Abstract
In infectious disease surveillance, public health data such as environmental, hospital, or census data have been extensively explored to create robust models of disease dynamics. However, this information is also subject to its own biases, including latency, high cost, contributor biases, and imprecise resolution. Simultaneously, new technologies, including Internet and mobile phone based tools, now enable information to be garnered directly from individuals at the point of care. Here, we consider how these crowdsourced data offer the opportunity to fill gaps in and augment current epidemiological models. Challenges and methods for overcoming limitations of the data are also reviewed. As more new information sources become mature, incorporating these novel data into epidemiological frameworks will enable us to learn more about infectious disease dynamics.
Keywords: Crowdsourcing, Surveillance, Technology, Bias
Global patterns of disease burden are constantly shifting. Recent studies of the emergence of novel infectious diseases have indicated numerous drivers, including the shift of populations to urban centers, increased mobility, and evolving human–animal interactions [1, 2]. Understanding disease dynamics in populations provides the best opportunity for understanding, controlling, and predicting disease spread. Spatiotemporal models based on public health surveillance data have been extensively explored for this purpose, elucidating patterns and processes by which infectious diseases diffuse across regions. These models traditionally rely on official or government sources, such as environmental, hospital, or census data [3, 4]. Although these data sets are robust and validated and attempt to report on entire populations and their collection is facilitated by intermediaries, they suffer from inherent limits resulting from latency, high cost, contributor biases, and imprecise demographic and geographic resolution [5, 6]. Additionally, studies have indicated areas of deficiency in traditional health systems, including timeliness and financial barriers to care [7].
Simultaneously, new technologies, including Internet tools such as social media or mobile devices, all coupled with global positioning systems, enable a new form of infectious disease information to be garnered directly from citizens. These crowdsourced data evade potentially constraining infrastructure costs and regulations, can be generated in real time, and can be used to fill in gaps in health information due to barriers in health-seeking behaviors through traditional systems [8–10]. Furthermore, these tools can now be deployed at scales that enable information to be garnered at a population level.
Generally, crowdsourcing is the process of obtaining services, ideas, or other information via a large group from the public, rather than a specific set of people (such as government institutions or hospitals). From crisis management to bioinformatics and ecology, information from individuals is providing disparate views and solutions, supplementing existing systems in normal or interrupted use [11–14]. In infectious disease surveillance, crowdsourcing offers the opportunity for collection of symptom and related information right from the point of care [15].
Although considered “gold standards,”, the prerequisite acquisition, aggregation, and validation steps in traditional clinical data sets naturally incur limitations. For example, the United States Centers for Disease Control and Prevention’s (CDC) influenza-like illness (ILI) surveillance system has been the primary metric for measuring national influenza activity. Yet because of differences in laboratory practices and patient populations seen by different providers, comparison of the CDC data between regions and across seasons is not straightforward [16]. Furthermore, temporal trends in the CDC data can be driven by multiple factors that are difficult to disentangle (Fig. 1); during holiday weeks, there could be a higher percentage of ILI visits based on increased disease activity and/or changes in health-seeking behavior, since there are fewer patient visits to sentinel sites overall at these times [17].
Fig. 1.
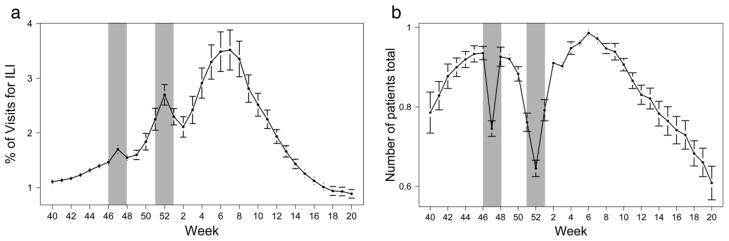
a Average percentage of visits to CDC sentinel sites for ILI by week. b Average number of patients seen at sentinel sites by week. Data are for seasons 2000–2011, pandemic seasons and those with 53 weeks excluded. Holiday weeks (shaded areas: 46–48, Thanksgiving and 51–1, New Years) show both an increase in %ILI visits and a decreased amount of patient visits
On an international scale, the World Health Organization (WHO) field reports of infectious disease outbreaks come from technical institutions and organizations that have the capacity to contribute to international outbreak alert and response. The WHO’s network provides some access to information from affected regions but is limited to organizationally obtained information and their reach [18]. Additionally, the data collection process can be affected by unequal selection whereby larger outbreaks are more likely to be detected, so that estimates of transmissibility may be biased upward [19]. Filling some of these gaps, news media have proven useful, in aggregate, for providing early information of epidemiological value for population-level disease surveillance and have decreased time to outbreak detection substantially [20]. More than 60% of all initial outbreak reports come from unofficial informal sources, such as news media [21]. However, Internet-based news is also subject to distinct limitations based on credibility, detection speed, reach to isolated populations, and geographic coverage of areas where media are restricted or limited. Figure 2 demonstrates the differences in these data sources, illustrating HealthMap [22] disease alerts by continent from 2006 to 2009, in contrast to WHO disease reports for the same time period. These pervasive limitations of current data sources hinder our understanding of disease dynamics. For instance, seasonality of infection risk in malaria is poorly understood [23], and domestically, we have weak understanding of temporal and spatial variation in influenza incidence as described above.
Fig. 2.
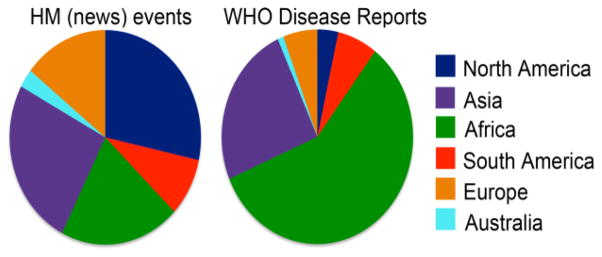
Disease events by continent via news reports 2006–2009, compared with WHO disease reports for the same time period
Crowdsourcing offers a real-time picture of disease by harnessing information as individuals are diagnosed or even before [8, 24]. These temporal advantages are especially vital since increased ease of mobility decreases the time for infectious diseases to spread globally to the scales of hours or minutes, much quicker than even the serial interval of many diseases [25]. Additionally, these tools can spatially augment information in places that current surveillance sites do not cover [9, 26]. Another benefit of working directly with the public is that it augments engagement and enables individuals to become more aware of and involved in their own health, as anecdotal evidence has shown [10]. Thus, this approach can provide an avenue for targeted health education and rapidly measuring responses to public health interventions. Finally, through crowdsourcing infectious disease information, we can learn about aspects of disease dynamics that are not accessible through traditional data, such as contact patterns and aspects of the social environment [27, 28].
Simultaneously, crowdsourced data present their own challenges. There are issues of validation, which current studies are addressing by bringing reported data together with diagnostic or other clinical measures, such as emergency room crowding [29]. Additionally, low specificity, 1– P(false alarm), can result from confounding factors such as media events [9, 30] or demographic biases [31, 32]. Although more work is needed, some studies have uncovered demographic or temporal factors shaping use of the tools [30–32].
Every data source includes biases and challenges that must be robustly understood before the data can be used to study disease dynamics. Further studies of crowdsourced data should continue to focus on addressing issues of population representativeness, reporting bias, and validation in order to demonstrate how the data can be used as a complement to existing epidemiological sources. As crowdsourcing data types and sources become more ubiquitous, we expect these data to serve as a vital component of global disease surveillance efforts.
Acknowledgments
Research reported in this publication was supported by grants from the National Library of Medicine of the National Institutes of Health under Award Numbers G08 LM009776, and R01 LM010812 and Google.org.
Footnotes
Compliance with Ethics Guidelines
Conflict of Interest Rumi Chunara, Mark S. Smolinski, and John S. Brownstein declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Morse SS, Mazet JA, Woolhouse M, Parrish CR, Carroll D, Karesh WB, et al. Prediction and prevention of the next pandemic zoonosis. The Lancet. 2012;380(9857):1956–65. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogich TL, Chunara R, Scales D, Chan E, Pinheiro LC, Chmura AA, et al. Preventing pandemics via international development: a systems approach. PLoS medicine. 2012;9(12):e1001354. doi: 10.1371/journal.pmed.1001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hay SI, Tatem AJ, Graham AJ, Goetz SJ, Rogers DJ. Global environmental data for mapping infectious disease distribution. Adv Parasitol. 2006;62:37–77. doi: 10.1016/S0065-308X(05)62002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reis BY, Mandl KD. Time series modeling for syndromic surveillance. BMC Med Inform Decis Mak. 2003;3 doi: 10.1186/1472-6947-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tatem AJ, Riley S. Effect of poor census data on population maps. Science. 2007;318(5847):43. doi: 10.1126/science.318.5847.43a. author reply. [DOI] [PubMed] [Google Scholar]
- 6.Tuite AR, Tien J, Eisenberg M, Earn DJ, Ma J, Fisman DN. Cholera epidemic in Haiti, 2010: using a transmission model to explain spatial spread of disease and identify optimal control interventions. Ann Intern Med. 2011;154(9):593–601. doi: 10.7326/0003-4819-154-9-201105030-00334. [DOI] [PubMed] [Google Scholar]
- 7.Basu S, Andrews J, Kishore S, Panjabi R, Stuckler D. Comparative Performance of Private and Public Healthcare Systems in Low- and Middle-Income Countries: A Systematic Review. PLoS Med. 2012;9(6):e1001244. doi: 10.1371/journal.pmed.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, Brilliant L. Detecting influenza epidemics using search engine query data. Nature. 2009;457(7232):1012–4. doi: 10.1038/nature07634. [DOI] [PubMed] [Google Scholar]
- 9.Chunara R, Andrews J, Brownstein J. Social and News Media Enable Estimation of Epidemiological Patterns Early in the 2010 Haitian Cholera Outbreak. American Journal of Tropical Medicine and Hygiene. 2011;86:39–45. doi: 10.4269/ajtmh.2012.11-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chunara R, Chhaya V, Bane S, Mekaru S, Chan E, Freifeld C, et al. Online reporting for malaria surveillance using micro-monetary incentives, in urban India 2010–2011. Malaria Journal. 2012;11(43) doi: 10.1186/1475-2875-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakhani KR, Boudreau KJ, Loh P-R, Backstrom L, Baldwin C, Lonstein E, et al. Prize-based contests can provide solutions to computational biology problems. Nature biotechnology. 2013;31(2):108–11. doi: 10.1038/nbt.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson DP, Cobb J, Korpela E, Lebofsky M, Werthimer D. SETI@ home: an experiment in public-resource computing. Communications of the ACM. 2002;45(11):56–61. [Google Scholar]
- 13.Meymaris K, Henderson S, Alaback P, Havens K, editors. AGU Fall Meeting Abstracts. 2008. Project BudBurst: Citizen Science for All Seasons. [Google Scholar]
- 14.Bengtsson L, Lu X, Thorson A, Garfield R, von Schreeb J. Improved response to disasters and outbreaks by tracking population movements with mobile phone network data: a post-earthquake geospatial study in Haiti. PLoS medicine. 2011;8(8):e1001083. doi: 10.1371/journal.pmed.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chunara R, Freifeld CC, Brownstein JS. New technologies for reporting real-time emergent infections. Parasitology. 2012;1(1):1–9. doi: 10.1017/S0031182012000923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Centers for Disease Control and Prevention. [Accessed March 13, 2012.];FluView. Available from: gis.cdc.gov/grasp/fluview/fluportaldashboard.html.
- 17.Copeland KR, Allen AE, editors. Proceedings of the Survey Research Methods Section. American Statistical Association; 2005. Basic Models for Mapping Prescription Drug Data. [Google Scholar]
- 18.The World Health Organization. [Accessed March 6, 2013.];Global Outbreak Alert & Response Network. Available from: http://www.who.int/csr/outbreaknetwork/en/%5D.
- 19.Cauchemez S, Epperson S, Biggerstaff M, Swerdlow D, Finelli L, Ferguson NM. Using Routine Surveillance Data to Estimate the Epidemic Potential of Emerging Zoonoses: Application to the Emergence of US Swine Origin Influenza A H3N2v Virus. PLoS Med. 2013;10(3):e1001399. doi: 10.1371/journal.pmed.1001399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan EH, Brewer TF, Madoff LC, Pollack MP, Sonricker AL, Keller M, et al. Global capacity for emerging infectious disease detection. Proc Natl Acad Sci U S A. 2010;107(50):21701–6. doi: 10.1073/pnas.1006219107. Epub 2010 Nov 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The World Health Organization. [Accessed March 6, 2013.];Global Alert and Response: Epidemic intelligence - systematic event detection. Available from: http://www.who.int/csr/alertresponse/epidemicintelligence/en/index.html.
- 22.Freifeld CC, Mandl KD, Reis BY, Brownstein JS. HealthMap: global infectious disease monitoring through automated classification and visualization of Internet media reports. Journal of American Medical Informatics Association. 2008;15(2):150–7. doi: 10.1197/jamia.M2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wesolowski A, Eagle N, Tatem AJ, Smith DL, Noor AM, Snow RW, et al. Quantifying the impact of human mobility on malaria. Science. 2012;338(6104):267–70. doi: 10.1126/science.1223467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tilston NL, Eames KT, Paolotti D, Ealden T, Edmunds WJ. Internet-based surveillance of Influenza-like-illness in the UK during the 2009 H1N1 influenza pandemic. BMC Public Health. 2010;10(1):650. doi: 10.1186/1471-2458-10-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hufnagel L, Brockmann D, Geisel T. Forecast and control of epidemics in a globalized world. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(42):15124–9. doi: 10.1073/pnas.0308344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfe ND, Heneine W, Carr JK, Garcia AD, Shanmugam V, Tamoufe U, et al. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proceedings of the National Academy of Sciences. 2005;102(22):7994–9. doi: 10.1073/pnas.0501734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Read JM, Edmunds WJ, Riley S, Lessler J, Cummings DA. Close encounters of the infectious kind: methods to measure social mixing behaviour. Epidemiol Infect. 2012;140(12):2117–30. doi: 10.1017/S0950268812000842. Epub 2012 Jun 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chunara R, Bouton L, Ayers JW, Brownstein JS. Assessing the online social environment for surveillance of obesity prevalence. 2013 doi: 10.1371/journal.pone.0061373. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dugas AF, Hsieh Y-H, Levin SR, Pines JM, Mareiniss DP, Mohareb A, et al. Google Flu Trends: correlation with emergency department influenza rates and crowding metrics. Clinical infectious diseases. 2012;54(4):463–9. doi: 10.1093/cid/cir883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan EH, Sahai V, Conrad C, Brownstein JS. Using Web Search Query Data to Monitor Dengue Epidemics: A New Model for Neglected Tropical Disease Surveillance. PLoS Negl Trop Dis. 2011;5(5):e1206. doi: 10.1371/journal.pntd.0001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chunara R, Aman S, Smolinski M, Brownstein JS. Flu Near You: An Online Self-reported Influenza Surveillance System in the USA [Google Scholar]
- 32.Wesolowski A, Eagle N, Noor AM, Snow RW, Buckee CO. Heterogeneous Mobile Phone Ownership and Usage Patterns in Kenya. PloS one. 2012;7(4):e35319. doi: 10.1371/journal.pone.0035319. [DOI] [PMC free article] [PubMed] [Google Scholar]


