Abstract
Background and Aims
Sexual reproduction is one of the most important moments in a life cycle, determining the genetic composition of individual offspring. Controlled pollination experiments often show high variation in the mating system at the individual level, suggesting a persistence of individual variation in natural populations. Individual variation in mating patterns may have significant adaptive implications for a population and for the entire species. Nevertheless, field data rarely address individual differences in mating patterns, focusing rather on averages. This study aimed to quantify individual variation in the different components of mating patterns.
Methods
Microsatellite data were used from 421 adult trees and 1911 seeds, structured in 72 half-sib families collected in a single mixed stand of Quercus robur and Q. petraea in northern Poland. Using a Bayesian approach, mating patterns were investigated, taking into account pollen dispersal, male fecundity, possible hybridization and heterogeneity in immigrant pollen pools.
Key Results
Pollen dispersal followed a heavy-tailed distribution (283 m on average). In spite of high pollen mobility, immigrant pollen pools showed strong genetic structuring among mothers. At the individual level, immigrant pollen pools showed highly variable divergence rates, revealing that sources of immigrant pollen can vary greatly among particular trees. Within the stand, the distribution of male fecundity appeared highly skewed, with a small number of dominant males, resulting in a ratio of census to effective density of pollen donors of 5·3. Male fecundity was not correlated with tree diameter but showed strong cline-like spatial variation. This pattern can be attributed to environmental variation. Quercus petraea revealed a greater preference (74 %) towards intraspecific mating than Q. robur (36 %), although mating preferences varied among trees.
Conclusions
Mating patterns can reveal great variation among individuals, even within a single even-age stand. The results show that trees can mate assortatively, with little respect for spatial proximity. Such selective mating may be a result of variable combining compatibility among trees due to genetic and/or environmental factors.
Keywords: Mating system, pollen dispersal, immigrant pollen structure, male fecundity, hybridization, microsatellite, Bayesian approach, oak, Quercus robur, Q. petraea
INTRODUCTION
Sexual reproduction is one of the most important moments in a life cycle, determining the genetic composition of individual offspring. The way that genetic information is transferred from generation to generation underlies not only individual genotypes but also the distribution of genetic diversity within and among populations. Furthermore, mating patterns have an effect on adaptability potential of populations and, ultimately, the rate of species evolution (Schoen and Brown, 1991; Barrett and Harder, 1996). In wind- and animal-pollinated plants, outcross mating encompasses the processes of gene dispersal within and gene flow among populations (Ward et al., 2005; Ashley, 2010).
Mating patterns within populations are typically far from random. The mechanics of pollen transport alone can constrain the probability of pollination between plants, probably regardless of the pollen vector used. Most often, the probability of pollen dispersal decreases with distance from a source plant (Levin and Kerster, 1974; Ashley, 2010). This implies the existence of a ‘mating neighbourhood’ (although not a discrete spatial sub-unit, cf. Rousset, 2007) with dimensions scaled by a pollen dispersal distribution. However, proximity between potential mates does not ensure mating because, for example, flowering asynchrony (involving timing of pollen shedding and female receptivity) can limit mating opportunities (El-Kassaby and Ritland, 1986). Furthermore, successful pollination itself also does not guarantee fertilization and thus mating. This is because individual plants may produce pollen whose abilities to germinate on different pistils differ because of incompatibility reactions or variable pollen tube growth rates (Harder and Routley, 2006). In animal-pollinated species, specific foraging behaviour can lead to non-random mating patterns, such as increased geitonogamy (Finer and Morgan, 2003). In the case of wind-pollinated plants, temporal directionality of wind flow can result in an anisotropic distribution of mating probabilities (Shen et al., 1981; Burczyk et al., 2004). Among other factors, differences in male fecundity strongly influence mating probabilities (Burczyk et al., 2004; Klein et al., 2008).
Controlled pollination experiments often show high variation in mating system parameters at the individual level (Aas, 1991; Steinhoff, 1998), suggesting a persistence of individual variation in natural populations. Individual variation in self-fertilization can arise from, for example, a variable amount of outcross pollen being available to different individuals (Mimura and Aitken, 2006; Restoux et al., 2008). This can be due to flowering asynchrony, which in turn may reveal seasonal fluctuations or be genetically determined lifetime stable characteristics. However, in many species, a genetically driven compatibility system may lead to important interindividual variation in mating system preferences. Individual variation may have significant implications for a population and for the entire species. For example, individual variation in self-fertilization can lead to unpredictable oscillations in inbreeding rates (Coelho and Vencovsky, 2003) and thus may be a basis for stronger (positive or negative) inbreeding effects than predicted by simple models. Therefore, identifying individual mating parameters is a first step towards understanding the diversity of mating system variation.
Mating patterns also have implications for gene flow, and thus for genetic structure at a landscape level. One useful measure of gene flow is the rate of pollen immigration (Ashley, 2010). It is often argued that high immigration rates are synonymous with high gene flow rates (e.g. Streiff et al., 1999). Existing methods regularly assume a random pool of pollen gametes immigrating from outside a local population (Burczyk et al., 2002; Klein et al., 2005; Chybicki and Burczyk, 2010a). However, in real populations, immigration patterns may differ individually for reasons similar to those described above. For example, a female with extremely early or late stigma receptivity may be exposed to a male pollen pool different from that of a female of an average phenology (El-Kassaby and Ritland, 1986). Thus, it may not be reasonable to summarize immigration levels and pools of immigrating pollen by a single parameter.
Contemporary mating patterns can be assessed through so-called ‘indirect’ or ‘direct’ approaches (Sork et al., 1999). Relying upon theoretical models, indirect methods provide summary parameters, such as inbreeding coefficients (Chybicki et al., 2009) or effective density of pollen donors (Austerlitz and Smouse, 2002), which are informative about mating patterns in general. Nonetheless, indirect methods do not allow for in-depth understanding of the processes behind the summary measures. Specifically, indirect methods are not well suited to study individual variation, and to disentangle the effects of different factors, such as pollen dispersal, fecundity and mate compatibility, among others. Therefore, mating patterns are usually studied using direct approaches often referred to as paternity analysis. In the simplest case, mating patterns are inferred from categorically assigned pollen donors (see review by Ashley, 2010). This detailed information should in theory provide a comprehensive network of mating patterns upon which to base inferences about the determinants of mating. While the information on individual paternity is usually of little interest, it is often used to frame mating patterns into a series of parameters describing different components of mating, such as self-fertilization level, mean pollen dispersal distance, dispersal kernel parameters, variance of male fertility or the covariance between male fertility and phenotypic characters. In an idealized situation, one could identify explicitly a male parent of any individual offspring. However, although highly polymorphic genetic markers (e.g. microsatellites) theoretically allow for categorical paternity inference, multiple (and therefore uncertain) paternity assignments are often problematic in the analysis of real data. Furthermore, extensive pollen dispersal in wind- and animal-pollinated plants means that many actual fathers are not found in the local population (Ashley, 2010). Genotyping problems (including null alleles, mutations or just genotyping mistakes) may also cause type I or type II errors in paternity assignments, unpredictably confounding the inference on mating patterns (Slavov et al., 2005). As a consequence, errors in paternity assignments bias inferences about mating parameters. On the other hand, the lack of certainty of paternity assignment is inherently accounted for in more advanced approaches, here called ‘paternity modelling’. These use fractional paternity assignments (Devlin et al., 1988) in the framework of a dedicated probability (mixture) model to estimate directly the parameters (components) of mating that are defined arbitrarily during model construction (Burczyk et al., 2002; Oddou-Muratorio et al., 2005; Klein et al., 2008). However, there is a need for further statistical developments because the existing implementations of ‘paternity modelling’ tools do not take into account individual variation in mating system parameters.
This study aimed to investigate comprehensively individual mating patterns within a mixed stand of Quercus robur and Q. petraea. Interspecific mating is possible in this species complex (Bacilieri et al., 1996; Curtu et al., 2007; Gugerli et al., 2007; Lepais and Gerber, 2011), challenging us to incorporate hybridization parameters into paternity modelling. In particular, we focused on inferring the distribution of individual selfing vs. outcrossing vs. pollen immigration rates as well as a differentiation of immigrating pollen pools at the individual female level. Male fecundity, pollen dispersal and individual intra- vs. interspecific mating preferences were also of interest. These permit detailed testing of the null model of random mating. To make the underlying model more realistic, we used a Bayesian approach that adopts the methods introduced previously by Burczyk et al. (2002) and Klein et al. (2008) to the system under study. The main refinements of the model are (1) accounting for typing errors; (2) a differentiation among immigrant pollen pools received by individual mother plants; and (3) a multispecies (or groups) scheme for different mating rates.
MATERIALS AND METHODS
The study site and genotyping
The study was conducted within a 5 ha mixed stand of Quercus robur and Q. petraea located in Jamy near Grudziądz, Poland (18°55′41″E, 53°32′8′'N). The stand was previously used for studying patterns of seed and pollen dispersal based on naturally regenerated seedlings (Chybicki and Burczyk, 2010a). The 135-year-old stand originated from planting, probably using locally collected seed material. The distribution of individuals is almost uniform, while the distribution of species is nearly random (Fig. 1). Taxonomical assignment was conducted based on leaf morphology (A. Boratyński, Institute of Dendrology, Polish Academy of Sciences, Poland, unpubl. res.), and no additional (e.g. genetic) tests were undertaken to verify species identity. Therefore, the term ‘pure species’ when used here means ‘putatively pure species’, allowing for some uncertainty about the actual taxonomic identity. The study site Jamy is located at the north-eastern margin of the natural range of Q. petraea. It is fenced to prevent free penetration of larger animals (such as wild boar, roe deer, etc.) to protect natural oak regeneration. The stand is a part of a forest mosaic composed of beech, oak and pine. Pine is the most abundant tree species in the closest neighbourhood.
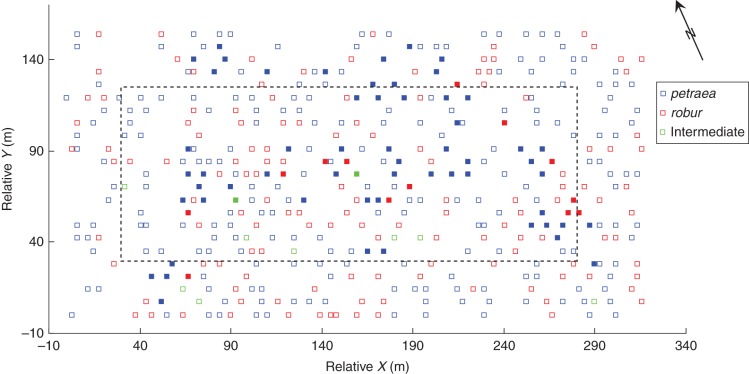
Fig. 1.
A map of the distribution of adult trees in the study site (Jamy, near Grudziądz, Poland). Mother trees are marked by filled squares.
Here we made use of data collected previously for all 421 adult individuals of both species. These include genotypes at five (out of six available in the original data set) unlinked, and highly polymorphic simple sequence repeat (SSR) loci (MSQ 4, ssrQrZAG 7, ssrQrZAG 20, ssrQpZAG 9 and ssrQpZAG 110), XY co-ordinates and diameter at breast height (DBH) measurements (Chybicki and Burczyk, 2010a). To study mating patterns, 72 trees were sampled for acorns. From these, 1911 seeds were successfully genotyped in total with 7–81 seeds per maternal family. The mother trees were selected to cover as much as possible of the total area of the stand (Fig. 1). However, due to variable female fecundity (the material was collected in a non-mast year), some regions remained under-represented. We arbitrarily classified mother trees as peripherally located (vs. centrally) if they were located <35 m from the border of the stand.
The model
Mating patterns were investigated with the aid of a probabilistic mating model (see Burczyk et al., 2002; Burczyk and Chybicki, 2004; Burczyk and Koralewski, 2005; Klein et al., 2011; Chybicki, 2013), originally referred to as the neighbourhood model (Adams and Birkes, 1991). In the neighbourhood model, in addition to genetic data, paternity is modelled through taking simultaneously into account spatial positions of trees as well as their fecundities, either in terms of fixed effects (Burczyk et al., 2002) or as random variables (Klein et al., 2008). These features make the neighbourhood model clearly different from the methods based on paternity inferred solely from genetic markers.
Total probability of an offspring genotype
The core of this model is the equation for total probability for an offspring genotype:
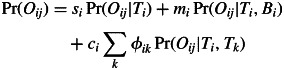 |
(1) |
where Oij is a multilocus genotype of the j-th offspring in a family group mothered by the i-th adult tree, Ti is a multilocus genotype of the i-th adult tree, Bi is the allele frequency in an immigrant pollen pool of the i-th adult tree, {si, mi, ci} are per-family mixture rates corresponding to self-fertilization, pollination with immigrant pollen and local outcrossing (note that si + mi + ci = 1), and φik is the rate of mating between the i-th and the k-th adult tree (note that Σk φik = 1). Conditional probabilities for offspring genotype Pr(Oij|.) follow Mendel's laws. As in the previous neighbourhood models (e.g. Burczyk et al., 2002; Klein et al., 2008), φik is not a true parameter but instead it refers to a function of parameters describing a process of pollen dispersal, a distribution of individual fecundity and a rate of inter- vs. intraspecific fertilization. Here, φik takes the form
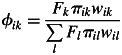 |
(2) |
where Fk is the fecundity of the k-th adult tree, πik is the probability of pollen dispersal at the distance between the k-th and the i-th adult tree, and wik is the probability of fertilization of an egg of the i-th plant with a sperm of the k-th plant as a function of species status of both parents.
Pollen dispersal
Here we assumed that pollen dispersal follows an exponential-power kernel (Austerlitz et al., 2004) of the form
| (3) |
where r is the distance of pollen dispersal (i.e. the distance between a point of pollen emission and the point of pollen deposition), a is a parameter of scale, b is a shape parameter of a dispersal kernel and Γ( … ) is the standard gamma function. An exponential-power kernel can take various shapes, which is useful to model various modes of dispersal. If b = 1, then dispersal follows simple exponential decay. If b > 1, then dispersal is concentrated around an emission point in a normal-like fashion (i.e. a light-tailed kernel). On the other hand, if b < 1, then the probability of dispersal decreases very slowly except for very short distances (i.e. a heavy-tailed kernel). Given the assumed dispersal kernel, then πik = f(dik; a, b) (eqn 2), where dik is the distance between the i-th and the k-th tree, and a and b are parameters of dispersal kernel. However, instead of a scale parameter, a theoretical average distance of (forward) pollen dispersal δ = a Γ(3/b)/Γ(2/b) was of direct interest, so the equation [eqn (3)] was rearranged appropriately to be a function of b and δ.
Fecundity
We used a model of individual fecundity distribution introduced by Klein et al. (2008). Briefly, fecundity of the i-th male Fi is assumed to follow log-normal distribution with variance Σ2 and the mean fixed arbitrarily at 1. Klein et al. (2008) showed that under the assumed distribution of fecundity, Σ2 + 1 equals a theoretical ratio of observed to effective density of pollen donors, i.e. Σ2 + 1 = dp/dep, which hereafter will be referred to as the relative effective density of pollen donors (REDP). In other words, REPD reflects the level of variation in pollen production within a population in terms of census to effective number of males.
Intra- vs. interspecific fertilization
In order to account for possible limitation of inter- vs. intraspecific mating, we used a vector of parameters corresponding to probabilities that the i-th individual can be successfully fertilized with pollen of the z-th species {ωiz} (z = 1, 2, … , Z; where Z is the number of taxonomic groups identified in the study plot). Note that all ωiz for the i-th individual sum up to 1. As a consequence, for a pair of individuals, of which the i-th is a mother tree and the k-th is a pollen donor belonging to the z-th species, a function wik = ωiz. Because three taxonomic types were identified in the study site, we arbitrarily set z = 1 for Q. petraea, z = 2 for Q. robur and z = 3 for the intermediate type.
Typing errors
The conditional probabilities for an offspring genotype [eqn (1)] were computed assuming a possibility of random typing errors, occurring at a locus-specific rate ɛl. Here, ɛl denotes the probability that a random allele at the l-th locus has been incorrectly typed as one of the remaining alleles at the locus. Thus, the model for typing errors resembles the K-Allele Model of mutation (Crow and Kimura, 1970). All conditional probabilities were derived in an unpublished manuscript (I. J. Chybicki and J. Burczyk, unpubl. res.) and are described in detail in the manual of NM+ software ver. 1.1 (Chybicki and Burczyk, 2010b) available at the web site http://www.ukw.edu.pl/pracownicy/strona/igor_chybicki/software_ukw/.
Immigrant pollen pool
Immigrant allele frequencies (Bi) were modelled individually for each half-sib family. Although it is possible that all mother trees receive pollen from a common immigrant pollen pool (Bi = Bj = … = B), here we assumed Bi to be subjected to random drift that causes differentiation of immigrant pollen captured by different mother trees. In the simplest scenario, the random drift can occur as an effect of non-random sampling of pollen gametes from a background population. For this purpose, we used the so-called F-model of allele frequencies (Gaggiotti and Foll, 2010), in which allele frequencies in different sub-populations follow a Dirichlet distribution with parameters {ptl1, … , ptlM} (1/θi – 1), where ptlm is the frequency of the m-th allele (out of M in total) at the l-th locus in the total population, and θi is the scaled variance of allele frequencies within a sub-population. Balding (2003) showed that, under Dirichlet distribution, θi can be interpreted as the probability that two genes drawn from a sub-population are identical by descent through shared ancestry within a sub-population. Thus, θi is an analogue of the classical Wright's FST. However, because here θi describes a rate of divergence of an immigrant pollen pool for the i-th mother, θi can be treated as an analogue of the Φft parameter used in the TwoGener method (Austerlitz and Smouse, 2001; Smouse et al., 2001). Therefore, the parameter θi is approximately the probability of identity of two random pollen gametes drawn from an immigrant pollen pool for the i-th mother tree. It is worth mentioning that recent simulation tests confirmed the usefulness of the F-model in the analysis of pollen pool structure based on the similar Bayesian framework (Chybicki, 2013).
Inferential procedures
Because the above model depends on a large number of parameters, to estimate them we used a Bayesian approach that is conceptually similar to that proposed by Klein et al. (2008). Due to non-zero typing error rates, every candidate parent (regardless of its genotype) has a non-zero probability of being a father. Also, because error rates {ɛ1, … ,ɛL} as well as allele frequencies in background populations {Bi, … ,BN} were set as estimable, transition probabilities [eqn (1)] were no longer constants but depended on current values of ɛl and Bi. These two technical issues severely slow the computation of the likelihood function, making the MCMC (Markov chain Monte Carlo) algorithm as proposed in Klein et al. (2008) practically unmanageable. Therefore, to address this issue, we made use of auxiliary (latent) variables that depend on the data and the current parameter values. This allows analyses to be carried out in a reasonable time using a personal computer, even for large samples (as in this study). As explained below, the auxiliary variables store the results of paternity assignments conducted cyclically based on eqn (1) and conditioned on the current values of the parameters (see ‘Algorithm’ section). Therefore, the algorithm used here is in part analogous to that used in Hadfield et al. (2006) and Moran and Clark (2011).
Auxiliary variables
We also introduced additional variables which were not of direct interest but which facilitated the estimation of the goal parameters. These auxiliary variables are somewhat redundant as they are all deduced from assigned paternities. However, to show clearly which information is used to estimate a given parameter, we decided to avoid more complex representation. First, we introduced nik to represent the number of offspring in the i-th family sired by the k-th adult individual. Additionally, xi, yi and zi were used to store the number of progeny in the i-th family which were a result of self-fertilization, outcrossing with locally produced pollen and outcrossing with immigrant pollen, respectively. To store the number of progeny in the i-th family sired by an adult belonging to the z-th species, we used giz. Finally, ailm was used to store a number of copies of the m-th allele at the l-th locus carried by immigrant pollen in the pool of the i-th family (i.e. in Bi).
For simplicity, we will use {nij} to denote multiple variables {ni1, … ,niK} falling into the same category. A set of {ni1, … ,niK} consists of variables which were assumed to follow a multinomial distribution with parameters {φik} [see eqn (1)], i.e. {nik} ∼ Multinomial({φik}). Similarly, {xi, yi, zi} ∼ Multinomial(si, ci, mi), {ailm} ∼ Multinomial({pilm}) and {gis} ∼ Multinomial({ωis}). Assuming independence across offspring genotypes, all the auxiliary variables were inferred given the current parameter values, using a Bernoulli scheme based on eqn (1). First, with probability
| (4) |
the j-th offspring in the i-th family was a result of self-fertilization, so that after a positive outcome of a Bernoulli scheme xi = xi + 1. Alternatively, yi = yi + 1 and zi = zi + 1 were results of a Bernoulli scheme with probability
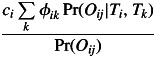 |
(5) |
and
| (6) |
respectively. Given an outcome of the above Bernoulli scheme, the remaining auxiliary variables were deduced in a nested scheme as follows. If the j-th offspring was deduced to be a result of outcrossing with locally produced pollen, then nik = nik + 1 with probability
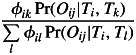 |
(7) |
Simultaneously it results in giz = giz + 1, with z corresponding to the taxonomic status of the k-th adult assigned as the father. If the j-th offspring was deduced to be a result of outcrossing with immigrant pollen, ailm = ailm + 1, where the m-th allele at the l-th locus was deduced at random under the assumption of Mendelian transmission given the maternal genotype and the current Bi.
Prior distributions
In the case of parameters referring to proportions that are directly related to the auxiliary variables, we used the Dirichlet distribution Dirichlet({αi}) as a prior distribution. In particular, (si, ci, mi) ∼ Dirichlet(αs, αc, αm) and {ωiz} ∼ Dirichlet({1})(i.e. a uniform distribution). As mentioned earlier, in the case of immigrant pollen allele frequencies at the l-th locus in the i-th family {pilm} ∼ Dirichlet({ptlm}[1/θi – 1]). Furthermore, because individual fecundity was assumed to follow log-normal distribution, it resulted in log(Fi) ∼ Normal(–σ2/2,σ 2), where σ2 = log(Σ2 + 1) was a (estimable) hyperparameter (Klein et al., 2008).
Typing error rates at the l-th locus ɛl as well as the hyperparameter θi were assumed to follow an exponential distribution truncated at 1, with the scale parameters βɛ and βθ, respectively. On the other hand, {ptlm} were assumed to follow Dirichlet({1}), i.e. the uniform distribution. The parameters taking real positive values, including parameters of dispersal kernel b and δ as well as hyperparameters σ, θi, αs, αc, αm, βɛ and βθ, were assumed (as in Klein et al., 2008) to follow an improper prior distribution proportional to 1/b, 1/δ, 1/σ, 1/θi, 1/αs, 1/αc, 1/αm, 1/βɛ and 1/βθ, respectively. In the case of the δ parameter, to avoid an uncontrolled drift towards extremely large values (Moran and Clark, 2011), we used a prior distribution truncated at 100 000 m.
Empirical estimates
Along with parameters of the model at any hierarchical level, we estimated additional quantities of interest. All these quantities were estimated in the same manner, i.e. by computing a specified function of values of current parameters during MCMC sampling, in order to obtain an empirical posterior distribution of this function. In this way, we estimated the empirical (observed) REDPO as a function of the variance in inferred fecundity (Klein et al., 2008). REPDO was of interest because the theoretical REDP can be biased because of the assumed prior distribution (Klein et al., 2011). To assess if there was any association between male fecundity and tree size, we estimated the empirical distribution of Spearman rank correlation between the DBH and Fi. In the same way, we assessed whether there was any relationship between the rate of local mating (ci) and a preference towards intraspecific mating (ωi1 and ωi2, for petraea- and robur-type, respectively). To compare mating preferences between the two pure species, we estimated the empirical posterior distribution of the mean difference of ωi1 between the two species:
| (8) |
where 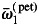 and
and 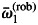 correspond to averages over individual ωi1 values, computed separately for petraea and robur sub-groups. The quantity di1 was expected to take values > 0, if there is positive overall skew towards intraspecific mating. Generally, similar procedures were employed whenever we wished to test a difference between two defined groups. Also, we estimated cumulative probability (CDF) of pollen dispersal distance based on the integral 0∫L 2πr f(r; a, b) dr, computed for a discrete set of upper limits (L). [Note that f(r; a, b) is defined in eqn (3).] Because the hyperparameters αs, αc and αm were set as estimable, we were able to assess the level of over-dispersion among individual proportions, i.e. whether si, ci, mi exhibit multinomial or extramultinomial variation among mother trees. To measure an overall over-dispersion, we used a single summary parameter γ = (1 + αs + αc + αm)−1, estimated during MCMC. The parameter γ takes 0 if proportions follow pure multinomial variation and >0 if there is some extramultinomial variation among individuals. Although there is a boundary issue, i.e. αs,αc,αm ∈ (0, + ∞), that makes it impossible to test formally for γ = 0, still the highest posterior density (HPD) set for γ can inform whether substantial over-dispersion occurs.
correspond to averages over individual ωi1 values, computed separately for petraea and robur sub-groups. The quantity di1 was expected to take values > 0, if there is positive overall skew towards intraspecific mating. Generally, similar procedures were employed whenever we wished to test a difference between two defined groups. Also, we estimated cumulative probability (CDF) of pollen dispersal distance based on the integral 0∫L 2πr f(r; a, b) dr, computed for a discrete set of upper limits (L). [Note that f(r; a, b) is defined in eqn (3).] Because the hyperparameters αs, αc and αm were set as estimable, we were able to assess the level of over-dispersion among individual proportions, i.e. whether si, ci, mi exhibit multinomial or extramultinomial variation among mother trees. To measure an overall over-dispersion, we used a single summary parameter γ = (1 + αs + αc + αm)−1, estimated during MCMC. The parameter γ takes 0 if proportions follow pure multinomial variation and >0 if there is some extramultinomial variation among individuals. Although there is a boundary issue, i.e. αs,αc,αm ∈ (0, + ∞), that makes it impossible to test formally for γ = 0, still the highest posterior density (HPD) set for γ can inform whether substantial over-dispersion occurs.
Algorithm
To approximate posterior distributions, a class of the MCMC approach was used called the Gibbs sampler. First, each cycle started by inferring new values for the auxiliary variables, given current parameter values. Then, for {si, ci, mi}, {ωis} and {pilm}, new values were sampled directly from the posterior multinomial–Dirichlet distribution (the Gibbs sampling), given auxiliary variables inferred in the current cycle. For the remaining parameters, the Metropolis–Hastings procedure was used, which needs to define a proposal distribution. For parameters and hyperparameters that take positive real numbers, i.e. b, δ, σ2, αs, αc, αm, βɛ and βθ, as in Klein et al. (2008), we used a proposal constructed by multiplying the current value by the random deviate drawn from log-normal distribution with a mean 1 and variance 0·1. In the case of parameters distributed continuously within the 0–1 interval, including {θi}, {ptjk} and {ɛl}, we propose a new value using a random walk algorithm with reflection, with the additive distortion uniformly distributed between –0·1 and 0·1. Because allele frequencies must sum up to unity, in the case of {ptjk} we used the approach of updating simultaneously frequencies for two randomly chosen alleles (cf. Falush et al., 2003). Estimation procedures were implemented in the NM2F computer program written in Pascal (the version running under Microsoft Windows is freely available at http://www.ukw.edu.pl/pracownicy/strona/igor_chybicki/software_ukw/). Estimates were derived with 50 000 MCMC samples, rejecting the first 10 000 updates. To weaken autocorrelation of subsequent updates due to Metropolis steps, only every 25th update was saved. Due to the large sample size as well as non-zero (and simultaneously estimable) genotyping errors, the estimation of all parameters in the full model took approx. 3 d on an average-speed PC unit (Intel Core 2 Duo E6750 @ 2·66 GHz).
RESULTS
Marker polymorphism
While adults had no missing genotypes (Chybicki and Burczyk, 2010a), 52 single-locus genotypes (0·5 %) were missing in progeny. The number of alleles per locus detected in progeny ranged from 24 to 30. However, due to apparent non-uniform allele frequency distributions, the effective number of alleles was low, ranging from 3·5 to 6·2. Observed heterozygosity in progeny was 78·4 % and did not deviate significantly from the panmictic expectation (77·8 %). The theoretical cumulative multilocus probability of paternity exclusion equalled EP = 0·9994, so that the probability of excluding all except the actual father (estimated as EP240) reached roughly 87 %. However, this proportion seemed to over-estimate the actual value, given that non-trivial typing error rates were found (see the next section).
Genotyping errors
Genotyping error rates were estimated individually for each locus assuming the exponential prior distribution. The posterior average error rates spanned from 1·4 to 8·1 %, with the grand average being 5·4 %. The scale hyperparameter of the prior distribution was slightly higher, reaching 0·066, with the 95 % credible interval (also known as Bayesian confidence interval, CIB) between 0·024 and 0·164.
Mating patterns
Composition of progeny
In total, average rates of self-fertilization, outcrossing with a local tree and outcrossing with immigrant pollen were 0·5 % (from 0 to 7·4 %), 49·8 % (from 11·9 to 87·5 %) and 49·7 % (from 12·4 to 88·0 %) (Table 1), respectively. Interestingly, only a single Q. petraea tree (Family 11) exhibited a significant proportion of selfed progeny (7·4 %), while the others were classified as pure outcrossers. On average, robur-like mother trees experienced slightly, but significantly (P < 0·05), more outcrossing with immigrant pollen (56·7 %) than petraea-like trees (48·6 %). We also noted that mother trees (regardless of taxonomic assignation) in the periphery of the stand received significantly greater proportions of immigrant pollen than centrally located trees (60·5 % vs. 45·1 %).
Table 1.
Characteristics of half-sib families
| Family | ni | Putative species | Location | X | Y | si | ci | mi | θi | Mating preference | Mating system |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 9 | petraea | Central | 89·9 | 70 | 0·002 | 0·738 | 0·260 | 0·091 | petraea | Outcrossing |
| 2 | 44 | petraea | Central | 72·5 | 70 | 0·001 | 0·420 | 0·579 | 0·036 | petraea | Outcrossing |
| 3 | 10 | petraea | Central | 66·7 | 91 | 0·002 | 0·668 | 0·329 | 0·061 | Any | Outcrossing |
| 4 | 7 | petraea | Central | 75·4 | 77 | 0·003 | 0·613 | 0·384 | 0·103 | Any | Outcrossing |
| 5 | 20 | petraea | Central | 66·7 | 77 | 0·002 | 0·327 | 0·672 | 0·046 | Any | Outcrossing |
| 6 | 24 | petraea | Peripheral | 81·2 | 133 | 0·001 | 0·313 | 0·686 | 0·082 | Any | Outcrossing |
| 7 | 14 | petraea | Peripheral | 142·1 | 133 | 0·003 | 0·475 | 0·523 | 0·038 | Any | Outcrossing |
| 8 | 16 | petraea | Central | 159·5 | 119 | 0·002 | 0·435 | 0·563 | 0·113 | Any | Outcrossing |
| 9 | 69 | petraea | Central | 150·8 | 91 | 0·001 | 0·854 | 0·146 | 0·184 | petraea | Outcrossing |
| 10 | 31 | petraea | Central | 147·9 | 77 | 0·001 | 0·875 | 0·124 | 0·106 | petraea | Outcrossing |
| 11 | 54 | petraea | Central | 121·8 | 91 | 0·074 | 0·691 | 0·235 | 0·046 | petraea | Mixed |
| 12 | 37 | petraea | Central | 110·2 | 77 | 0·001 | 0·682 | 0·317 | 0·060 | petraea | Outcrossing |
| 13 | 7 | petraea | Peripheral | 168·2 | 126 | 0·030 | 0·515 | 0·456 | 0·102 | Any | Outcrossing |
| 14 | 7 | petraea | Peripheral | 205·9 | 140 | 0·007 | 0·352 | 0·641 | 0·062 | Any | Outcrossing |
| 15 | 10 | petraea | Peripheral | 179·8 | 126 | 0·002 | 0·629 | 0·369 | 0·054 | Any | Outcrossing |
| 16 | 9 | petraea | Central | 185·6 | 119 | 0·003 | 0·753 | 0·244 | 0·098 | petraea | Outcrossing |
| 17 | 8 | petraea | Peripheral | 203 | 133 | 0·003 | 0·353 | 0·644 | 0·125 | Any | Outcrossing |
| 18 | 7 | petraea | Central | 171·1 | 119 | 0·003 | 0·540 | 0·457 | 0·222 | Any | Outcrossing |
| 19 | 23 | petraea | Central | 208·8 | 119 | 0·001 | 0·293 | 0·705 | 0·092 | Any | Outcrossing |
| 20 | 46 | petraea | Central | 220·4 | 119 | 0·001 | 0·580 | 0·419 | 0·150 | petraea | Outcrossing |
| 21 | 12 | petraea | Central | 214·6 | 105 | 0·003 | 0·199 | 0·799 | 0·228 | Any | Outcrossing |
| 22 | 19 | petraea | Central | 208·8 | 91 | 0·001 | 0·410 | 0·588 | 0·075 | Any | Outcrossing |
| 23 | 12 | petraea | Central | 220·4 | 84 | 0·002 | 0·520 | 0·477 | 0·200 | Any | Outcrossing |
| 24 | 14 | petraea | Central | 200·1 | 77 | 0·002 | 0·744 | 0·254 | 0·210 | petraea | Outcrossing |
| 25 | 79 | petraea | Central | 220·4 | 77 | 0·036 | 0·581 | 0·383 | 0·070 | petraea | Outcrossing |
| 26 | 25 | petraea | Central | 176·9 | 91 | 0·001 | 0·144 | 0·855 | 0·428 | Any | Outcrossing |
| 27 | 73 | petraea | Central | 179·8 | 77 | 0·000 | 0·842 | 0·157 | 0·058 | petraea | Outcrossing |
| 28 | 20 | petraea | Central | 182·7 | 84 | 0·002 | 0·686 | 0·313 | 0·105 | petraea | Outcrossing |
| 29 | 21 | petraea | Central | 211·7 | 77 | 0·002 | 0·581 | 0·417 | 0·045 | robur | Outcrossing |
| 30 | 64 | petraea | Central | 263·9 | 49 | 0·001 | 0·558 | 0·441 | 0·160 | petraea | Outcrossing |
| 31 | 14 | petraea | Central | 255·2 | 49 | 0·002 | 0·691 | 0·307 | 0·071 | Any | Outcrossing |
| 32 | 37 | petraea | Central | 272·6 | 49 | 0·001 | 0·730 | 0·269 | 0·048 | petraea | Outcrossing |
| 33 | 11 | petraea | Central | 261 | 56 | 0·002 | 0·659 | 0·339 | 0·068 | petraea | Outcrossing |
| 34 | 39 | petraea | Central | 255·2 | 84 | 0·001 | 0·354 | 0·645 | 0·076 | petraea | Outcrossing |
| 35 | 17 | petraea | Central | 269·7 | 42 | 0·002 | 0·401 | 0·597 | 0·077 | Any | Outcrossing |
| 36 | 77 | petraea | Central | 249·4 | 91 | 0·000 | 0·570 | 0·430 | 0·058 | petraea | Outcrossing |
| 37 | 21 | petraea | Central | 261 | 77 | 0·002 | 0·624 | 0·375 | 0·089 | petraea | Outcrossing |
| 38 | 19 | petraea | Peripheral | 287·1 | 49 | 0·002 | 0·607 | 0·391 | 0·057 | Any | Outcrossing |
| 39 | 12 | petraea | Peripheral | 290 | 28 | 0·003 | 0·404 | 0·593 | 0·061 | Any | Outcrossing |
| 40 | 11 | petraea | Central | 165·3 | 35 | 0·002 | 0·338 | 0·660 | 0·169 | Any | Outcrossing |
| 41 | 81 | petraea | Central | 174 | 35 | 0·001 | 0·530 | 0·470 | 0·053 | petraea | Outcrossing |
| 42 | 41 | petraea | Central | 171·1 | 63 | 0·001 | 0·707 | 0·293 | 0·044 | petraea | Outcrossing |
| 43 | 7 | petraea | Central | 165·3 | 63 | 0·002 | 0·498 | 0·500 | 0·069 | Any | Outcrossing |
| 44 | 9 | petraea | Peripheral | 52·2 | 7 | 0·002 | 0·188 | 0·810 | 0·114 | Any | Outcrossing |
| 45 | 22 | petraea | Peripheral | 58 | 28 | 0·003 | 0·301 | 0·696 | 0·051 | Any | Outcrossing |
| 46 | 19 | petraea | Central | 63·8 | 63 | 0·021 | 0·355 | 0·624 | 0·050 | Any | Outcrossing |
| 47 | 16 | petraea | Central | 75·4 | 63 | 0·002 | 0·426 | 0·572 | 0·115 | Any | Outcrossing |
| 48 | 30 | petraea | Peripheral | 69·6 | 140 | 0·002 | 0·526 | 0·472 | 0·064 | petraea | Outcrossing |
| 49 | 34 | petraea | Central | 130·5 | 63 | 0·001 | 0·672 | 0·327 | 0·059 | petraea | Outcrossing |
| 50 | 31 | petraea | Peripheral | 87 | 140 | 0·002 | 0·451 | 0·547 | 0·061 | petraea | Outcrossing |
| 51 | 13 | petraea | Peripheral | 110·2 | 133 | 0·002 | 0·465 | 0·533 | 0·050 | Any | Outcrossing |
| 52 | 13 | petraea | Peripheral | 84·1 | 147 | 0·002 | 0·456 | 0·542 | 0·125 | Any | Outcrossing |
| 53 | 8 | petraea | Peripheral | 188·5 | 147 | 0·003 | 0·416 | 0·581 | 0·055 | Any | Outcrossing |
| 54 | 15 | petraea | Peripheral | 174 | 140 | 0·002 | 0·543 | 0·455 | 0·075 | Any | Outcrossing |
| 55 | 16 | petraea | Peripheral | 46·4 | 21 | 0·002 | 0·119 | 0·880 | 0·057 | Any | Outcrossing |
| 56 | 41 | petraea | Peripheral | 55·1 | 21 | 0·001 | 0·238 | 0·761 | 0·195 | Any | Outcrossing |
| 57 | 7 | petraea | Central | 261 | 91 | 0·003 | 0·422 | 0·575 | 0·126 | Any | Outcrossing |
| 58 | 68 | robur | Central | 153·7 | 84 | 0·001 | 0·476 | 0·524 | 0·358 | petraea | Outcrossing |
| 59 | 26 | robur | Central | 142·1 | 84 | 0·001 | 0·597 | 0·402 | 0·094 | petraea | Outcrossing |
| 60 | 30 | robur | Central | 118·9 | 77 | 0·023 | 0·628 | 0·349 | 0·045 | Any | Outcrossing |
| 61 | 7 | robur | Peripheral | 214·6 | 126 | 0·003 | 0·275 | 0·723 | 0·162 | Any | Outcrossing |
| 62 | 12 | robur | Central | 240·7 | 105 | 0·002 | 0·186 | 0·812 | 0·195 | Any | Outcrossing |
| 63 | 46 | robur | Central | 188·5 | 70 | 0·001 | 0·735 | 0·264 | 0·034 | petraea | Outcrossing |
| 64 | 21 | robur | Central | 278·4 | 63 | 0·001 | 0·483 | 0·516 | 0·039 | Any | Outcrossing |
| 65 | 25 | robur | Central | 266·8 | 84 | 0·001 | 0·530 | 0·469 | 0·180 | Any | Outcrossing |
| 66 | 40 | robur | Peripheral | 281·3 | 56 | 0·001 | 0·295 | 0·705 | 0·029 | robur | Outcrossing |
| 67 | 40 | robur | Central | 275·5 | 56 | 0·001 | 0·365 | 0·634 | 0·048 | petraea | Outcrossing |
| 68 | 23 | robur | Central | 176·9 | 63 | 0·026 | 0·296 | 0·678 | 0·193 | robur | Outcrossing |
| 69 | 11 | robur | Central | 66·7 | 56 | 0·003 | 0·422 | 0·575 | 0·050 | Any | Outcrossing |
| 70 | 7 | robur | Peripheral | 66·7 | 21 | 0·003 | 0·274 | 0·723 | 0·034 | Any | Outcrossing |
| 71 | 29 | intermediate | Central | 159·5 | 77 | 0·001 | 0·443 | 0·556 | 0·068 | Any | Outcrossing |
| 72 | 74 | intermediate | Central | 92·8 | 63 | 0·001 | 0·798 | 0·201 | 0·182 | petraea | Outcrossing |
ni, number of progeny in the i-th family; putative species, species assignation of the mother tree; X and Y, relative co-ordinates (in metres); si, posterior average self-fertilization rate; ci, posterior average outcrossing rate with local trees; mi, posterior average outcrossing with an immigrant pollen; θi, divergence of an immigrant pollen pool of the i-th mother tree from the total background population (alternative paternity identity within immigrant pollen); mating preference, preference towards mating with trees of a given (if any) species, inferred based on a test of whether the rate of acceptance of conspecific pollen is >0·5 (see text for details); mating system, preference towards purely outcross pollination or mixed self- and outcross pollination, inferred based on a test of whether a rate of self-fertiliztion is >0 (see text for details).
In addition to model parameters, the hyperparameters of the Dirichlet prior distribution were also estimated (Table 2). The posterior averages (95 % CIBs) were rather low, reaching 0·034 (0·006–0·082), 2·307 (1·406–3·681) and 2·322 (1·399–3·634) for αs, αc and αm, respectively. The posterior average of the over-dispersion parameter γ equalled 0·183 (median = 0·182, mode = 0·181), with 95 % HPD between 0·116 and 0·251. Thus, the results provided strong support for significant over-dispersion in individual rates {si, ci, mi} among mother trees.
Table 2.
Estimates of empirical averages of individual rates of self-fertilization (s), outcrossing with local pollen (c) and outcrossing with immigrant pollen (m) together with estimates of hyperparameters of the Dirichlet distribution (see text)
| Parameter | s | c | m | αs | αc | αm |
|---|---|---|---|---|---|---|
| Mean | 0·005 | 0·498 | 0·497 | 0·034 | 2·307 | 2·322 |
| Median | 0·004 | 0·498 | 0·497 | 0·030 | 2·233 | 2·251 |
| Mode | 0·003 | 0·496 | 0·498 | 0·021 | 2·037 | 2·094 |
| Q2·5 % | 0·001 | 0·462 | 0·462 | 0·006 | 1·406 | 1·399 |
| Q97·5 % | 0·010 | 0·534 | 0·534 | 0·082 | 3·681 | 3·634 |
Mean, median and mode are point estimates given the posterior distribution, while Q values are quantiles of the posterior distribution that limits the 95 % credible interval.
Pollen dispersal kernel
Joint estimation of pollen dispersal kernel parameters with the remaining model parameters proved difficult. In particular, we found the estimate of average pollen dispersal distance (δ) to float occasionally towards extremely large values. Hence, the posterior distribution for δ was strongly right-skewed (Fig. 2A), resulting in substantial differences between posterior mean and median estimates, 3366 and 283 m, respectively. Using a default kernel density estimation procedure available in the R package (CRAN), we obtained (after log-transformation) a rough assessment of the mode equal to 121 m. In spite of a large number of offspring, we obtained a very wide 95 % HPD interval for δ, ranging between 38 and 20 189 m. The posterior mean and median for shape parameter b equalled 0·166 and 0·143, respectively. Similarly to δ, the posterior distribution of b was apparently right-skewed (Fig. 2B), resulting in the mode estimate (approximate) of 0·106. The 95 % HPD around b ranged between 0·020 and 0·366. Thus, the observed dispersal kernel was classified as ‘heavy-tailed’ with strong confidence. Based on the estimated CDF (Fig. 2C), the expected mean rate of pollen flow from outside 150 m (the mother-edge average distance) was equal to 33 % (CIB 9–65 %), and thus did not deviate significantly from the estimated average pollen immigration rate.
Fig. 2.
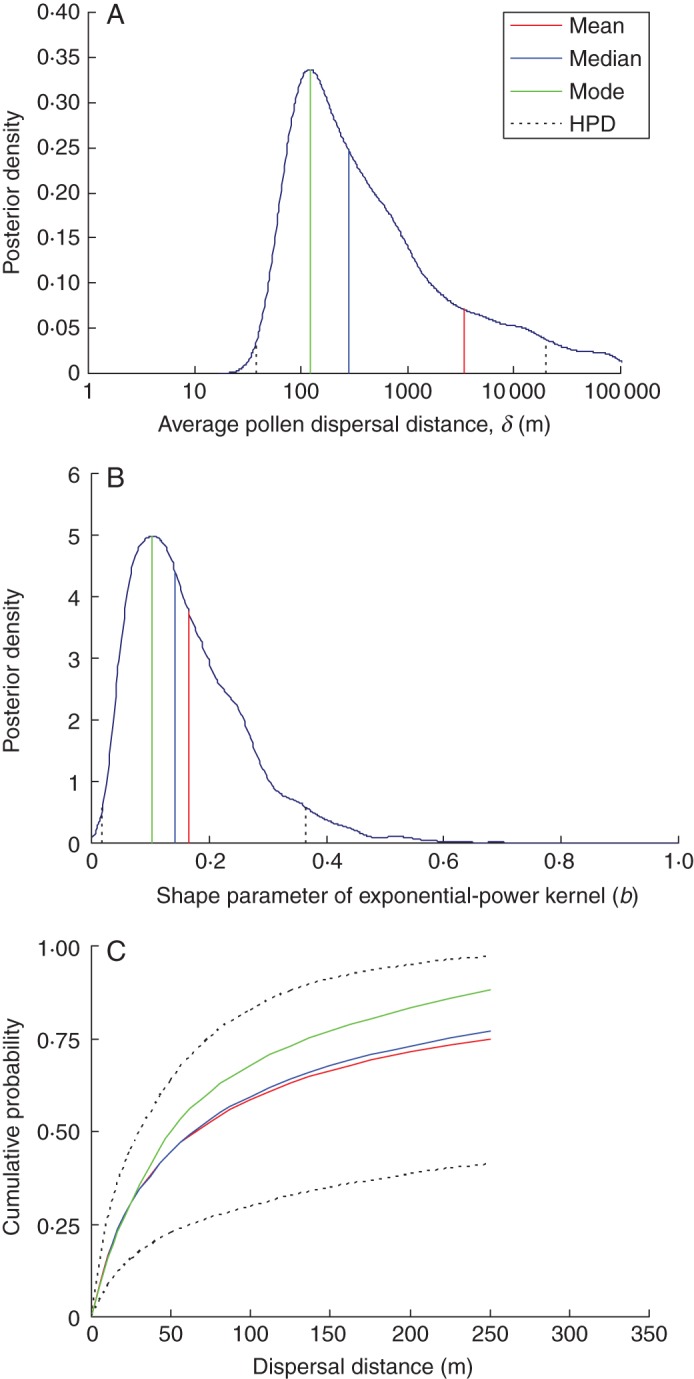
The posterior distributions of parameters of the exponential power kernel for pollen dispersal (A, B) together with corresponding pollen dispersal cumulative probability distribution (C). The three curves in (C) correspond to three point estimates of dispersal kernel parameters, i.e. mean, median and mode (see text). Note that the x-axis in (A) is given in a logarithmic scale.
Intra- and interspecific mating preferences
Generally, mother trees revealed great variability in mating preferences (Fig. 3). In order to summarize the results for pure species, we focused on rates of accepting conspecific pollen gametes. In the case of petraea-type mother trees, the average ωi1 equalled 0·735 (95 % CIB 0·698–0·772), spanning from 0·158 to 0·961. In the case of robur-type mother trees, the average ωi2 was apparently lower, reaching 0·363 (95 % CIB 0·278–457), and spanned between 0·043 and 0·783. The mean difference (di1) of ωi1 between the two species equalled 0·217, with a 95 % CIB of between 0·120 and 0·311. Thus, on average, petraea mother trees had a significantly higher preference for mating with petraea trees rather than robur mother trees. We employed CIBs that were significantly greater than 0·5 for ωi1 and ωi2 (for petraea- and robur-types, respectively) as criteria for specificity of the mating process at the individual level. Using these criteria, 23 out of 57 (40 %) petraea-type mother trees were classified as preferring to mate intraspecifically. On the other hand, only two out of 13 (15%) robur-type mother trees appeared to prefer receiving pollen gametes of the same species. Interestingly, among the remaining robur-type mother trees, four preferred to mate interspecifically (i.e. with petraea-type trees), while seven had no apparent mating preferences. In comparison, only one petraea-type mother tree was found to mate preferentially with robur-type trees, while the remaining 33 mother trees had no specific preferences. It is worth mentioning that both intermediate mother trees revealed a preference towards backcrossing with Q. petraea, with one exhibiting ωi1 close to 1 (and significantly >0·5).
Fig. 3.
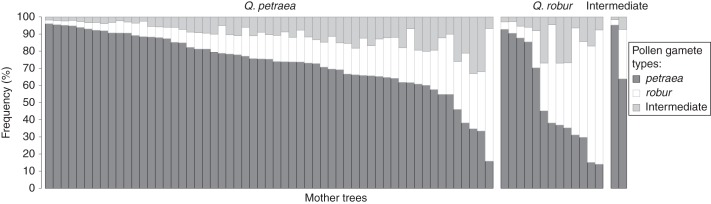
Individual per-mother tree distributions of the probability of accepting a pollen gamete of petraea, robur or intermediate type. Each bar represents a single mother tree. Mother trees were grouped according to putative species assignation.
Using the Spearman correlation coefficient (r), we also measured the relationship between the rate of local mating (ci) and the preference towards intraspecific mating (ωi1 and ωi2, for petraea- and robur-type trees, respectively). In the case of petraea-type trees, we observed a positive and highly significant relationship. In this case, the normal-like posterior distribution was centred on an average r = 0·42, with a confidence interval (CI) between 0·24 and 0·58. On the other hand, for robur-type trees, the relationship was negative yet insignificant, with an average r = –0·23 and 95 % CI between –0·60 and 0·17.
Male fecundity
The hyperparameter σ of log-normal prior distribution for fecundity was concentrated on a 95 % CIB between 1·080 and 1·504, with a mean of 1·272 and median 1·264. It resulted in the theoretical REDP being equal to 5·32, with a 95 % CIB between 3·21 and 9·60. The empirical REDPO appeared slightly lower than the REDP, with a posterior mean and median equal to 4·25 and 4·11, respectively. The 95 % CIB was between 3·01 and 6·19. At the individual level, male fecundy showed high variance, with individual posterior averages ranging from 0·079 to 14·71. Because fecundity was assumed to follow a log-normal distribution with a mean equal to 1, to examine how many outliers were in the stand due to extreme fecundity values we searched for individual values significantly different from 1. We found that six trees had fecundity significantly greater than 1, while 22 had fecundity significantly lower than 1. The average fecundity of those sub-sets of superior and inferior trees was equal to 8·84 and 0·25, respectively. We found that the correlation between DBH and fecundity was positive but not significant (r = 0·072, 95 % CIB –0·008–0·153).
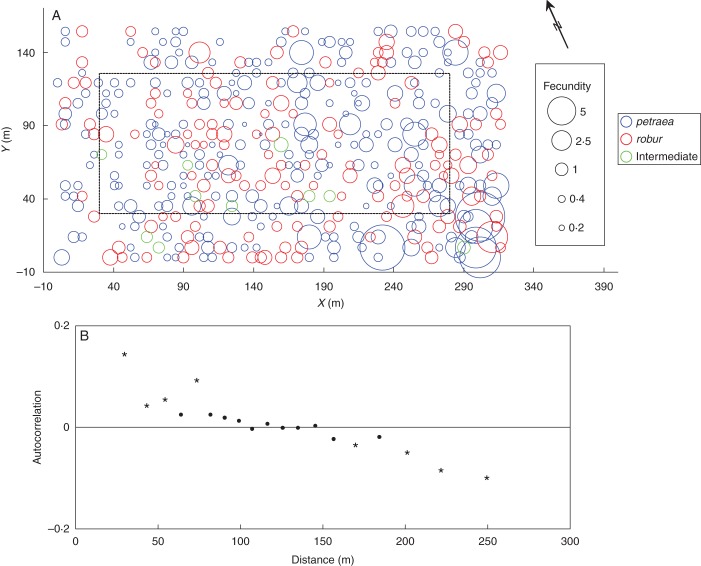
At the taxonomic level, fecundity did not differ between the three groups, taking on average 0·95 (95 % CIB 0·76–1·16), 0·91 (95 % CIB 0·70–1·17) and 0·85 (95 % CIB 0·29–1·98) for petraea-type, robur-type and intermediate trees, respectively. However, inspection of the spatial distribution of individual male fecundity revealed that the most fecund trees were located in the southern corner of the stand (Fig. 4A). Using a spatial autocorrelogram (Moran's I), with posterior means as data, we revealed significant clustering (P < 0·001) of trees having a similar fecundity up to 70 m (the extent of positive autocorrelation), and significant repulsion of trees located >160 m apart (the extent of negative autocorrelation) (Fig. 4B). Thus, the autocorrelogram confirmed the presence of a strong cline-like distribution of fecundity within the stand.
Fig. 4.
Relative male fecundity in the stand. (A) Distribution of fecundity in the stand: Q. petraea, Q. robur and intermediate trees as indicated in the key. The dashed line shows the border of the central zone. (B) Spatial autocorrelogram summarizing a spatial structure of fecundity: significant autocorrelation coefficients are marked with asterisks.
The genetic structure of immigrant pollen
The average differentiation (θi) over all mother trees equalled 0·101, with a 95 % CI between 0·085 and 0·119. Individual values spanned from 0·029 to 0·428. On average, petraea-type mother trees had slightly lower θi than robur-type trees (0·098 vs. 0·112). However, the difference appeared not to be significant. Also, peripheral mother trees had lower average θi than central trees (0·079 vs. 0·111), but, again, the difference was not significant. We also did not observe significant correlation between mi and θi.
DISCUSSION
The present study concentrated on mating patterns by using comprehensive analysis of paternity. We attempted to account for all major factors influencing paternity, including non-random pollen dispersal, uneven male fecundity and non-random interspecific mating, as well as some more technical issues such as genotyping errors or uncertainty about immigrant pollen pools. Our approach allowed us to identify detailed patterns present at the level of individual females, which would remain cryptic if mating patterns were assessed with the use of the classical neighbourhood model (Adams and Birkes, 1991; Burczyk et al., 2002).
Mating system
Oaks are mostly allogamous species (Bacilieri et al., 1993; Dow and Ashley, 1998a; Streiff et al., 1999; Nakanishi et al., 2004; Curtu et al., 2009; Chybicki and Burczyk, 2010a; Lepais and Gerber, 2011). Nonetheless, experiments based on controlled pollination showed that there might be a significant variation in ability to self-fertilize among oak trees (Aas, 1991; Steinhoff, 1993, 1998). Possible causal mechanisms may include genetically driven self-incompatibility that inhibits fertilization with selfed pollen (Hagman, 1975; Yacine and Bouras, 1997; Buschbom et al., 2011). In natural conditions, poor flowering synchrony between male and female flowers of the same individual (Bacilieri et al., 1995) and biased sex allocation (see Ducousso et al., 1993) can additionally influence the chance of self-fertilization. Our results seem to support the above opinion, as significant over-dispersion was identified. However, detailed examination revealed that only a single tree (out of 72) showed occasional (and significant) self-fertilization, with 7·4 % or about four seeds being produced through self-fertilization. Thus, to a great extent, the statistical effect of over-dispersion in selfing rates can be attributable to the exceptional behaviour of a single tree. Moreover, our previous study (Chybicki and Burczyk, 2010a) in the same stand showed some preference towards self-fertilization in another tree (Family 68), that is no longer the case here. Therefore, any furhter in-depth conclusions should be drawn with caution.
Pollen flow
In wind-pollinated tree species, pollen dispersal usually follows a leptokurtic, heavy-tailed distribution (Streiff et al., 1999; Austerlitz et al., 2004; Goto et al., 2006; Shimatani et al., 2007; Chybicki and Burczyk, 2010a). Our results conform to this pattern. In particular, the probability distribution of dispersal was apparently more heavy-tailed than a simple exponential kernel. In other words, the dispersal probability decreases very slowly at long distances, suggesting a relatively high likelihood of long-distance dispersal. These findings are concordant with the previous estimates (Dow and Ashley, 1998a; Streiff et al., 1999; Nakanishi et al., 2004; Chybicki and Burczyk, 2010a; Craft and Ashley, 2010), which generally suggest a great potential for long-distance pollen dispersal in oaks. Unfortunately, the average and the cumulative probability distribution of dispersal distance estimated in this study showed unsatisfactorily wide confidence intervals, preventing the precise predictions of patterns of pollen dispersal. In our opinion, this was mainly due to the inability to capture nuances of different heavy-tailed models (within a range of confidence intervals for b) at the spatial scale of the study plot (<300 m). A solution in this case would be to link immigration rates with the dispersal kernel (e.g. Shimatani et al., 2007; Chybicki and Burczyk, 2010a; Moran and Clark, 2011). However, it requires a reasonable model of forest landscape at least in the neighbourhood of the study plot, which is not readily available in our case. Nonetheless, the cumulative probability distribution is strongly dependent on the assumed dispersal kernel (see Gérard et al., 2006; Goto et al., 2006), and thus it has low overall confidence. The same warning is also valid for the estimates of the average (forward) dispersal distance obtained in this study. However, although this information is generally interesting, the estimate of the average dispersal distance is less important for predicting essential features of gene flow patterns (Klein et al., 2006).
Estimates of pollen immigration rates are generally very sensitive to the presence of typing errors (Gérard et al., 2006), the assumed frequencies in the pollen source background population (Burczyk and Chybicki, 2004) and the presence of unsampled local fathers. In our study, all these issues were accounted for, so it is our belief that the estimated immigration rates were nearly unbiased. Interestingly, they appeared to be about 20 % lower than our previous estimates for the same stand based on procedures ignoring typing errors (m = 0·623; Chybicki and Burczyk, 2010a); however, in that case, the analyses were conducted based on established seedlings. Moreover, if we assume that the edge effect, as detected in this study, did not affect our previous results (due to the apparently central location of potential mother trees), the difference may increase even further (i.e. 45·1 % vs. 62·3 %). It seems that the present approach effectively removed the upward bias, expected under naïve transition probabilities. Nonetheless, still about 50 % of pollination events were due to immigrant pollen, which is quite comparable with previous studies on oaks in general (Dow and Ashley, 1998a; Streiff et al., 1999; Nakanishi et al., 2004; Chybicki and Burczyk, 2010a; Craft and Ashley, 2010). Surprisingly, the mean pollen immigration rate corresponded quite well to the prediction based on the estimated (posterior average) CDF for pollen dispersal. This might suggest that there is no obvious heterogeneity between the modes of local vs. long-distance pollen dispersal, i.e. that pollen dispersal follows roughly a single monotonic probability function. This would be in opposition to the opinion that, for wind-borne pollen, long-distance and local movements are based on different aerodynamic processes (Di-Giovanni et al., 1996; Williams, 2008). However, our conclusion should be treated with caution, because estimates of dispersal kernel parameters were characterized by low precision.
Generally, the probability of paternal identity decreases together with the extent of pollen flow (Austerlitz and Smouse, 2001). Therefore, one might expect θi to decrease together with pollen immigration (Nakanishi et al., 2004, 2005; Craft and Ashley, 2010), because immigrant pollen can be assumed to come from a large background population. However, in our study, some trees that received a lot of pollen from outside the stand also revealed a very high level of homogeneity in immigrant pollen (e.g. Family 25, Table 1). Given that 2θi approximates the proportion of full-sibs among offspring produced with immigrant pollen (Hardy et al., 2004), our results indicated the presence of correlated mating among trees located further than 150 m apart. In the case of animal-pollinated species, such a correlation can be readily explained (Schoen and Clegg, 1984) as an effect of long-distance transportation of a pollen load from one plant to another (Hardy et al., 2004). However, in the case of wind-pollinated species, it is not obvious how this phenomenon could actually occur. Here, correlated mating over long distances seems extremely unlikely, because it requires that pollen gametes can travel between two particular trees structured as a homogeneous cloud. In our opinion, a possible explanation, that would lead to high genetic identity among immigrant pollen gametes, could be high mating compatibility (e.g. flowering synchronization) between particular oak trees located within the stand and near to the border of the stand. Alternatively, our estimates of θi may have been subject to positive bias. Although the simulation studies did not suggest any strong bias in indirect estimates of paternal identity probability (Hardy et al., 2004; Chybicki, 2013), Abraham et al. (2011) warned that the probability of paternal identity can be over-estimated (up to 4-fold) with indirect approaches when the proportion of full-sibs is very low. Nonetheless, even if this is the case in our study, the proportion of full-sibs among half-immigrant progeny would be quite high in the most extreme cases (see Table 1). Despite this speculation, our results demonstrated that a high proportion of immigrant pollen in the total pollen cloud of a single tree does not necessarily imply high pollen gene flow in certain cases, although this can be generally true (Nakanishi et al., 2004, 2005; Craft and Ashley, 2010).
Intra- vs. interspecific mating
Multispecies oak populations are often the scene of interspecific mating that leads to hybridization and introgression (Whittemore and Schaal, 1991; Bacilieri et al., 1996; Curtu et al., 2007; Gugerli et al., 2007; Peñaloza-Ramirez et al., 2010; Lepais and Gerber, 2011). Our study revealed non-random interspecific mating between Q. robur and Q. petraea. In particular, we detected asymmetric mating proportions, with Q. petraea acting generally as a donor while Q. robur acted as a recipient of interspecific pollen. However, we need to recall here that the taxonomic status of some trees in the study stand may have been incorrectly assigned. Problems with taxonomic discrimination between the studied oaks have been discussed often (e.g. Kremer et al., 2002; Curtu et al., 2007). Generally, using a multivariate approach, one is able to discriminate between the two species quite easily. However, because hybrids can manifest the phenotypes of a pure species (Gugerli et al., 2007), genetic markers are needed to improve discrimination. Unfortunately, our marker set has poor discrimination power for this purpose (as a comparison, see, for example, Lepais et al., 2009; Neophytou et al., 2010; Chybicki et al., 2012). Nevertheless, the observed asymmetric mating proportions could be due, hypothetically, to Q. petraea being a more invasive species (Petit et al., 2004). Yet controlled crosses did reveal asymmetric rates of interspecific fertilization, with Q. robur being more successful as female than Q. petraea (Steinhoff, 1998). These findings were confirmed in a few field studies (Bacilieri et al., 1996; Curtu et al., 2009; Lepais and Gerber, 2011). A hypothesis of species succession through genome invasion (Petit et al., 2004) seems very attractive in our case, especially because in the study area Q. petraea reaches the north-eastern limit of the natural distribution. However, because the stand originated artificially from planting, any in-depth interpretation should be made with caution.
Also, we detected a positive relationship between the rate of local mating (ci) and the preference towards intraspecific mating in Q. petraea. A possible explanation could be that flowering times of the two oak species are not synchronized (Williams et al., 2001). If so, those Q. petraea trees which are more synchronized with Q. robur, as compared with the species average, could receive local pollen mostly from Q. robur. However, because the overall rate of successful interspecific fertilization (in a direction from Q. robur to Q. petraea) is low, much of the fertilization is due to immigrant, putatively conspecific, pollen. Unfortunately, we have no data on flowering phenology in the stand. Scarce published data do not support the hypothesis about significantly asynchronous flowering between Q. robur and Q. petraea (Bacilieri et al., 1995; Chesnoiu et al., 2009). However, because a similar hypothesis has already been formulated (Curtu et al., 2009), it would be an attractive subject for future studies.
Fecundity
Plant size is often considered as a proxy of fecundity. In the simplest case, fecundity (sexual function) increases allometrically with size, because larger individuals have more branches and therefore more reproductive organs (Enquist and Niklas, 2002). Also, larger plants have a larger energetic budget than smaller plants. Theoretically, in wind-pollinated hermaphrodites, such as oaks, larger plants tend to be more male-biased than smaller plants (Sakai and Sakai, 2003). The prediction holds true especially when the fecundity of larger plants in relation to smaller plants is higher than their relative seed dispersal area. This evolutionary prediction stems from the fact that larger (taller) wind-pollinated plants can disperse their pollen more extensively, achieving higher male reproductive success when compared with smaller plants. However, in natural conditions, microhabitat quality is a proximate factor of fecundity because plants that grow in a richer microhabitat are potentially larger and more fecund. However, in real-world studies, tree size was found to be unrelated to male reproductive success in the wind-pollinated tree species Quercus macrocarpa (Dow and Ashley, 1998b) and Fagus crenata (Shimatani et al., 2007). In contrast, Asuka et al. (2005) found a positive correlation between the proportion of fertilizations and the tree diameter in one of the two studied populations of F. crenata. In our case, estimated male fecundity was spatially structured, with a clinal distribution. This was not dependent on species identity and did not co-vary with tree size. Thus, it seems that male fecundity was related to resources that do not influence tree size. Better exposure to wind and/or insolation seems a possible cause because the southern corner of the stand is slightly elevated. Another explanation could be that, among size-related variables other than the tree diameter, the crown volume is a better predictor of male fecundity (Dow and Ashley, 1998b). However, in the area of high male fecundity (see Fig. 4A), trees grow in high density, so that their crown volumes probably do not deviate from the stand average.
Our study has demonstrated that the relative effective density of pollen donors is about 5. In other words, if there are approx. 100 individuals ha−1, the estimated distribution of male fecundity translates into about 20 pollen donors ha−1 effectively contributing to the local pollen production. One possible consequence of the uneven contribution of individuals to the next generation is a decrease of the effective population size resulting in genetic drift within populations (Wilson and Levin, 1986). Obviously, the present results describe contemporary mating patterns and thus they provide little evidence of the overall reduction of effective population size during the life time of the population. However, a previous study revealed that effective population size can be relatively stable across different seed collection years (Dering and Chybicki, 2012). Paternity data for future seasons would help in reaching a final conclusion.
In order to capture the finest details of mating patterns from the perspective of individual females, we have developed a complex mating model and used a Bayesian approach to estimate parameters of interest. Consequently, we have demonstrated that mating patterns in oaks are far from being random. This applies to all mating components: self-fertilization; outcross mating within a local population; and mating with distant males through pollen immigration. On the other hand, male features such as proximity to focal females or male size (here DBH) appeared to be poor proxies of male mating success. Therefore, the question remains: why is a given female more likely to mate with a particular male? Is the number of mates per female genetically or environmentally determined? As sexual selection by female choice has not been excluded in oaks (Dow and Ashley, 1998a), besides flower phenology, there might be additional cryptic mechanisms determining the potential of females to mate with few or many males. Temporal data on mating patterns should shed more light on these issues. Nevertheless, the long-term evolutionary implications of assortative vs. random mating in oaks are yet to be understood (Soularue and Kremer, 2012).
ACKNOWLEDGEMENTS
We thank Katarzyna Meyza, Ewa Sztupecka and Artur Dzialuk for their help during collection of the material in the field (K.M., E.S. and A.D.) and in the laboratory work (K.M. and E.S.). We also thank Jane Sampson for the thorough reading and comments on the first version of the manuscript. The study was supported by the Polish Ministry of Science and Higher Education (the IUVENTUS Plus programme) [grant no. IP2010 005470].
LITERATURE CITED
- Aas G. Crossing experiments with pedunculate and sessile oak (Quercus robur and Q. petraea) Allgemeine Forst- und Jagdzeitung. 1991;162:141–145. [Google Scholar]
- Abraham ST, Zaya DN, Koenig WD, Ashley MV. Interspecific and intraspecific pollination patterns of Valley Oak, Quercus lobata, in a mixed stand in coastal central California. International Journal of Plant Sciences. 2011;172:691–699. [Google Scholar]
- Adams WT, Birkes DS. Estimating mating patterns in forest tree populations. In: Fineschi S, Malvoti ME, Cannata F, Hattemer HH, editors. Biochemical markers in the population genetics of forest trees. The Hague: SPB Academic Publishing; 1991. pp. 157–172. [Google Scholar]
- Ashley MV. Plant parentage, pollination, and dispersal: how DNA microsatellites have altered the landscape. Critical Reviews in Plant Sciences. 2010;29:148–161. [Google Scholar]
- Asuka Y, Tomaru N, Munehara Y, Tani N, Tsumura Y, Yamamoto S. Half-sib family structure of Fagus crenata saplings in an old-growth beech–dwarf bamboo forest. Molecular Ecology. 2005;14:2565–2575. doi: 10.1111/j.1365-294x.2005.02585.x. [DOI] [PubMed] [Google Scholar]
- Austerlitz F, Smouse PE. Two-generation analysis of pollen flow across a landscape. IV. Estimating the dispersal parameter. Genetics. 2002;161:355–363. doi: 10.1093/genetics/161.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austerlitz F, Smouse PE. Two-generation analysis of pollen flow across a landscape. II. Relation between Fft, pollen dispersal and interfemale distance. Genetics. 2001;157:851–857. doi: 10.1093/genetics/157.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austerlitz F, Dick CW, Dutech C, et al. Using genetic markers to estimate the pollen dispersal curve. Molecular Ecology. 2004;13:937–954. doi: 10.1111/j.1365-294x.2004.02100.x. [DOI] [PubMed] [Google Scholar]
- Bacilieri R, Roussel G, Ducousso A. Hybridization and mating system in a mixed stand of sessile and pedunculate oak. Annales des Sciences Forestières. 1993;50:122–127. [Google Scholar]
- Bacilieri R, Ducousso A, Kremer A. Genetic, morphological, ecological and phenological differentiation between Quercus petraea (Matt.) Liebl. and Quercus robur L. in a mixed stand of northwest of France. Silvae Genetica. 1995;44:1–10. [Google Scholar]
- Bacilieri R, Ducousso A, Petit RJ, Kremer A. Mating system and asymmetric hybridization in a mixed stand of European oaks. Evolution. 1996;50:900–908. doi: 10.1111/j.1558-5646.1996.tb03898.x. [DOI] [PubMed] [Google Scholar]
- Balding DJ. Likelihood-based inference for genetic correlation coefficients. Theoretical Population Biology. 2003;63:221–230. doi: 10.1016/s0040-5809(03)00007-8. [DOI] [PubMed] [Google Scholar]
- Barrett SCH, Harder LD. Ecology and evolution of plant mating. Trends in Ecology and Evolution. 1996;11:73–79. doi: 10.1016/0169-5347(96)81046-9. [DOI] [PubMed] [Google Scholar]
- Burczyk J, Chybicki I. Cautions on direct gene flow estimation in plant populations. Evolution. 2004;58:956–963. doi: 10.1111/j.0014-3820.2004.tb00430.x. [DOI] [PubMed] [Google Scholar]
- Burczyk J, Koralewski TE. Parentage versus two-generation analyses for estimating pollen-mediated gene flow in plant populations. Molecular Ecology. 2005;14:2525–2537. doi: 10.1111/j.1365-294X.2005.02593.x. [DOI] [PubMed] [Google Scholar]
- Burczyk J, Adams WT, Moran GF, Griffin AR. Complex patterns of mating revealed in a Eucalyptus regnans seed orchard using allozyme markers and the neighborhood model. Molecular Ecology. 2002;11:2379–2391. doi: 10.1046/j.1365-294x.2002.01603.x. [DOI] [PubMed] [Google Scholar]
- Burczyk J, Lewandowski A, Chalupka W. Local pollen dispersal and distant gene flow in Norway spruce (Picea abies [L.] Karst.) Forest Ecology and Management. 2004;197:39–48. [Google Scholar]
- Buschbom J, Yanbaev Y, Degen B. Efficient long-distance gene flow into an isolated relict oak stand. Journal of Heredity. 2011;102:464–472. doi: 10.1093/jhered/esr023. [DOI] [PubMed] [Google Scholar]
- Chesnoiu EN, Sofletea N, Curtu AL, Toader A, Radu R, Enescu M. Bud burst and flowering phenology in a mixed oak forest from Eastern Romania. Annales des Sciences Forestières. 2009;52(199- 206) [Google Scholar]
- Chybicki IJ. Note on the applicability of the F-model in analysis of pollen pool heterogeneity. Journal of Heredity. 2013 doi: 10.1093/jhered/est029. in press. http://dx.doi.org/10.1093/jhered/est029 . [DOI] [PubMed] [Google Scholar]
- Chybicki IJ, Burczyk J. Realized gene flow within mixed stands of Quercus robur L. and Q. petraea (Matt.) Liebl. revealed at the stage of naturally established seedling. Molecular Ecology. 2010a;19:2137–2151. doi: 10.1111/j.1365-294X.2010.04632.x. [DOI] [PubMed] [Google Scholar]
- Chybicki IJ, Burczyk J. NM + : software implementing parentage-based models for estimating gene dispersal and mating patterns in plants. Molecular Ecology Resources. 2010b;10:1071–1075. doi: 10.1111/j.1755-0998.2010.02849.x. [DOI] [PubMed] [Google Scholar]
- Chybicki IJ, Trojankiewicz M, Oleksa A, Dzialuk A, Burczyk J. Isolation-by-distance within naturally established populations of European beech (Fagus sylvatica) Botany. 2009;87:791–798. [Google Scholar]
- Chybicki IJ, Oleksa A, Kowalkowska K, Burczyk J. Genetic evidence of reproductive isolation in a remote enclave of Quercus pubescens in presence of cross-fertile species. Plant Systematics and Evolution. 2012;298:1045–1056. [Google Scholar]
- Coelho ASG, Vencovsky R. Intrapopulation fixation index dynamics in finite populations with variable outcrossing rates. Scientia Agricola. 2003;60:305–313. [Google Scholar]
- Craft KJ, Ashley MV. Pollen-mediated gene flow in isolated and continuous stands of bur oak, Quercus macrocarpa (Fagaceae) Annals of Botany. 2010;97:1999–2006. doi: 10.3732/ajb.0900390. [DOI] [PubMed] [Google Scholar]
- Crow JF, Kimura M. An introduction to population genetics theory. New York: Harper and Row; 1970. [Google Scholar]
- Curtu AL, Gailing O, Finkeldey R. Evidence for hybridization and introgression within a species-rich oak (Quercus spp.) community. BMC Evolutionary Biology. 2007;7:218. doi: 10.1186/1471-2148-7-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtu AL, Gailing O, Finkeldey R. Patterns of contemporary hybridization inferred from paternity analysis in a four-species forest. BMC Evolutionary Biology. 2009;9:284. doi: 10.1186/1471-2148-9-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dering M, Chybicki IJ. Assessment of genetic diversity in two-species oak seed stands and their progeny populations. Scandinavian Journal of Forest Research. 2012;27:2–9. [Google Scholar]
- Devlin B, Roeder K, Ellstrand NC. Fractional paternity assignment – theoretical development and comparison to other methods. Theoretical and Applied Genetics. 1988;76:369–380. doi: 10.1007/BF00265336. [DOI] [PubMed] [Google Scholar]
- Di-Giovanni F, Kevan PG, Arnold J. Lower planetary boundary layer profiles of atmospheric conifer pollen above a seed orchard in northern Ontario, Canada. Forest Ecology and Management. 1996;83:87–97. [Google Scholar]
- Dow BD, Ashley MV. High levels of gene flow in bur oak revealed by paternity analysis using microsatellites. Journal of Heredity. 1998a;89:62–70. [Google Scholar]
- Dow BD, Ashley MV. Factors influencing male mating success in bur oak, Quercus macrocarpa. New Forests. 1998b;15:161–180. [Google Scholar]
- Ducousso A, Michaud H, Lumaret R. Reproduction and gene flow in the genus Quercus. Annales des Sciences Forestières. 1993;50:91s–106s. [Google Scholar]
- El-Kassaby YA, Ritland K. The relationship of outcrossing rate to reproductive phenology and supplemental mass pollination in a Douglas-fir seed orchard. Silvae Genetica. 1986;35:240–244. [Google Scholar]
- Enquist BJ, Niklas KJ. Global allocation rules for patterns of biomass partitioning in seed plants. Science. 2002;295:1517–1520. doi: 10.1126/science.1066360. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer MS, Morgan MT. Effects of natural rates of geitonogamy on fruit set in Asclepias speciosa (Apocynaceae): evidence favoring the plant's dilemma. American Journal of Botany. 2003;90:1746–1750. doi: 10.3732/ajb.90.12.1746. [DOI] [PubMed] [Google Scholar]
- Gaggiotti OE, Foll M. Quantifying population structure using the F-model. Molecular Ecology Resources. 2010;10:821–830. doi: 10.1111/j.1755-0998.2010.02873.x. [DOI] [PubMed] [Google Scholar]
- Gérard PR, Klein EK, Austerlitz F, Fernández-Manjarrés JF, Frascaria-Lacoste N. Assortative mating and differential male mating success in an ash hybrid zone population. BMC Evolutionary Biology. 2006;6:96. doi: 10.1186/1471-2148-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S, Shimatani K, Yoshimaru H, Takahashi Y. Fat-tailed gene flow in the dioecious canopy tree species Fraxinus mandshurica var. japonica revealed by microsatellites. Molecular Ecology. 2006;15:2985–2996. doi: 10.1111/j.1365-294X.2006.02976.x. [DOI] [PubMed] [Google Scholar]
- Gugerli F, Walser J-C, Dounvai K, Holderegger R, Finkeldey R. Coincidence of small-scale spatial discontinuities in leaf morphology and nuclear microsatellite variation of Quercus petraea and Q. robur in a mixed forest. Annals of Botany. 2007;99:713–722. doi: 10.1093/aob/mcm006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield JD, Richardson DS, Burke T. Towards unbiased parentage assignment: combining genetic, behavioural and spatial data in a Bayesian framework. Molecular Ecology. 2006;15:3715–3730. doi: 10.1111/j.1365-294X.2006.03050.x. [DOI] [PubMed] [Google Scholar]
- Hagman M. Incompatibility in forest trees. Proceedings of the Royal Society B: Biological Sciences. 1975;188:313–326. [Google Scholar]
- Harder LD, Routley MR. Pollen and ovule fates and reproductive performance by flowering plants. In: Harder LD, Barret SCH, editors. Ecology and evolution of flowers. Oxford: Oxford University Press; 2006. pp. 61–80. [Google Scholar]
- Hardy OJ, González-Martínez SC, Colas B, Fréville H, Mignot A, Olivieri I. Fine-scale genetic structure and gene dispersal in Centaurea corymbosa (Asteraceae). II. Correlated paternity within and among sibships. Genetics. 2004;168:1601–1614. doi: 10.1534/genetics.104.027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein EK, Lavigne C, Picault H, Renard M, Gouyon P-H. Pollen dispersal of oilseed rape: estimation of the dispersal function and effects of field dimension. Journal of Applied Ecology. 2006;43:141–151. [Google Scholar]
- Klein EK, Desassis N, Oddou-Muratorio S. Pollen flow in the wildservice tree, Sorbus torminalis (L.) Crantz. IV. Whole inter-individual variance of male fecundity estimated jointly with dispersal kernel. Molecular Ecology. 2008;17:3323–3336. doi: 10.1111/j.1365-294X.2008.03809.x. [DOI] [PubMed] [Google Scholar]
- Klein EK, Carpentier FH, Oddou-Muratorio S. Estimating the variance of male fecundity from genotypes of progeny arrays: evaluation of the Bayesian forward approach. Methods in Ecology and Evolution. 2011;2:349–361. [Google Scholar]
- Kremer A, Dupouey JL, Deans JD, et al. Leaf morphological differentiation between Quercus robur and Quercus petraea is stable across western European mixed oak stands. Annals of Forest Science. 2002;59:777–787. [Google Scholar]
- Lepais O, Gerber S. Reproductive patterns shape introgression dynamics and species succession within the European white oak species complex. Evolution. 2011;65:156–170. doi: 10.1111/j.1558-5646.2010.01101.x. [DOI] [PubMed] [Google Scholar]
- Lepais O, Petit RJ, Guichoux E, et al. Species relative abundance and direction of introgression in oaks. Molecular Ecology. 2009;18:2228–2242. doi: 10.1111/j.1365-294X.2009.04137.x. [DOI] [PubMed] [Google Scholar]
- Levin D, Kerster H. Gene flow in seed plants. Evolutionary Biology. 1974;7:139–220. [Google Scholar]
- Mimura M, Aitken SN. Increased selfing and decreased effective pollen donor number in peripheral relative to central populations in Picea sitchensis (Pinaceae) American Journal of Botany. 2007;94:991–998. doi: 10.3732/ajb.94.6.991. [DOI] [PubMed] [Google Scholar]
- Moran EV, Clark JS. Estimating seed and pollen movement in a monoecious plant: a hierarchical Bayesian approach integrating genetic and ecological data. Molecular Ecology. 2011;20:1248–1262. doi: 10.1111/j.1365-294X.2011.05019.x. [DOI] [PubMed] [Google Scholar]
- Nakanishi A, Tomaru N, Yoshimaru H, Kawahara T, Manabe T, Yamamoto S. Patterns of pollen flow and genetic differentiation among pollen pools in Quercus salicina in a warm temperate old-growth evergreen broad-leaved forest. Silvae Genetica. 2004;53:258–264. [Google Scholar]
- Nakanishi A, Tomaru N, Yoshimaru H, Manabe T, Yamamoto S. Interannual genetic heterogeneity of pollen pools accepted by Quercus salicina individuals. Molecular Ecology. 2005;11:4469–4478. doi: 10.1111/j.1365-294X.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- Neophytou C, Aravanopoulos F, Fink S, Dounavi A. Detecting interspecific and geographic differentiation patterns in two interfertile oak species (Quercus petraea (Matt.) Liebl. and Q. robur L.) using small sets of microsatellite markers. Forest Ecology and Management. 2010;259:2026–2035. [Google Scholar]
- Oddou-Muratorio S, Klein EK, Austerlitz F. Pollen flow in the wildservice tree, Sorbus torminalis (L.) Crantz. II. Pollen dispersal and heterogeneity in mating success inferred from parent–offspring analysis. Molecular Ecology. 2005;14:4441–4452. doi: 10.1111/j.1365-294X.2005.02720.x. [DOI] [PubMed] [Google Scholar]
- Peñaloza-Ramírez JM, González-Rodríguez A, Mendoza-Cuenca L, Caron H, Kremer A, Oyama K. Interspecific gene flow in a multispecies oak hybrid zone in the Sierra Tarahumara of Mexico. Annals of Botany. 2010;105:389–399. doi: 10.1093/aob/mcp301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Bodénès C, Ducousso A, Roussel G, Kremer A. Hybridization as a mechanism of invasion in oaks. New Phytologist. 2004;161:151–164. [Google Scholar]
- Restoux G, Silva DE, Sagnard F, Torre F, Klein E, Fady B. Life at the margin: the mating system of Mediterranean conifers. Web Ecology. 2008;8:94–102. [Google Scholar]
- Rousset F. Inferences from spatial population genetics. In: Balding D, Bishop M, Cannings C, editors. Handbook of statistical genetics. 3rd edn. John Wiley & Sons; 2007. pp. 945–979. [Google Scholar]
- Sakai A, Sakai S. Size-dependent ESS sex allocation in wind-pollinated cosexual plants: fecundity vs. stature effects. Journal of Theoretical Biology. 2003;222:283–295. doi: 10.1016/s0022-5193(03)00024-9. [DOI] [PubMed] [Google Scholar]
- Shen H, Rudin D, Lindgren D. Study of the pollination pattern in a Scots pine seed orchard by means of isoenzyme analysis. Silvae Genetica. 1981;30:7–15. [Google Scholar]
- Schoen DJ, Brown AHD. Intraspecific variation in population gene diversity and effective population size correlates with the mating system in plants. Proceedings of the National Academy of Sciences, USA. 1991;88:4494–4497. doi: 10.1073/pnas.88.10.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen DJ, Clegg MT. Estimation of mating system parameters when outcrossing events are correlated. Proceedings of the National Academy of Sciences, USA. 1984;81:5258–5262. doi: 10.1073/pnas.81.16.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimatani K, Kimura M, Kitamura K, Suyama Y, Isagi Y, Sugita H. Determining the location of a deceased mother tree and estimating forest regeneration variables by use of microsatellites and spatial genetic models. Population Ecology. 2007;49:317–330. [Google Scholar]
- Slavov GT, Howe GT, Gyaourova AV, Birkes DS, Adams WT. Estimating pollen flow using SSR markers and paternity exclusion: accounting for mistyping. Molecular Ecology. 2005;14:3109–3121. doi: 10.1111/j.1365-294x.2005.02620.x. [DOI] [PubMed] [Google Scholar]
- Smouse PE, Dyer RJ, Westfall RD, Sork VL. Two-generation analysis of pollen flow across a landscape. I. Male gamete heterogeneity among females. Evolution. 2001;55:260–271. doi: 10.1111/j.0014-3820.2001.tb01291.x. [DOI] [PubMed] [Google Scholar]
- Sork VL, Nason J, Campbell DR, Fernandez J. Landscape approaches to contemporary gene flow in plants. Trends in Ecology and Evolution. 1999;14:219–224. doi: 10.1016/s0169-5347(98)01585-7. [DOI] [PubMed] [Google Scholar]
- Soularue J-P, Kremer A. Assortative mating and gene flow generate clinal phenological variation in trees. BMC Evolutionary Biology. 2012;12:79. doi: 10.1186/1471-2148-12-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhoff S. Results of species hybridization with Quercus robur L. and Quercus petraea (Matt.) Liebl. Annales des Sciences Forestieres. 1993;50:137s–143s. [Google Scholar]
- Steinhoff S. Controlled crosses between pedunculate and sessile oak: results and conclusion. Allgemeine Forst- und Jagdzeitung. 1998;169:163–168. [Google Scholar]
- Streiff R, Ducousso A, Lexer C, Steinkellner H, Gloessl J, Kremer A. Pollen dispersal inferred from paternity analysis in a mixed oak stand of Quercus robur L and Q. petraea (Matt.) Liebl. Molecular Ecology. 1999;8:831–841. [Google Scholar]
- Ward M, Dick CW, Gribel R, Lowe AJ. To self, or not to self … A review of outcrossing and pollen-mediated gene flow in neotropical trees. Heredity. 2005;95:246–254. doi: 10.1038/sj.hdy.6800712. [DOI] [PubMed] [Google Scholar]
- Whittemore AT, Schaal BA. Interspecific gene flow in sympatric oaks. Proceedings of the National Academy of Sciences, USA. 1991;88:2540–2544. doi: 10.1073/pnas.88.6.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CG. Aerobiology of Pinus taeda pollen clouds. Canadian Journal of Forest Research. 2008;38:2177–2188. [Google Scholar]
- Williams JH, Boecklen WJ, Howard DJ. Reproductive processes in two oak (Quercus) contact zones with different levels of hybridization. Heredity. 2001;87:680–690. doi: 10.1046/j.1365-2540.2001.00968.x. [DOI] [PubMed] [Google Scholar]
- Wilson JB, Levin DA. Some genetic consequences of skewed fecundity distributions in plants. Theoretical and Applied Genetics. 1986;73:113–121. doi: 10.1007/BF00273727. [DOI] [PubMed] [Google Scholar]
- Yacine A, Bouras F. Self- and cross-pollination effects on pollen tube growth and seed set in holm oak Quercus ilex L (Fagaceae) Annales des Sciences Forestières. 1997;54:447–462. [Google Scholar]