7738–7745. doi: 10.1021/jo9602736. [DOI] [PubMed] [Google Scholar]
- [33].Turek-Etienne TC, Strickland CL, Distefano MD. Biochemical and structural studies with prenyl diphosphate analogues provide insights into isoprenoid recognition by protein farnesyl transferase. Biochemistry. 2003;42:3716–3724. doi: 10.1021/bi0266838. [DOI] [PubMed] [Google Scholar]
- [34].Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- [35].Rowland MM. Chemistry. University of Tennessee; Knoxville: 2011. Chemical Tools to Characterize Membrane-Protein Binding Interactions Using Synthetic Lipid Probes; p. 125. vol. Ph.D. [Google Scholar]
- [36].Ballell L, Alink KJ, Slijper M, Versluis C, Liskamp RM, Pieters RJ. A new chemical probe for proteomics of carbohydrate-binding proteins. Chembiochem. 2005;6:291–295. doi: 10.1002/cbic.200400209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.