Abstract
Correlated chemical imaging is an emerging strategy for acquisition of images by combining information from multiplexed measurement platforms to track, visualize, and interpret in situ changes in the structure, organization, and activities of interesting chemical systems, frequently spanning multiple decades in space and time. Acquiring and correlating information from complementary imaging experiments has the potential to expose complex chemical behavior in ways that are simply not available from single methods applied in isolation, thereby greatly amplifying the information gathering power of imaging experiments. However, in order to correlate image information across platforms, a number of issues must be addressed. First, signals are obtained from disparate experiments with fundamentally different figures of merit, including pixel size, spatial resolution, dynamic range, and acquisition rates. In addition, images are often acquired on different instruments in different locations, so the sample must be registered spatially so that the same area of the sample landscape is addressed. The signals acquired must be correlated in both spatial and temporal domains, and the resulting information has to be presented in a way that is readily understood. These requirements pose special challenges for image cross-correlation that go well beyond those posed in single technique imaging approaches. The special opportunities and challenges that attend correlated imaging are explored by specific reference to correlated mass spectrometric and Raman imaging, a topic of substantial and growing interest.
Overview
Chemical imaging can usefully be defined as “the spatial (and temporal) identification and characterization of the molecular chemical composition, structure, and dynamics of any given sample.”1 Ideally this encompasses the ability to detect, identify, and visualize the spatial distribution of molecules, known or unknown, over multiple size scales, with arbitrary time resolution, and with single-molecule detection limits. Scientists use chemical imaging to address a wide variety of problems – visualizing the arrangement of atoms patterned on a surface at nanometer scale,2 the molecular contents of single cells at sub-micrometer scale,3 and colorless gases released into Earth's atmosphere by cities on the kilometer scale.4 While none of these imaging applications comes close to fulfilling the above ideal, analytical techniques have been developed in imaging modes that permit access to a considerable fraction of this “chemical imaging space”. Now researchers are beginning to invest substantial effort in learning how to combine different imaging modalities, thus amplifying the information gathering power of imaging experiments.
Chemical imaging involves probing a sample, then detecting a signal that provides spatial and temporal information about the chemical state. Images are usually acquired either by an encoding/decoding operation, as in optical imaging, or by rastering sample and probe past one another. Imaging probes come in a stunning array of interactions – bombarding the sample with photons, charged particles, atoms, or sound waves, for example. Readout can involve detection of these agents, whether they are similar to the incident probe (fluorescence microscopy), or not (photoacoustic microscopy5), or reading out the interaction of proximal probes, as in atomic force microscopy, AFM. Chemical imaging can also be accomplished by addition of labels or contrast agents which either generate a detectable event independently, e.g. positron emitters for tomography, PET, and radionuclides for autoradiography, or enhance information from an external probe, e.g. fluorescent tags in fluorescence imaging and microbubbles for ultrasound.6 The specific combination of probe and readout define the capabilities, limitations, and therefore useful applications of chemical imaging techniques. From these underlying principles emerge important secondary characteristics such as spatial and temporal resolution, sample penetration depth, sensitivity, multiplex capability, sample preparation requirements, destructiveness, chemical specificity, and information content. One of the principal aims of correlated imaging is to carefully mix and match the imaging tools so that these secondary characteristics can be optimized across the combined imaging platforms.
Since all possible combinations of chemical imaging techniques and applications constitute a vast subject area, here the focus is limited to correlated chemical imaging of molecules in bioanalytical research. Emphasis is placed on how combining suitable, complementary imaging techniques circumvents specific limitations and provides advantages to the researcher. General themes in correlated imaging will be illustrated by reference to correlated mass spectrometric and confocal Raman imaging.
Specific approaches to biomolecular imaging
Biological organisms are chemically, spatially, and temporally complex, exhibiting diversity in size, structure, shape, and concentration spanning orders of magnitude. Chemical and structural features change on time scales ranging from years of organismal life to the millisecond time-frame of a nerve cell's action potential.7 Understanding how biological systems function and malfunction ultimately requires integration of information across all of these scales, a challenging task where chemical imaging plays a critical role, especially at the microscopic level.
One of the first applications of light microscopy was by Anton van Leeuwenhoek who examined single cells in the first biomolecular microscopic imaging experiment.8 Probing a sample with incoherent white light and observing what is transmitted or reflected reports principally on morphology, but adaptations of the basic principle can yield chemical information. For example, in fluorescence microscopy, a molecule of interest is tagged with an organic dye, protein, or quantum dot, which can then be excited to yield the spatial distribution of the probe. Because fluorophores can be engineered for specific excitation and emission wavelengths and also designed with affinity for particular biomolecules, fluorescence microscopy has become a powerful method for visualizing specific molecular species. Many other imaging methods, such as autoradiography,9 positron emission tomography (PET),10 and ultrasound biomicroscopy (UBM),11 achieve chemical specificity in a similar targeted manner.
How can molecular imaging be performed on biological systems de novo, or when a reliable tagging approach does not exist?12 In this case imaging techniques must deliver information to allow for broad initial chemical survey, while retaining chemical specificity and structural detail for identification of unknowns. Two techniques – mass spectrometry and Raman spectroscopy – serve as excellent imaging platforms for this purpose. Functioning on distinct principles, they provide uniquely information-rich chemical images and complementary advantages. For example, Raman microscopy is non-invasive and non-destructive, so the integrity of the sample is preserved for the MS imaging, thus accessing the high chemical specificity inherent in mass measurement with MS, which however, is inherently destructive. We will use these two techniques to illustrate the challenges and opportunities in correlated imaging.
Mass Spectrometric Imaging
In contemporary practice, even high molecular weight biomolecules can be volatilized, ionized, and detected, making MS a robust technique for non-targeted biochemical analysis of tissue samples and single cells.13 Furthermore, highly-accurate mass assignments and tandem MS (fragmentation) experiments can greatly amplify the information about the identity and structure of compounds, such as peptides. Performing MS with a microprobe, such as a focused laser or primary ion beam, allows different regions of a chemically heterogeneous sample to be chemically profiled.14 Rastering the microprobe automatically across an area of interest, i.e. microprobe-mode mass spectrometric imaging (MSI), yields a chemical map of the region. Each position in the array constitutes a “pixel” in the resulting image and contains a full mass spectrum. Alternatively, microscope-mode MSI involves probing the entire field of view simultaneously with a defocused probe, then preserving spatial information by transmitting the ions to a position-sensitive detector via a stigmatic mass spectrometer.15, 16 Optimal representation of the data remains a significant challenge. It is typically visualized by filtering the spectral data to display the localization of specific ions, resulting in an ion image. A single MSI experiment can generate a separate ion image from each distinct ion within the scanned mass range, and typically tens to hundreds of ions are detected depending on sample complexity and the resolving power of the mass analyzer(s). Nevertheless, the richness of the data, especially when tandem MSn experiments are performed, allows more powerful means to visualize the data.
Probe choice has a profound effect on MSI data content and quality, especially in terms of ions observed, sensitivity, and spatial resolution. The most common probes used are ultraviolet lasers for laser desorption ionization (LDI)17, 18 or matrix-assisted LDI (MALDI)13, 19-21 and focused ion beams for secondary ion mass spectrometry (SIMS),22-27 though many other viable probe types exist including desorption electrospray ionization (DESI),28-32 capillary-controlled liquid microjunctions (LMJ),29, 33-36 laser-ablation electrospray ionization (LAESI),29, 37-40 and laser ablation with post-ionization by inductively-coupled plasma (LA-ICP).41-43 An overview and comparison of these MSI probe types is given in Table 1.
Table 1.
Performance comparison of some common MSI microprobes.
| Probe | Typical lateral resolution (μm) | Max. lateral resolution (μm) | Accessible chemical information | Advantages | Disadvantages |
|---|---|---|---|---|---|
| SIMS | 1-526, 27 | 0.0247 | Lipids,25 peptides,26, 27 metabolites,150 characteristic molecular fragments,151 elements,24 isotopes23 | Highest attainable lateral MSI resolution, ion beam etching capability allows high resolution depth profiling, 3D imaging | Harder ionization (vs MALDI, ESI), mass range limited to <2.5 kDa27, expensive instrumentation, requires vacuum-compatible samples |
| MALDI | 25-20072 | 0.693 | Lipids,19 metabolites,13 peptides,13, 21 proteins20 | Excellent high mass range, broadly applicable across many biomolecule classes | Matrix application complicates low-mass spectrum (< 500 Da), may reduce lateral resolution |
| LDI | 25-10017 | 0.2152 | Metabolites, characteristic ion fragments,18 elements, isotopes | No sample pretreatment, performed on standard MALDI instrumentation | <1 kDa mass range limit, harder ionization (vs MALDI, ESI) |
| LA-ICP | 12-16043 | 442 | Elements, isotopes42 | Atmospheric pressure sampling, excellent sampling efficiency | Hard ionization limits chemical information |
| LA-ESI | 20029 | 10153 | Lipids, metabolites, small drug molecules29 | Atmospheric pressure sampling, no sample pretreatment | Mass range limited to <1 kDa by laser ablation process |
| DESI | 20029 | 4032 | Lipids, metabolites, small drug molecules29 | Atmospheric pressure sampling, no sample pretreatment | Low spatial resolution |
| LMJ | 400-70034, 154/100 (non-contact mode35) | 1533 | Lipids,33 metabolites, small drug molecules34, 36 | Atmospheric pressure sampling, no sample pretreatment, potential for in-line sample processing36 | Low typical spatial resolution, microjunction stability varies with tissue type & condition |
MALDI is a relatively soft (non-fragmenting) ion generation scheme which offers excellent mass range (to MDa),44 high sensitivity,45 and μm-scale probe diameters in custom instrumentation.46 SIMS employs probes with diameters as small as 20 nm 47 suitable for cell- and subcellular imaging, albeit with a harsher ionization mechanism leading to lower sensitivity (higher detection limits), particularly for intact biomolecules. Conveniently, ion beams can also serve to etch away surface layers, such as the contamination from culture medium with cultured cells, 48, 49 making it possible to produce three-dimensional images.50-52 DESI involves bombarding a sample surface with charged solvent droplets. In comparison to MALDI and SIMS, it provides lower lateral resolution (>35 μm)53 in exchange for a softer ionization mechanism, which is more amenable to ionization of intact biomolecules. DESI is also performed at ambient conditions rather than in vacuum, a useful feature enabling direct analysis of live biological samples.54
Raman Microscopy
In vibrational Raman spectroscopy the frequency spectrum of inelastically-scattered light is measured to obtain information about the functional groups present in a sample.55 Because a large number of cellular constituents are Raman-active to some degree, Raman spectroscopy offers broad, non-targeted detection of biomolecules in complex matrices, e.g. cells, tissues, and biofilms, with chemical specificity relating to structure – as opposed to molecular identity in MS. Moreover, by employing resonance Raman spectroscopy (RRS) and surface-enhanced Raman spectroscopy (SERS), sensitivity can be enhanced dramatically, even yielding single-molecule detection under ideal circumstances.56
Similar to MSI, a focused laser is raster scanned across a sample to produce chemical images where each pixel is composed of a full Raman spectrum. Both Raman and IR images provide information on the spatial distribution of the components of a sample, with the intensity of each component related to its abundance. This makes it possible to elucidate quantitative sample component distributions.57 In contrast with MSI, Raman spectroscopy is both nondestructive and amenable to ambient conditions. Thus, it is well-suited to live cell imaging.58 Furthermore, carrying out confocal Raman microscopy (CRM) offers many additional advantages such as 3D and subsurface sample imaging capability, sub-μm (diffraction-limited) lateral resolution, and reduced autofluorescence background by confining the region of excitation and analysis to a small volume.55 Alternatively, the development of spatially offset Raman spectroscopy (SORS) has afforded subsurface analysis of turbid materials at greater depths than CRM. Based on acquiring Raman spectra at spatial positions offset from the incident laser, the acquired spectra allow signal contributions from the surface and the subsurface layers to be separated, thereby producing pure Raman spectra from the subsurface layers.59, 60 Because the overpowering Raman and fluorescence signals that arise from the surface can be suppressed, it is feasible to image at depths in the mm range in SORS.
Scanning vs. Fourier Image Formation
2D images showing the spatial distributions of analytes can be acquired in two different ways. In traditional optical microscopy, images are obtained by focusing light onto an object, measuring the field that is scattered or diffracted, and then processing the information to obtain a spatial map of the distribution of components. Alternatively, the sample and source may be rastered relative one another to produce point by point maps. In the first method, Fourier imaging, the entire object is illuminated, and the resultant scattered/transmitted light is collected by a lens system and transferred to a detector. The collection lens system performs a Fourier transform on the characteristic radiation from the sample to extract spatial frequency information from the object after which it undergoes an inverse Fourier transform to form the image.61, 62 This method is maximally efficient, because the collected light contains information from each point on the sample.
In scanning based imaging, light is tightly focused to a small spot on the surface. Spectral information is acquired from each spatial location, and images are constructed by rastering, while recording spectra at each position. After collection, the multispectral data can be reconstructed in various ways to form an image.63 Scanned images usually require longer acquisition times, since the time required is proportional to the number of pixels in the image.
Introduction to Correlated Imaging
Because all individual imaging approaches have natural limitations, correlating information acquired from complementary experiments, such as MS (LDI or SIMS) and vibrational spectroscopy (Raman or IR), has the potential to provide more complete information about complex spatial distributions of chemical and molecular components than that available from either technique in isolation.
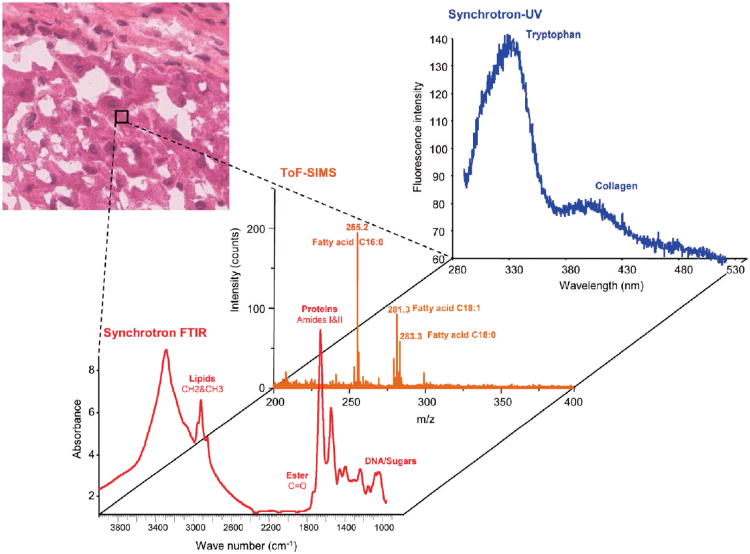
While single-technique chemical imaging is well developed, experiments correlating information from independent imaging approaches are just beginning to appear. For example, synchrotron FTIR and ToF-SIMS microspectroscopies were coupled in a recent study of steatotic liver tissue.64 Images acquired from combining these two techniques showed the distribution of lipids and other tissue components and distinctively revealed differences between normal and steatotic tissue. In another study, images of tissue from a cirrhosis liver were acquired using a multi-modal platform consisting of ToF-SIMS coupled to both Synchrotron FTIR and synchrotron UV- microspectroscopies, viz. Fig. 1.65 In both studies, the images provide multimodal chemical information along with the spatial distribution of cellular components, which holds great promise for early diagnosis. In another report, a multi-modal imaging system, comprised of LDI-MS, Raman and fluorescence microspectroscopies, was used to explore a single-cell algae (Euglena gracilis), yielding detailed information about the internal structure and chemical composition of cells. Images of individual algal cells provided information on the content and distribution of photosynthetic molecules and phospholipids.66 Raman and mass spectrometric imaging have also been applied in environmental/geological research. CRM and SIMS images from the same sample at the same location were successfully acquired in a study of Akilia supracrustal rocks, allowing the presence and physical and molecular composition of apatite-based graphitic inclusions in the rocks to be confirmed.67 In an art conservation application, FTIR, SIMS were combined with x-ray (SEM-EDX) imaging to identify the mineral content and map their spatial distribution on fragments of African wood art.68 Finally, in our laboratories, CRM and SIMS-MS imaging were combined to study processed samples of Miscanthus × gigenteus. These images provide detailed information on the spatial distribution of cell wall components, and correlating Raman and mass spectra from specific spatial locations allow assignment of intracellular globular structures to hemicellulose-rich lignin complexes, an assignment which could not be made definitively from either image alone.17
Figure 1.
Illustration of the combination of multimodal microspectroscopies from a single small region of liver tissue. The spectra represent multimodal data – synchrotron FTIR, TOF-SIMS, and synchrotron UV absorption – from a single region, pixel, of a liver sample. Multimodal spectra such as these presage heterocorrelated images in which every pixel in the image contains multiple spectra spatially registered and optimally scaled for high value added post-processing. Reprinted with permission. Adapted with permission from ref. 65, Copyright 2010 American Chemical Society.
Outline of the Article
The remainder of this article begins by focusing on MS and Raman imaging, identifying their characteristic capabilities and weaknesses. Using these as examples, the generic challenges in multispectral image correlation are described. Finally, opportunities for advancing the state-of-the-art are identified and discussed.
II. Challenges and Opportunities in Mass Spectrometric and Raman Imaging
Challenges in Mass Spectrometric Imaging
MSI is unique in providing comprehensive molecular maps as a basis to study biological systems. In practice, however, a number of challenges present themselves, many relating to the central problem of ionizing and detecting low-abundance analytes in complex chemical environments, which often interfere with measurement.
Sample Preparation
MSI techniques generally impose stringent sample preparation requirements, which must be met while preserving as much of the original spatial chemical information as possible. Flat samples avoid topographically-induced signal artifacts;69 thus tissues are typically frozen, thin-sectioned (<20 μm thickness), and thaw-mounted to a flat substrate for analysis. In the case of vacuum MALDI and SIMS sources, ion extraction prefers a conductive substrate; often a metal plate or indium tin oxide-coated microscope slide, the latter permitting transmission light microscopy as well. The vacuum requirement of conventional MALDI and SIMS presents an additional challenge in high-spatial resolution work, since biological samples with high water content may collapse and disrupt native structure upon vacuum desiccation. Chemical fixation,70, 71 stabilization with glycerol,71, 72 and cryogenic methods such as freeze-drying and frozen-hydrated preparation 52, 73 have all been successfully used to address this issue.
Since MSI is a form of surface analysis, sample preparation may also include steps to uncover subsurface features of interest, especially the case for SIMS, which probes only the top few nanometers of the sample.74 Freeze-fracturing cells is one effective way to accomplish this,75 and in-source manipulation methods offer effective alternatives. Polyatomic “cluster” primary ion beams such as C60 or Ar2000 excel at removing material while causing little sample damage,76, 77 thus serving as effective etching tools to interrogate cell contents.78 Similarly, orthogonal fast ion beam milling can shave off nanometer-scale layers of material between imaging scans.79
Sensitivity and Ion Suppression
MS provides excellent sensitivity, down to zeptomole LODs with optimized sample preparation.45 However, since MSI is necessarily performed in situ to preserve spatiochemical features, optimization is more difficult and cannot include means to simplify a complex mixture – although notably, a liquid microjunction probe does allow separation prior to electrospray ionization.36 As a result, MSI detection limits are decreased in practice due to ion suppression,80 arising from co-desorption of compounds such as inorganic salts or easily-ionized molecules such as glycerophospholipids,81 potentially producing false negatives in MSI. It also complicates quantitation in MSI, since ion intensity may not accurately report analyte concentration in this case. One way to reduce ion suppression is to remove interfering compounds while taking care to leave analytes of interest undisturbed, and various chemical rinses, including ammonium salt solutions,82 water and organic solvent rinses,83 have been devised to accomplish this. Care must be taken to avoid analyte loss or delocalization during rinsing, and additional method optimization, such as pH adjustment, is helpful in some cases.84
LDI benefits greatly from addition of a matrix which assists in desorption and/or ionization of the sample, and this can be true for SIMS as well.21, 27 The best known and most versatile examples are the organic laser-absorbing compounds for MALDI, but a similar effect can be achieved with metal coatings,18 derivatized nanoparticles,85 and nanostructured substrates.86, 87 Selecting the most effective matrix for a given specimen is certainly a challenge, but in MSI it is equally important to optimize extraction (high sensitivity) without significant chemical delocalization (loss of spatial information). Unfortunately these two goals compete; pneumatic spray of organic matrices 72 offers superior sensitivity while drier applications such as sublimation preserve small spatial features by generating a uniform coat of μm-sized crystals.88 Preparation can be improved by separating extraction and crystallization steps,89, 90 but optimizing both sensitivity and spatial resolution remains a challenge.
Spatial Resolution
SIMS is capable of high resolution (<100 nm) chemical imaging with monatomic ion sources, but these typically yield few intact molecular ions. In contrast, polyatomic cluster ion sources, including C60, Bi3, and Au400, greatly enhance molecular ion yields and now rival monatomic sources in focus as well.21, 47, 91, 92 Likewise, laser ablation probes can reach essentially diffraction-limited focus 46, 93 and near-field enhancement approaches can surpass this limit.94 Given the substantial progress in MSI microprobe quality, spatial resolution is now limited largely by the ability to detect sufficient ion counts,95 a challenge related closely to sample preparation and detection limits. Also, increasing spatial resolution leads to longer acquisition times and larger data volumes, so at high resolution, datasets become prohibitively large, unless the imaged region is decreased accordingly.
Chemical Specificity: Mass Accuracy and Resolution
MS is a highly chemically-selective detection platform, but it is not without limits. One such limit is mass accuracy; nominal (integer) or low-accuracy mass assignments leave the identity of the detected ion highly ambiguous, whereas a small mass error (e.g. ±0.0001 Da) provides an accurate mass from which the exact elemental composition of the ion can be inferred.96 The accuracy of mass measurements depends on the performance of the MS analyzer used, the observed mass range, and even sampling conditions. Thus, unambiguous elemental formulae are often not obtainable. Chemical interference, a second limit to the chemical specificity of MS detection, arises when peaks from two or more ions overlap and cannot be distinguished. The consequence of chemical interference in MSI experiments is that an ion filter will sum of the overlapping peaks and display them as a single image, thus concealing chemical complexity. This problem can be mitigated by MS instrumentation, i.e. higher resolution analyzers such as the Orbitrap and the Fourier-transformion cyclotron resonance (FT-ICR) instrumentation provide higher resolution than other analyzers and hence can observe subtle mass differences. “Adequate” mass resolution is difficult to define, since this depends on the masses and mass differences of the analytes of interest, but several recent publications have demonstrated the importance of resolving nearly-isobaric species in MSI experiments, especially among lipids, metabolites, and other small molecules.78, 97 In some cases, several distinct biomolecules are perfectly isobaric, and in this case identification depends on additional steps to separate and identify the ions, such as ion mobility chromatography 98 or tandem MS.99
Data Processing and Analysis
MS images can produce up to hundreds of gigabytes of data, and continuing improvements to effective spatial resolution as well as three-dimensional imaging capability stand to further increase dataset size. Thus, one challenge is to compress the data without losing useful spectral information, and another is to automate data processing to efficiently discern significant spatiochemical features against a rich chemical background.100 Workflows typically involve spectral refinement (smoothing, baseline correction, peak alignment or binning), image refinement (pixel normalization or spatial denoising), and classification of image regions into anatomically- or chemically-distinct regions.101 Classification is achieved by clustering or multivariate approaches, such as principal component analysis, k-means clustering, and maximum autocorrelation factorization,102-104 which allows informative segmentation maps of the analyzed region to be constructed that may reveal distinct chemical regions and colocalization of chemical species. Despite this progress, MS image processing remains complex, time-consuming, computationally intensive, and prone to image artifacts,105 so additional work is needed to build an efficient and reliable pipeline for it.
Challenges in Raman Imaging
Because vibrational Raman spectroscopy provides chemical functional group information, it does not require labeling to generate image contrast. Thus, it non-destructive and requires little or no sample preparation. Raman signals are not affected by water, which scatters only weakly, making it well suited to study biological samples. Performing Raman spectroscopy in a confocal microscope allows image acquisition at high spatial resolution in all three dimensions. Confocal Raman microscopy (CRM), thus, is a tool with applications in both plant106 and biomedical107 research. An overview and comparison of different approaches to vibrational imaging is given in Table 2.
Table 2. Comparison of Vibrational Imaging Techniques.
| Technique | Lateral resolution | Chemical information | Advantages | Limitations | References |
|---|---|---|---|---|---|
| IR | Diffraction limited ∼λ/2 (2.5-25μm) | Molecular | Label free, non-invasive imaging | Low resolution due to long IR wavelengths; Strong interference from water absorption | 155, 156 |
| Raman | Diffraction limited ∼ λ/2 | Molecular | Label free, non-invasive imaging; Raman signals are not affected by water therefore is suitable for biological samples; Better spatial resolution than IR due to the use of shorter wavelengths | Low sensitivity; Long experimental acquisition times; Experiments require high powers that can lead to thermal damage | 63, 106, 107 |
| SERS | Diffraction limited ∼ λ/2 | Molecular | Higher sensitivity leading to faster image acquisition; Metal nanoparticles can quench autofluorescence that is common with biological samples | Poor reproducibility; SERS is substrate dependent therefore not applicable to all samples | 112-114 |
| TERS | Molecular | Higher sensitivity than native Raman and is not limited to a metal substrate; Label free analysis; Sub-diffraction limited spatial resolution due to the size of the probe | Area of enhanced signal is very small | 119, 124 | |
| CARS | Lateral0.40μm-0.30μm axial ∼1.5 μm |
molecular and cellular structural | Label free, non-invasive imaging; High sensitivity compared to native Raman accesses video-rate imaging at modest laser powers; 3-D sectioning capabilities enable depth imaging of thick tissues and cells | High costs of implementing the set-up | 157-159 |
SERS Imaging
While Raman microscopy is rich in information about the composition and spatial distribution of analytes in heterogeneous materials, Raman scattering is an inherently weak process (ca. 1 scattered photon in 109 incident photons), resulting in long imaging times and limiting its application to sample abundant components. Thus, there is strong interest in methods to enhance sensitivity, such as SERS, where the apparent Raman cross-section is enhanced by many orders of magnitude by placing it in close proximity to a nanoparticle or a roughened noble metal surface.108, 109 The signal enhancement is attributed, in part, to the strong electromagnetic fields that are generated upon excitation of local surface plasmon resonances (LSPR). Population-averaged SERS enhancements are typically in the range of 103-106, while individual molecule enhancements as high as 1014 can be achieved under the right conditions, rendering Raman comparable to fluorescence in sensitivity.110 SERS can be exploited in imaging experiments,111 and, being non-invasive, it is attractive for biological and biomedical applications. Over the years SERS imaging has been applied to cancer research studies,107, 112 in vivo imaging studies,113 medical diagnostics,114 bacteria and biofilms110 and biological processes in cells.89 SERS has achieved great success at probing intracellular components and processes, however, SERS has not yet proven capable of probing the nucleus, because the nuclear membrane pores are too small to admit nanoparticles. In addition to its sensitivity advantages, the metallic features responsible for SERS can quench the autofluorescence ubiquitous in biological experiments. However, because SERS intensities vary strongly with nanoscopic details of the molecular environment, it is difficult to make quantitative comparisons of SERS signals between sample sites.115 Another challenge to employing SERS imaging is the need for a metallic substrate. Great effort has been invested in extending SERS beyond metallic nanoparticle substrates.116 One advance is tip-enhanced Raman scattering (TERS),117, 118 in which the Raman signal is enhanced by the intense fields generated at a tip that can be rastered over the sample. TERS experiments combine scanning probe microscopy features with SERS to provide spatial, structural, and chemical information. The major advantage of TERS is that the enhanced Raman signals are confined to a small area immediately surrounding the tip, which is typically much smaller than a diffraction-limited laser focal spot, thus enabling imaging with nm-scale lateral resolution.119, 120 However, the Raman signal in TERS is still weak, primarily because the sampled region (20-50 nm diameter) is small. To circumvent these difficulties, Tian et al. recently developed a novel set of enhancement media based on SiO2-shell-Au/Ag core nanoparticles. These structures show great promise, since they combine the spatial localization available from TERS with the ability to bring the enhancement medium to the sample under a wide range of environmental conditions.121
Spatial Resolution
Spatial resolution in Raman microscopy is primarily determined by the wavelength of light, λ, numerical aperture of the objective, NA = n sin θ, and the refractive index of the medium, n. Abbe theory gives the minimum distance Δx between two adjacent points that can be resolved by a microscope. When the objective and condenser NAs are identical, it is defined by the two-point Rayleigh resolution criterion, 122, 123
| (1) |
Raman imaging is primarily implemented in a scanning confocal configuration, which rejects signal contributions out of the focal plane, resulting in higher axial resolution compared to conventional microscopy. The depth resolution of a confocal microscope is,123
| (2) |
thus being determined by the physical properties of the sample and the efficiency of the collection and imaging optics. In practice, axial resolution is degraded by spherical aberration, and the actual resolution realized is typically less than the theoretical value. Clearly, high NA objectives, imaging in a high index medium, e.g. oil, and employing shorter wavelengths all improve spatial resolution, with best results being on the order of ∼ λ/2 (∼200 nm). Enhanced spatial resolution in Raman imaging can also be obtained by TERS; in a recent communication, a λ/60 resolution was reported in TERS imaging study of carbon nanotubes.124 Near-field scanning optical microscopy and Raman spectroscopy have also been combined to achieve high spatial resolution imaging. For example, 100 nm resolution was reported in a DNA imaging study.125 In addition, standard Raman experiments can be modified by spatial oversampling followed by deconvolution to enhance the spatial resolution.126
Information content
Image formation in CRM entails collecting spectra at a spatial location (pixel) on a sample, which upon processing yields a map of chemical functional groups identified by their characteristic fingerprint vibrations.127-129 Unfortunately, while this is an efficient method to analyze small datasets, it is not suitable for large image datasets having subtle molecular variability within the image. These datasets require much more sophisticated chemometric tools, in order to extract all the information present in the CRM image.
Chemometrics – essentially “the entire process whereby data, e.g. numbers in a table, are transformed into information used for decision making,”130 – is a powerful adjunct in Raman imaging, since each pixel represents an entire spectrum. Chemometric tools can extract subtle relationships hidden in the complex chemical and physical phenomena represented in spectral datasets.131, 132 Pattern recognition tools, which dominate usage in chemical analysis, can further be grouped into unsupervised and supervised learning approaches. Unsupervised learning techniques, such as principal component analysis (PCA) and hierarchal cluster analysis (HCA), work well for initial analysis of Raman images, as they seek to identify data clustering without a priori conditions.
In PCA, the variation present in a data matrix is decomposed and represented using a small number of factors - the principal components – chosen to expose the underlying basis for the observed behavior. Frequently, PCA is implemented, and the principal component values for a large number of samples are plotted in an n-dimensional space in order to identify common responses. HCA, on the other hand, examines abstract inter-point distances between samples and represents that information in a two-dimensional dendrogram, in which clusters of data can be identified by eye. The dendrograms are created through an iterative process of sample-specific cluster joining, which is repeated until only one cluster remains. The distances between clusters give information on variations in the data, thereby identifying those data subsets that are most alike, i.e. are clustered. PCA has become a staple in Raman spectroscopy and imaging generally,131, 133-135 while HCA is a common approach used in biological Raman imaging.136-138
Supervised learning approaches are designed to construct models, which are used to classify samples. Unlike unsupervised models, these approaches excel when there is significant a priori information about the sample. A set of known samples, the training set, is used to establish the number of classes and how the different classes are distinguished from each other. Validation diagnostics are critical in order to assess the reliability and quality of the model and the sensitivity to the various parameters within the model. Sample tools are designed to determine the relationship between different samples and identify any unusual samples, while the variable tools are used to also determine the relationship between the different variables and identify any outlying variables.130 Two frequently used examples of supervised learning techniques are K-Nearest neighbor (KNN) and Soft Independent Modeling of Class Analogies (SIMCA). In KNN, the class of the unknown sample is considered to be the class of the samples that are found nearest to it in multi-dimensional space.130 KNN's ability to identify samples is a powerful tool that has been used in the identification of counterfeit drugs.139 SIMCA, models are designed based on the shape and position of an object formed by the samples within an abstract row space to define the class. PCA is used for modeling the object formed by an individual class. Classes are represented in multi-dimensional space, and samples are classified by determining the spatial region in which the samples belong.
III. Correlating MSI and CRM
Preliminaries – Sample Requirements
Additional challenges arise when correlating two spectral imaging modes; we will illustrate the generic problems by considering the specific issues raised when CRM and MSI are combined. These begin with sample preparation, where fortunately the requirements are not mutually exclusive. CRM and MSI both perform best with thin (<20 μm), flat specimens; tissue samples must be sectioned and mounted to a substrate for analysis. Glass microscope slides are convenient, since they allow additional optical microscopy to be performed, e.g. to map the specimen morphologically with stains, afterwards. Because MALDI and SIMS ion sources both depend on a uniform electric field for ion extraction, conductive indium tin oxide (ITO)-coated slides are typically used, and fortuitously ITO does not interfere with Raman measurements.140 Since the samples are introduced to vacuum for MS analysis, biological specimens must be fixed and dried or otherwise stabilized, e.g. by frozen hydration or glycerol addition, beforehand. Chemical fixation methods, such as ethanol or formaldehyde treatment, are Raman-compatible for some samples, although care must be exercised, especially for protein components where chemical cross-linking can affect α-helix and β-sheet specific vibrations.141 MSI may benefit from additional chemical treatments depending on the experiment. Typically these involve either chemical washes to reduce ion suppression or chemical coatings, e.g. organic matrices, metal plasma, or nanoparticles, to enhance sensitivity. Many such treatments have yet to be tested for compatibility with Raman imaging, though we have previously shown that in some cases they are not only compatible but in fact mutually beneficial.140
General Technical Challenges
To fully exploit the possibilities inherent in correlated imaging, a number of technical challenges must be addressed. These stem from the fact that the experiments are performed sequentially and because experimental capabilities and conditions differ between MSI and CRM. Three major experimental concerns for the particular case of MS-Raman imaging are spatial registry, sample integrity, and dynamic range differences.
Digital image correlation142 is typically applied to pairs of images before and after application of a small perturbation, thus yielding information about the differential response to the perturbation. When the perturbation is in the temporal domain, raster-scan image correlation spectroscopy can be used to produce image data in which each differential pixel represents information offset in time by the raster period.143 Of more utility are ideas borrowed from generalized image cross-correlation spectroscopy (ICCS).144 In ICCS, fluctuations between two differently-labeled species are correlated in both space and time using the generalized correlation function, , where δIa,b are the intensity fluctuations corresponding to labels a and b, which might be mass and vibrational frequency in MS-CRM correlations. Of course, ICCS is optimally applied only when the labels can be observed simultaneously under exactly the same conditions, a requirement which clearly cannot be met in the MS-Raman experiment. To address this problem, Todd and coworkers developed a semiautomated analytical image correlation approach and specifically addressed the correlation of optical (i.e. morphological) and SIMS data in images presenting both regular and irregular features, differentially tagged according to their chemical (atomic) composition.145 They addressed both the image registration problem, by devising a relative positioning scheme, and the differential (between SIMS and optical refraction) sensitivity issue to produce a semi-automated system to identify complementary optical and MS image features.
In order to implement these approaches to image correlation, spatial registry must be achieved between the two image acquisition modes. A method that is adaptable to both MSI and CRM identifies the exact regions of interest (ROIs) on the sample,17 and a structural landmark can be used to achieve spatial registry. In our previous work on lignocellulosic materials, spatial landmarks were established with a bright field microscopy image, and based on the optical image, a grid of 50-100 μm pitch (typical) for LDI-MS was defined and used as a fiducial reference to establish a small number (ROIs) for study by SIMS and CRM imaging.
While great emphasis is placed on the spatial registry, it is just as important to ensure that the orientation of the sample is maintained to minimize scaling issues arising from sample rotation. This is an issue that can be solved by an index notation along with the spatial registry feature to ensure the sample is placed on the same scale while imaging using both techniques. While the obvious concern from an imaging perspective would be that the images would not be identical, rotation has the ability to produce results that produce apparent chemical differences based on sample orientation. An example of this arises in Raman experiments with functional groups presenting signals that are excitation laser polarization-dependent.146 Finally, different techniques invariably produce images with different characteristic pixel sizes, leading to image dilation uncertainties. For images obtained with comparable, but not equal, pixel sizes, such as are obtained by SIMS and CRM, the image pattern classification scheme described below offers the potential to harmonize information. Unfortunately when the pixel sizes are very different, as they are in MALDI-MSI and CRM, there is little that can be done ex post facto. In this case it is better to work to make the pixel sizes more closely commensurate, for example by reducing the focal diameter of the MALDI laser.
Another area of concern is the differing dynamic ranges of the image acquisition techniques. For example, mass spectrometry can be sensitive, especially when compared to unenhanced Raman experiments. As a result, data acquired from MS and native Raman can present significant variations in feature acquisition, rendering correlation challenging. Features which may lie in the middle of the dynamic range of one technique might not be detectable by the other. In the MSI-CRM example, increasing the sensitivity of CRM, for example by using SERS, could alleviate the problem, but thorough cross-validation experiments with painstakingly fabricated serially diluted reference standards would be needed.
Sequential vs. Parallel Imaging
Heterocorrelated imaging is, of necessity, nearly always sequential, which raises the problem of sample registration and regions of interest (ROIs). Accurately locating and addressing the ROI for the downstream imaging experiment (MSI, in our case) can be achieved through the use of mutually-detectable fiducial marks in the specimen or coordinate landmarks printed on the substrate, as described above.147 Any misalignment of ROIs between the two imaging systems must be corrected post-acquisition, and of course misidentification essentially dictates re-imaging. Thus, although sequential imaging is the straightforward approach, in the absence of highly specialized instrumentation it introduces extra challenges and potential sources of error, which must be addressed.
Parallel image acquisition with a hybrid Raman/mass spectrometer is another solution to address the issues inherent in sequential imaging. In fact, some of the challenges associated with incorporating a confocal Raman microscope (CRM) into a mass spectrometer have already been addressed, at least indirectly. Methods for delivering focused laser radiation into the source for LDI while minimally perturbing ion extraction optics critical to MS sensitivity have been developed, the relevant result being that two geometries – transmission 46 and reflection,93 both orthogonal to the surface – allow nearly diffraction-limited focus on a sample surface. The high-numerical aperture objective lens required for these configurations is also suitable for both delivering the excitation laser and also efficiently collecting Raman scattered light from a sample, thus, a single lens with proper transmission and aberration characteristics could fulfill all three roles.
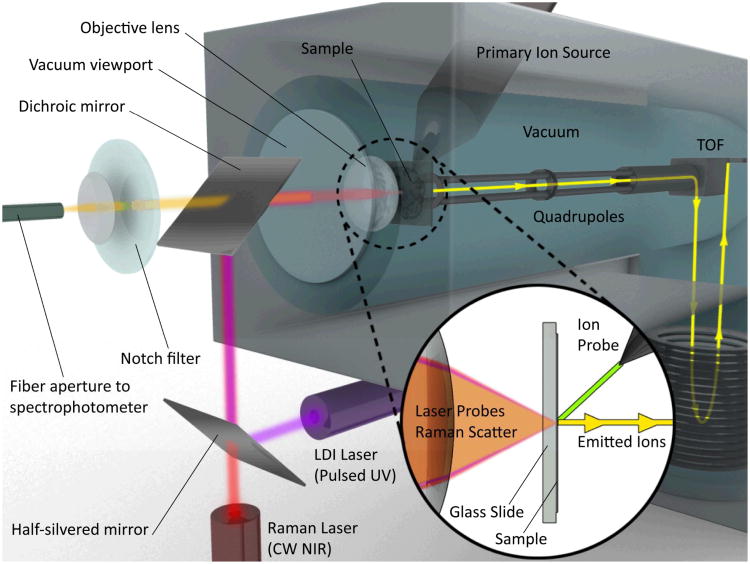
One possible configuration for such a transmission-mode hybrid CRM/MS instrument is shown in Fig. 2. In this system, MS is performed by either UV LDI or C60 SIMS depending on sample and imaging requirements, e.g. lateral and depth resolution. LDI is accomplished coaxially in transmission-mode through a vacuum window and transparent sample substrate, in a geometry originally demonstrated by Hillencamp et al. 46 and more recently implemented by the Caprioli group for tissue imaging.148 C60 primary ions for SIMS are delivered at 45° to the sample surface. Probes are aligned and fixed in position, and imaging is accomplished by rastering the sample on an X/Y translational stage with sub-μm precision in either continuous motion or discrete point mode, depending on sensitivity and time requirements. Ions generated from either source are extracted with a low voltage electric field into a (QTOF) system capable of tandem MS with CID, similar to the dual-source QTOF design reported by Carado et al.149 In a novel extension of this previous work, CRM can be performed here in backscatter mode using the same objective lens employed to focus the UV LDI laser, allowing both excitation lasers to be focused to <1 μm at the sample. Z-axis piezoelectric devices could then be used to allow confocal access to various depths for 3D imaging.
Figure 2.
Concept schematic of a hybrid CRM- QTOF LDI/C60-SIMS chemical imaging instrument showing one potential arrangement of the sample, optics and mass analyzer. While not truly simultaneous, this instrument provides the ability to probe the sample with both imaging modalities without moving the sample between instruments and hence provides enhanced temporal and spatial registration between the imaging approaches.
Although data acquisition would still be sequential in the sense that the Raman spectrum would be acquired at each pixel before (destructive) probing by MS, properly aligned probes would yield Raman and MS data automatically spatially co-registered at each pixel without the need for physical markers or post-acquisition correction. Previous heterocorrelated imaging involved manual comparison of images, but with such automatically registered CRM/MSI one might think about combining mass and Raman spectra into a single hybrid spectrum for each pixel. This would open new possibilities for statistical analysis and image or sample classification based on the complete coherent assembly of mass spectrometric and light scattering information, potentially a powerful new tool in tissue, cell, or organelle-level profiling. Another advantage offered by such an instrument is that CRM could be used to conduct a relatively rapid (100 msec/pixel) chemical survey scan to identify particularly appealing ROIs before a lengthier (>1 sec/pixel) MSI scan is initiated, supplementing the reflected visible light image typically used for sample positioning within an MS source.
Heterocorrelated Imaging of Lignocellulosic Materials
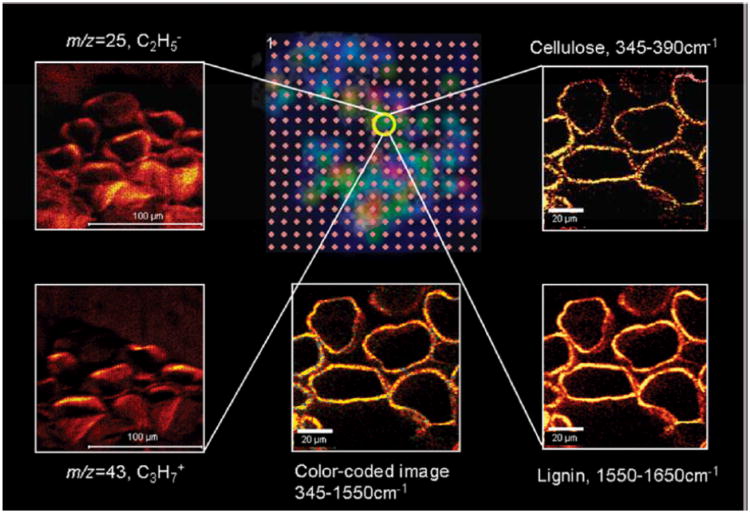
We have recently applied a heterocorrelated chemical imaging approach utilizing CRM and MSI in order to characterize biofuel feedstock at subcellular spatial resolution, as shown in Fig. 3.17Miscanthus × giganteus is a fast-growing grass that generates high mass yield at low cost, so it is an appealing alternative to corn as a biofuel source. However, polysaccharides, cellulose, and hemicellulose must first be freed from the lignin matrix of the cell walls before they can be hydrolyzed and fermented. Thus, elucidating how these molecules are distributed within the plant's cells, and how various chemical treatments extract them, are critical questions for optimizing Miscanthus biofuel processing methods. To accomplish this, LDI, SIMS, and CRM were incorporated into a combined study of processed Miscanthus. After optimizing the methods individually18, 84 in order to visualize lignin and saccharide distributions within cross-sections at μm-scale spatial resolution, LDI, SIMS, and CRM were performed sequentially on a common vascular bundle region of a processed plant. ROIs were identified and registered using fiducial landmarks on the sample, allowing precise alignment of the images. As a result, the compounds of interest could be detected by both Raman scattering, with lignin-related bands at 1607 and 1630 cm−1, and SIMS, with characteristic ions at m/z 95, corresponding to a C6H5OH2+ fragment ions of lignin, for cross-validating chemical images.
Figure 3.
Overview of LDI/SIMS/CRM heterocorrelated imaging applied to lignocellulosic materials. The LDI-MS grid (center top) is color-coded, corresponding to the intensity of m/z = 45 ions obtained by laser desorption-ionization excitation spots on 100 μm centers. The yellow circle highlights the spot where high resolution imaging was performed by both negative (m/z = 25, C2H-, top left) and positive (m/z = 43, C3H7+, bottom left) ion SIMS, as well as CRM, characterized by the cellulose band, 345 – 390 cm−1 (top right), and the lignin band, 1550 – 1650 cm−1 (bottom right). (Bottom center) Composite CRM image combining information from both cellulose (green) and lignin (yellow) bands. Adapted with permission from ref. 17, Copyright 2010 American Chemical Society.
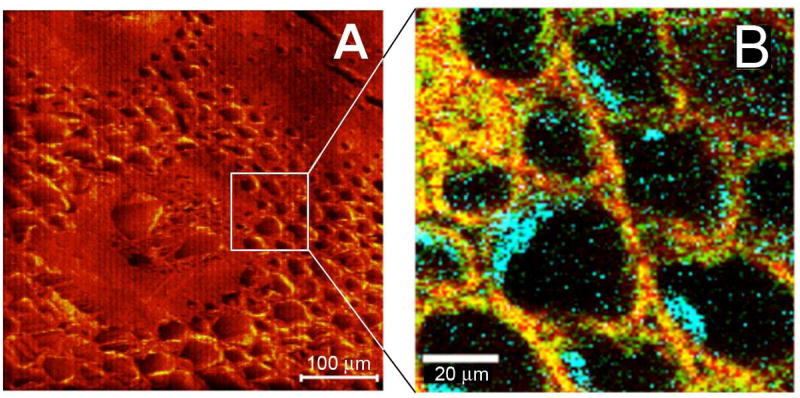
Interestingly, completely new information was available from the correlated imaging experiments. As shown in Fig. 4,17 the processed Miscanthus samples exhibit a globular mass associated with the interior cell walls. Examination of the Raman spectrum in these regions revealed a band at 478 cm−1, characteristic of lignin-hemicellulose complexes. While the cell walls show characteristic cellulose and lignin bands where these components are colocated, Raman bands characteristic of hemicelluloses, another major component of cell walls, are not visible, either because the hemicelluloses in Miscanthus cell walls exhibit intrinsically weak Raman scattering, or because their abundance is low. To probe this further, SIMS images were acquired at the same positions. The distribution maps of fragment ions corresponding to lignin (m/z 95, C6H5OH2+) and cellulose (m/z 105, C4H9O3+) were found to overlap in the cell wall regions, consistent with the CRM images. Detailed analysis of the mass spectra from the globular mass region shows that the intensities of two ions increase significantly: m/z 133 (C5H9O4+), a fragment ion from pentose, and m/z 181 (C6H12O6H+), assigned to either a hexose fragment ion or a pentose cluster ion. Because pentose is the scaffold of hemicellulose, the increased signal intensity of pentose fragment ions from the wall-associated globular structures confirms the tentative assignment made from the CRM image. In this manner, hemicellulose was found to be localized primarily with lignin as globular structures within the cells, and CRM was subsequently used to show that NaOH treatment delignifies cells from the inside first without disturbing the cellulose.106
Figure 4.
Correlation of the negative ion SIMS image (A) and CRM image (B) from the vascular bundle region of processed Miscanthus. Color-coded CRM image; red = lignin, 1550 – 1650 cm−1, green = cellulose, 345 – 390 cm−1, and blue = lignin-hemicellulose (460 – 500 cm−1) complex. Adapted with permission from ref. 17, Copyright 2010 American Chemical Society.
Critical Challenges for the Future
It is clear that combining information acquired from two complementary experiments, such as mass spectrometric imaging (LDI/SIMS) and Raman microscopy, has the potential to provide chemical information that may not be available from either method alone. MS and Raman experiments are performed in different experimental environments and the performance levels of the experiments differ significantly in quantitative analysis. Therefore, realizing the full potential of correlating disparate imaging tools requires effort in two stages. First, the performance of each technique must be optimized in isolation. Then, careful design of an image correlation strategy must take into account sample registration, differences in instrumental operating characteristics, such as dynamic range, spatial resolution, and depth probed, and chemometric strategies for extracting maximal chemical information from the images acquired.
Raman and MS imaging experiments provide complementary information, with Raman experiments providing functional group information, and mass spectrometry giving accurate mass information that can enable identification of molecular ion species. However, Raman imaging can achieve spatial resolution ∼1 μm, while LDI is typically implemented at ≥ 25 μm laser spot sizes, making image cross-correlation difficult. SIMS has better spatial resolution (< 1 μm under optimal conditions), so on the basis of image resolution alone, SIMS is a better imaging partner for CRM. However, SIMS sacrifices access to high mass ions that are so informative in biological samples. In static mode, where MS and Raman images are acquired asynchronously, the image data can be correlated off-line by mask-pattern cross-correlation,125 using, for example, the LDI mass spectrum as a mask for the development of Raman and SIMS cross-correlated images. Considering a mask consisting of Nx × Ny pixels, the cross-correlation can be obtained by moving the mask over the image, calculating the cross-correlation coefficient, Sij, as a function of the spatial position in the image,
| (3) |
To validate these derived images, three formal spaces: the pattern space, P, mask space, M, and classification space, C, must be constructed and analyzed in such a way that all of the mathematical distortions which exist between MS and CRM images can be corrected computationally allowing accurate mathematical cross-correlation. These distortions are (a) dilation (change of scale), (b) rotation and translation, which can potentially be addressed by the landmark registration approach described above, and (c) dynamic range, which can be addressed by varying the relative CRM/MS signal intensities, vide infra. Although one can manually correlate the image features, the capacity to do this over large sample spaces is compromised in complex biological samples. For these more challenging situations, a classification algorithm can be implemented. Identifying the subset, P′, of P which most closely matches the mask, permits a multidimensional classification space to be defined by eqn. (5). Minimizing the classification matrix,
| (4) |
identifies the location of the feature encoded in the mask, M.
Finally, one of the major advantages of MS techniques is the high sensitivity they afford while Raman techniques are greatly hampered by low sensitivity, thus Raman techniques are not efficient tools for the study of systems with low concentration. The weaker sensitivity of Raman scattering compared to MS can be improved by the use of nanoparticle enhanced SERS and the SHINERS technique121 which has been shown to afford superior performance compared regular SERS, but again quantitative comparisons will require careful validation with serially diluted samples.
IV. Conclusions
Correlating information from independent image acquisition platforms constitutes a grand challenge problem in modern chemical analysis. In this article we have considered some of the generic technical challenges associated with correlated imaging and illustrated them with specific reference to MS-Raman correlated imaging. The range of possible operating modes and specific optimization procedures is large, meaning that achieving optimal experimental design is critical. For maximal utilization of multimodal imaging data it is crucial to develop efficient solutions for cross-platform sample registry, to address image distortion effects (dilation, rotation/translation, dynamic range effect) and to implement optimal chemometric strategies for post-acquisition processing. Generically, these issues can be addressed either by fusing disparate image data sets across space and time or by building instruments that allow true simultaneous image acquisition. The effort to do either is significant, yet, it is sure to be handsomely repaid, as the information that can be extracted from correlated images can greatly exceed what it is possible to learn from single imaging tools used in isolation.
Acknowledgments
For work carried out in the authors' laboratories, they acknowledge the Department of Energy Office of Biological and Environmental Research through grant SC0006642 for support of RM and EL and NIH through P30 DA018310 for partial support of the construction of the new hybrid mass spectrometer.
Table of Abbreviations
- AFM
Atomic force microscopy
- CRM
Confocal Raman microscopy
- DESI
Desorption electrospray ionization
- DNA
Deoxyribonucleic acid
- FT-ICR
Fourier-transform ion cyclotron resonance
- FTIR
Fourier-transform infrared
- HCA
Hierarchical cluster analysis
- ICCS
Image cross-correlation spectroscopy
- IR
Infrared
- ITO
Indium tin oxide
- KNN
K-nearest neighbor
- LAESI
Laser-ablation electrospray ionization
- LA-ICP
Laser-ablation inductively coupled plasma ionization
- LDI
Laser desorption/ionization
- LOD
Limit of detection
- LMJ
Liquid microjunction
- LSPR
Local surface plasmon resonances
- MALDI
Matrix-assisted laser desorption/ionization
- MDa
Megadalton(s)
- MS
Mass spectrometry
- MSI
Mass spectrometry imaging
- NA
Numerical aperture
- PCA
Principal component analysis
- PET
Positron emission tomography
- QTOF
Quadrupole time-of-flight
- ROI
Region of interest
- RRS
Resonance Raman spectroscopy
- SEM-EDX
Scanning electron microscopy/energy-dispersive X-ray spectroscopy
- SERS
Surface-enhanced Raman spectroscopy
- SHINERS
Shell-isolated nanoparticle-enhanced Raman spectroscopy
- SIMCA
Soft independent modeling of class analogies
- SIMS
Secondary ion mass spectrometry
- SORS
Spatially offset Raman spectroscopy
- TERS
Tip-enhanced Raman scattering
- ToF
Time-of-flight
- UBM
Ultrasound biomicroscopy
References
- 1.Jackson NB. Visualizing Chemistry: The Progress and Promise of Advanced Chemical Imaging. The National Academies Press; Washington, DC: 2006. [PubMed] [Google Scholar]
- 2.Eigler DM, Schweizer EK. Nature. 1990;344:524–526. [Google Scholar]
- 3.Monroe EB, Jurchen JC, Lee J, Rubakhin SS, Sweedler JV. J Am Chem Soc. 2005;127:12152–12153. doi: 10.1021/ja051223y. [DOI] [PubMed] [Google Scholar]
- 4.Mays KL, Shepson PB, Stirm BH, Karion A, Sweeney C, Gurney KR. Environ Sci Technol. 2009;43:7816–7823. doi: 10.1021/es901326b. [DOI] [PubMed] [Google Scholar]
- 5.Wang LV, Hu S. Science. 2012;335:1458–1462. doi: 10.1126/science.1216210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greco A, Mancini M, Gargiulo S, Gramanzini M, Claudio PP, Brunetti A, Salvatore M. J Biomed Biotechnol. 2012;2012 doi: 10.1155/2012/519238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Squire L. Fundamental neuroscience. Elsevier/Academic Press; 2008. [Google Scholar]
- 8.Leewenhoeck A. Philosophical Transactions. 1684;14:568–574. [Google Scholar]
- 9.Solon EG, Schweitzer A, Stoeckli M, Prideaux B. AAPS Journal. 2010;12:11–26. doi: 10.1208/s12248-009-9158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson TE, Manning HC. J Nucl Med Technol. 2009;37:151–161. doi: 10.2967/jnmt.109.062729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pysz MA, Gambhir SS, Willmann JK. Clin Radiol. 2010;65:500–516. doi: 10.1016/j.crad.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw JE, Epand RF, Epand RM, Li Z, Bittman R, Yip CM. Biophys J. 2006;90:2170–2178. doi: 10.1529/biophysj.105.073510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubakhin SS, Romanova EV, Nemes P, Sweedler JV. Nat Meth. 2011;8:S20–S29. doi: 10.1038/nmeth.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmerman T, Rubakhin S, Sweedler J. J Am Soc Mass Spectrom. 2011;22:828–836. 836. doi: 10.1007/s13361-011-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luxembourg SL, Mize TH, McDonnell LA, Heeren RMA. Analyt Chem. 2004;76:5339–5344. doi: 10.1021/ac049692q. [DOI] [PubMed] [Google Scholar]
- 16.Altelaar AFM, Taban IM, McDonnell LA, Verhaert PDEM, de Lange RPJ, Adan RAH, Mooi WJ, Heeren RMA, Piersma SR. Int J Mass Spectrom. 2007;260:203–211. [Google Scholar]
- 17.Li Z, Chu LQ, Sweedler JV, Bohn PW. Analyt Chem. 2010;82:2608–2611. doi: 10.1021/ac100026r. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Bohn PW, Sweedler JV. Bioresour Technol. 2010;101:5578–5585. doi: 10.1016/j.biortech.2010.01.136. [DOI] [PubMed] [Google Scholar]
- 19.Goto-Inoue N, Hayasaka T, Zaima N, Setou M. BBA Molec Cell Biol Lipids. 2011;1811:961–969. doi: 10.1016/j.bbalip.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Seeley EH, Schwamborn K, Caprioli RM. J Biol Chem. 2011;286:25459–25466. doi: 10.1074/jbc.R111.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heile A, Lipinsky D, Wehbe N, Delcorte A, Bertrand P, Felten A, Houssiau L, Pireaux JJ, De Mondt R, Van Vaeck L, Arlinghaus HF. Appl Surf Sci. 2008;255:941–943. [Google Scholar]
- 22.Vickerman JC. Analyst. 2011;136:2199–2217. doi: 10.1039/c1an00008j. [DOI] [PubMed] [Google Scholar]
- 23.Steinhauser ML, Bailey AP, Senyo SE, Guillermier C, Perlstein TS, Gould AP, Lee RT, Lechene CP. Nature. 2012;481:516–519. doi: 10.1038/nature10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wedlock LE, Wedlock MRK, Cliff JB, Filgueira L, Saunders M, Berners-Price SJ. Metallomics. 2011;3:917–925. doi: 10.1039/c1mt00053e. [DOI] [PubMed] [Google Scholar]
- 25.Passarelli MK, Winograd N. BBA Molec Cell Biol Lipids. 2011;1811:976–990. doi: 10.1016/j.bbalip.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nygren H, Malmberg P. Proteomics. 2010;10:1694–1698. doi: 10.1002/pmic.200900782. [DOI] [PubMed] [Google Scholar]
- 27.McDonnell LA, Piersma SR, Altelaar AFM, Mize TH, Luxembourg SL, Verhaert PDEM, van Minnen J, Heeren RMA. J Mass Spectrom. 2005;40:160–168. doi: 10.1002/jms.735. [DOI] [PubMed] [Google Scholar]
- 28.Girod M, Shi Y, Cheng JX, Cooks RG. J Am Soc Mass Spectrom. 2010;21:1177–1189. doi: 10.1016/j.jasms.2010.03.028. [DOI] [PubMed] [Google Scholar]
- 29.Wu C, Dill AL, Eberlin LS, Cooks RG, Ifa DR. Mass Spectrom Rev. 2012 doi: 10.1002/mas.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu C, Ifa DR, Manicke NE, Cooks RG. Analyst. 2010;135:28–32. doi: 10.1039/b919816d. [DOI] [PubMed] [Google Scholar]
- 31.Wiseman JM, Ifa DR, Zhu Y, Kissinger CB, Manicke NE, Kissinger PT, Cooks RG. Proc Nat Acad Sci. 2008;105:18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kertesz V, Van Berkel GJ. Rapid Commun Mass Spectrom. 2008;22:2639–2644. doi: 10.1002/rcm.3662. [DOI] [PubMed] [Google Scholar]
- 33.Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Analyt Chem. 2011;84:141–148. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Berkel GJ, Kertesz V, Koeplinger KA, Vavrek M, Kong ANT. J Mass Spectrom. 2008;43:500–508. doi: 10.1002/jms.1340. [DOI] [PubMed] [Google Scholar]
- 35.Ovchinnikova OS, Kertesz V, Van Berkel GJ. Rapid Commun Mass Spectrom. 2011;25:3735–3740. doi: 10.1002/rcm.5285. [DOI] [PubMed] [Google Scholar]
- 36.Kertesz V, Van Berkel GJ. Analyt Chem. 2010;82:5917–5921. doi: 10.1021/ac100954p. [DOI] [PubMed] [Google Scholar]
- 37.Nemes P, Vertes A. J Vis Exper. 2010:1–4. doi: 10.3791/2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nemes P, Woods AS, Vertes A. Analyt Chem. 2010;82:982–988. doi: 10.1021/ac902245p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nemes P, Barton AA, Li Y, Vertes A. Analyt Chem. 2008;80:4575–4582. doi: 10.1021/ac8004082. [DOI] [PubMed] [Google Scholar]
- 40.Nemes P, Vertes A. Analyt Chem. 2007;79:8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 41.Wu B, Becker JS. Int J Mass Spectrom. 2011;307:85–91. [Google Scholar]
- 42.Wu B, Becker JS. Int J Mass Spectrom. 2012;323–324:34–40. [Google Scholar]
- 43.Becker JS, Niehren S, Matusch A, Wu B, Hsieh HF, Kumtabtim U, Hamester M, Plaschke-Schlütter A, Salber D. Int J Mass Spectrom. 2010;294:1–6. [Google Scholar]
- 44.Aksenov A, Bier M. J Am Soc Mass Spectrom. 2008;19:219–230. doi: 10.1016/j.jasms.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 45.Sjödahl J, Kempka M, Hermansson K, Thorsén A, Roeraade J. Analyt Chem. 2005;77:827–832. doi: 10.1021/ac0400966. [DOI] [PubMed] [Google Scholar]
- 46.Hillenkamp F, Unsöld E, Kaufmann R, Nitsche R. Nature. 1975;256:119–120. doi: 10.1038/256119a0. [DOI] [PubMed] [Google Scholar]
- 47.Kollmer F, Paul W, Krehl M, Niehuis E. Surf Interf Anal. 2012 in press. [Google Scholar]
- 48.Brison J, Benoit DSW, Muramoto S, Robinson M, Stayton PS, Castner DG. Surf Interf Anal. 2011;43:354–357. doi: 10.1002/sia.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurczy ME, Piehowsky PD, Willingham D, Molyneaux KA, Heien ML, Winograd N, Ewing AG. J Am Soc Mass Spectrom. 2010;21:833–836. doi: 10.1016/j.jasms.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fletcher JS, Lockyer NP, Vaidyanathan S, Vickerman JC. Analyt Chem. 2007;29:2199–2206. doi: 10.1021/ac061370u. [DOI] [PubMed] [Google Scholar]
- 51.Mao D, Wucher A, Brenes DA, Lu C, Winograd N. Analyt Chem. 2012;84:3981–3989. doi: 10.1021/ac2032589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MA, Graham DJ, Castner DG. Analyt Chem. 2012;84:4880–4885. doi: 10.1021/ac300480g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campbell DI, Ferreira CR, Eberlin LS, Cooks RG. Analyt Bioanalyt Chem. 2012:1–10. doi: 10.1007/s00216-012-6173-6. [DOI] [PubMed] [Google Scholar]
- 54.Zhang JI, Talaty N, Costa AB, Xia Y, Tao WA, Bell R, Callahan JH, Cooks RG. Int J Mass Spectrom. 2011;301:37–44. [Google Scholar]
- 55.Dieing T. Confocal Raman Microscopy. Springer; Berlin: 2011. [Google Scholar]
- 56.Nie S, Emory SR. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 57.Mendelsohn R, Chen HC, Rerek ME, Moore DJ. J Biomed Opt. 2003;8:185–190. doi: 10.1117/1.1560645. [DOI] [PubMed] [Google Scholar]
- 58.Klein K, Gigler Alexander M, Aschenbrenner T, Monetti R, Bunk W, Jamitzky F, Morfill G, Stark Robert W, Schlegel J. Biophys J. 2012;102:360–368. doi: 10.1016/j.bpj.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stone N, Faulds K, Graham D, Matousek P. Analyt Chem. 2010;82:3969–3973. doi: 10.1021/ac100039c. [DOI] [PubMed] [Google Scholar]
- 60.Stone N, Kerssens M, Lloyd GR, Faulds K, Graham D, Matousek P. Chemical Science. 2011;2:776–780. [Google Scholar]
- 61.Caprihan A, Fukushima E, Rosato AD, Kos M. Rev Sci Instrum. 1997;68:4217–4220. [Google Scholar]
- 62.Yelleswarapu CS, Kothapalli SR, Rao D. Optics Communications. 2008;281:1876–1888. doi: 10.1016/j.optcom.2007.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schaeberle MD, Morris HR, Turner JF, Treado PJ. Analyt Chem. 1999;71:175A–181A. doi: 10.1021/ac990251u. [DOI] [PubMed] [Google Scholar]
- 64.Le Naour F, Bralet MP, Debois D, Sandt C, Guettier C, Dumas P, Brunelle A, Laprévote O. Plos One. 2009;4:1–10. doi: 10.1371/journal.pone.0007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petit VW, Réfrégiers M, Guettier C, Jamme F, Sebanayakam K, Brunelle A, Laprévote O, Dumas P, Le Naour F. Analyt Chem. 2010;82:3963–3968. doi: 10.1021/ac100581y. [DOI] [PubMed] [Google Scholar]
- 66.Urban PL, Schmid T, Amantonico A, Zenobi R. Analyt Chem. 2011;83:1843–1849. doi: 10.1021/ac102702m. [DOI] [PubMed] [Google Scholar]
- 67.McKeegan KD, Kudryavtsev AB, Schopf JW. Geology. 2007;35:591–594. [Google Scholar]
- 68.Mazel V, Richardin P, Touboul D, Brunelle A, Walter P, Laprévote O. Analyt Chim Acta. 2006;570:34–40. [Google Scholar]
- 69.Lee JLS, Gilmore IS, Seah MP, Levick AP, Shard AG. Surf Interf Anal. 2012;44:238–245. [Google Scholar]
- 70.Malm J, Giannaras D, Riehle MO, Gadegaard N, Sjövall P. Analyt Chem. 2009;81:7197–7205. doi: 10.1021/ac900636v. [DOI] [PubMed] [Google Scholar]
- 71.Tucker KR, Li Z, Rubakhin S, Sweedler JV. J Am Soc Mass Spectrom. 2012 doi: 10.1007/s13361-012-0472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Monroe EB, Annangudi SP, Hatcher NG, Gutstein HB, Rubakhin SS, Sweedler JV. Proteomics. 2008;8:3746–3754. doi: 10.1002/pmic.200800127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chandra S, Bernius MT, Morrison GH. Analyt Chem. 1986;58:493–496. doi: 10.1021/ac00293a053. [DOI] [PubMed] [Google Scholar]
- 74.Ryan KE, Smiley EJ, Winograd N, Garrison BJ. Appl Surf Sci. 2008;255:844–846. [Google Scholar]
- 75.Chandra S, Morrison GH, Wolcott CC. J Microsc. 1986;144:15–37. doi: 10.1111/j.1365-2818.1986.tb04670.x. [DOI] [PubMed] [Google Scholar]
- 76.Fletcher JS, Lockyer NP, Vickerman JC. Surf Interf Anal. 2006;38:1393–1400. [Google Scholar]
- 77.Fletcher JS, Rabbani S, Barber AM, Lockyer NP, Vickerman JC. Surf Interf Anal. 2012 in press. [Google Scholar]
- 78.Rabbani S, Fletcher JS, Lockyer NP, Vickerman JC. Surf Interf Anal. 2011;43:380–384. [Google Scholar]
- 79.Szakal C, Narayan K, Fu J, Lefman J, Subramaniam S. Analyt Chem. 2011;83:1207–1213. doi: 10.1021/ac1030607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jones EA, Lockyer NP, Kordys J, Vickerman JC. J Am Soc Mass Spectrom. 2007;18:1559–1567. doi: 10.1016/j.jasms.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 81.Lay JO, Gidden J, Liyanage R, Emerson B, Durham B. Lipid Technol. 2012;24:36–40. [Google Scholar]
- 82.Angel PM, Spraggins JM, Baldwin HS, Caprioli R. Analyt Chem. 2012;84:1557–1564. doi: 10.1021/ac202383m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lemaire R, Wisztorski M, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. Analyt Chem. 2006;78:7145–7153. doi: 10.1021/ac060565z. [DOI] [PubMed] [Google Scholar]
- 84.Shariatgorji M, Kallback P, Gustavsson L, Schintu N, Svenningsson P, Goodwin RJA, Andren PE. Analyt Chem. 2012;84:4603–4607. doi: 10.1021/ac203322q. [DOI] [PubMed] [Google Scholar]
- 85.Taira S, Sugiura Y, Moritake S, Shimma S, Ichiyanagi Y, Setou M. Analyt Chem. 2008;80:4761–4766. doi: 10.1021/ac800081z. [DOI] [PubMed] [Google Scholar]
- 86.Calavia R, Annanouch FE, Correig X, Yanes O. J Proteom. 2012;75:5061–5068. doi: 10.1016/j.jprot.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 87.Walker BN, Stolee JA, Pickel DL, Retterer ST, Vertes A. J Phys Chem C. 2010;114:4835–4840. [Google Scholar]
- 88.Hankin J, Barkley R, Murphy R. J Am Soc Mass Spectrom. 2007;18:1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang J, Caprioli RM. Analyt Chem. 2011;83:5728–5734. doi: 10.1021/ac200998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Monroe EB, Koszczuk BA, Losh JL, Jurchen JC, Sweedler JV. Int J Mass Spectrom. 2007;260:237–242. [Google Scholar]
- 91.Weibel D, Wong S, Lockyer N, Blenkinsopp P, Hill R, Vickerman JC. Analyt Chem. 2003;75:1754–1764. doi: 10.1021/ac026338o. [DOI] [PubMed] [Google Scholar]
- 92.Dubey M, Brison J, Grainger DW, Castner DG. Surf Interf Anal. 2011;43:261–264. doi: 10.1002/sia.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spengler B, Hubert M. J Am Soc Mass Spectrom. 2002;13:735–748. doi: 10.1016/S1044-0305(02)00376-8. [DOI] [PubMed] [Google Scholar]
- 94.Dutoit B, Zeisel D, Deckert V, Zenobi R. J Phys Chem B. 1997;101:6955–6959. [Google Scholar]
- 95.Piehowski PD, Davey AM, Kurczy ME, Sheets ED, Winograd N, Ewing AG, Heien ML. Analyt Chem. 2009;81:5593–5602. doi: 10.1021/ac901065s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Godfrey A, Brenton A. Analyt Bioanalyt Chem. 2012;404:1159–1164. doi: 10.1007/s00216-012-6136-y. [DOI] [PubMed] [Google Scholar]
- 97.Smith D, Aizikov K, Duursma M, Giskes F, Spaanderman DJ, McDonnell L, O'Connor P, Heeren R. J Am Soc Mass Spectrom. 2011;22:130–137. doi: 10.1007/s13361-010-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xuan Y, Creese AJ, Horner JA, Cooper HJ. Rapid Commun Mass Spectrom. 2009;23:1963–1969. doi: 10.1002/rcm.4101. [DOI] [PubMed] [Google Scholar]
- 99.Bai L, Romanova EV, Sweedler JV. Analyt Chem. 2011;83:2794–2800. doi: 10.1021/ac200142m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Watrous JD, Alexandrov T, Dorrestein PC. J Mass Spectrom. 2011;46:209–222. doi: 10.1002/jms.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jones EA, Deininger SO, Hogendoorn PCW, Deelder AM, McDonnell LA. J Proteom. 2012;75:4962–4989. doi: 10.1016/j.jprot.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 102.Henderson A, Fletcher JS, Vickerman JC. Surf Interf Anal. 2009;41:666–674. [Google Scholar]
- 103.McCombie G, Staab D, Stoeckli M, Knochenmuss R. Analyt Chem. 2005;77:6118–6124. doi: 10.1021/ac051081q. [DOI] [PubMed] [Google Scholar]
- 104.Alexandrov T, Becker M, Deininger SO, Ernst Gn, Wehder L, Grasmair M, von Eggeling F, Thiele H, Maass P. J Proteome Res. 2010;9:6535–6546. doi: 10.1021/pr100734z. [DOI] [PubMed] [Google Scholar]
- 105.Deininger SO, Cornett D, Paape R, Becker M, Pineau C, Rauser S, Walch A, Wolski E. Analyt Bioanalyt Chem. 2011;401:167–181. doi: 10.1007/s00216-011-4929-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chu LQ, Masyuko R, Sweedler JV, Bohn PW. Bioresour Technol. 2010;101:4919–4925. doi: 10.1016/j.biortech.2009.10.096. [DOI] [PubMed] [Google Scholar]
- 107.Schaeberle MD, Kalasinsky VF. Analyt Chem. 1996;68:1829. doi: 10.1021/ac951245a. [DOI] [PubMed] [Google Scholar]
- 108.Jeanmaire DL, Vanduyne RP. J Electroanal Chem. 1977;84:1–20. [Google Scholar]
- 109.Haynes CL, McFarland AD, van Duyne RP. Analyt Chem. 2005;77:338A–346A. [Google Scholar]
- 110.Ivleva NP, Wagner M, Horn H, Niessner R, Haisch C. Analyt Chem. 2008;80:8538–8544. doi: 10.1021/ac801426m. [DOI] [PubMed] [Google Scholar]
- 111.Kneipp K, Kneipp H. Chem Rev. 1999;99:2957. doi: 10.1021/cr980133r. [DOI] [PubMed] [Google Scholar]
- 112.Lee S, Chon H, Lee M, Choo J, Shin SY, Lee YH, Rhyu IJ, Son SW, Oh CH. Biosens Bioelectron. 2009;24:2260–2263. doi: 10.1016/j.bios.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 113.Liu R, Liu JF, Zhou Xx, Jiang GB. Trends Anal Chem. 2011;30:1462–1476. [Google Scholar]
- 114.Vo-Dinh T, Yan F, Wabuyele MB. Surface-Enhanced Raman Scattering: Physics and Applications. 2006;103:409–426. [Google Scholar]
- 115.Hankus ME, Honggang L, Gibson GJ, Cullum BM. Analyt Chem. 2006;78:7535–7546. doi: 10.1021/ac061125a. [DOI] [PubMed] [Google Scholar]
- 116.Zhong-Qun T, Bin R, Jian-Feng L, Zhi-Lin Y. Chem Commun. 2007;2007:3514–3534. [Google Scholar]
- 117.Stockle RM, Suh YD, Deckert V, Zenobi R. Chem Phys Lett. 2000;318:131–136. [Google Scholar]
- 118.Sungho P, Pengxiang Y, Corredor P, Weaver MJ. J Am Chem Soc. 2002;124:2428. doi: 10.1021/ja017406b. [DOI] [PubMed] [Google Scholar]
- 119.Carrier SL, Kownacki CM, Schultz ZD. Chem Commun. 2011;47:2065–2067. doi: 10.1039/c0cc05059h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pettinger B, Picardi G, Schuster R, Ertl G. Single Mol. 2002;3:285–294. [Google Scholar]
- 121.Li JF, Huang YF, Ding Y, Yang ZL, Li SB, Zhou XS, Fan FR, Zhang W, Zhou ZY, Wu DY, Ren B, Wang ZL, Tian ZQ. Nature. 2010;464:392–395. doi: 10.1038/nature08907. [DOI] [PubMed] [Google Scholar]
- 122.Everall NJ. Analyst. 2010;135:2512–2522. doi: 10.1039/c0an00371a. [DOI] [PubMed] [Google Scholar]
- 123.Juang CB, Finzi L, Bustamante CJ. Rev Sci Instrum. 1988;59:2399–2408. [Google Scholar]
- 124.Cancado LG, Hartschuh A, Novotny L. J Raman Spec. 2009;40:1420–1426. [Google Scholar]
- 125.Deckert V, Zeisel D, Zenobi R, Vo-Dinh T. Analyt Chem. 1998;70:2646–2650. doi: 10.1021/ac971304f. [DOI] [PubMed] [Google Scholar]
- 126.Schlucker S, Huffman SW, Levin IW. Biomedical Vibrational Spectroscopy and Biohazard Detection Technologies. 2004;5321:157–163. [Google Scholar]
- 127.Colthup NB, Daly LH, Wiberley SE. Introduction to infrared and Raman spectroscopy. Boston: Academic Press; 1990. [Google Scholar]
- 128.Larkin P. Infrared and Raman spectroscopy: principles and spectral interpretation. Amsterdam; Boston: Elsevier; 2011. [Google Scholar]
- 129.Lin-Vien D. The Handbook of infrared and raman characteristic frequencies of organic molecules. Boston: Academic Press; 1991. [Google Scholar]
- 130.Beebe KR, Pell RJ, Seasholtz MB. Chemometrics: a practical guide. New York: Wiley; 1998. [Google Scholar]
- 131.Shinzawa H, Awa K, Kanematsu W, Ozaki Y. J Raman Spec. 2009;40:1720–1725. [Google Scholar]
- 132.Wei-Qi L, Jian-Hui J, Hai-Feng Y, Ozaki V, Guo-Li S, Ru-Qin Y. Analyt Chem. 2006;78:6003–6011. [Google Scholar]
- 133.Eliasson C, Engelbrektsson J, Lorén A, Abrahamsson J, Abrahamsson K, Josefson M. Chemometr Intell Lab Syst. 2006;81:13–20. [Google Scholar]
- 134.Hayden CA, Morris MD. Appl Spectrosc. 1996;50:708–714. [Google Scholar]
- 135.Sasic S. Appl Spectrosc. 2007;61:239–250. doi: 10.1366/000370207780220769. [DOI] [PubMed] [Google Scholar]
- 136.Kniggendorf AK, Gaul TW, Meinhardt-Wollweber M. Appl Spectrosc. 2011;65:165–173. [Google Scholar]
- 137.Draux F, Gobinet C, Sulé-Suso J, Trussardi A, Manfait M, Jeannesson P, Sockalingum GD. Anal Bioanal Chem. 2010;397:2727–2737. doi: 10.1007/s00216-010-3759-8. [DOI] [PubMed] [Google Scholar]
- 138.Tfayli A, Gobinet C, Vrabie V, Huez R, Manfait M, Piot O. Appl Spectrosc. 2009;63:564–570. doi: 10.1366/000370209788347048. [DOI] [PubMed] [Google Scholar]
- 139.Dégardin K, Roggo Y, Been F, Margot P. Analyt Chim Acta. 2011;705:334–341. doi: 10.1016/j.aca.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 140.Nie B, Masyuko RN, Bohn PW. Analyst. 2012;137:1421–1427. doi: 10.1039/c2an15790j. [DOI] [PubMed] [Google Scholar]
- 141.Kniggendorf AK, Gaul TW, Meinhardt-Wollweber M. Microsc Res Tech. 2011;74:177–183. doi: 10.1002/jemt.20889. [DOI] [PubMed] [Google Scholar]
- 142.Kasper A, Bartsch E, Sillescu H. Langmuir. 1998;14:5004–5010. [Google Scholar]
- 143.Gielen E, Smisdom N, De Clercq B, Vandeven M, Gijsbers R, Debyser Z, Rigo JM, Hofkens J, Engelborghs Y, Ameloot M. J Fluoresc. 2008;18:813–819. doi: 10.1007/s10895-007-0308-8. [DOI] [PubMed] [Google Scholar]
- 144.Nohe A, Keating E, Loh C, Underhill MT, Petersen NO. Faraday Discuss. 2004;126:185–195. doi: 10.1039/b304943d. [DOI] [PubMed] [Google Scholar]
- 145.Schaaff TG, McMahon JM, Todd PJ. Analyt Chem. 2002;74:4361–4369. doi: 10.1021/ac025693b. [DOI] [PubMed] [Google Scholar]
- 146.Fischer S, Schenzel K, Fischer K, Diepenbrock W. Macromolecular Symposia. 2005;223:41–56. [Google Scholar]
- 147.Lau KH, Christlieb M, SchrÖder M, Sheldon H, Harris AL, Grovenor CRM. J Microsc. 2010;240:21–31. doi: 10.1111/j.1365-2818.2010.03380.x. [DOI] [PubMed] [Google Scholar]
- 148.Zavalin A, Todd EM, Rawhouser PD, Yang J, Norris JL, Caprioli RM. J Mass Spectrom. 2012;47:1473–1481. doi: 10.1002/jms.3108. [DOI] [PubMed] [Google Scholar]
- 149.Carado A, Passarelli MK, Kozole J, Wingate JE, Winograd N, Loboda AV. Analyt Chem. 2008;80:7921–7929. doi: 10.1021/ac801712s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kotze HL, Armitage EG, Fletcher JS, Henderson A, Williams KJ, Lockyer NP, Vickerman JC. Surf Interf Anal. 2012 [Google Scholar]
- 151.Piehowski PD, Carado AJ, Kurczy ME, Ostrowski SG, Heien ML, Winograd N, Ewing AG. Analyt Chem. 2008;80:8662–8667. doi: 10.1021/ac801591r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Stöckle R, Setz P, Deckert V, Lippert T, Wokaun A, Zenobi R. Analyt Chem. 2001;73:1399–1402. doi: 10.1021/ac001440b. [DOI] [PubMed] [Google Scholar]
- 153.Coello Y, Jones AD, Gunaratne TC, Dantus M. Analyt Chem. 2010;82:2753–2758. doi: 10.1021/ac9026466. [DOI] [PubMed] [Google Scholar]
- 154.Van Berkel GJ, Kertesz V. Analyt Chem. 2006;78:4938–4944. doi: 10.1021/ac060690a. [DOI] [PubMed] [Google Scholar]
- 155.Offroy M, Roggo Y, Milanfar P, Duponchel L. Analyt Chim Acta. 2010;674:220–226. doi: 10.1016/j.aca.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 156.Colley CS, Kazarian SG, Weinberg PD, Lever MJ. Biopolymers. 2004;74:328–335. doi: 10.1002/bip.20069. [DOI] [PubMed] [Google Scholar]
- 157.Evans CL, Potma EO, Puoris'haag M, Cote D, Lin CP, Xie XS. Proc Natl Acad Sci USA. 2005;102:16807–16812. doi: 10.1073/pnas.0508282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cheng JX, Xie XS. J Phys Chem B. 2004;108:827–840. [Google Scholar]
- 159.Duncan MD, Reintjes J, Manuccia TJ. Opt Lett. 1982;7:350–352. doi: 10.1364/ol.7.000350. [DOI] [PubMed] [Google Scholar]