Abstract
The microbiota plays a critical, weight-promoting role in diet-induced obesity (DIO), but the pathways that cause the microbiota to induce weight gain are unknown. We report that mice deficient in lymphotoxin (LT), a key molecule in gut immunity, were resistant to DIO. Ltbr−/− mice differed in microbial community composition compared to their heterozygous littermates, including an overgrowth of segmented filamentous bacteria (SFB). Furthermore, cecal transplantation conferred leanness to germ-free recipients. Housing Ltbr−/− mice with their obese siblings rescued weight gain, demonstrating the communicability of the obese phenotype. Ltbr−/− animals lacked interleukin 23 (IL-23) and IL-22 that can regulate SFB. Mice deficient in these pathways also resisted DIO, demonstrating that intact mucosal immunity guides diet-induced changes to the microbiota to enable obesity.
Over two-thirds of adults in the United States are overweight or obese, and studies suggest that the incidence of obesity will continue to rise in coming decades1–3. Although early twin-studies suggested an important role for host genetics in obesity4,5, recent evidence demonstrates that obesity is associated with a change in the intestinal microbiota6,7. Germ-free rodent models have demonstrated that the commensal microbiota contribute substantially to overall body weight, and even obesity induced by high-fat diet (HFD) is enervated in the absence of commensal bacteria8,9. Several studies have suggested that dietary components are the primary determinants of the composition of the microbiota10,11, but how the microbiota responds to changes in diet and ultimately impacts diet-induced obesity (DIO) is unknown.
Mucosal immunity lies in a delicate balance with the microbiota; a consequence of this symbiosis is a reciprocal relationship between host and microbe that contribute to host health12. As a result, there has been great speculation into the role of mucosal homeostasis in host illness, especially in the case of obesity. Recently it has been argued that in the absence of inflammasome signaling pathogenic microbes penetrate the mucosa and result in tumor necrosis factor (TNF) production, which causes non-alcoholic steatohepatitis in diets deficient for essential amino acids13. That study is similar to what has been argued for Toll-like receptor 5 (TLR5) in metabolic syndrome, where the absence of TLR5 results in a dysbiosis that induces weight gain14. However, although both of these studies and others have been informative of host-microbe dynamics in metabolic disease, it remains to be definitively demonstrated which mucosal immune pathways regulate the microbiota to induce normal weight gain after exposure to HFD in wild-type animals or humans.
Several epidemiological studies have linked polymorphisms in the TNF–Lymphotoxin (LT) locus to obesity and type II diabetes15,16. While the role for TNF in obesity has been extensively studied, the role that LT might play in this disease has been largely ignored. LT is essential for normal mucosal immunity17–19. LTα forms a membrane bound heterotrimer with LTβ and binds LTβ receptor (LTβR). Deficiency in either LTα or LTβR results in complete secondary lymphoid aplasia including a lack of mesenteric lymph nodes, Peyer’s patches and isolated lymphoid follicles20. The LTβR pathway directly regulates expression of interleukin 23 (IL-23), a heterodimeric cytokine composed of p19 and p40 subunits, and results in production of IL-22 from RORγt+ innate lymphocytes; furthermore, Ltbr−/− mice fail to express sufficient IL-23 and IL-22 to eliminate mucosal pathogens21,22. Thus, intact agonism of the LT pathway is a critical for normal mucosal defense.
Lta, the gene encoding LTα, is located 1.3 kb away from Tnf16,20. Mechanistic studies in rodents have revealed that in the absence of TNF signaling, animals are less susceptible to insulin resistance provoked by DIO23. However, deficiency in TNF signaling only has a modest impact on adiposity in DIO, arguing that the TNF pathway alone does not explain changes in body mass. As a result, the polymorphisms linking TNF to obesity may be informative of an actual mechanistic connection between LT and obesity. This notion is best illustrated by the fact that some polymorphisms linking TNF to obesity actually lie within coding exons of Lta16. A consequence of the work on the “obesity-associated microbiome” is that environmental exposure, either in the form of HFD or through colonization of obesity-inducing microbiota, may actually play a critical role in the pathogenesis of obesity and associated diseases7. The microbiota lie in a delicate homeostasis with host immunity, and given the epidemiological data linking LT to obesity and the key role of LT in regulating mucosal defense, we hypothesized that LT-driven pathways regulate changes to the microbiota that promote weight gain in DIO.
Results
LTβR and LTα are essential for weight gain in DIO
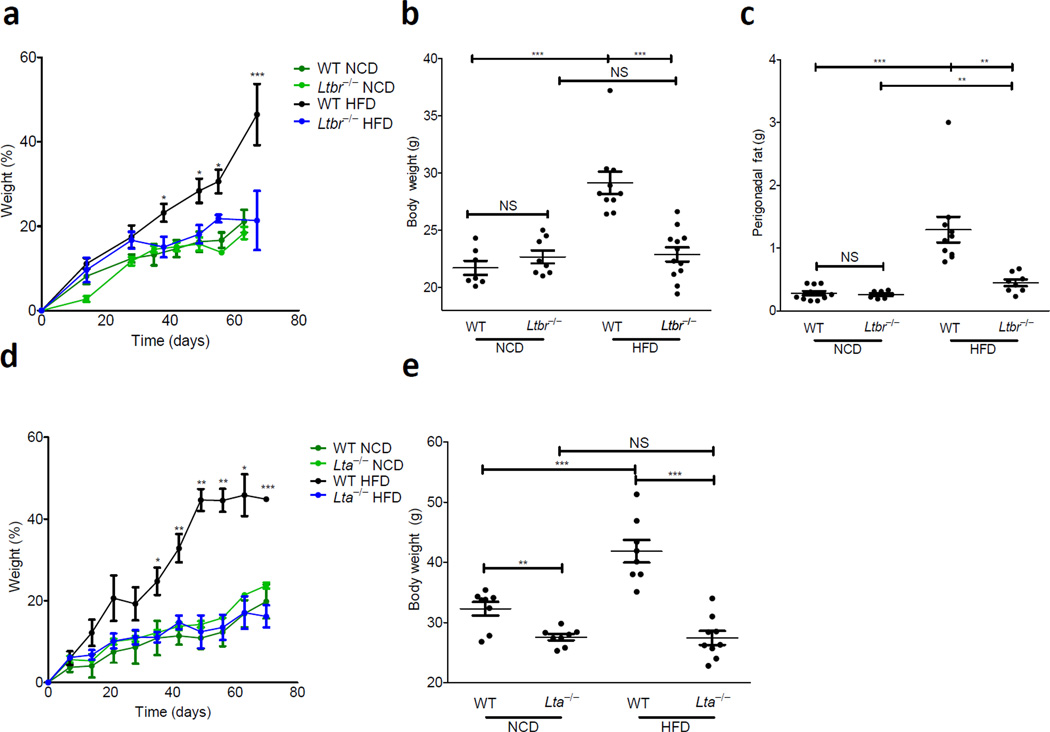
To address the role of the LT pathway in DIO, we challenged wild-type and Ltbr−/− adult animals with HFD. Animals were kept on normal chow diet (NCD) in our vivarium until 9 weeks of age where they were either switched onto HFD or maintained on NCD (for composition of all diets see Supplementary Table 1). While there was no difference in growth between wild-type and Ltbr−/− mice on NCD, wild-type mice on HFD gained significantly more weight than Ltbr−/− animals, which were resistant to DIO (Fig. 1a). There was no difference in weight after 9 weeks of dietary challenge between wild-type and Ltbr−/− animals maintained on NCD; wild-type and Ltbr−/− animals weighed 21.70 ± 0.60 g and 22.66 ± 0.56 g at the end of NCD, respectively (Fig. 1b). However, at the end of HFD, wild-type and Ltbr−/− groups were significantly different, weighing 29.13 ± 0.99 g and 22.87 ± 0.62 g, respectively (Fig. 1b). In contrast to wild-type mice, Ltbr−/− animals did not gain additional weight after prolonged HFD, suggesting a role for the LT pathway in controlling excess weight gain induced by HFD.
Figure 1. LTβR is essential for weight gain in response to HFD.
(a–e) 9 week old WT (C57BL6), Ltbr−/−, or Lta−/− animals were subject to a HFD or NCD for 9 weeks. (a) Weight gain as a percentage over starting weight is plotted (b) Absolute weight in grams at the end of diet. (c) Perigonadal fat was removed and weighed at the end of diet; weight of fat is plotted. (d) Weight gain as a percentage of starting weight is plotted against days on diet. (e) Weight at the end of diet for mice in E. (Data is reflective of 2–3 independent experiments per genotype, with n=5–12 mice in all groups; statistics demonstrate differences between HFD groups; Student’s t-test for individual points along growth curves; 1-Way Anova with Bonferonni post-test for dot plots: *P <0.05, ** P <0.01, *** P <0.001)
At the time of sacrifice it was clear that changes in weight gain corresponded with changes in adiposity. Necropsy revealed the perigonadal fat pad of wild-type animals was much larger than that of Ltbr−/− mice (Supplementary Fig. 1). To quantify these results, the perigonadal fat pad of wild-type and Ltbr−/− mice was dissected out and weighed at the end of diet. Both in absolute terms and as a percentage of body weight, the perigonadal fat pad of wild-type mice had expanded much more than that of Ltbr−/− mice on HFD. This finding was in stark contrast to their relative adiposity on NCD, where wild-type and Ltbr−/− animals weighed similarly at the end of diet and had similar body composition (Fig. 1c and Supplementary Fig. 1). Together, this data demonstrates that LTβR is essential for excess weight gain and adiposity induced by HFD.
LTα forms part of a membrane bound heterotrimer that binds to LTβR, and polymorphisms in coding exons of LTα have been linked to obesity16. We therefore challenged wild-type and Lta−/− mice with HFD to determine whether this ligand was essential for weight gain. Consistent with our results in Ltbr−/− animals, Lta−/− animals resisted DIO and showed similar growth on HFD to both wild-type and Lta−/− mice on NCD (Fig. 1d); these growth patterns reflected stark differences in body composition between wild-type and Lta−/− animals on HFD, with the latter being much leaner than the former (Fig. 1e and Supplementary Fig. 1). There was a modest, but detectable difference in weight between Lta−/− and wild-type animals on NCD, and this could be attributed to additional agonism of the TNFR pathway as LTα can form a soluble homotrimer that binds to TNFR (Fig. 1f). However unlike wild-type animals and similar to Ltbr−/− mice, Lta−/− animals did not appear to increase body weight on HFD, contextualizing the significance of the LT pathway in DIO. Furthermore, Ltb−/− animals also resisted weight gain compared to wild-type animals on HFD (Supplementary Fig. 2). Together, the data for Lta−/−, Ltb−/− and Ltbr−/− animals demonstrates the importance of the intact membrane bound LT pathway in DIO.
LTβR regulates the microbiota to induce weight gain
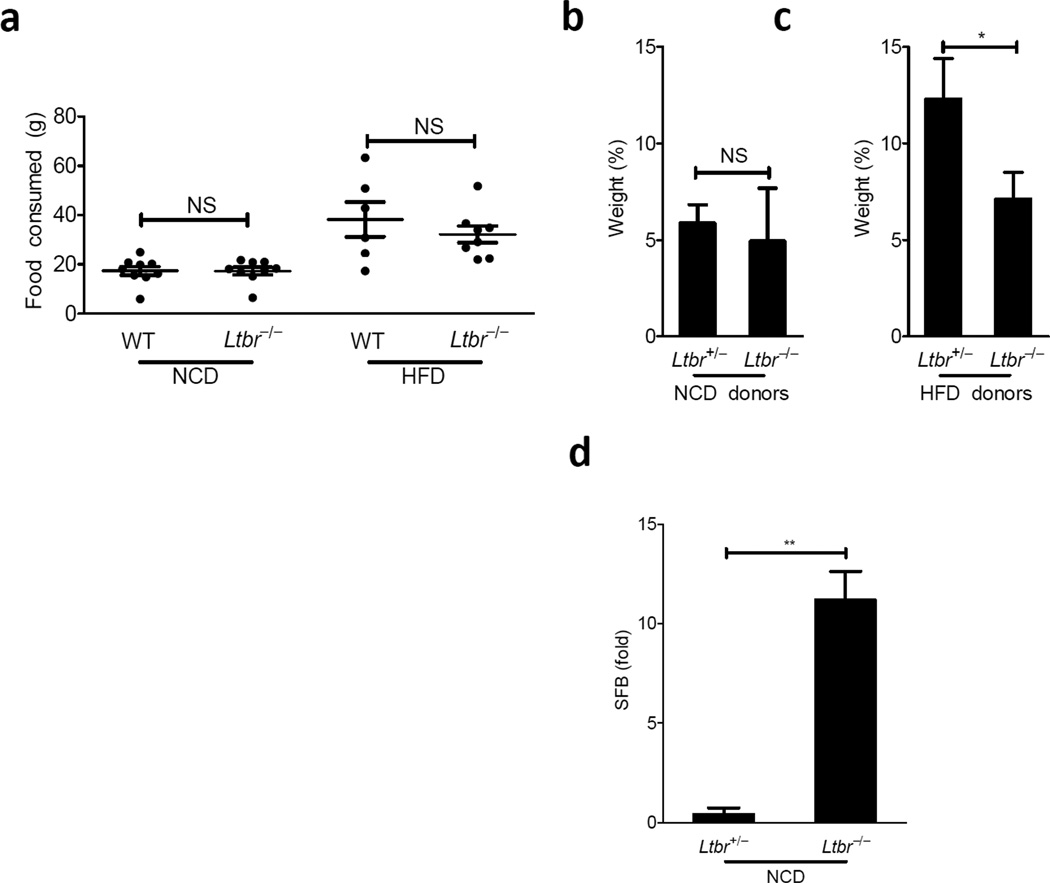
To better understand the mechanism by which the LT pathway promoted weight gain in DIO, we addressed the food intake of wild-type and Ltbr−/− animals on NCD and HFD. There were no obvious changes in feeding behavior between both groups on NCD or on HFD (Fig. 2a), suggesting that differences in weight gain were occurring despite similar consumption patterns. Studies in axenic mice have revealed that the intestinal microbiota enable access to greater caloric intake, and as a result germ-free mice weigh substantially less than their conventionalized littermates9. Because the LT-signaling pathways plays such a prominent role in normal mucosal defense and because feeding behavior was similar between groups, we wondered if the LT-pathway influenced changes in the microbiota that promoted weight gain.
Figure 2. LTβR influences weight gain through changes in the microbiota.
(a) Food consumed represents the weight difference of food between n and n+1 days of a given cage of mice over the course of the first two weeks on diet (b–c) Germ free mice were gavaged with cecal contents from Ltbr+/− or Ltbr−/− littermates maintained on NCD or HFD for 9–10 weeks starting at 9 weeks of age. Cecal contents from two donors was pooled. Recipients were kept on diets of similar compositions to donors. (b–c) Weight gain as a percentage of starting body weight is shown 20 days after gavage of germ free recipients from the NCD (b) and HFD (c) groups. (D) RT-PCR for SFB on DNA from stool collected from Ltbr+/− and Ltbr−/− mice 4 weeks after NCD start (n=4 mice per group, representative of 3 independent experiments for a and d; n=3–5 germ free mice/group, representative of 2 independent experiments Summary of p-values: * P <0.05, ** P <0.001, student’s t test for a and d, paired-t test for b and c)
To address this issue, we amplified the V1-V2 tags of 16S rRNA encoding genes from stool samples obtained from Ltbr+/− and Ltbr−/− animals on NCD and HFD and subjected the resulting PCR products to 454 Pyrosequencing. We performed Principle Coordinate Analysis (PCA) to spatially discriminate the variable region tag sequences of 16S rRNA encoding genes from Ltbr+/− and Ltbr−/− stool DNA. PCA revealed genotype- and diet-specific clustering dependent on the two largest components of variation (Supplementary Fig. 3). Intriguingly, PCA1 (52.18% of variation) strongly separated NCD and HFD groups and was consistent with a HFD-induced expansion of the Firmicute phyla observed in both genotypes (Supplementary Fig. 3,4); PCA2 (17.96% of variation) separated null and heterozygous animals, and demonstrated differences not explained by diet alone.
In addition to Firmicute expansion, a hallmark of the “obese microbiome” in human stool is a loss of commensal diversity. Even though total bacterial content as addressed by bacterial DNA per gram of stool was similar between either genotype and diet, Ltbr+/− animals experienced reduced commensal diversity after the start of HFD, while Ltbr−/− animals maintained a similarly diverse community to either group at the start of diet (Supplementary Fig. 4).
Although there were no differences at the phyla level between genotypes, we observed an overgrowth of the Erysopelotrichi class after HFD in Ltbr+/− (obese) animals that was completely absent in Ltbr−/− (lean) animals after HFD (Supplementary Table 2); this underrepresentation of Erysopelotrichi may have been occurring due to the overabundance of the Cytophagia class of the Bacteroidetes phyla (Supplementary Table 2). Importantly, the relative abundance of both of these classes was similar between genotypes on NCD (data not shown). This is a significant finding given that the Erysipelotrichi class has previously been reported as overabundant in the obese state24, and its ability to overgrow and monopolize the gastrointestinal niche after HFD has implicated it as a species that may influence metabolic disease in the host and whose colonization status after HFD appears to depend on LTβR.
To test how the changes to the microbiota contributed to weight gain, we transplanted the cecal contents of Ltbr+/− and Ltbr−/− mice into wild-type germ-free recipients. Recipients were maintained on a diet of similar composition to their donors (Supplementary Table 1). Consistent with our results in SPF mice, there was no difference in weight gain between recipients that received cecal contents from Ltbr+/− or Ltbr−/− donors on NCD after 20 days of diet (Fig. 2b), suggesting that although there were detectable differences in the microbial communities at this point, in and of themselves, these differences seen on NCD were unable to explain differential weight gain between genotypes. In contrast, the cecal contents of Ltbr+/− animals conferred greater weight gain than that of Ltbr−/− animals when both donor and recipient groups were kept on HFD (Fig. 2c). Although recipient groups weighed differently at and prior to 20 days after transplant, Ltbr−/− recipients caught up in weight gain after this time point (data not shown); however, this data could be due to the fact that recipient animals are wild-type and thus have an intact mucosal immune response. Wild-type gnotobiotic animals may be achieving normalization of their microbial communities after 3 weeks of HFD, a time period for maturation of ILFs and other key elements of mucosal immunity. Altogether these data demonstrate that changes in the microbial communities colonizing Ltbr+/− and Ltbr−/− animals after HFD are at least transiently causative of excess weight gain in the heterozygous group after HFD.
The fecal stream is composed of allochthonous (transient) and autochthonous (permanent resident) microbes and is informative of microbiota living throughout the gastrointestinal tract25. Further analysis of stool revealed changes in specific operational taxonomic units (OTUs) between heterozygous and knockout animals 4 weeks of HFD that extended beyond the class level to that of changes in specific genera. There were several OTUs over- and under- represented in in Ltbr−/− mice after HFD; we focused on overrepresented species because such species appear to require an LTβR-mediated immune response for clearance from the microbiota in response to HFD. Such clearance could contribute to the loss of commensal diversity experienced by heterozygous animals after HFD was initiated (Supplementary Fig. 4). One OTU significantly overrepresented in Ltbr−/− animals was not classifiable beyond the Clostridiales order (Supplementary Table 2). The OTU detected in our analysis had high sequence homology with the V1-V2 region of the 16S rRNA encoding gene of segmented filamentous bacteria (SFB) (Supplementary Fig. 5), an autochthonous, unclassified Clostridiales order member that is able to induce a potent TH17 cytokine-based immune response in mice26–28. SFB is detectable in the fecal stream of mice throughout adulthood but lives within the mucus layer of the ileum29,30. Quantitative PCR with primers specific for SFB demonstrated that SFB experienced a moderate overgrowth in Ltbr−/− which was detectable both in the feces and terminal ileum on NCD and HFD (Fig. 2d and Supplementary Fig. 6). Therefore, the LTβR pathway regulates changes in the microbiota, including loss of commensal diversity, after the initiation of HFD.
Sibling cohousing rescued weight gain and SFB regulation
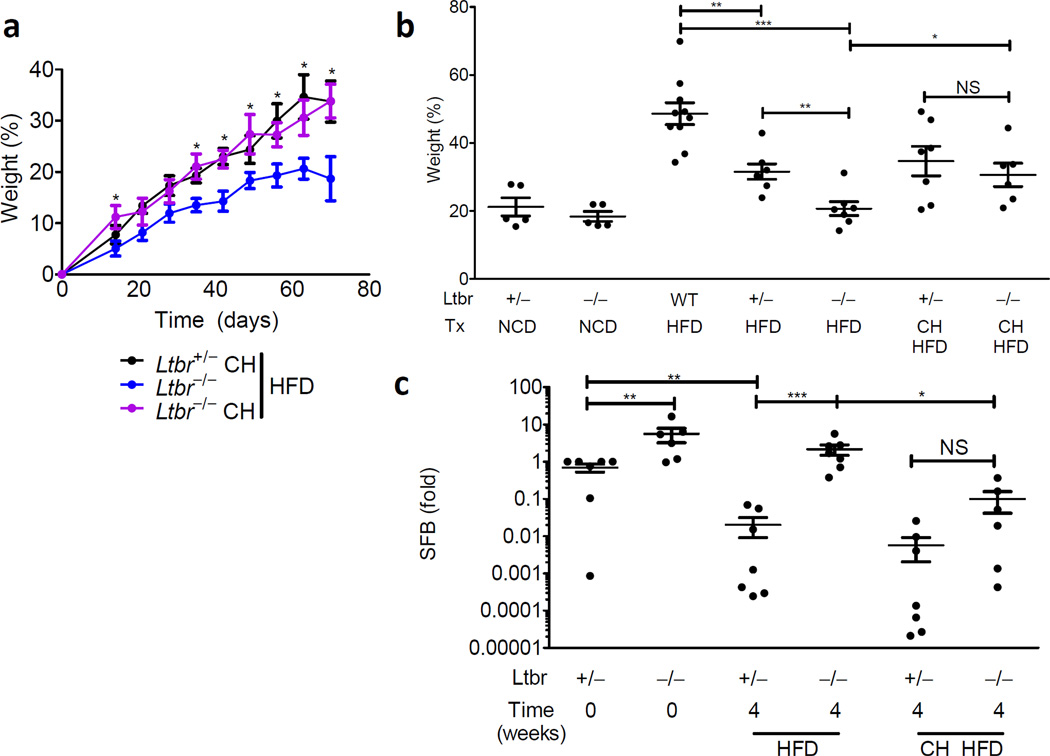
Given the conflicting viewpoints presented by various studies regarding genetic and environmental causes for obesity4,5,7, we wondered whether environmental manipulation would influence the phenotype of LTβR-deficient animals. To explore this, Ltbr+/− and Ltbr−/− littermates were weaned into cages separated by genotype or into cages where genotypes were mixed. Mice are coprophagic and fecal consumption is a mechanism by which mice housed in the same cage constantly colonize one another; cohousing is a commonly exploited experimental technique to facilitate microbiota exposure28,31. Separately housed Ltbr−/− mice resisted excess body weight deposition induced by diet but Ltbr−/− mice cohoused with their Ltbr+/− littermates experienced a rescued capacity for excess weight gain in response to HFD (Fig. 3a,b). These data suggest that Ltbr+/− littermates, which maintain intact regulation of their own microbiota, maybe constantly exposing Ltbr−/− mice to obesity-inducing microbes and supplementing growth. Although both mice are exposed to the other’s microbiota, the phenotype of heterozygous animal, which maintains an intact mucosal immune response, is dominant.
Figure 3. Environmental exposure reveals horizontal transmissibility of the obese phenotype.
(a–c) Ltbr+/− or Ltbr−/− were genotyped and weaned either separately or together (CH) at 3 weeks of age. (a) Weight gain as a percentage over starting weight is plotted for adult animals started on HFD at 9 weeks of age. (b) Weight gain as apercentage over starting weight after 9 weeks of diet; Tx indicates combination of diet and housing conditions. (c) RT-PCR for SFB in stool relative to Ltbr+/− littermates stool at diet start. (n=5–12 mice per group; growth curve is reflective of 3 independent experiments and statistics demonstrate differences between Ltbr−/− groups; Student’s t-test for individual points along growth curves and dot-plots: * P <0.05, ** P <0.01, *** P <0.001)
Because species diversity loss is a hallmark of the obese microbiome in humans, and because SFB was overrepresented in Ltbr−/− animals, we used SFB as a representative marker species for changes to the microbiota. Intriguingly, the proportion of SFB was substantially reduced after heterozygous animals were placed on HFD, suggesting that HFD creates environmental changes that are unfavorable to SFB (Fig. 3c). Strictly speaking, these changes could be either directly through changes in nutrition or mediated by indirect mechanisms. However, Ltbr−/− animals separately housed from their Ltbr+/− littermates experienced very modest, if any decreases in SFB after HFD, arguing that although both animals change nutritional source, this species-specific change cannot occur without intact host immunity (Fig. 3c). It is exciting to note that Ltbr−/− animals housed with their Ltbr+/− littermates experienced a rescued ability to reduce SFB abundance in their stool; this regained ability to clear SFB in response to HFD coincided with rescue of weight gain in the co-housed animals (Fig. 3c). These data demonstrate that exposing Ltbr−/− mice with their LTβR-replete siblings not only rescued weight gain, but rescued changes in the microbiota normally induced by exposure to HFD. The transmissibility of the obese phenotype tracked with changes in the microbiota normally associated with the obese state.
LTβR regulates IL-23/IL-22
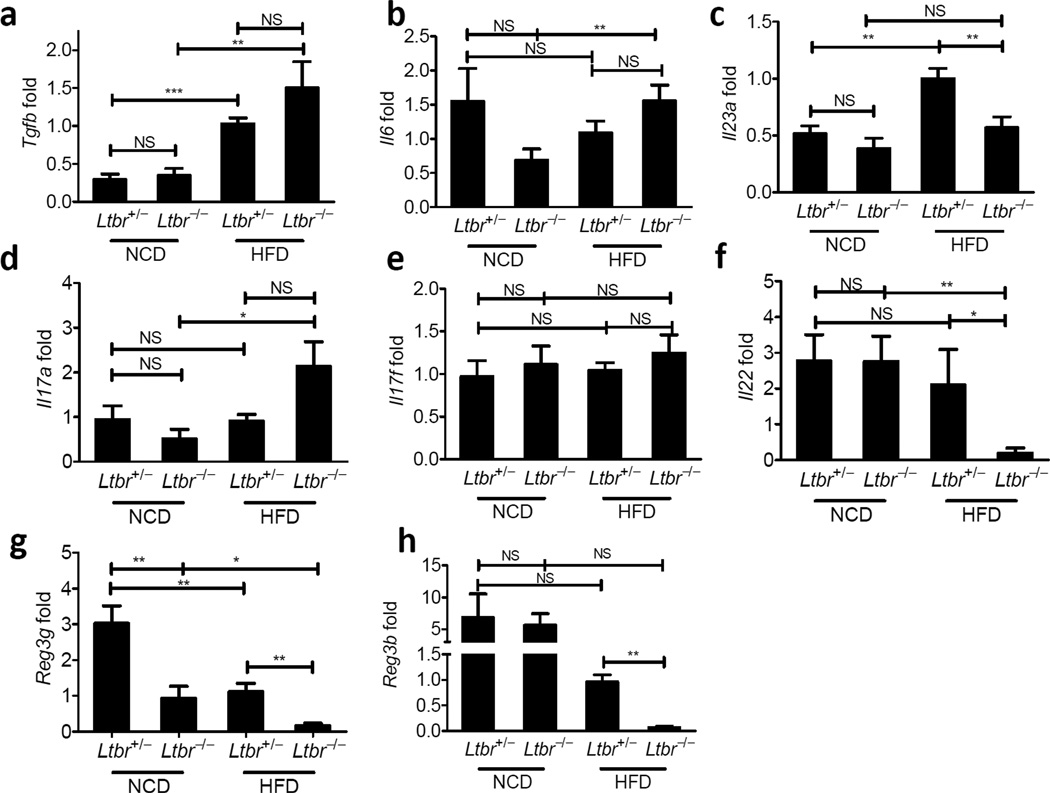
The behavior of SFB prompted us to consider elements of the TH17 cytokine pathway that might be regulated by LTβR, because this particular immune response relies on SFB for induction26,28. The abundance of transcripts encoding transforming growth factor-β (TGF-β), IL-6, IL-17A, and IL-17F were similar between Ltbr+/− and Ltbr−/− groups after HFD and between groups on NCD (Fig. 4). However, transcripts encoding IL-23p19 and IL-22, a key downstream cytokine regulated by IL-23, were reduced in Ltbr−/− mice (Fig. 4c,f). Furthermore, IL-23p19 was induced by HFD as compared to the NCD state. Additionally, members of the RegIII antimicrobial peptide family, which are downstream of the IL-23–IL-22 signaling pathway were also greatly reduced by HFD, but their expression after HFD was partially dependent on LTβR, as evidenced by the observation that Ltbr−/− animals had little to no expression of these antimicrobial peptides after HFD (Fig. 4g,h). The selective loss of transcripts in the IL-23–IL-22 pathway and not the IL-17A/F pathway in LTβR-deficient mice suggested preferential involvement of this signaling axis in regulating the microbiota and DIO.
Figure 4. LTβR agonizes the innate IL23/IL22 axis.
Ltbr+/− and Ltbr−/− animals were fed HFD for 10 weeks starting at 9 weeks of age. After challenge, PCR for targets was performed on cDNA from whole colon. Data is plotted relative to Ltbr+/− animals on NCD and normalized to HPRT. (a) Tgfb (b) Il6 (c) Il23a (d) Il17a (e) Il17f (f) Il22 (g) Reg3g (h) Reg3b: (n=3–9 mice per group, representative of 2 independent experiments, * P <0.05, ** P <0.01, student’s t test)
IL-23 is regulated by LTβR and necessary for DIO
To confirm the importance of the LT-signaling pathway in IL-23 production, we cultured colons of wild-type, Ltbr+/−, and Ltbr−/− animals after HFD and measured IL-23p19p40 in the supernatants by ELISA. We observed that there was no difference in IL-23 expression between Ltbr+/− and Ltbr−/− groups on NCD; however, we observed that IL-23 was induced in Ltbr+/− animals after HFD but this induction did not occur in Ltbr−/− animals fed HFD (Fig. 5a). This is an intriguing observation because LTβR has previously been shown to impact IL-23 production in models of Citrobacter rodentium infection but not in the naïve state21,22. This finding suggests that similar to mucosal pathogenic challenge, HFD stimulus was sufficient to evoke an immune response dependent on LTβR, which resulted in IL-23 expression. To address the significance of IL-23 in weight gain, we challenged Il23a−/− animals with HFD; Il23a−/− animals resisted HFD induced weight gain and excess adiposity (Fig. 5b–d). Because HFD-induced IL-23 expression was dependent on LTβR, the phenotype of p19−/− animals is consistent with the necessity of the LT pathway in inducing IL-23 for DIO.
Figure 5. HFD induces LTβR-dependent IL23 which is essential for DIO.
(a) WT (C57BL/6), Ltbr+/−, and Ltbr−/− animals were fed HFD for 10 weeks. At the end of diet, animals were sacrificed and colons were removed and cultured overnight. Supernatants were subjected to ELISA for IL23p19p40 and resulting data was normalized per milligram of colon cultured. (b–d) WT (C57BL/6) mice or Il23a−/− animals were challenged with HFD starting at 9 weeks of age or 9 weeks. (b) Weight as a percentage over starting weight is plotted. (c) Perigonadal fat was removed and weighed at the end of diet; weight of fat is plotted. (d) Fat from (c) is plotted as a percentage of body weight. Weight gain as a percentage of starting weight is plotted. (n=5–9 mice per group; growth curve is reflective of 2 independent experiments and statistics demonstrate differences between HFD groups; Student’s t-test for individual points along growth curves; 1-Way Anova with Bonferonni post-test for dot plots: * P <0.05, **P <0.01)
RORγt+ cells are essential for weight gain after HFD
The LT pathway is essential to enable RORγt+ innate lymphoid cells to produce IL-22 after acute bacterial infection21,22. To study whether IL-22 regulated by the LT pathway after HFD is essential for DIO, Rorc−/− mice were selected because it has previously been shown that Ltbr−/− mice fail to evoke IL-22 production from RORγt+ lymphocytes in response to acute bacterial infection32. Rorc−/− mice were challenged with HFD. Rorc+/− mice gained significantly greater weight after HFD than their Rorc−/− littermates (Fig. 6a). One could argue that Rorc−/− mice may resist weight gain due to a lack in IL-23p19p40 due to the lack of lymphoid structure in these animals, but it is important to note that IL-23p19p40 abundance was similar from colons of Rorc+/− and Rorc−/− mice (data not shown). The LT–IL-23 axis is known to be essential in regulating IL-22 production from innate RORγt+ cells, and the results of Rorc−/− mice are consistent with the involvement of this LT-mediated axis in DIO.
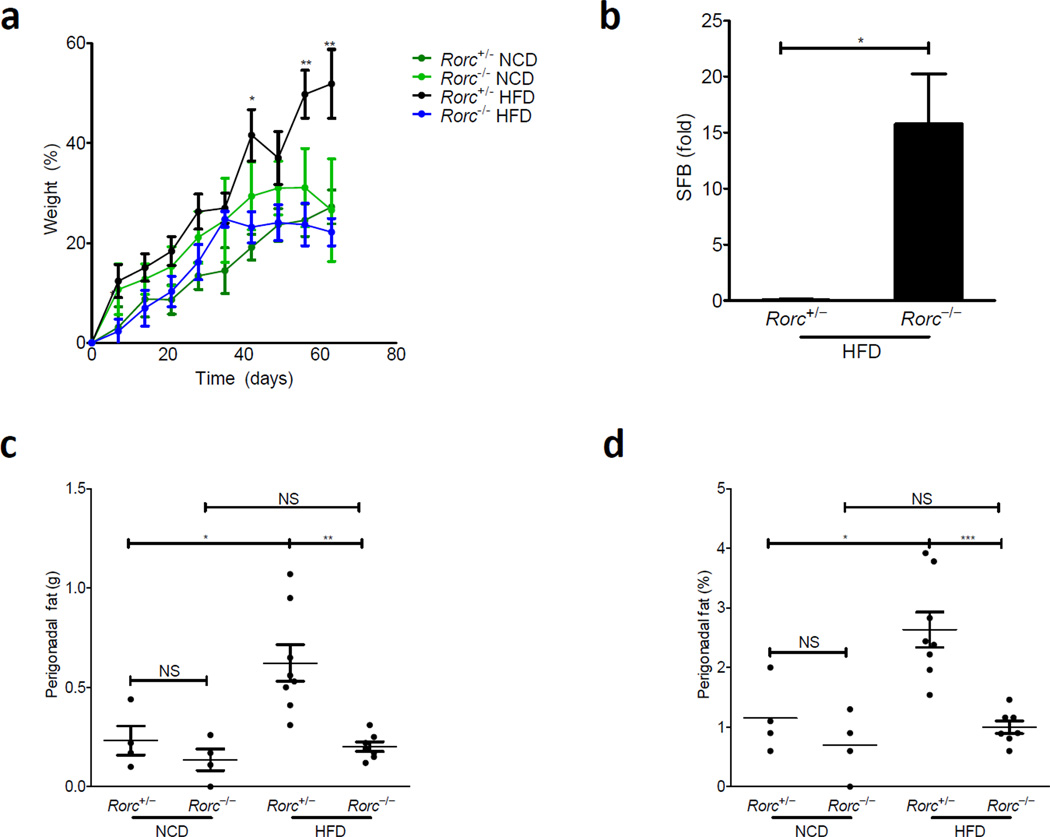
Figure 6. The transcription factor, RORγt, is required for weight gain and SFB homeostasis in DIO.
Rorc+/− or Rorc−/− littermates were challenged with HFD for 9 weeks starting at 5 weeks of age. (a) Weight as a percentage over starting weight is plotted. (b) RT-PCR for SFB in stool relative to Rorc+/− littermates after 4 weeks of HFD normalized to levels at diet start (n=4 mice in each group). (c) Perigonadal fat was removed and weighed at the end of diet; weight of fat is plotted. (d) Fat from (c) is plotted as a percentage of body weight. Weight gain as a percentage of starting weight is plotted (n=7–8 mice per group; growth curve is reflective of 3 independent experiments and statistics demonstrate differences between HFD groups; Student’s t-test for individual points along growth curves and SFB levels; 1-Way Anova with Bonferonni post-test for dot plots: * P <0.05, ** P <0.01, *** P <0.001)
Consistent with our results in Ltbr−/− animals, Rorc−/− animals also sustained higher representation of SFB after HFD (Fig. 6b). This observation suggests that the upstream defects in immunity are leading to a consistent downstream overgrowth in the microbiota. Similar to Ltbr−/− animals, in the absence of RORγt+ cells, the perigonadal fat pad did not expand in response to HFD (Fig. 6c,d).
IL-22 rescues the impact of HFD on SFB and DIO
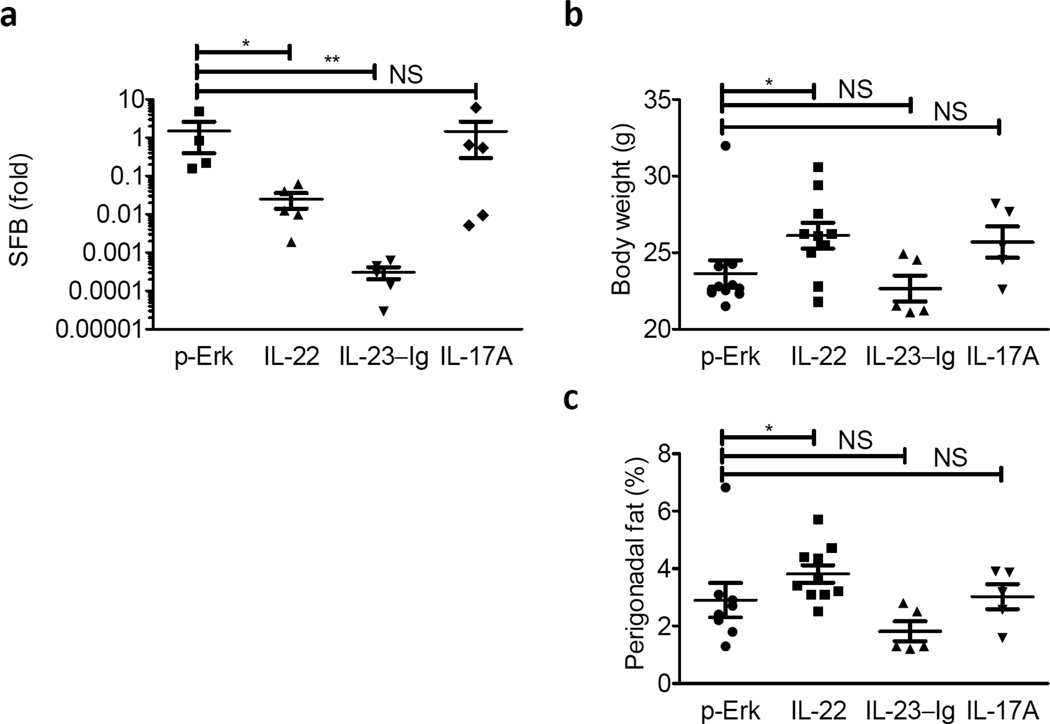
Given that HFD appeared to induce an LTβR-dependent agonism of the IL-23–IL-22 cytokine axis, we wondered whether restoring elements of this axis would rescue commensal homeostasis or weight gain in Ltbr−/− hosts. To address this question, we delivered IL-22, IL-23-Ig and IL-17A via hydrodynamic injection to Ltbr−/− adults at the start of diet and challenged them with diet for 9 weeks. Hydrodynamic injection resulted expression of IL-22, IL-23-Ig, or IL-17A detectable in the serum; notably, IL-22 was detectable in IL-23-Ig treated animals although IL-17A was not (data not shown). Both IL-22 and IL-23-Ig, but not IL-17A, were able to reduce the colonization of SFB after HFD initiation in Ltbr−/− hosts (Fig. 7a). Although IL-22 and IL-23-Ig treated groups had reduced abundance of SFB in the stool, total body size and perigonadal fat pads depot expansion occurred most extensively in Ltbr−/− animals after delivery of IL-22 and not IL-23-Ig or IL-17A (Fig. 7b, c). These data suggest a role for IL-22 downstream of LTβR in DIO.
Figure 7. IL22 Restores SFB homeostasis and perigonadal fat pad expansion in LTβR−/− mice.
LTβR−/− females were treated with 10 µg of plasmid encoding empty vector (p-Erk), IL-22, IL-23-Ig, or IL-17A at the start of diet. (a) RT-PCR for SFB normalized to Day 0 of diet are plotted. (b–c) (B) total body weight and (c) perigonadal fat pads were weighed and are plotted as a percentage of final weight. (n=5–10 mice per group; Student’s t-test for log-transformed data for SFB; Two-tailed Mann-Whitney test for body weight and perigonadal fat due to outlier in p-ERK group; * P <0.05, ** P <0.01)
DISCUSSION
While it has been argued that diet influences the microbiota independently of host genotype10, the possibility that innate immune responses serve as a critical pivot for species specific responses to HFD and microbiota provides a potential link between host responses to diet, the intestinal microbiota, and obesity. We have observed that HFD initiates two changes in the organism – namely a change in the composition of the commensal microbiota and an LTβR-dependent immune response within the host. We argue that these two changes in host and microbiota are not unrelated, and in fact, changes in host immunity induced by diet actually cultivate changes in the microbial community of the distal gut. Our study has now demonstrated that the LT–IL-23–IL-22-pathway, essential for innate immune defense against gut pathogens, is also essential for regulation of specific commensal responses to HFD.
Broadly speaking, it is thought that changes in the composition of the commensal microbiota occur as a result of altered nutritional quality of diet; in this model, the host is a static entity, not influencing the overgrowth or clearance of organisms in response to a change in diet. One study is particularly illustrative of this hypothesis and its limitations. It was determined that unrelated mammals, which consume similar diets (herbivores versus omnivores versus carnivores) maintain similar microbial communities33. A notable exception within the study was pandas and bears; pandas and bears are a unique group because although they are closely related, dietary composition within the group varies greatly. Intriguingly, the microbial communities of bears regardless of dietary habits is similar; this exceptional group provides some evidence to support a role for the host in regulating its microbiota34. Further support for the role of the host in shaping its microbial community comes from studies utilizing reciprocal transplantation between zebrafish and mice. Mice that were colonized with zebrafish microbiota fostered growth of the foreign bacteria, but microbes from zebrafish could only grow to abundance levels of closely related microbes that naturally grew in mice35. This data suggests that selective pressures in the host play some deterministic role in aspects of the microbiota, such as abundance of specific species.
HFD induced host response would be part of the selective pressure of the gut as an ecological niche. In our model, production of antimicrobial peptides through the IL-22 (antibacterial peptides) pathway would directly antagonize the growth of some microbes, such as SFB. Species overgrowth, such as that exhibited by members of the Erysopelotrichi class, could occur in place of organisms that are eliminated by the host. In the LTβR−/− model system, where the host lacks some elements of mucosal immunity, a change in nutrient composition was not enough to induce reductions for some bacteria, specifically SFB. Intriguingly, when the IL22 was restored in LTβR-deficient hosts, SFB was once again cleared and body size normalized. Therefore, some bacteria can become important biomarkers tracking DIO in given species.
This model presupposes the existence of an inflammatory state in the host gut induced by HFD; while many groups have focused on inflammation in host adipose tissue, the possibility that inflammation is not restricted to fat depots alone has had quiet support in recent years. This was initially hinted at by the finding that HFD induces NF-κB expression in the colon early after the start of HFD36. Given the important symbiosis shared between the intestinal microbiota and mucosal inflammatory responses, it is logical to consider how changes in immunity influence the microbiota and in turn, how those changes to the microbiota feedback to influence not only local immunity but systemic host health.
A major “defect” in the microbiome of LTβR-dependent animals after HFD is that the microbial communities of Ltbr−/− animals after HFD are not less diverse. It is entirely possible that the enhanced immune response of the obese host exerts greater pressure on the microbial community of the distal gut, preventing the survival of species that could otherwise normally populate a unique niche. SFB serves as a useful biomarker, but the role of SFB and other species whose reduction from the gut relies on mucosal immunity for DIO remain to be determined and will benefit from gnotobiotic models and reconstitution of selective gut flora.
Even though some reports argue that genes play a large role in obesity 4,5, the consistent dysbiosis present in obese individuals suggests a strong role for environmental contribution to this disease 7. Polymorphisms in the Lta gene locus have been linked to obesity, but the role of the LTβR signaling in DIO appears to rely on changes within the commensal microbiota. Moreover, the importance of this immune response on weight gain can be subverted by changes in housing. We feel that the viewpoints regarding the importance of genetics and environment are not at odds when it comes to obesity. We propose the possibility that the host response induced by HFD may actually help provide inertia for the obese state by facilitating occupation of an obese microbiome; the intestinal microbiota can thus serve as agents to transmit beneficial energy harvest to immunocompromised hosts. In the mammalian population this may be a mechanism by which mothers that are colonized with microbiota that is more efficient at energy harvest may colonize their offspring at the time of birth and after when their immune systems are not completely developed; from this perspective, the microbiota would facilitate more efficient utilization of scarce food resources.
Population-wide implications for this argument are interesting because this model suggests a potential to manipulate weight gain – either to promote or inhibit it – by regulating the microbiota through antibiotic/probiotic regimens, regulating the host through vaccination, or complementary strategies them employ both vaccination and direct modification of the microbiota. Even so, the precise microbes that promote such weight gain and the specific host responses that foster their growth need to be better established in order to create useful strategies in manipulating host-microbe interactions to influence weight gain.
Online Methods
Mice
WT C57BL/6 mice were obtained from Jackson Laboratories, Harlan Laboratories, or the National Cancer Institute (NCI). Lta−/−, Ltb−/−, Ltbr−/−,Il23a−/−, and Rorc−/− mice were bred in our vivarium at the University of Chicago. In cases of all heterozygous animals, breedings were set up where one parent was a null and the other was a heterozygous animal (usually the father). Mice were genotyped by PCR and weaned as early as 21 days and as late as 28 days after birth. Cohousing experiments were performed with 3 heterozygous and 2 homozygous null animals in a cage. Germ-free C57BL/6 mice were maintained in the gnotobiotic facility at the University of Chicago. Mice were maintained according to the standards set by the University of Chicago’s IACUC (Protocol #71866 and #58771).
HFD and NCD Challenge Experiments
All SPF mice were maintained on Harlan Teklad 2918 until the start of diet where they were either switched onto 88137 or maintained on 2918 for the duration of the experiment. Mice were weighed every 7–10 days after the start of diet. At the end of diet (63–70 days after initiation), mice were sacrificed by CO2 euthanasia and cervical dislocation. Mice were weighed again after sacrifice and perigonadal fat (periuterine or epididymal fat in the case of female and male mice, respectively) was dissected and weighed.
Food Consumption
Mice were started on either NCD or HFD at day zero and food was measured daily. Successive weights were subtracted from the previous day measured (day n – day n+1) and data is plotted adjusted for days between measurements (1–3). 5 mice were housed per cage.
Cecal and Stool DNA Extraction
Cecal samples were collected at the time of sacrifice (at the end of NCD or HFD respectively) and frozen at −80 °C until time of processing. Stool samples were collected freshly in our vivarium at 0, 4, and 9 weeks after the start of diet and frozen at −20 °C. All extraction was done utilizing the QIAamp ® DNA Stool Mini Kit (Qiagen) from Qiagen. Briefly, samples were lysed in a detergent solution and mechanically dissociated using a Mini-beadbeater from Biospec products (BioSpec Products) for 90 s at maximum setting. Samples were treated with InhibitEX matrix to prevent DNA damage and to inhibit PCR disrupting agents. Subsequently, proteins were digested with Proteinase K, samples were bound to a column, washed twice, and eluted in the supplied buffer. Quality of DNA and concentrations were determined utilizing Nanodrop.
PCR Amplification and 454 Pyrosequencing of 16S rDNA
Sequences were done from two different litters of animals in two different facilities to ensure thorough understanding of microbial communities (one set of samples from one litter with V1-V2 regions prepared at the Univeristy of Chicago (primers listed at end) listed as T1; another set of samples from another litter amplified for V3-V4 regions Trial 2 listed as T2 in Supplementary Table 2). For T1, sequencing and analysis were done as described previously37. V1-V2 regions of 16S rDNA from stool or cecal samples were amplified with TaKaRa Ex Taq PCR mixture (TAKARA Bio USA). The PCR program was set at 95 °C 10 min, 30 cycles of 95 °C 1 min, 50 °C 1 min, 72 °C 1.5 min, followed by 72 °C for 10 min. PCR products were purified using the AMPure Kit (Agentcourt Bioscience). The resulting product was analyzed on a 2% agarose gel and by nanodrop. Products were then pooled at equal concentrations and sequenced on a GS Titanium 70 × 75 picotitre plate according to the manufactuerer’s protocols for GS FLX (Roche Applied Science) at the Roy J Carver Center at the University of Illinois at Urbana Champaign. For “T2” PCR primers used were specific for the V3-V5 region of 16s rRNA. DNA was prepared and submitted to Research and Testing Laboratories for amplification, barcoding and sequencing.
Analysis of Pyrosequencing
16S rRNA sequence analysis was performed via MOTHUR suite of programs, version 1.17.0 (ref. 38). Low-quality sequences were trimmed, and redundant sequences were removed to create a simplified dataset. Sequences were aligned to the SILVA reference database, chimeric sequences were removed and operational taxonomical unit (OTU) clustering was performed via average neighbor methodology. Simpson diversity index (a measure of biodiversity within a habitat) was calculated using table of OTU abundances. Principal coordinate analysis was performed using matrix of Yue and Clayton similarity measures. To minimize complications from unclassified OTUs, phylotype-based analysis was also preformed. High quality sequences were taxonomically annotated via the RDP classifier tool38. Differentially abundant features were determined via Metastats39
Sequence Alignment
OTU alignment to the known 16s SFB rRNA encoding V1-V2 region was performed utilizing ClustalW2 available from the European Bioinformatics Institute.
Germ Free Experiments
Germ free NCD and HFD were described in Supplementary Table 1. WT C57BL/6 germ-free mice were gavaged with fresh cecal contents from Ltbr+/− and Ltbr−/− donors maintained on similar diets.
Colon Culture and ELISA
Proximal colon pieces weighing less than 50 mg were cut in small pieces and incubated in 0.4 ml of RPMI 1640× containing 10% FBS, amphotericin, gentamicin, penicillin, and streptomycin for 48 h in tissue plates, as previously described by Zheng et al.40 IL-23p19p40 in supernatants was measured by ELISA (eBiosciences) according to the manufacturer’s recommendations.
Hydrodynamic Injection
Hydrodynamic injection was performed as described by Tumanov et al. by placing mice in a conical restraining device with an attached heating element. 10 µg of a plasmid vector expressing IL-23-Ig (B. Becher, Switzerland), IL-17A (C. Dong, USA), IL-22 (IL-22, graciously provided by Genentech) or empty vector (p-ERK, graciously provided by Genentech) were injected one or two days prior to the start of HFD in 1.8 ml TransIT-EE Hydrodynamic Delivery Solution (MIR 5340, Mirus Bio LLC) over a period that lasted less than five seconds22.
Real-Time PCR
RNA was extracted from colon samples frozen at −80 °C in RNALater (Life Technologies). Briefly, samples were homogenized in TRIzol Reagent ® (Invitrogen) and underwent phenol-chloroform extraction. The product was treated with Amplification Grade DNAse I available from Sigma Aldrich (Sigma Aldrich Corporation). Product integrity was verified by running samples on 2% agarose gels. 2 µg of RNA was utilized to make cDNA using M-MLTV Reverse Transcriptase and associated buffers, dNTPs, and oligo-dT primer from Promega (Promega). Samples were amplified on an ABI 7900 instrument (Applied Biosystems Inc.) using SsoFast™ EvaGreen® Supermix (Bio-Rad Laboratories); primer concentrations were 0.5 µM in the final reaction. Correct melting temperatures for all products were verified after amplification. For all products, amplification in all samples resulted in correct melting temperatures. For IL-22 and RegIIIβ targets, amplification often resulted in multiple products and reactions with the resulted in multiple products are excluded from both groups. For IL-22, no Ltbr−/− animal produced a product with the correct melting temperature, likely due to the low transcript abundance for this product in these mice. Amplification data for all PCR reactions was submitted to Real-Time PCR Miner for accurate Ct value calculation and Primer Efficiency assessment 41. Fold relative to WT normalized to HPRT was calculated utilizing the Pfaffl method.
Statistical Methods
Statistical analysis for Supplementary Table 2 was described in “Analysis of Pyrosequencing” and is the outputted p-value from Metastats. For all other statistical tests, raw data was input into GraphPad Prism v5 and analyzed through statistical tests available in the software.
Primers:
| Primer | Sequence | Reference |
|---|---|---|
| HPRT | HPRTF: TGAAGAGCTACTGTAATGATCAGTCAAC HPRTR: AGCAAGCTTGCAACCTTAACCA |
22 |
| IL23p19 | IL23p19f: GGT GGC TCA GGG AAA TGT IL23p19R:GAC AGA GCA GGC AGG TAC AG |
40 |
| TGFβ | TGFβF: CACTGATACGCCTGAGTG TGFβR: GTGAGCGCTGAATCGAAA |
42 |
| IL-6 | IL6F:TCC AAT GCT CTC CTA ACA GAT AAG IL6R:CAA GAT GAA TTG GAT GGT CTT G |
40 |
| IL-17A | IL-17A Ex2F ctccagaaggccctcagactac IL-17A Ex3R agctttccctccgcattgacacag |
43 |
| IL-17F | IL-17F Ex1F gaggataacactgtgagagttgac IL-17F Ex2R2 gagttcatggtgctgtcttcc |
43 |
| IL-22 | IL22F: TCC GAG GAG TCA GTG CTA AA IL22R: AGA ACG TCT TCC AGG GTG AA |
40 |
| RegIIIγ | RegIIIgF: ATG GCT CCT ATT GCT ATG CC RegIIIgR: GAT GTC CTG AGG GCC TCT T-3’ |
40 |
| RegIIIβ | RegIIIbF: ATG GCT CCT ACT GCT ATG CC RegIIIbR: GTG TCC TCC AGG CCT CTT T |
40 |
| EUA | EUAF: ACTCCTACGGGAGGCAGCAGT EUAR: ATTACCGCGGCTGCTGGC |
44 |
| SFB | SFBF: GACGCTGAGGCATGAGAGCAT SFBR: GACGGCACGGATTGTTATTCA |
44 |
Sequencing Primers/Adaptors for T1 only:
Primers
TA-27FMID1 CGTATCGCCTCCCTCGCGCCATCAGACGAGTGCGTAGAGTTTGATCCTGGCTCAG
TA-27FMID2 CGTATCGCCTCCCTCGCGCCATCAGACGCTCGACAAGAGTTTGATCCTGGCTCAG
TA-27FMID3 CGTATCGCCTCCCTCGCGCCATCAGAGACGCACTCAGAGTTTGATCCTGGCTCAG
TA-27FMID4 CGTATCGCCTCCCTCGCGCCATCAGAGCACTGTAGAGAGTTTGATCCTGGCTCAG
TA-27FMID5 CGTATCGCCTCCCTCGCGCCATCAGATCAGACACGAGAGTTTGATCCTGGCTCAG
TA-27FMID6 CGTATCGCCTCCCTCGCGCCATCAGATATCGCGAGAGAGTTTGATCCTGGCTCAG
TA-27FMID7 CGTATCGCCTCCCTCGCGCCATCAGCGTGTCTCTAAGAGTTTGATCCTGGCTCAG
TA-27FMID8 CGTATCGCCTCCCTCGCGCCATCAGCTCGCGTGTCAGAGTTTGATCCTGGCTCAG
TA-27FMID9 CGTATCGCCTCCCTCGCGCCATCAGTAGTATCAGCAGAGTTTGATCCTGGCTCAG
TA-27FMID10 CGTATCGCCTCCCTCGCGCCATCAGTCTCTATGCGAGAGTTTGATCCTGGCTCAG
TA-27FMID11 CGTATCGCCTCCCTCGCGCCATCAGTGATACGTCTAGAGTTTGATCCTGGCTCAG
TA-27FMID13 CGTATCGCCTCCCTCGCGCCATCAGCATAGTAGTGAGAGTTTGATCCTGGCTCAG
TA-27FMID14 CGTATCGCCTCCCTCGCGCCATCAGCGAGAGATACAGAGTTTGATCCTGGCTCAG
TB-338R CTATGCGCCTTGCCAGCCCGCTCAGTGCTGCCTCCCGTAGGAGT
Supplementary Material
Acknowledgements
The authors would like to thank B. Becher from University of Zurich for IL-23-Ig, C. Dong from MD Anderson (Huston, TX) for IL-17, O. Wenjun from Genentech (South San Francisco, CA) for IL-22. V.U. was supported by the American Heart Association (AHA Predoctoral 11PRE7320015) and the NIH Medical Scientist Training Program grant GM007281 to the University of Chicago Pritzker School of Medicine. This research was supported by pilot grants from the DDRCC (p30 DK42086), the University of Chicago Institutional Translational Medicine (UL1 RR024999), and US National Institutes of Health grants, AI090392 and CA134563 to Y.X.F.
Footnotes
Author Contributions
V.U. partook in the performance of all experiments. T.K., A.T.V., E.P.K. contributed in Figure 1, 4, and 5. S.D. and S.F. contributed to Figure 2. V.U. and V.P. performed bioinformatic analyses. L.D. contributed in Figure 7. D.L., C.N., H.T., and E.C. contributed to experimental design, interpretation of results, and critical assessment of the manuscript. V.U. and Y.X.F. designed all experiments and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Wang Y, Beydoun MA, Liang L, Caballero B, Kumanyika SK. Will All Americans Become Overweight or Obese? Estimating the Progression and Cost of the US Obesity Epidemic. Obesity. 2008;16:2323–2330. doi: 10.1038/oby.2008.351. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2010 < http://www.cdc.gov/obesity/data/trends.html#State>.
- 3.Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and Trends in Obesity Among US Adults, 1999–2008. JAMA. 2010;303:235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- 4.Stunkard AJ, Foch TT, Hrubec Z. A Twin Study of Human Obesity. JAMA. 1986;256:51–54. [PubMed] [Google Scholar]
- 5.Stunkard AJ, et al. An Adoption Study of Human Obesity. NEJM. 1986;314:193–198. doi: 10.1056/NEJM198601233140401. [DOI] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-Induced Obesity Is Linked to Marked but Reversible Alterations in the Mouse Distal Gut Microbiome. Cell Host & Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muegge BD, et al. Diet Drives Convergence in Gut Microbiome Functions Across Mammalian Phylogeny and Within Humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a Human Gut Microbiota’s Response to Diet in Gnotobiotic Mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberl G. A new vision of immunity: homeostasis of the superorganism. Mucosal Immunol. 2010 doi: 10.1038/mi.2010.20. [DOI] [PubMed] [Google Scholar]
- 13.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. doi: http://www.nature.com/nature/journal/v482/n7384/abs/nature10809.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vijay-Kumar M, et al. Metabolic Syndrome and Altered Gut Microbiota in Mice Lacking Toll-Like Receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norman RBC, Ravussin E. Linkage Between Obesity and a Marker Near the Tumor Necrosis Factor-Alpha Locus in Pima Indians. J. Clin. Invest. 1995;96:158–162. doi: 10.1172/JCI118016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahajan A, et al. Obesity-dependent association of the TNF/LTA locus with type 2 diabetes in North Indians. J. Mol. Med. 2010;88:515–522. doi: 10.1007/s00109-010-0594-5. [DOI] [PubMed] [Google Scholar]
- 17.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 18.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J. Clin. Invest. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pamir N, McMillen TS, Edgel KA, Kim F, LeBoeuf RC. Deficiency of Lymphotoxin-alpha does not exacerbate high fat diet induced obesity but does enhance inflammation in mice. Amer.J. Phys. Endo. Metab. 2012 doi: 10.1152/ajpendo.00447.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu Y-X, Chaplin DD. Development Maturation of Secondary Lymphoid Tissues. Annu. Rev. Immunol. 2003;17:399–433. doi: 10.1146/annurev.immunol.17.1.399. [DOI] [PubMed] [Google Scholar]
- 21.Ota N, et al. IL-22 bridges the lymphotoxin pathway with the maintenance of colonic lymphoid structures during infection with Citrobacter rodentium. Nat Immunol. 2011;12:941–948. doi: 10.1038/ni.2089. [DOI] [PubMed] [Google Scholar]
- 22.Tumanov Alexei V, et al. Lymphotoxin Controls the IL-22 Protection Pathway in Gut Innate Lymphoid Cells during Mucosal Pathogen Challenge. Cell Host & Microbe. 2011;10:44–53. doi: 10.1016/j.chom.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uysal KT, Wiesbrock SM, Marino MW, Hotamisligil GS. Protection from obesity-induced insulin resistance in mice lacking TNF-[alpha] function. Nature. 1997;389:610–614. doi: 10.1038/39335. [DOI] [PubMed] [Google Scholar]
- 24.Turnbaugh PJ, et al. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Sci. Trans. Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savage DC. Microbial Ecology of the Gastrointestinal Tract. Annu. Rev.f Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 26.Wu H-J, et al. Gut-Residing Segmented Filamentous Bacteria Drive Autoimmune Arthritis via T Helper 17 Cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klaasen HL, et al. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect. Immun. 1993;61:303–306. doi: 10.1128/iai.61.1.303-306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanov II, et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sczesnak A, et al. The Genome of Th17 Cell-Inducing Segmented Filamentous Bacteria Reveals Extensive Auxotrophy and Adaptations to the Intestinal Environment. Cell Host & Microbe. 2011;10:260–272. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prakash T, et al. Complete Genome Sequences of Rat and Mouse Segmented Filamentous Bacteria, a Potent Inducer of Th17 Cell Differentiation. Cell Host & Microbe. 2011;10:273–284. doi: 10.1016/j.chom.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. doi: http://www.nature.com/nature/journal/v478/n7368/abs/nature10434.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cella M, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ley RE, et al. Evolution of Mammals and Their Gut Microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaishnava S, et al. The Antibacterial Lectin RegIIIγ Promotes the Spatial Segregation of Microbiota and Host in the Intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal Gut Microbiota Transplants from Zebrafish and Mice to Germ-free Recipients Reveal Host Habitat Selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding S, et al. High-Fat Diet: Bacteria Interactions Promote Intestinal Inflammation Which Precedes and Correlates with Obesity and Insulin Resistance in Mouse. PLoS ONE. 2010;5:e12191. doi: 10.1371/journal.pone.0012191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poroyko V, et al. Gut Microbial Gene Expression in Mother-Fed and Formula-Fed Piglets. PLoS ONE. 2010;5:e12459. doi: 10.1371/journal.pone.0012459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schloss PD, et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 41.Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Firan M, Dhillon S, Estess P, Siegelman MH. Suppressor activity and potency among regulatory T cells is discriminated by functionally active CD44. Blood. 2006;107:619–627. doi: 10.1182/blood-2005-06-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivanov II, et al. The Orphan Nuclear Receptor ROR[gamma]t Directs the Differentiation Program of Proinflammatory IL-17+ T Helper Cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 44.Barman M, et al. Enteric Salmonellosis Disrupts the Microbial Ecology of the Murine Gastrointestinal Tract Infect. Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.