Abstract
Genetic interactions reveal the functional relationships between pairs of genes. In this study, we describe a method for the systematic generation and quantitation of triple mutants, termed Triple Mutant Analysis (TMA). We have used this approach to interrogate partially redundant pairs of genes in S. cerevisiae, including ASF1 and CAC1, two histone chaperones. After subjecting asf1Δ cac1Δ to TMA, we found that the Swi/Snf Rdh54 protein, compensates for the absence of Asf1 and Cac1. Rdh54 more strongly associates with the chromatin apparatus and the pericentromeric region in the double mutant. Moreover, Asf1 is responsible for the synthetic lethality observed in cac1Δ strains lacking the HIRA-like proteins. A similar TMA was carried out after deleting both CLB5 and CLB6, cyclins that regulate DNA replication, revealing a strong functional connection to chromosome segregation. This approach can reveal functional redundancies that cannot be uncovered using traditional double mutant analyses.
Introduction
The systematic study of genetic interactions by high-throughput methods in numerous organisms has provided insight into a variety of complicated biological processes (Beltrao et al., 2010). For example, in S. cerevisiae double mutants obtained by crossing a given gene knockout with a large number of other viable single gene ablations or modifications can reveal either synthetic reductions in growth or they can exhibit epistasis (Collins et al., 2010; Schuldiner et al., 2005; Tong et al., 2004). Synthetic lethality or synthetic sickness occurs when two genes have overlapping, important functions so that the absence of both genes results in a severe growth defect that is only seen when both genes are absent. In the same fashion, double mutant combinations can reveal sensitivity to environmental conditions or to antibiotics. In contrast, the combination of two knockouts that remove elements of the same pathway shows no additional phenotype (epistasis). Epistatic interactions often correspond to gene products that participate in protein-protein interactions and/or function in the same biological pathway (Beltrao et al., 2010; Collins et al., 2007).
More recently, this type of systematic genetic analysis has been extended to other organisms, including S. pombe (Roguev et al., 2008; Roguev et al., 2007; Ryan et al., 2012), C. elegans (Lehner et al., 2006), D. melanogaster (Horn et al., 2011) and mammalian cells (Bassik et al., 2013; Lin et al., 2012; Roguev et al., 2013) and has also been carried out in the presence of specific exogenous stresses (Ideker and Krogan, 2012) and using specific point mutants of multifunctional genes (unpublished data). To date, almost all genetic interaction data collected in any organism has been generated using systems that create mutants in a pair-wise fashion, even though great mechanistic insight could be uncovered through higher perturbation studies. In this study, we describe an approach, termed Triple Mutant Analysis (TMA), which facilitates higher-level genetic interaction analysis by allowing for the generation and quantitative analysis of triple mutants in budding yeast.
In budding yeast, Asf1 and the CAF-1 complex, comprising Cac1, Cac2 and Cac3, are the two known histone H3-H4 chaperones (De Koning et al., 2007). Deletion of ASF1 results in slower growth, with an accumulation of cells in G2, as well as a marked increase in gross chromosomal rearrangements (GCR) (Kats et al., 2006; Ramey et al., 2004). Asf1, interacting with histones H3-H4, also recruits Rtt109, an enzyme responsible for histone H3-K56 acetylation (Collins et al., 2007; Driscoll et al., 2007; Han et al., 2007b). Furthermore, Asf1 plays a role in the monitoring of replication and chromatin assembly, through interaction with the checkpoint kinase Rad53. Similarly, deletion of any of the three subunits of CAF-1 (Cac1, Cac2 or Cac3) results in elevated GCR (Myung et al., 2003) and mutation of CAC1, also known as RLF2, affects the structure of telomeric chromatin and the transcriptional silencing of HML and HMR (Enomoto and Berman, 1998). As expected, the double mutant asf1Δ cac1Δ is more seriously compromised with cells having a doubling time about twice that of the wild type cells, with a long delay in the G2/M phase of the cell cycle (Kats et al., 2006). Moreover, Asf1 and Cac1 have also been shown to have overlapping roles in deactivating the DNA damage checkpoint after cells have repaired a double-strand break (Kim and Haber, 2009). Despite this, asf1Δ cac1Δ strains are surprisingly robust (Kats et al., 2006; Ramey et al., 2004). Therefore, we hypothesized that there could be additional chromatin remodelers acting as functional “back-ups” to Asf1 and CAF-1, and hence we subjected the double mutant to TMA.
Similar to Asf1 and CAF-1, two of yeast six cyclin B homologs, Clb5 and Clb6, act redundantly in regulating the timing of DNA replication in mitosis and meiosis (Decesare and Stuart, 2012; Donaldson et al., 1998); however a clb5Δ clb6Δ double mutant is not particularly sick, suggesting an additional pathway exists for this process. In both of these instances, we show novel genetic outcomes derived from the TMA approach that provide mechanistic insight into the pathways being interrogated that could not have been gleaned from the analysis of single mutants, illustrating the utility of this approach. We propose that the experimental and computational framework described here could be used to interrogate higher order relationships in various biological processes in many organisms, including in mammalian cells.
Results and Discussion
In some instances, eliminating redundant genes results in cell death (e.g. deleting the three cyclin A homologs (CLN1, CLN2 and CLN3) or deleting 2 cyclin B homologs (CLB1 and CLB2) are lethal (Schneider et al., 1996; Surana et al., 1991)). Similarly, deleting components of parallel pathways can result in cell death (e.g. ablating pairs of genes involved in chromosome transmission, such as CTF18 and CTF4, are synthetically lethal (Xu et al., 2007)). However, in other situations, removing two parallel processes leaves the double mutant slow-growing, but still viable. One such case involves the absence of two known histone H3-H4 chaperone complexes – Asf1 and the CAF-1 complex. If Asf1 and CAF-1 are the only histone H3-H4 chaperones, how do double mutant cells remain viable? In mammalian cells, the HIRA complex also acts as an independent histone chaperone (Ray-Gallet et al., 2002); in budding yeast, the components of this complex (Hir1, Hir2, Hir3 and Hpc2) appear to be involved in regulating histone transcription, but are also involved in histone deposition working in association with Rtt106 (Green et al., 2005; Krawitz et al., 2002; Sharp et al., 2002; Silva et al., 2012). To address this question, we have used the SGA technique (Tong et al., 2004) and the quantitative scoring system associated with the E-MAP approach (Schuldiner et al., 2005) to develop a quantitative strategy to analyze triple mutants, which we term Triple Mutant Analysis (TMA). Normally, double mutant analysis in S. cerveisiae involves the crossing of two differently marked mutant strains to generate a double mutant strain. TMA involves the use of a double mutant strain that is crossed to a panel of mutant strains, allowing for the systematic creation of triple mutants. In this study, a haploid MATα asf1Δ::HPH cac1Δ::URA3 strain was crossed against MATa haploids with knockouts of non-essential genes or hypomorphic alleles of essential genes, marked by KAN.
We anticipated three scenarios that might emerge from the selection of Ura+ HPHR KANR triple mutants. First, the triple mutant might exhibit an even more severe growth defect, revealing a third gene whose product might act as an additional histone H3-H4 chaperone. Second, mutations that caused more severe growth defects might be uncovered if, for example, the absence of both chaperones exacerbates the phenotype of another mutant not involved in chromatin remodeling (e.g. by altering the expression of other genes). A third scenario, which would represent a positive interaction, could correspond to a suppression of the intrinsically poor growth of asf1Δ cac1Δ via removal of a gene whose mis-regulated product was responsible for the poor growth associated with the double mutant.
TMA of asf1Δ cac1Δ
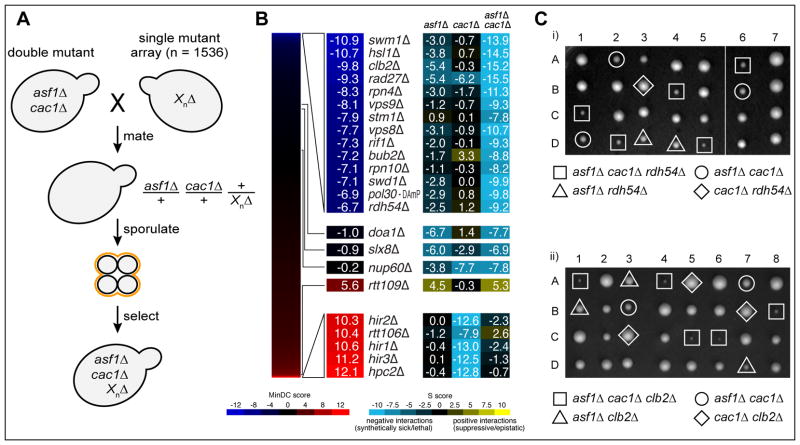
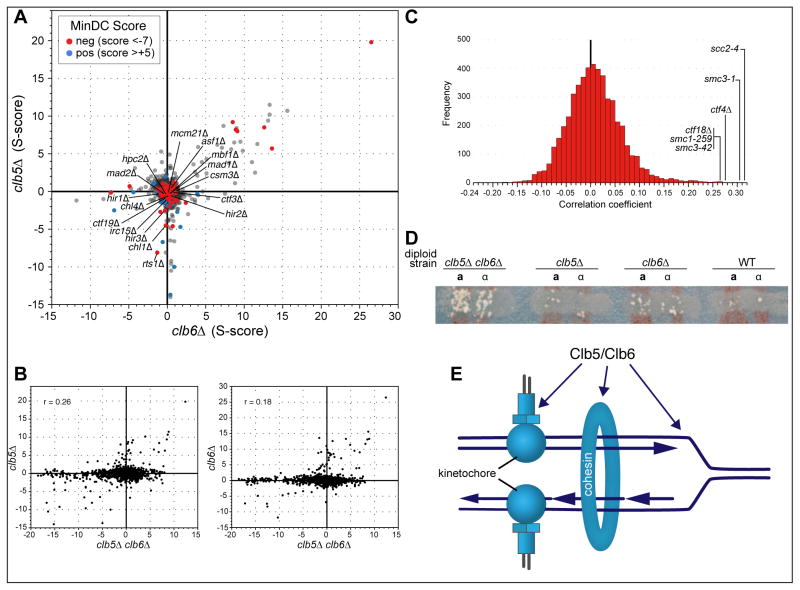
In the genetic cross, we can select for two different sets of double mutants (asf1Δ geneXΔ; cac1Δ geneXΔ) as well as for the triple mutants (asf1Δ cac1Δ geneXΔ) using the appropriate antibiotic resistant markers (Figure 1A; Supp. Data 1). Analysis of this data using previously described analytical tools can provide genetic scores (or S-scores) (Collins et al., 2010) for all three sets of mutants that range from negative (i.e. synthetic sickness) (blue) to positive (suppression) (yellow) (Figure 1B). However, inspection of the triple mutant scores alone can be deceiving if not compared to the scores derived from each corresponding double mutant. For example, there are interactions of a gene knockout with both asf1Δ and cac1Δ (i.e. the triple mutant) that were not significantly different from a deletion of the gene with either asf1Δ or cac1Δ. Indeed, the S-score from the asf1Δ slx8Δ double mutant (−6.7) is similar to what is observed with the asf1Δ cac1Δ slx8Δ triple mutant (−6.9) (Figure 1B, middle). Other examples include asf1Δ doa1Δ and cac1Δ nup60Δ (Figure 1B), suggesting that only one of the two starting mutants is contributing to the phenotype observed in the triple mutant. To separate these cases from situations where the triple mutant score is strong when each double mutant has little effect, we performed a minimum difference comparison (MinDC) between the score derived from the triple mutant compared to the more significant score of the two double mutant combinations. The resulting MinDC scores, which also range from negative (dark blue) to positive (red) (Figure 1B; Supp. Data 2), reveal that the most significant negative interactions found with both ASF1 and CAC1 absent are with swm1Δ (−10.9), hsl1Δ (−10.7), clb2Δ (−9.8), rad27Δ (−9.3), and rpn4Δ (−8.3). Hsl1 is a Nim1-like kinase that acts on the cell cycle regulator, Swe1 (Booher et al., 1993). Clb2, a B-type cyclin (Surana et al., 1991) and Swm1, a component of the Anaphase Promoting Complex (APC) (Hall et al., 2003) are implicated in cell cycle progression, as is Rpn4, a component of the 19S proteasome (Xie and Varshavsky, 2001). Rad27 is a 5′ flap endonuclease involved in DNA repair (Reagan et al., 1995). Among the genes that appeared to act redundantly with Asf1 and Cac1 is Radh54, also known as Tid1, a Swi/SNF homolog that has been implicated in chromatin remodeling (Kwon et al., 2008; Prasad et al., 2007). Deletion of RDH54 resulted in the 14th lowest MinDC score (−6.7) (Figure 1B).
Figure 1. Triple mutant analysis of asf1Δ cac1Δ.
A) TMA using a strain deleted for both ASF1 and CAC1 crossed to a library of 1536 different mutants. The mutants represent all major biological processes, with a particular emphasis on chromatin biology (Ryan et al., 2012). Following mating, the diploid cells are sporulated and the triple mutant haploid strains are selected. B) Double and triple mutant S-scores range from positive (yellow) to negative (blue). A minimum difference comparison was obtained by subtracting the triple mutant S-score from the S-score of the more severe of the two double mutant combinations. MinDC scores range from positive (red) to negative (dark blue). C) Meiotic tetrad dissection yields the triple mutants asf1Δ cac1Δ radh54Δ and asf1Δ cac1Δ clb2Δ, as well as all corresponding double mutants.
To confirm these results, we carried out tetrad analysis after sporulating diploids heterozygous for asf1Δ cac1Δ and either clb2Δ or rdh54Δ (Figure 1C), as well as hsl1Δ (data not shown). In each case, asf1Δ cac1Δ segregants are slow-growing, but the phenotype of the triple mutants are far more severe. Hsl1 was originally identified as a synthetic lethal mutation (histone synthetic lethality) in cells lacking the N-terminal tail of histone H3 (Ma et al., 1996) and it appears that asf1Δ cac1Δ phenocopies the H3 tail mutation. Hsl1 is required for degradation of the Swe1 kinase and therefore in a hsl1Δ mutant background, Swe1 is constitutively active and cells are prevented from completing the cell cycle (Sia et al., 1996). Consistent with this interpretation, we showed that deleting SWE1 in the asf1Δ cac1Δ hsl1Δ background suppressed its lethality (data not shown). Interestingly, the cyclin Clb2 is also genetically linked to the Swe1-Hsl1 network (Simpson-Lavy et al., 2009). We also confirmed the TMA negative interaction with Rdh54 (Figure 1C). Radh54 plays important roles in meiotic recombination, where it partners with the meiosis-specific recombination protein Dmc1, but has relatively minor roles in mitotic recombination (Dresser et al., 1997; Klein, 1997). In mitotic cells, its role in recombination is less evident, but rdh54Δ cells have defects in DNA damage checkpoint adaptation after induction of a single, unrepaired DSB (Lee et al., 2001). Rdh54 also appears to act redundantly with Rad54 and Uls1, two other Swi/Snf homologs that work to free Rad51 from dsDNA to enable it to repair DSBs (Chi et al., 2011). However, neither rad54Δ nor uls1Δ exhibit any significant genetic interaction with asf1Δ cac1Δ (Supp. Data 1, 2).
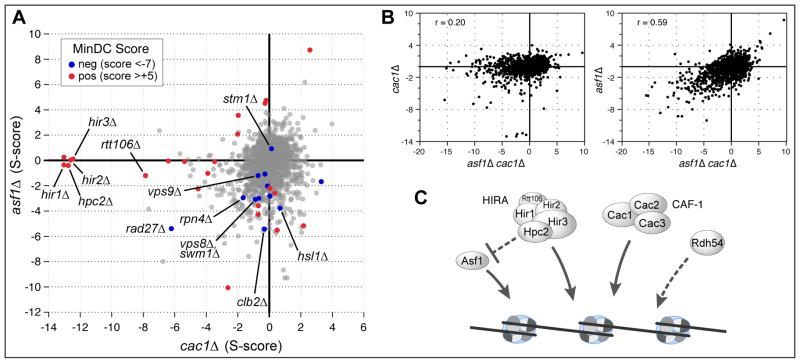
On the positive end of the MinDC scores, we again see cases where one of the double mutant combinations is similar to what is seen in the triple mutant; for example the S-score for asf1Δ cac1Δ rtt109Δ (+5.3) is similar to that observed for rtt109Δ asf1Δ (+4.5) (Figure 1B). This is an example of classic epistasis, where the Rtt109 histone acetyltransferase is known to be dependent on Asf1 for its activity (Collins et al., 2007; Driscoll et al., 2007; Han et al., 2007a). However, when the MinDC analysis was applied, we found all five members of the HIRA-like complex (HIR1, HIR2, HIR3, HPC2 and RTT106) to be the genes with largest MinDC scores (Figure 1B). Double mutants lacking cac1Δ and one of the HIR complex genes display severe growth defects (Sharp et al., 2002), a finding that is recapitulated in our study. However, this severe defect is dramatically suppressed by deleting ASF1. The entire dataset are displayed in a more comprehensive fashion in Figure 2. The S-scores for the asf1Δ and cac1Δ double mutants are displayed along two axes. For each gene, the MinDC score for the triple mutant is displayed by node color. Dark blue indicates cases where the triple mutant grew better than the single mutants (MinDC scores >+5); red shows where the triple mutant was more severely impaired (MinDC scores <−7)). Overall, the genetic profile obtained from the double mutant is more similar to that seen with asf1Δ than cac1Δ (Figure 2B), suggesting that defects in asf1Δ cac1Δ are largely a reflection of those that arise when Asf1 is absent.
Figure 2. Comparison of the S- and MinDC scores from the asf1Δ cac1Δ TMA.
A) Scatter plot of the S-scores derived from asf1Δ and cac1Δ double mutants with the corresponding MinDC scores highlighted in red (positive: >+5) and dark blue (negative: <−7). B) Scatter plot of S-scores from asf1Δ and cac1Δ double mutants compared to asf1Δ cac1Δ triple mutants. C) Model of how chromatin regulators, Asf1, HIR-C, CAF-1 and Rdh54, functionally interact to ensure efficient chromatin regulation (see text for details).
In summary, our data suggest that Rdh54 is potentially acting in a parallel pathway to Asf1 and CAF-1, one that may only function when the other pathways are disabled (Figure 2C) - a conclusion supported by additional observations presented later. Moreover, that deleting ASF1 apparently suppresses double deletions of HIRA and CAF-1 is surprising because one might have anticipated that components of the HIRA complex would be the functionally substitute for Asf1 and CAF-1 and hence have strongly negative MinDC scores. The utility of the TMA scoring system is that it reveals very strong suppression even though the triple mutants are not generally much better growing than average triple mutant strains and hence would not provide significant S-scores. These suppressive relationships are consistent with the observation that HIRA/Rtt106 proteins normally act to regulate Asf1. When HIRA/Rtt106 is absent, Asf1 apparently acts in an abnormal fashion, causing a severe defect in growth when CAF-1 is also absent (Figure 2C). Thus deleting ASF1 rescues the extreme defect of cac1Δ hir/rtt106/hpc2Δ mutants. For example, in the absence of Cac1 and HIRA/Rtt106, the level of histones may fall too low, but asf1Δ may ensure adequate histone abundance.
Functional comparison of Asf1 and Rtt109 using triple mutant analysis
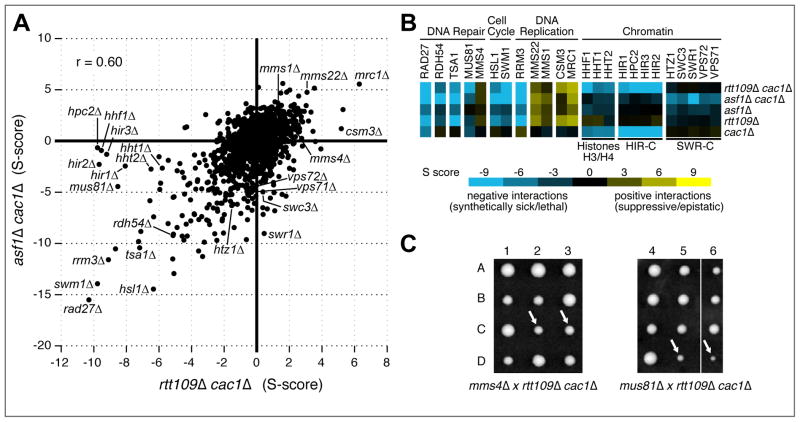
One major role of Asf1 is to promote the acetylation of histone H3-K56 by the histone acetyltransferase, Rtt109 (Collins et al., 2007; Driscoll et al., 2007; Han et al., 2007a). To determine what degree the defects associated with asf1Δ are related to Rtt109 function, we carried a similar TMA using a strain deleted for both RTT109 and CAC1 (Supp. Data 3). Overall, there is a strong correlation of the double mutant scores from asf1Δ cac1Δ and rtt109Δ cac1Δ (r=0.60) (Figure 3A) (for scatter plots using MinDC scores, see Supp. Figure 1). For example, deletion of RAD27, SWM1, HSL1 or RDH54, in combination with rtt109Δ cac1Δ, all provided negative S-scores, as was also observed with asf1Δ cac1Δ. Furthermore, negative interactions were also seen with both sets of triple mutants when Rrm3, a DNA helicase (Ivessa et al., 2002), or Tsa1, a thioredoxin peroxidase involved in DNA replication stress (Inoue et al., 1999), were removed (Figure 3A,B). Similarly, positive interactions were observed with both pairs of double mutants combined with deletions of MMS22, MMS1, CSM3 and MRC1, genes involved in monitoring DNA replication progression (Mohanty et al., 2006; Vaisica et al., 2011). These data support the idea that Asf1 and Rtt109 primarily work together in regulating chromatin function.
Figure 3. Comparison of the S-scores from asf1Δ cac1Δ and rtt109Δ cac1Δ.
A) Scatter plot of the triple mutant S-scores from asf1Δ cac1Δ versus rtt109Δ cac1Δ double mutants. B) A selection of genetic interactions (S-scores) derived from the triple and double mutants analyses. Yellow and blue correspond to positive and negative genetic interactions, respectively. Note the cac1Δ double mutant scores were averaged from data obtained from both asf1Δ cac1Δ and rtt109Δ cac1Δ starter strains. C) Tetrad analysis shows a difference in the viability of mms4Δ rtt109Δ cac1Δ and mus81Δ rtt109Δ cac1Δ segregants, marked by white arrows.
Nonetheless, several interesting differences between asf1Δ cac1Δ and rtt109Δ cac1Δ hint at functional differences between Asf1 and Rtt109. For example, the negative interactions when cac1Δ is combined with hir1Δ, hir2Δ, hir3Δ or hpc2Δ are not suppressed by rtt109Δ (Figure 3A,B), unlike what is seen with asf1Δ. This suggests that Asf1 has at least some roles related to CAF-1 and HIRA functions that are independent of Rtt109. Similar results were observed with deletions of one of the two genes encoding for histones H3 and H4 (HHT1, HHT2 or HHF1) (Figure 3A,B), consistent with the notion that the suppressive activity is related to histone levels. Furthermore, only asf1Δ cac1Δ, but not rtt109Δ cac1Δ, displayed strong negative interactions when combined with deletions of components of the SWR-C complex (SWR1, VPS71, VPS72, SWC3 and HTZ1), which replaces histone H2A with the variant Htz1 in chromatin (Kobor et al., 2004; Krogan et al., 2003; Mizuguchi et al., 2004). Again, these data imply that the Asf1 functions associated with these genetic interactions are not related to Rtt109.
Finally, we observed a striking different genetic interaction pattern with rtt109Δ cac1Δ versus asf1Δ cac1Δ double and triple mutants with deletions of MUS81 and MMS4, encoding an endonuclease complex that promotes crossovers during homologous recombination (de los Santos et al., 2003; Ho et al., 2010). Only mus81Δ, and not mms4Δ, displays a strong negative interaction with rtt109Δ cac1Δ (Figure 3B), as confirmed by tetrad analysis (Figure 3C), even though Mus81 and Mms4 are thought to function exclusively together. These data suggest that Mus81 works independently of Mms4 in certain instances. It is interesting to note that in mammals, Mus81 has been shown to pair with two distinct partners, Eme1 and Eme2 (Shin et al., 2012). Although no other partner as been found in budding yeast, our results raise the possibility of a second partner or that in some circumstances, Mus81 can act alone. This result is an example where Rtt109 seemingly functions outside of its role with Asf1 since asf1Δ does not provide the same pattern of interactions (Figure 3B). We suggest that the Rtt109 acetyltransferase has substrates other than histone H3-K56 that are unrelated to Asf1. Collectively, these data demonstrate the insight that can be gained from higher-level genetic analysis that could not be gleaned from information derived from double mutants, as it teases out important differences between factors that were thought to work in the same functional pathway.
Rad54 plays a role in chromatin regulation in the absence of Asf1 and CAF-1
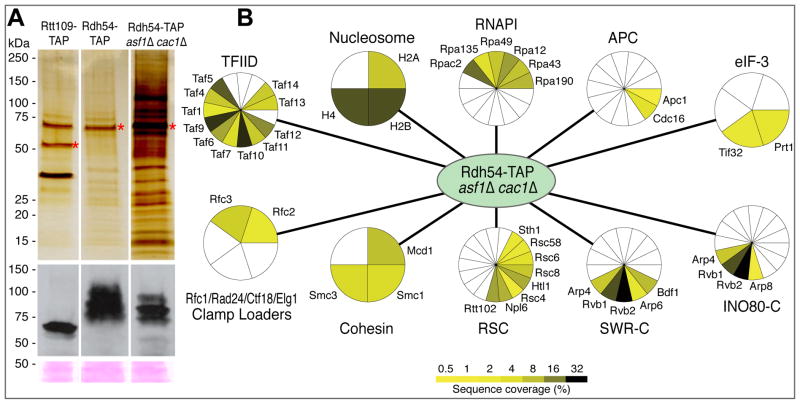
The fact that deleting RDH54 results in a marked growth defect in an asf1Δ cac1Δ or rtt109Δ cac1Δ background suggests that Rdh54 plays a more major role in chromatin regulation in the absence of the two chaperones. To test this idea, we TAP-tagged and purified Rdh54 from both wild type and asf1Δ cac1Δ strains (Krogan et al., 2006). As shown in Figure 4A, there was a marked increased in the number and quantity of co-purifying proteins when the purification was carried out in a strain lacking Asf1 and Cac1. This increase did not reflect a difference in the amount of Rdh54-TAP that was in vivo or purified from the two strains (Figure 4A). Mass spectrometry analysis revealed that several of the additional factors seen specifically co-purifying with Rad54-TAP in the double deletion background are components of chromatin modifying enzymes, including Ino80 and RSC, as well as the core histones H2A, H2B and H4 (Figure 4B, Supp. Data 4). We also recovered one member of the SWR-C complex, Arp6, as well as several proteins that are part of both Ino80 and SWR-C. Interestingly, we had identified the catalytic subunit of the SWR-C, Swr1, another member of the SNF2 family of ATPases, as having a MinDC score of −6.4 (Supp. Data 2), suggesting that SWR-C also functions in a redundant pathway to Rdh54. Furthermore, we identified 11/15 subunits of the general transcription factor, TFIID, 6 proteins of RNA polymerase I and Rfc2 and Rfc3, components of multiple RFC complexes involved in DNA repair and replication. We also recovered several components of the Anaphase Promoting Complex (APC), again consistent with a strong negative TMA score with deletion of SWM1, a component of the APC, in the asf1Δ cac1Δ background. One explanation for the increased association of these proteins with Rdh54 is that they could be more highly expressed in the double mutant background. To test this, we globally assessed the gene expression changes in asf1Δ cac1Δ and found that the majority of the genes corresponding to these proteins were actually down-regulated (Supp. Figure 2; Supp. Data 5). That there are so many complexes involved in chromatin regulation found specifically associated with Rdh54-TAP asf1Δ cac1Δ strongly suggests that Rdh54 plays a more central role in overall chromatin regulation in the absence of the well-characterized histone chaperones.
Figure 4. Affinity tag purifications of Rdh54-TAP and Rad54-TAP asf1Δ cac1Δ.
A) Rad54-TAP was purified from strains in the presence and absence of Asf1 and Cac1. The purified material was subjected to SDS-PAGE and stained with silver (Top). As a control, Rtt109-TAP was purified in the same manner. Red asterisks mark the tagged proteins. (Bottom) Western blot analysis was carried out using extracts prior to purification. Bands from a Ponceau stain serve as a loading control. B) Network of interactions derived from Rdh54-TAP asf1Δ cac1Δ using affinity tag purification-mass spectrometry. Proteins found to interact with either Rdh54-TAP or Rtt109-TAP were removed. Complexes or pathways are labeled and the sequence coverage (%) from the mass spectrometry analysis is shown using a yellow/green color scheme.
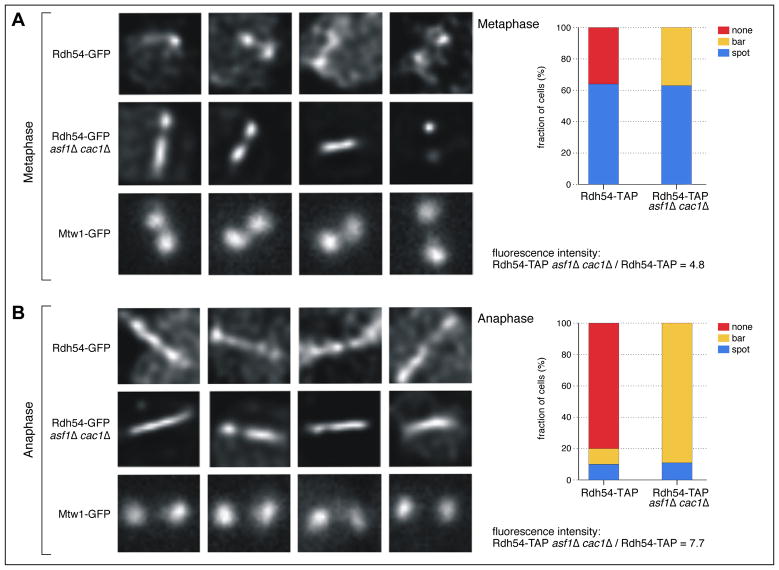
Interestingly, we also found that components of cohesion (Mcd1/Scc1, Smc1 and Smc3) preferentially associated with Rdh54-TAP in the asf1Δ cac1Δ mutant background (Figure 4), suggesting that Rdh54 plays a role in aspects of chromosome segregation. To further explore this connection, we carried out a cytological analysis of Rdh54-GFP in the presence and absence of Asf1 and Cac1. In wild-type cells, Rdh54 is frequently localized to the metaphase spindle in mitosis (64% of cells). Specifically, Rdh54 is concentrated at centromeres and flanking pericentric chromatin and exists exclusively as “spots” (Figure 5A). Unlike kinetochore proteins, including Mtw1, Rdh54 does not always appear as two spots of equal intensity representing sister kinetochores. Instead, one or two foci are seen (e.g. Rdh54-GFP, Metaphase: first and second panel, respectively). In the absence of Asf1 and Cac1 in metaphase, there is 4.8-fold increase in the concentration of Rdh54-GFP along the pericentric chromatin and this localization is apparent in all cells; 63% have foci spots whereas 37% now have elongated “bars”, or linear extensions between the spindle poles (>2 microns) (Figure 5A). Upon anaphase onset, only 20% of the Rad54-GFP wild-type cells display fluorescence, with an equal amount showing focal spots and bars (Figure 5B). In asf1Δ cac1Δ cells, there is a substantial increase of overall fluorescence (7.7 fold) and it is seen in all cells, 89% as bars. In general, the strong prevalence of this pattern is reminiscent of condensin, which is concentrated in the pericentric chromatin and lies proximal to the spindle axis in metaphase (Stephens et al., 2011). This suggests that Rad54 plays a role in chromatin remodeling and/or chromosome condensation.
Figure 5. Rdh54-GFP is redistributed to pericentric chromatin in asf1Δ cac1Δ mutants during mitosis.
Representative images of Rdh54-GFP (C-terminal fusion) in wild-type (WT) and asf1Δ cac1Δ cells. A) Metaphase: Rdh54-GFP localization is compared to a core component of the yeast kinetochore (Mtw1-GFP). The concentration of Rdh54-GFP along the spindle is ~ 5-fold greater in the mutant vs. WT. B) Anaphase: In the absence of Asf1 and Cac1, Rdh54 appears as a bar along the metaphase spindle in nearly 90% of cells. The concentration of Rdh54-GFP along the spindle is approximately 7.7-fold greater in the mutant vs. WT. Foci are defined as diffraction-limited (or slightly larger) fluorescent spots. For the kinetochore protein Mtw1 (bottom row) spots are approximately spherical, with sister kinetochores exhibiting similar shape and intensity (Haase et al., 2012). Bars are defined as linear extensions of fluorescence (ex. bars in 1st and 3rd panel of Rdh54-GFP asf1Δ cac1Δ Metaphase). In instances where a bar and a foci are observed (ex. 2nd panel Rdh54-GFP, asf1Δ cac1Δ Anaphase) the cell is scored as containing a bar.
The pericentromere surrounding the point centromere is functionally distinct from the bulk chromosome arms during mitosis (Verdaasdonk et al., 2012). Several chromatin remodeling complexes, including RSC and ISW2, are required for histone stability and chromatin packaging within the pericentromere as this region experiences spindle tension force. The enrichment of Rdh54 in the absence of the Asf1 and CAF-1 histone chaperones suggests it plays a role in the maintenance of chromatin under tension. Its localization along the spindle axis also places Rdh54 in a position to regulate chromatin loops formed by the action of condensin (Stephens et al., 2011). Condensin and cohesin compact the pericentromere into axial loops that constitute the chromatin spring. The combined function of pericentric compaction, together with chaperones to regulate histone dynamics, reflect a critical function in building a chromatin spring that resists microtubule-based pulling forces throughout mitosis. The recruitment of Rdh54 to this region in the absence of histone chaperones reflects the ability of the cell to respond to changes in the stoichiometry of components that regulate chromatin stiffness. Collectively, this localization data is consistent with the protein-protein interaction data (Figure 4) that suggests Rdh54 is more functionally relevant in the absence of the two chaperones.
TMA of CLB5 and CLB6 reveals a connection of the cyclins to chromosome segregation
In a similar fashion to Asf1 and CAF-1, we analyzed another situation of apparent redundancy. The two cyclin B proteins, Clb5 and Clb6, function in the regulation of the timing of DNA replication, although they are not essential individually or when both are absent. Among B-type cyclins, Clb5 and Clb6 play distinct roles in regulating the initiation of S phase. Either Clb5 or Clb6 can activate early-firing replication origins, but late origin firing is dependent only on Clb5 (Donaldson, 2000). However, neither CLB5 nor CLB6 is essential and even the clb5Δ clb6Δ double mutant is viable. Moreover, early expression of Clb2 during S phase does not restore normal replication timing in the absence of Clb5 and Clb6 (Donaldson, 2000). Intriguingly, the clb5Δ clb6Δ double mutant is unable to promote DNA replication in meiosis, most likely by the absence of CLB2 expression (Mai and Breeden, 2000). Clb5 has also been shown to play an important role in mitotic spindle positioning, a function that is not replaced by overexpressing Clb2 (Segal et al., 1998). Clb5 and Clb6 has also been implicated in the establishment of sister-chromatid cohesion in cells under replication stress; cells lacking Clb5, Clb6 and the securin, Pds1 are inviable (Hsu et al., 2011).
To establish if there were novel gene functions that maintained the viability of the clb5Δ clb6Δ double mutant, we performed TMA as described above, crossing clb5Δ::URA3 clb6Δ::HPH against KAN-marked gene knockouts (Figure 6A; Supp. Data 6 and 7). Again, there were a number of synthetically sick interactions where there was little or no defect for either single mutant. Unlike the situation with asf1Δ and cac1Δ, there were very few instances where the synthetic lethality of clb5Δ clb6Δ was also seen in one or the other single mutant (Figure 6B). However, mtc1Δ is synthetically sick with clb5Δ clb6Δ (−16.4) and clb5Δ (−10.5) but not clb6Δ (0.5); conversely vps53Δ is synthetically sick with clb5Δ clb6Δ (−10.3) and clb6Δ (−11.8) but not at all with clb5Δ (1.2). Furthermore, there were only very weak correlations between the pattern of genetic interactions between clb5Δ clb6Δ and either clb5Δ (r = 0.26) or clb6Δ (r = 0.18) (Figure 6B). In many cases, the basis of this lethality is not evident. Mbf1 is required for gene expression in the G1 to S transition and it is possible that some other early S phase functions are essential without Clb5 and Clb6. One striking set of genes with common genetic interactions was the HIR complex where hir1Δ (−14.6), hir2Δ (−11.8), hir3Δ (−12.8), and hpc2Δ (−14.6) all show a common strongly negative interaction in clb5Δ clb6Δ but not with either single mutant (−0.3, −0.5, −2.5, −0.5 for clb5Δ and −0.9, 0.3, −0.3, −0.1 for clb6Δ, respectively).
Figure 6. Triple mutant analysis of clb5Δ clb6Δ.
A) Scatter plot of the S-scores derived from clb5Δ and clb5Δ double mutants with the corresponding MinDC scores highlighted in red (positive: >+5) and dark blue (negative: <−7). B) Scatter plot of S-scores from clb5Δ or clb6Δ double mutants compared to clb5Δ clb6Δ triple mutants. C) Comparison of genetic profile generated from clb5Δ clb6Δ to profiles from large genetic interaction dataset (Data Set S4 from (Ryan et al., 2012)). The most highly correlated profiles belong to strains harboring mutations in SCC2, SMC3, CTF4, CTF18 and SMC1. D) Diploids homozygous for the indicated genotype and for ade2 were cross-streaked with haploid MATa and MATα ade5 cells. Chromosome loss (or mitotic crossing-over) was scored as papillae that grow when the matings are replica-plated to minimal medium (see Methods). E) Model of how Clb5 and Clb6 function to ensure efficient chromosome segregation (see text for details).
To characterize more generally the phenotype of clb5Δ clb6Δ mutants we compared interactions across all tested gene knockouts with the hierarchically clustered quantitative genetic interaction data from previous studies (Ryan et al., 2012). We found that clb5Δ clb6Δ most strongly correlated with mutations affecting the establishment of sister-chromatid cohesion, including point mutants in cohesin, smc1-259, smc3-1, smc3-42 and in the cohesin loading factor Scc2 (scc2-4) as well as with deletions in CTF4 and CTF18, (Stirling et al., 2012; Yuen et al., 2007) (Figure 6C; Supp. Data 8). clb5Δ clb6Δ showed especially strong negative interactions with ctf3Δ (−12.1), ctf19Δ (−13.3), irc15Δ (−13.9), mcm21Δ (−13.8) and chl1Δ (−15.1) (Figure 6a). The set of strongly negative genes is strongly reminiscent of genes that were synthetically lethal with hypomorphic mutations of three cohesion proteins (McLellan et al., 2012). We confirmed that clb5Δ clb6Δ, but not the single mutants, exhibit a chromosome loss phenotype, consistent with a defect in cohesion (Figure 6D).
The idea that Clb5 and Clb6 might play an important role in chromosome segregation was further strengthened by our finding that the double mutant exhibited strong synthetic sickness with the spindle assembly checkpoint mutants mad1Δ (S-score = −10.7) and mad2Δ (S-score = −13.3), however there was little negative interaction with mad3Δ (S-score = −1.4). To verify these interactions, we carried out tetrad analysis, scoring spore colony size. We confirmed that both mad1Δ and mad2Δ, but not mad3Δ showed strong synthetic lethality (Supp. Table 1). In addition, tetrad analysis showed that bub1Δ, but not bub2Δ, also showed strong negative interactions. Despite the fact that Mad2 and Mad3 often appear to function together, there are other observations that suggest they have separate functions; for example the phosphorylation of Mad1 when the mitotic checkpoint is activated depends on Mad2 and Bub1 but not on Bub2 and Mad3 (Hardwick and Murray, 1995). Similarly, accurate meiotic chromosome segregation depends on Mad2 but not Mad3 (Tsuchiya et al., 2011). Moreover, a screen of synthetic genetic interactions with various chromosome transmission mutations found a number of instances where mad1Δ and mad2Δ, but not mad3Δ, exhibited synthetic lethality (Daniel et al., 2006). We suggest that the absence of both Clb5 and Clb6 establishes a delay in kinetochore assembly and/or spindle assembly that requires Mad1, Mad2 and Bub1 to delay mitosis, but not Mad3 or Bub2.
We summarize these findings in the scheme shown in Figure 6E. Especially around centromeres, DNA replication is coupled to the establishment of kinetochore attachment to microtubules and to extensive sister-chromatid cohesion in the pericentromeric region. In the absence of Clb5 and Clb6, replication is delayed and these critical steps may not be completed by the time mitosis is normally initiated. Consequently in clb5Δ clb6Δ, cells become dependent on the action of Mad1, Mad2 and Bub1 in establishing a spindle-assembly checkpoint and cannot tolerate defects in the establishment of cohesion. It is also possible that the chromosome loss phenotype is a reflection of a more general consequence of replication stress similar to the increased genome instability seen in mammalian cells with activated oncogenes (Halazonetis et al., 2008). However, clb5Δ clb6Δ does not show strong synthetic interactions with deletions either of DNA repair genes or components of the DNA damage checkpoint. Hence, we believe the effects are more directly linked to kinetochore assembly and the recruitment of cohesins in pericentromeric regions.
Perspective
Great insight into a variety of different biological processes has been extracted from genetic interaction maps in a variety of different organisms, in both budding and fission yeast (Collins et al., 2007; Fiedler et al., 2009; Roguev et al., 2008; Schuldiner et al., 2005; Wilmes et al., 2008) and bacteria (Butland et al., 2008; Typas et al., 2008), as well as more complex organisms such as D. melanogaster (Horn et al., 2011) and C. elegans (Lehner et al., 2006). More recently, quantitative genetic interaction mapping have been developed for mammalian cell lines (Bassik et al., 2013; Lin et al., 2012; Roguev et al., 2013). However, to date, essentially all of these data have been generated in a systematic, pair-wise fashion even though a deeper understanding of functional pathways could be gleaned from analyzing more than two genetic perturbations at a time. There have been genetic studies involving triple mutants in budding yeast (Tong et al., 2004; Zou et al., 2009), however these were qualitative in nature. Here we describe a quantitative approach, termed TMA (triple mutant analysis), that allows for higher-order interactions. We interrogated two pairs of genes known to act redundantly in budding yeast: histone chaperones Asf1 and Cac1 and the cyclins, Clb5 and Clb6. This has led to several discoveries. First, Rdh54/Tid1 functionally “backs-up” Asf1 and CAF-1 and interacts much more strongly with chromatin complexes when the two major chaperones are absent. Second, loss of Asf1 suppresses the defect seen when HIR-A proteins are removed in combination with cac1Δ. Such a complex relationship could not have been gleaned simply from double mutant analysis. Third, we found Clb5 and Clb6 play an important role in the regulation chromosome segregation. Finally, comparison of the TMA profiles of asf1Δ cac1Δ and rtt109Δ cac1Δ revealed functional differences between Asf1 and Rtt109 that, again, could not have been uncovered using more traditional genetic analysis. We suggest this approach will be a powerful way to uncover subtle functional differences between factors that were thought to exclusively function in the same pathway.
Combined with genetic interaction mapping under different conditions (Ideker and Krogan, 2012) and analysis that allows for genetic data to be collected at sub-protein resolution (unpublished data), quantitative TMA allows for the probing into a previously unexplored interactome space. As many genes in budding yeast do not have strong genetic interaction profiles (e.g. RDH54), more systematic TMA will be crucial in assigning their functions. Furthermore, extending this analysis into other organisms that have tools for genetic interaction mapping, will be important to further understand various processes in these organisms and will facilitate evolutionarily analysis of higher order genetic interactions. Finally, as platforms are developed in mammalian cells that are amenable for higher order genetic interactions using combinatorial RNAi approaches (Roguev et al., 2013), TMA will be crucial to identify pathway organization in normal conditions and in specific disease states. Ultimately, we argue work of this nature will be key for identifying targets for polypharmacy therapeutic intervention.
Methods
Detailed methods are presented in Supplemental Information. E-MAP analysis was conducted as described in (Schuldiner et al., 2006). TAP-tagged proteins were purified as previously described (Krogan et al., 2006).
Supplementary Material
Highlights.
High-throughput quantitative genetic analysis of triple mutants in budding yeast
Swi/Snf ATPase Rdh54 can compensate for absence of histone chaperones Asf1 and Cac1
Absence of both CLB5 and CLB6 creates kinetochore and cohesin-like defects
Genetic differences between ASF1 and RTT109 suggest Rtt109 has unidentified functions
Acknowledgments
We thank members of the Krogan and Haber groups for helpful discussion. This work was supported by grants from NIH (GM084448, GM084279, GM081879 and GM098101 to NJK; GM61766 and GM20056 to JEH and R37 GM32238 to KB). NJK is a Searle Scholar and a Keck Young Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, et al. A systematic Mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P, Cagney G, Krogan NJ. Quantitative genetic interactions reveal biological modularity. Cell. 2010;141:739–745. doi: 10.1016/j.cell.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher RN, Deshaies RJ, Kirschner MW. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butland G, Babu M, Diaz-Mejia JJ, Bohdana F, Phanse S, Gold B, Yang W, Li J, Gagarinova AG, Pogoutse O, et al. eSGA: E. coli synthetic genetic array analysis. Nat Methods. 2008;5:789–795. doi: 10.1038/nmeth.1239. [DOI] [PubMed] [Google Scholar]
- Chi P, Kwon Y, Visnapuu ML, Lam I, Santa Maria SR, Zheng X, Epshtein A, Greene EC, Sung P, Klein HL. Analyses of the yeast Rad51 recombinase A265V mutant reveal different in vivo roles of Swi2-like factors. Nucleic Acids Res. 2011;39:6511–6522. doi: 10.1093/nar/gkr297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Collins SR, Roguev A, Krogan NJ. Quantitative genetic interaction mapping using the E-MAP approach. Methods Enzymol. 2010;470:205–231. doi: 10.1016/S0076-6879(10)70009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Keyes BE, Ng YP, Freeman CO, Burke DJ. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics. 2006;172:53–65. doi: 10.1534/genetics.105.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decesare JM, Stuart DT. Among B-Type Cyclins Only CLB5 and CLB6 Promote Premeiotic S Phase in Saccharomyces cerevisiae. Genetics. 2012;190:1001–1016. doi: 10.1534/genetics.111.134684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD. The yeast mitotic cyclin Clb2 cannot substitute for S phase cyclins in replication origin firing. EMBO Rep. 2000;1:507–512. doi: 10.1093/embo-reports/kvd108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, Fangman WL. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell. 1998;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- Dresser ME, Ewing DJ, Conrad MN, Dominguez AM, Barstead R, Jiang H, Kodadek T. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, Silva AC, Shales M, Collins SR, van Wageningen S, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR, 3rd, Kaufman PD. Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J, Stephens A, Verdaasdonk J, Yeh E, Bloom K. Bub1 kinase and Sgo1 modulate pericentric chromatin in response to altered microtubule dynamics. Curr Biol. 2012;22:471–481. doi: 10.1016/j.cub.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hall MC, Torres MP, Schroeder GK, Borchers CH. Mnd2 and Swm1 are core subunits of the Saccharomyces cerevisiae anaphase-promoting complex. J Biol Chem. 2003;278:16698–16705. doi: 10.1074/jbc.M213109200. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007a;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem. 2007b;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Mazon G, Lam AF, Symington LS. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell. 2010;40:988–1000. doi: 10.1016/j.molcel.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn T, Sandmann T, Fischer B, Axelsson E, Huber W, Boutros M. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nat Methods. 2011;8:341–346. doi: 10.1038/nmeth.1581. [DOI] [PubMed] [Google Scholar]
- Hsu WS, Erickson SL, Tsai HJ, Andrews CA, Vas AC, Clarke DJ. S-phase cyclin-dependent kinases promote sister chromatid cohesion in budding yeast. Mol Cell Biol. 2011;31:2470–2483. doi: 10.1128/MCB.05323-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Krogan NJ. Differential network biology. Mol Syst Biol. 2012;8:565. doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Matsuda T, Sugiyama K, Izawa S, Kimura A. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J Biol Chem. 1999;274:27002–27009. doi: 10.1074/jbc.274.38.27002. [DOI] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kats ES, Albuquerque CP, Zhou H, Kolodner RD. Checkpoint functions are required for normal S-phase progression in Saccharomyces cerevisiae RCAF- and CAF-I-defective mutants. Proc Natl Acad Sci U S A. 2006;103:3710–3715. doi: 10.1073/pnas.0511102103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Haber JE. Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proc Natl Acad Sci U S A. 2009;106:1151–1156. doi: 10.1073/pnas.0812578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz DC, Kama T, Kaufman PD. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol Cell Biol. 2002;22:614–625. doi: 10.1128/MCB.22.2.614-625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Kwon Y, Seong C, Chi P, Greene EC, Klein H, Sung P. ATP-dependent chromatin remodeling by the Saccharomyces cerevisiae homologous recombination factor Rdh54. J Biol Chem. 2008;283:10445–10452. doi: 10.1074/jbc.M800082200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Pellicioli A, Malkova A, Foiani M, Haber JE. The Saccharomyces recombination protein Tid1p is required for adaptation from G2/M arrest induced by a double-strand break. Curr Biol. 2001;11:1053–1057. doi: 10.1016/s0960-9822(01)00296-2. [DOI] [PubMed] [Google Scholar]
- Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- Lin YY, Kiihl S, Suhail Y, Liu SY, Chou YH, Kuang Z, Lu JY, Khor CN, Lin CL, Bader JS, et al. Functional dissection of lysine deacetylases reveals that HDAC1 and p300 regulate AMPK. Nature. 2012;482:251–255. doi: 10.1038/nature10804. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ma XJ, Lu Q, Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- Mai B, Breeden L. CLN1 and its repression by Xbp1 are important for efficient sporulation in budding yeast. Mol Cell Biol. 2000;20:478–487. doi: 10.1128/mcb.20.2.478-487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JL, O’Neil NJ, Barrett I, Ferree E, van Pel DM, Ushey K, Sipahimalani P, Bryan J, Rose AM, Hieter P. Synthetic Lethality of Cohesins with PARPs and Replication Fork Mediators. PLoS Genetics. 2012;8:e1002574. doi: 10.1371/journal.pgen.1002574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Mohanty BK, Bairwa NK, Bastia D. The Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2006;103:897–902. doi: 10.1073/pnas.0506540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Pennaneach V, Kats ES, Kolodner RD. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc Natl Acad Sci U S A. 2003;100:6640–6645. doi: 10.1073/pnas.1232239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TK, Robertson RB, Visnapuu ML, Chi P, Sung P, Greene EC. A DNA-translocating Snf2 molecular motor: Saccharomyces cerevisiae Rdh54 displays processive translocation and extrudes DNA loops. J Mol Biol. 2007;369:940–953. doi: 10.1016/j.jmb.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey CJ, Howar S, Adkins M, Linger J, Spicer J, Tyler JK. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol Cell Biol. 2004;24:10313–10327. doi: 10.1128/MCB.24.23.10313-10327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D, Quivy JP, Scamps C, Martini EM, Lipinski M, Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- Reagan MS, Pittenger C, Siede W, Friedberg EC. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Talbot D, Negri GL, Shales M, Cagney G, Bandyopadhyay S, Panning B, Krogan NJ. Quantitative genetic-interaction mapping in mammalian cells. Nat Methods. 2013 doi: 10.1038/nmeth.2398. advance online publication 2013/02/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Methods. 2007;4:861–866. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- Ryan CJ, Roguev A, Patrick K, Xu J, Jahari H, Tong Z, Beltrao P, Shales M, Qu H, Collins SR, et al. Hierarchical Modularity and the Evolution of Genetic Interactomes across Species. Mol Cell. 2012;46:691–704. doi: 10.1016/j.molcel.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Yang QH, Futcher AB. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Weissman JS, Krogan NJ. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods. 2006;40:344–352. doi: 10.1016/j.ymeth.2006.07.034. [DOI] [PubMed] [Google Scholar]
- Segal M, Clarke DJ, Reed SI. Clb5-associated kinase activity is required early in the spindle pathway for correct preanaphase nuclear positioning in Saccharomyces cerevisiae. J Cell Biol. 1998;143:135–145. doi: 10.1083/jcb.143.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp JA, Franco AA, Osley MA, Kaufman PD. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 2002;16:85–100. doi: 10.1101/gad.925302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YK, Amangyeld T, Nguyen TA, Munashingha PR, Seo YS. Human MUS81 complexes stimulate flap endonuclease 1. FEBS J. 2012;279:2412–2430. doi: 10.1111/j.1742-4658.2012.08620.x. [DOI] [PubMed] [Google Scholar]
- Sia RA, Herald HA, Lew DJ. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol Biol Cell. 1996;7:1657–1666. doi: 10.1091/mbc.7.11.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AC, Xu X, Kim HS, Fillingham J, Kislinger T, Mennella TA, Keogh MC. The replication-independent histone H3-H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J Biol Chem. 2012;287:1709–1718. doi: 10.1074/jbc.M111.316489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Lavy KJ, Sajman J, Zenvirth D, Brandeis M. APC/CCdh1 specific degradation of Hsl1 and Clb2 is required for proper stress responses of S. cerevisiae. Cell Cycle. 2009;8:3003–3009. [PubMed] [Google Scholar]
- Stephens AD, Haase J, Vicci L, Taylor RM, 2nd, Bloom K. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol. 2011;193:1167–1180. doi: 10.1083/jcb.201103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Crisp MJ, Basrai MA, Tucker CM, Dunham MJ, Spencer FA, Hieter P. Mutability and mutational spectrum of chromosome transmission fidelity genes. Chromosoma. 2012;121:263–275. doi: 10.1007/s00412-011-0356-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher AB, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tsuchiya D, Gonzalez C, Lacefield S. The spindle checkpoint protein Mad2 regulates APC/C activity during prometaphase and metaphase of meiosis I in Saccharomyces cerevisiae. Mol Biol Cell. 2011;22:2848–2861. doi: 10.1091/mbc.E11-04-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Nichols RJ, Siegele DA, Shales M, Collins SR, Lim B, Braberg H, Yamamoto N, Takeuchi R, Wanner BL, et al. High-throughput, quantitative analyses of genetic interactions in E. coli. Nat Methods. 2008;5:781–787. doi: 10.1038/nmeth.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisica JA, Baryshnikova A, Costanzo M, Boone C, Brown GW. Mms1 and Mms22 stabilize the replisome during replication stress. Mol Biol Cell. 2011;22:2396–2408. doi: 10.1091/mbc.E10-10-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaasdonk JS, Gardner R, Stephens AD, Yeh E, Bloom K. Tension-dependent nucleosome remodeling at the pericentromere in yeast. Mol Biol Cell. 2012;23:2560–2570. doi: 10.1091/mbc.E11-07-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci U S A. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Boone C, Brown GW. Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics. 2007;176:1417–1429. doi: 10.1534/genetics.107.072876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen KW, Warren CD, Chen O, Kwok T, Hieter P, Spencer FA. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc Natl Acad Sci U S A. 2007;104:3925–3930. doi: 10.1073/pnas.0610642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Friesen H, Larson J, Huang D, Cox M, Tatchell K, Andrews B. Regulation of cell polarity through phosphorylation of Bni4 by Pho85 G1 cyclin-dependent kinases in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:3239–3250. doi: 10.1091/mbc.E08-12-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.