Abstract
MUC1 is an integral membrane glycoprotein expressed on epithelial and hematopoietic cells with a COOH-terminus (CT) that mediates intracellular signal transduction. To better understand MUC1-dependent signaling, we searched for proteins binding to its CT using the yeast two-hybrid system with the MUC1 CT as bait and a human epithelial cell cDNA library as prey. Of the six positive clones identified, all encoded calcium-modulating cyclophilin ligand (CAML). The MUC1 CT interacted with CAML in transformed yeast cells as revealed by growth on selective media and in situ X-α-galactosidase activity. Binding of the MUC1 CT to CAML in human epithelial cells was confirmed by reciprocal coimmunoprecipitations, confocal microscopy, protein crosslinking, and coupled transcription/translation analyses. By deletion mutagenesis, the NH2-terminus of CAML was responsible for binding to the MUC1 CT. Finally, transfection of cells with plasmids encoding MUC1 and CAML increased intracellular calcium levels compared with cells transfected with either plasmid alone, suggesting a possible biological significance of the MUC1-CAML interaction.
Keywords: Signal transduction, Oncogenesis, Yeast Two-Hybrid
1. Introduction
MUC1 is a membrane-tethered mucin that was originally cloned from human mammary and pancreatic tumor cells (Gendler et al., 1990; Lan et al., 1990). Based upon the protein structure predicted from its genomic organization, MUC1 possesses an NH2-terminal extracellular region, a single pass transmembrane domain, and an intracellular COOH-terminus (CT) (for reviews see Hattrup & Gendler, 2008; Voynow et al., 2006). Its ectodomain is divided into a distal region containing O-glycosylated 20-amino acid tandem repeats (TR) and a juxtamembrane SEA (sperm protein, enterokinase, agrin) domain that contains a unique site of autocatalytic cleavage (Levitin et al., 2005). The MUC1 CT possesses multiple amino acid phosphorylation sites that are located within consensus sequence motifs for signaling kinases and adaptor proteins involved in cell growth and differentiation (Wang et al., 2003; Zrihan-Licht et al., 1994). Indeed, previous studies have implicated a role for MUC1 in cell adhesion, proliferation, apoptosis, and oncogenesis (Chervenak & Illsley, 2000; Li et al., 2003; Quin & McGuckin, 2000; Raina et al., 2004). Some of these effects are thought to be mediated through the activation of signaling pathways and, in particular, MUC1 was demonstrated to initiate a calcium signal after ligation by intercellular adhesion molecule-1 (ICAM-1) (Rahn et al., 2004). The relationship between MUC1 and ICAM-1 is further strengthened by the fact that the expression of both proteins was up-regulated by inflammatory mediators such as TNF-α(Krunkosky et al., 2000; Koga et al., 2007). However, the physiologic function of MUC1 in the context of signal transduction remains to be clarified. In an attempt to further elucidate MUC1 signaling, we searched for proteins binding to its CT region using the yeast two-hybrid system. Of the positive clones encoding proteins that interacted with the MUC1 CT, we identified calcium-modulating cyclophilin ligand (CAML), a cytosolic protein previously recognized for its ability to interact with a variety of membrane receptors involved in signal transduction. This is the first report demonstrating that the MUC1 CT can bind to CAML. The potential role of MUC1-CAML interaction during signaling and oncogenesis is discussed.
2. Materials and methods
2.1. cDNA library construction and yeast two-hybrid screening
mRNA was purified from the ZR-75-1 breast cancer cell line (ATCC, Manassas, VA) using the Oligotex Direct mRNA kit (Qiagen, Valencia, CA) and 100 ng was used to synthesize first-strand cDNA using oligo(dT) as primer. A cDNA library was prepared using the Matchmaker Two-Hybrid System 3 kit (Clontech, Mountain View, CA) by 24 cycles of PCR amplification with specific primers supplied by the vendor. The amplified cDNA was transformed in Saccharomyces cerevisiae strain AH109 together with predigested pGADT7 vector containing a leucine-selectable marker, transformants were plate amplified on synthetic defined dropout medium lacking leucine (SD/-Leu), aliquoted, and stored as a pretransformed yeast library. The 72-amino acid MUC1 CT was cloned by PCR using a MUC1 cDNA template (kindly provided by Dr. Sandra Gendler, Mayo Clinic, Scottsdale, AZ) and ligated into pGBKT7 with EcoR I and BamH I (New England Biolabs, Ipswich, MA). pGBKT7 bait and pGADT7 prey plasmids encode c-Myc and hemagglutinin (HA) epitope-tagged proteins, respectively. The MUC1 CT plasmid was transformed into S. cerevisiae strain Y187 and mated with the pretransformed yeast AH109 cDNA library. Progeny were plated on SD/-Leu/-Trp medium containing 2.0 mg/ml of X-α-gal (5-bromo-4-chloro-3-indolyl-α-galactopyranoside, Clontech) as chromogenic substrate. Growing colonies were replicated on SD/-Ade/-His/-Leu/-Trp medium containing X-α-gal and positive yeast diploids were identified as blue colonies. Colonies were picked and individual plasmids were amplified, purified, and analyzed by restriction mapping of PCR products of insert DNAs. Automated di-deoxy DNA sequencing followed by analysis using the NCBI Blast search program revealed the identity of insert DNA in positive clones.
2.2. Identification of interacting protein domains by yeast two-hybrid mating
The human MUC1 FL (amino acids 1–475) and fragments containing the TR/SEA (1–375), TR (1–260), SEA (261–375), and CT (404–475) regions were cloned by PCR and ligated into pGBKT7 as above. The human CAML FL (amino acids 1–296), NH2-terminal (NT) (1–189) and CT (190–296) were obtained using the CAML construct cloned by yeast two-hybrid system described above and ligated into pGADT7 with EcoR I and BamH I. The MUC1 constructs were transformed into strain Y187 and the CAML constructs were transformed into strain AH109 for small scale matings to identify the interacting domains of the two proteins. Positive yeast diploids were identified by growth on SD/-Ade/-His/-Leu/-Trp medium containing X-α-gal.
2.3. Subcloning, transient transfection, and coimmunoprecipitation (coIP)
cMyc1-tagged MUC1 FL, TR/SEA, TR, SEA, or CT constructs were subcloned into pcDNA3.1 (Invitrogen, Carlsbad, CA) using Xho I and BamH I. HA-tagged CAML FL, NT, and CT constructs were cloned into pcDNA3.1 with EcoR V and Hind III. In some experiments, the cMyc-MUC1 and HA-CAML were subcloned into pEGFP-C1 or pEGFP-N1 (Clontech) to generate constructs with green fluorescent protein (GFP) tags. HEK293 cells (ATCC) at approximately 80% confluence in DMEM, 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) were cotransfected with plasmid DNAs using Lipofectamine (Invitrogen) according to the manufacturer’s instructions. Detergent lysates (1.0 mg) of transfected HEK293 cells or confluent ZR-75-1 cells were prepared and IPed with 1.0 μg of anti-cMyc, anti-HA, anti-MUC1, or anti-CAML Abs (Sigma, St. Louis, MO; Biomeda, Burlingame, CA; Santa Cruz Biotechnology, Santa Cruz, CA) as described (Wang et al., 2003). Precipitated proteins were resolved by SDS-PAGE and analyzed by Western blotting with primary Abs (anti-HA or anti-cMyc Abs for transfected HEK293 cells, or anti-CAML or anti-MUC1 Abs for ZR-75-1 cells) (1/1,000) and horseradish peroxidase-conjugated goat anti-mouse IgG or goat anti-rabbit IgG secondary Abs (1/10,000; KPL, Gaithersburg, MD).
2.4. Laser scanning confocal microscopy
Transfected HEK293 cells were cultured in 8-chamber slides (Nunc, Naperville, IL) precoated with poly-D-lysine, the cells were fixed with 4% paraformaldehyde for 15 min, and blocked with 1% bovine serum albumin and 5% normal goat serum for 30 min at 22°C. The slides were incubated for 1 hr with anti-cMyc or anti-HA Abs (1/100), washed with PBS containing 0.05% Tween 20, incubated for 45 min at 22°C with Alexa Fluor 488-conjugated goat anti-rabbit IgG or Alexa Fluor 594-conjugated goat anti-mouse IgG Abs (1/1,000; Invitrogen), mounted with Vectashield (Vector Laboratories, Burlingame, CA), and visualized using a Zeiss LSM410 confocal microscope (Carl Zeiss, Oberkochen, Germany). Images were recorded under a 63x NA1.4 objective with 3x zoom and analyzed with Adobe Photoshop (Adobe Systems, San Jose, CA).
2.5. Intracellular protein crosslinking
HEK293 cells stably expressing a CD8/MUC1 plasmid containing the CD8 ectodomain and the MUC1 CT (Wang et al., 2003) were transfected with HA-CAML FL, cultured in DMEM-LM supplemented with 10% dialyzed FBS, 4.0 mM photo-leucine, and 2.0 mM photo-methionine (Pierce, Rockford, IL), and exposed to ultraviolet irradiation (365 nm) for 14 minutes. Cell lysates were prepared and crosslinked proteins were analyzed by Western blotting as described above.
2.6. In vitro transcription/translation
Forty μl of the TnT Quick Master Mix (Promega, Madison, WI), 2.0 μl of plasmid DNA (0.5 μg/μl) encoding the PCR amplified MUC1 or CAML genes, and 8.0 μl of nuclease-free H2O were mixed and incubated for 90 min at 30°C. The TnT Master Mix contains all of the components needed for coupled in vitro transcription/translation (RNA polymerase, RNase inhibitor, rabbit reticulocyte lysate, amino acid mixture, and optimized reaction buffer). Synthesis of the individual MUC1 and CAML proteins was verified by Western blotting. Aliquots of the in vitro synthesized MUC1 and CAML proteins were mixed together and analyzed by coIP.
2.7. Intracellular Ca2+ measurements
HEK293 cells were nontransfected or transfected with plasmids encoding the MUC1 FL, CAML FL, or MUC1 plus CAML constructs. Total DNA was kept constant by adding pcDNA3.1 empty vector. At 48 hr post-transfection, the cells were transferred to 96-well plates at a density of 1.25 × 105 cells/well and loaded with 1.0 μM of Fluo-4 AM in loading buffer for 30 min at 37°C as described by the manufacturer (Invitrogen). The cells were washed twice with measurement buffer (loading buffer without albumin) and fluorescence was quantified using a Synergy HT plate reader (BioTek, Winooski, VT) with an excitation wavelength of 485 nm and measuring the emission at 528 nm.
3. Results
3.1. MUC1-CAML interaction by yeast two-hybrid screening
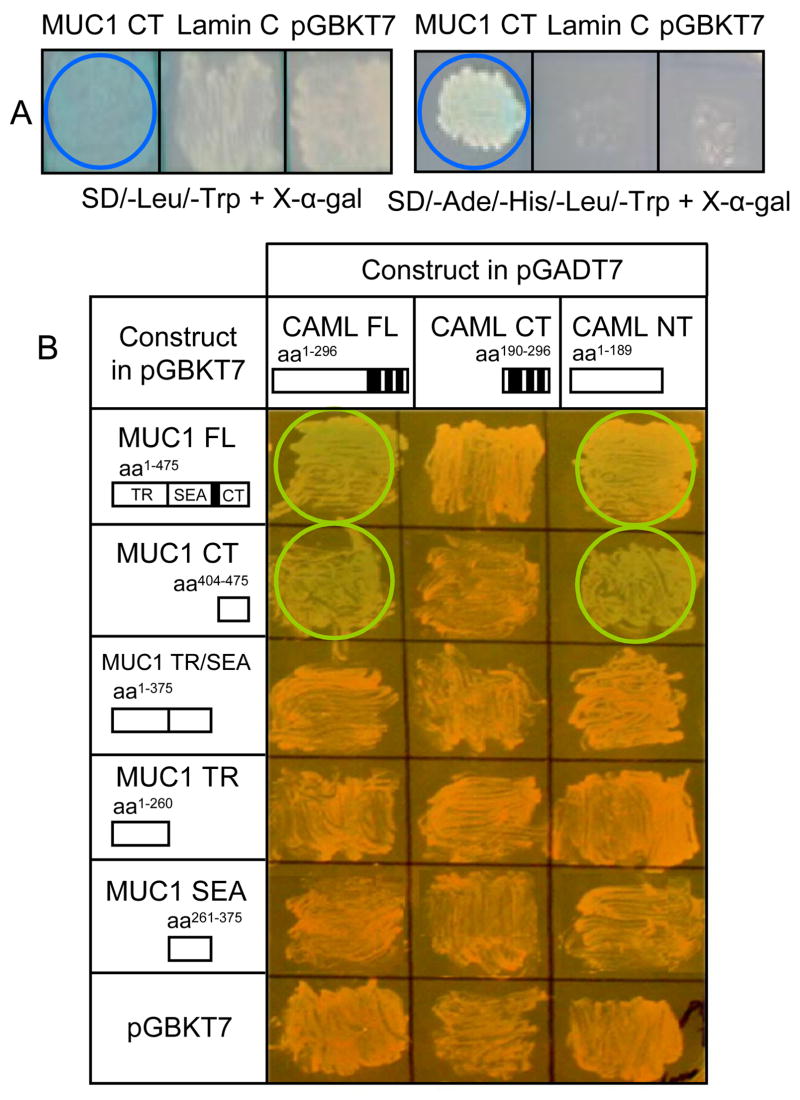
To identify MUC1-interacting proteins that might provide clues to its function, we performed 2 separate yeast two-hybrid screening analyses using a pretransformed human breast cancer cell cDNA library, the first with the entire 475-amino acid MUC1 FL cDNA reported by Gendler et al. (1990) and the second with the 72-amino acid CT (residues 404–475). Six clones were isolated using the MUC1 CT that were verified by repetitive selection as blue colonies on high-stringency quadruple drop-out medium (SD/-Ade/-His/-Leu/-Trp) containing X-α-gal. All of these clones coded for CAML by restriction enzyme mapping and sequence analysis (Supplement Fig. S1). One hundred and ten clones were isolated using the MUC1 FL clone on low-stringency medium (SD/-Leu/-Trp) containing X-α-gal. However, small scale matings revealed that only CAML interacted with both the MUC1 FL and CT regions (Supplement Fig. S2). Therefore, we focused on CAML as a possible binding partner with the MUC1 CT.
The largest CAML cDNA clone was 882 nucleotides in length, of which 849 nucleotides encoded the CAML open reading frame (residues 14–296) and matched the published sequence by 99.5% (Bram & Crabtree, 1994). Interaction of the MUC1 CT with CAML was verified by mating yeast strain Y187 containing the MUC1 CT cDNA in bait plasmid pGBKT7 with strain AH109 containing the CAML FL cDNA in prey plasmid pGADT7 (Fig. 1A). The MUC1 CT interacted with CAML as indicated by growth on high-stringency medium and in situ X-α-galactosidase activity (blue colonies). To confirm that the interaction of MUC1 with CAML was solely due to its CT, and to identify the region of CAML that was required for binding to MUC1, deletion mutants of both proteins were assayed in the yeast two-hybrid system on SD/-Leu/-Trp medium plus X-α-gal. Based in situ X-α-galactosidase activity, the MUC1 CT and CAML NT regions were needed for interaction (Fig. 1B).
Fig. 1.
MUC1-CAML interaction by yeast two-hybrid analysis. (A) Results of matings of yeast strains containing the MUC1 CT and CAML FL constructs on the indicated culture media. Positive interactions are indicated by blue colored circles. An irrelevant cDNA (lamin C) or empty pGBKT7 in place of the MUC1 CT failed to generate positive interaction. (B) Combinations of MUC1 and CAML deletion mutants were analyzed to delineate the domains of each protein required for interaction. Black boxes indicate transmembrane regions. Positive interactions are indicated by colored circles.
3.2. MUC1-CAML interaction by coIP
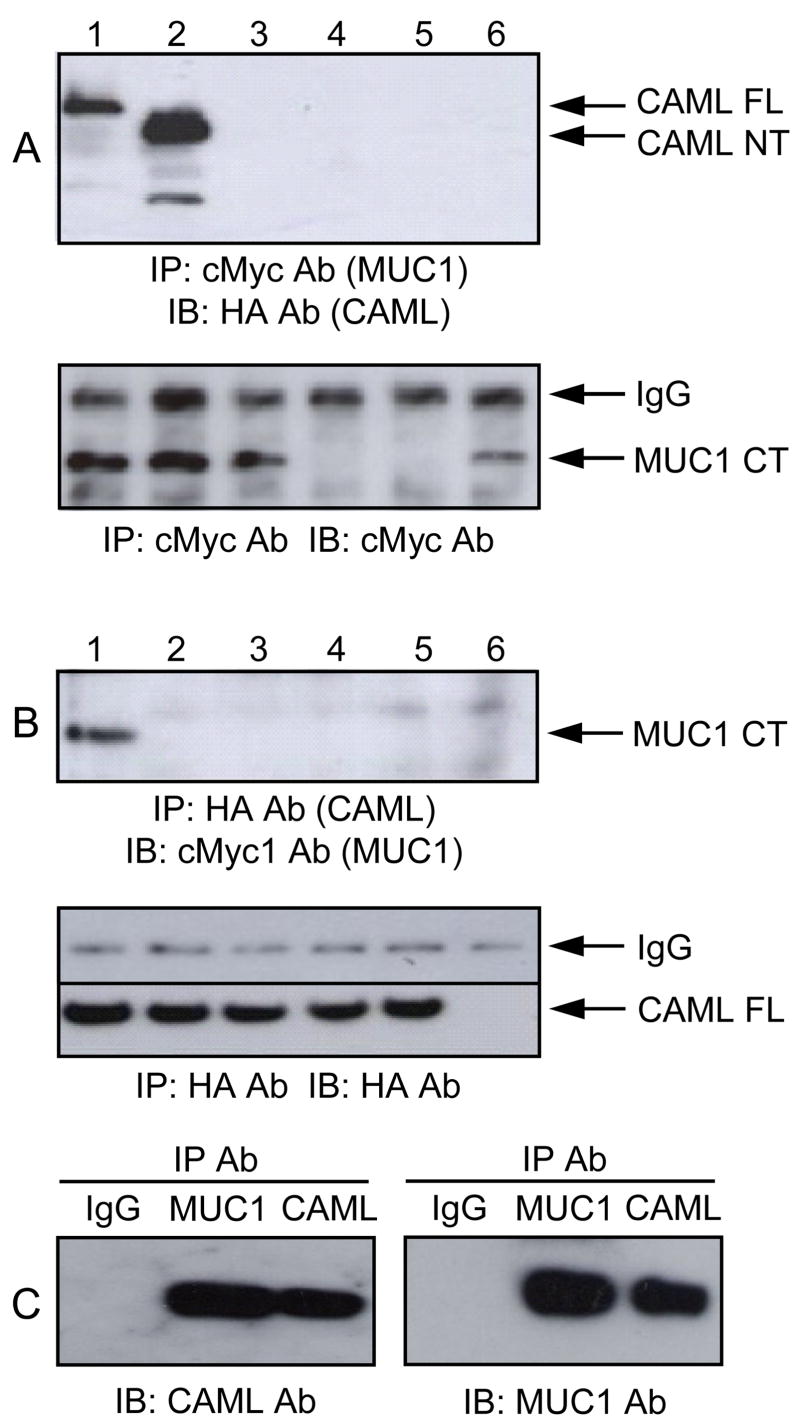
To confirm the results of the yeast two-hybrid analysis, we examined the association between MUC1 and CAML in HEK293 cells that do not endogenously express MUC1 or CAML proteins detectable by Western blotting. Mammalian expression plasmids coding for FL and deletion mutants of both proteins (see Fig. 1B) were generated containing epitope tags. These included MUC1 constructs with cMyc and GFP tags, and CAML constructs with HA and GFP tags. HEK293 cells transiently transfected with the individual plasmids expressed all of the constructs based upon GFP fluorescence (Supplement Fig. S3). Next, HEK293 cells were transiently transfected with the cMyc1-MUC1 CT plus the HA-CAML FL, NT, or CT clones and cell lysates were analyzed by coIP. The MUC1 CT coIPed only with the CAML FL and NT constructs (Fig. 2A). As controls, cells transfected with plasmids encoding cMyc-MUC1 CT alone, GFP alone, or empty vector did not reveal MUC1 CT-CAML coIP. In the reciprocal procedure, the CAML FL clone only coIPed with the MUC1 CT, and not constructs containing the MUC1 TR/SEA, TR, or SEA regions (Fig. 2B). Finally, to verify that a similar interaction occurred in cells endogenously expressing MUC1 and CAML, reciprocal coIPs were performed using ZR-75-1 cell lysates and Abs against the coding sequences of the two proteins. As shown in Fig. 2C, binding interaction between MUC1 and CAML was detected in both cases.
Fig. 2.
MUC1-CAML interaction by coIP. (A) HEK293 cells were cotransfected with plasmids encoding (1) cMyc-MUC1 CT + HA-CAML FL, (2) cMyc-MUC1 CT + HA-CAML NT, (3) cMyc-MUC1 CT + HA-CAML CT, (4) GFP only, (5) pcDNA empty vector, or (6) cMyc-MUC1 CT alone. Lysates were subjected to anti-cMyc Ab IP and anti-HA Ab IB. (B) HEK293 cells were cotransfected with plasmids encoding (1) HA-CAML FL + cMyc-MUC1 CT, (2) HA-CAML FL + cMyc-MUC1 TR/SEA, (3) HA-CAML FL + cMyc-MUC1 TR, (4) HA-CAML FL + cMyc-MUC1 SEA, (5) HA-CAML FL alone, or (6) pcDNA empty vector. Lysates were subjected to anti-HA Ab IP and anti-cMyc Ab IB. As controls, the lower panels in (A) and (B) illustrate the blots that were stripped and reprobed with the IP Ab. (C) ZR-75-1 cell lysates were IPed with normal IgG, anti-MUC1 Ab, or anti-CAML Ab and subjected to anti-CAML Ab or anti-MUC1 Ab IB.
3.3. MUC1-CAML interaction by confocal microscopy
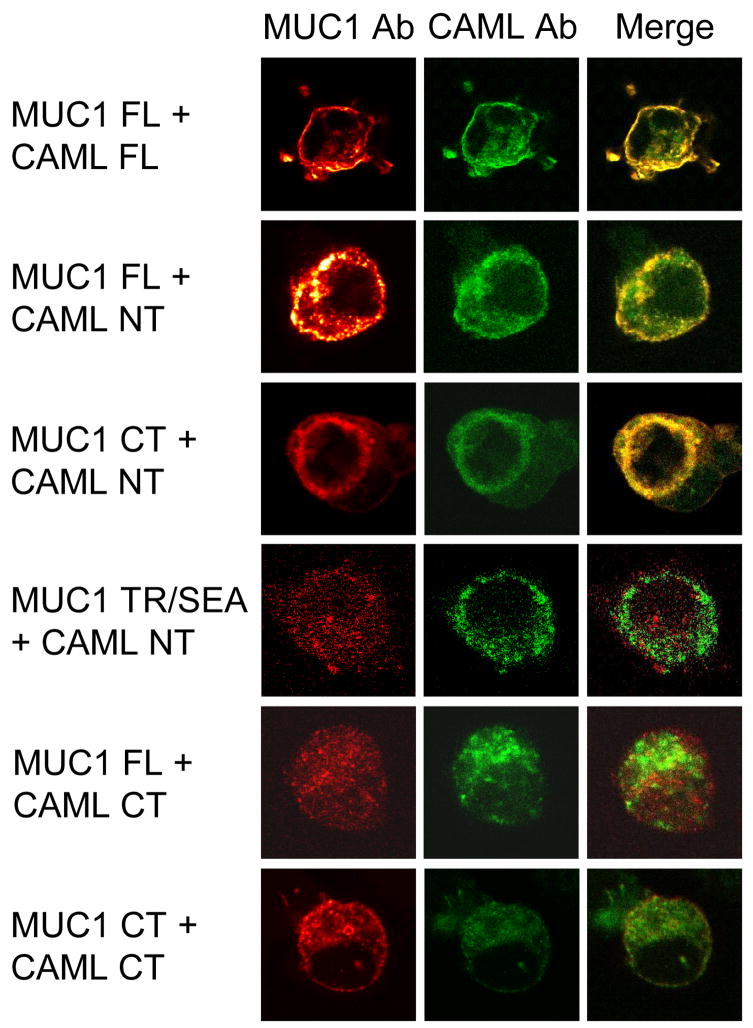
HEK293 cells were cotransfected with the cMyc1-MUC1 FL or deletion mutants lacking the TR/SEA or CT regions plus the HA-CAML FL, NT, or CT constructs, the cells were stained with anti-cMyc and anti-HA Abs plus Alexa Fluor 488- and Alexa Fluor 594-conjugated secondary Abs, and examined by laser scanning confocal microscopy. Colocalization of proteins, as revealed by yellow immunostaining of merged images, was evident only when cells expressed combinations of the MUC1 CT and CAML NT regions (Fig. 3).
Fig. 3.
MUC1-CAML interaction by confocal microscopy. HEK293 cells were cotransfected with plasmids encoding the indicated MUC1 and CAML constructs, stained with MUC1 (red) and CAML (green) Abs, and examined by confocal microscopy.
3.4. MUC1-CAML interaction by intracellular protein crosslinking
Metabolic incorporation of photoreactive amino acid analogs during protein translation followed by UV activation allows for covalent crosslinking within protein-protein interaction domains in their native environment within live cells (Suchanek et al., 2005). CD8/MUC1 CT-HEK293 cells were transfected with HA-CAML FL, the cells were untreated or exposed to UV irradiation, and cell lysates were analyzed by Western blotting with anti-CD8 or anti-HA Abs. (HEK293 cells expressing a CD8 ectodomain/MUC1 CT fusion protein [Wang et al., 2003] were used here due to destruction of the cMyc epitope in cMyc-MUC1 CT by the UV-crosslinking procedure [data not shown].) Following UV exposure, a new band reactive with both Abs was detected at ~70 kDa, the predicted size of a dimeric complex between the crosslinked CD8/MUC1 CT (34 kDa) and HA-CAML FL (37 kDa) monomers (Fig. 4A). As a control HEK293 cells expressing CD8 alone did not exhibit crosslinking with CAML. The upper band in CD8/MUC1 CT-HEK293 lysates reactive with CD8 Ab represents a CT tyrosine phosphorylated isoform while the lower band represent the non-phosphorylated isoform (Meerzaman et al., 2000). Both forms appeared to be crosslinked with CAML.
Fig. 4.
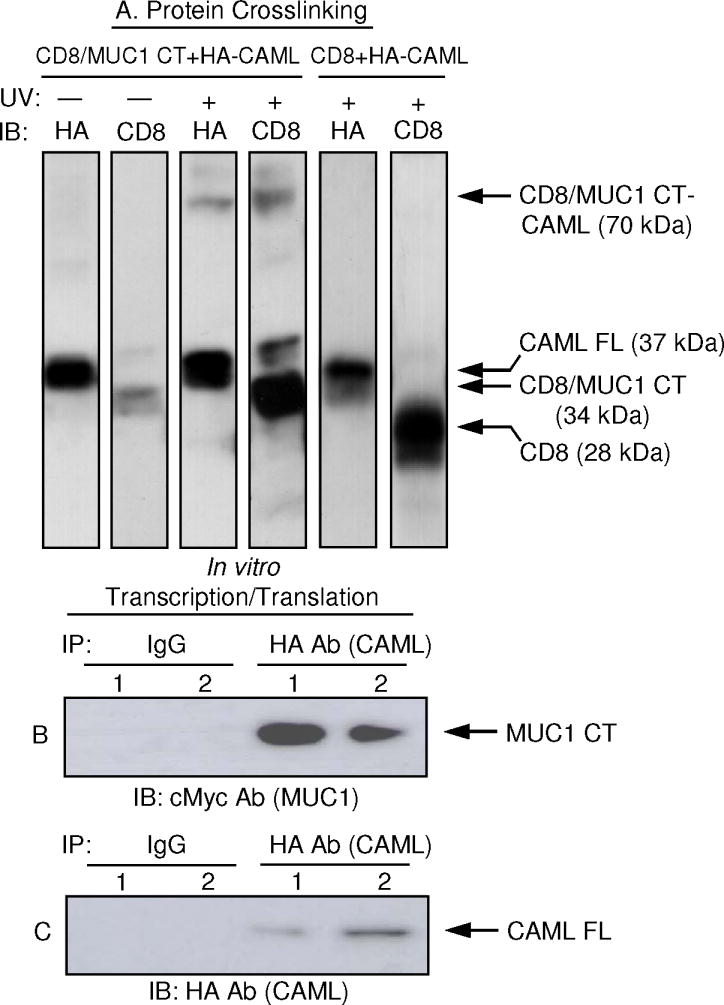
MUC1-CAML interaction by protein crosslinking and coupled in vitro transcription/translation. (A) HEK293 cells stably expressing CD8/MUC1 or CD8 alone were transfected with HA-CAML FL, the cells were metabolically labeled with photo-methionine and photo-leucine, untreated (−) or exposed to UV (+), and cell lysates were analyzed by immunoblotting with CD8 or HA Abs. (B) Lane 1, cMyc-MUC1 CT and HA-CAML FL cDNAs were separately subjected to transcription/translation, the proteins were mixed, and analyzed by coIP as indicated. Lane 2, cMyc-MUC1 CT and HA-CAML FL cDNAs were mixed prior to transcription/translation and analyzed by coIP. (C) The blot in (B) was stripped and reprobed with the IP Ab as a control.
3.5. MUC1-CAML interaction by coupled in vitro transcription/translation
Because the observed size of crosslinked MUC1-CAML was equal to the sum of the individual constituents, the results in Fig. 4A suggested that the MUC1 CT directly interacted with CAML without the involvement of intermediary components. To further test this hypothesis, the cMyc-MUC1 CT and HA-CAML FL cDNAs were amplified by PCR and subjected to coupled transcription/translation to prepare essentially pure recombinant polypeptides in the absence of additional intracellular proteins. The first experiment utilized separate reactions with the MUC1 CT and CAML FL cDNA templates and the individually synthesized proteins were subsequently mixed together and analyzed by coIP. In the second experiment, the two cDNA were simultaneously subjected to transcription/translation and the reaction products directly coIPed. As shown in Fig. 4B, coIP complexes between the MUC1 CT and CAML were detected in both cases.
3.6. Coexpression of MUC1 and CAML increases intracellular Ca2+ levels
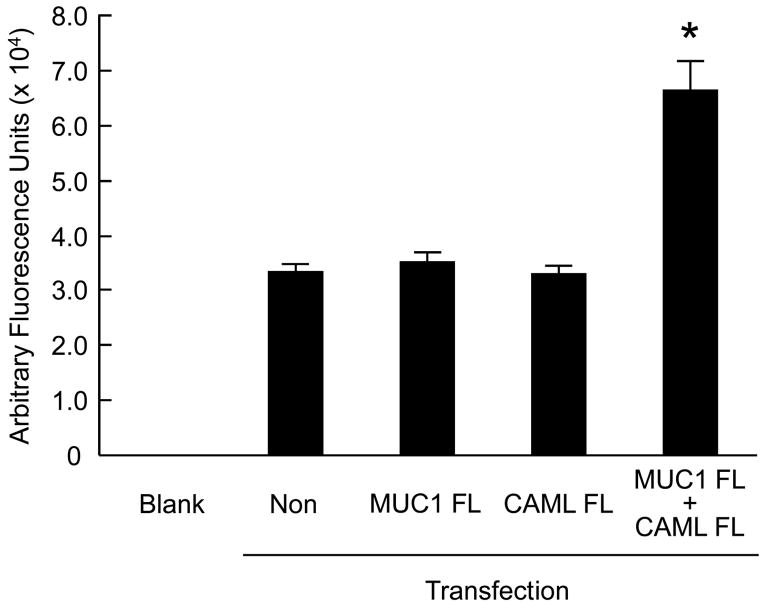
Several reports have demonstrated that interaction of CAML with some of its protein binding partners altered intracellular Ca2+ levels (Bram & Crabtree, 1994; Tovey et al., 2000; Grant et al., 2007). Therefore, HEK293 cells were transiently transfected with plasmids encoding the MUC1 FL or CAML FL constructs, or were cotransfected with MUC1 plus CAML, and intracellular Ca2+ levels were measured using the Fluo-4 fluorescence indicator. As shown in Fig. 5, coexpression of both proteins resulted in approximately 2-fold increased Ca2+ levels compared with cells expressing either protein alone.
Fig. 5.
Coexpression of MUC1 and CAML increases intracellular Ca2+ levels. HEK293 cells were nontransfected (non) or transiently transfected with plasmids encoding MUC1 FL, CAML FL, or MUC1 FL plus CAML FL constructs and intracellular Ca2+ levels were measured using the Fluo-4 fluorescence indicator. Blank refers to fluorescence generated in the absence of added cells (i.e. assay reagents only). Values are means ± SEM (N = 3). *, p < 0.05 comparing MUC1 plus CAML cotransfection with MUC1 or CAML single transfections.
4. Discussion
In this study, we identified CAML as a binding partner of the MUC1 CT. Interaction of the proteins was demonstrated by yeast two-hybrid analysis as revealed by growth on high-stringency selective media and in situ X-α-galactosidase activity. By deletion mutagenesis, the CAML NT was responsible for binding to the MUC1 CT. MUC1-CAML binding was confirmed by reciprocal coIPs in transfected HEK293 cells as well as ZR-75-1 breast cancer cells endogenously expressing both proteins. Intracellular colocalization of the two proteins by confocal microscopy, covalent protein crosslinking in live cells, and coIP of the proteins prepared by coupled in vitro transcription/translation provided convincing evidence for a stable and specific binding interaction between MUC1 and CAML. Finally, coexpression of MUC1 and CAML increased intracellular Ca2+ levels compared with cells expressing either protein alone, suggesting a possible functional significance for this protein-protein interaction.
CAML was originally identified in a yeast two-hybrid screen of human T cells as a protein interacting with cyclophilin B (Bram & Crabtree, 1994). It is a 296-amino acid polypeptide that is located in the endoplasmic reticulum and ubiquitously expressed by most tissues and organs. CAML possesses a hydrophilic NT and a CT with 3 predicted membrane-spanning domains. CAML is therefore predicted to be an integral membrane protein with the majority of the polypeptide on the cytoplasmic face where it may be available to interact with the intracellular regions of membrane-bound receptors. CAML has been shown to interact with an ever-expanding and assorted array of membrane proteins, including transmembrane activator and CAML interactor (TACI), EGFR, ATRAP, fibrocystin, GABAA receptor, Kaposi’s sarcoma-associated herpes virus protein K7, adenovirus E3-6.7K protein, and HIV-1 Vpu protein (Feng et al., 2002; Grant et al., 2007; Guo et al., 2005; Nagano et al., 2005; Tran et al., 2003; Varthakavi et al., 2008; von Bülow & Bram, 1997; Yuan et al., 2008).
In spite of the fact that multiple proteins bind to CAML, amino acid sequence alignments have failed to reveal a common sequence motif that is responsible for this interaction. In two instances, however, CAML-binding domains have been discovered or presumptively identified. In the case of the adenovirus E3-6.7K protein, the critical region was localized between residues 25 and 46 (Grant et al., 2007) and, as pointed out by these investigators, the combined results of von Bülow and Bram (1997) and Xia et al. (2000) suggested that the CAML-binding domain of TACI was located between residues 162 and 212. Our amino acid sequence alignments using the Genestream network server (http://xylian.igh.cnrs.fr/bin/align-guess.cgi) (Pearson et al., 1997) revealed that the COOH-terminal 15 residues of MUC1 (LSYTNPAVAATSANL) share 40.0% and 53.3% sequence homology with the CAML-binding domains of TACI and E3-6.7K, respectively. Interestingly, this region of MUC1 contains a Grb-2 binding motif that has been implicated in activation of the ERK1/2 MAP kinase (Wang et al., 2004). It is tempting to speculate that binding of CAML to this motif may competitively inhibit MUC1 → Grb-2 → ERK1/2 signal transduction.
Whereas CAML has been implicated in binding to multiple types of membrane-bound proteins, its exact function remains unclear. Two proposed functions have received noteworthy attention, stimulation of intracellular Ca2+ signal transduction and control of cell survival. CAML was initially described as a mediator of Ca2+ signaling in T cells in the context of a T cell receptor → NF-AT pathway (Bram & Crabtree, 1994). The interaction between CAML and adenovirus E3-6.7K, a protein that modulates intracellular Ca2+ homeostasis (Grant et al., 2007), and the fact that ATP-induced Ca2+ waves were altered in CAML expressing cells (Tovey et al., 2000), strengthens the validity of this concept. MUC1 mucin also has been shown to play a role in regulating cytosolic Ca2+ levels. Rahn et al. (2004) reported that ICAM-1 is a ligand for the MUC1 ectodomain on the surface of breast cancer cell lines or transfected HEK293 cells, activating outside-in signaling via the MUC1 CT to initiate Ca2+ signaling. The authors speculated that because Ca2+ signaling has been associated with cytoskeletal changes and motility, it is possible MUC1 promotes heterotypic cell-cell adhesion through ICAM-1 followed by a Ca2+-based pro-migratory signal within tumor cells, thus facilitating metastasis. Our demonstration that coexpression of MUC1 plus CAML results in increased intracellular Ca2+ levels compared with cells expressing MUC1 or CAML alone now provides a functional significance for the MUC1-CAML interaction and offers a possible mechanistic explanation to support the previous observations of Rahn et al. (2004).
Ca2+ signaling, possibly mediated through the action of CAML, also plays a role in cell survival. Two CAML-interacting virus proteins, adenovirus E3-6.7K and Kaposi’s sarcoma-virus protein K7, are known to inhibit apoptosis by altering cellular Ca2+ levels (Grant et al., 2007). Furthermore, expression of CAML appears to be an essential mediator of T cell survival (Tran et al., 2005). In B cells, a pro-survival role for the CAML-interacting TACI receptor has been documented. The TNF superfamily ligands, B-cell-activating factor (BAFF) and a proliferation-inducing ligand (APRIL), activate anti-apoptotic pathways, including phosphatidylinositol-3 kinase (PI3K)/Akt, through binding to TACI, BAFF receptor, and B cell maturation protein A (BCMA) and are major B cell survival factors, some of which play an important role in multiple myeloma (Klein et al., 2003). However, it is currently unclear how, if at all, the interaction between TACI and CAML generates pro-survival signals. It is nevertheless intriguing to note that expression of MUC1 also was recently reported to promote the growth and survival of human multiple myeloma cells (Kawano et al., 2008), in line with several studies suggesting that MUC1 exerts an anti-apoptotic effect. In the latter case, over-expression of human MUC1 in MUC1 knockout mice was correlated with decreased apoptosis in the mammary gland (Schroeder et al., 2004). Also, MUC1 expression attenuated DNA damage-induced apoptosis in HCT116 colon carcinoma cells and down-regulation of MUC1 expression in A549 lung and ZR-75-1 breast carcinoma cells by RNA interference was associated with increased sensitivity to genotoxic drugs through a mechanism involving its CT region (Ren et al., 2004). Finally, MUC1 expression in rat 3Y1 fibroblasts activated the anti-apoptotic PI3K/Akt and Bcl-xL pathways (Li et al., 2003).
In summary, we discovered a novel protein interaction between MUC1 and CAML that may be relevant to the proposed roles of this mucin in Ca2+ signaling and/or tumorigenesis. Future studies are needed to elucidate the molecular relationship, if any, between MUC1 and CAML in the context of these cellular processes.
Supplementary Material
Acknowledgments
This work was supported by grants from NIH (R21 ES013483 and R21 AI072291), the Cystic Fibrosis Foundation, the American Lung Association, and the Maryland Cigarette Restitution Fund.
Abbreviations
- Ab
antibody
- CAML
calcium-modulating cyclophilin ligand
- CT
COOH-terminus
- FL
full-length
- GFP
green fluorescent protein
- HA
hemagglutinin
- IB
immunoblot
- ICAM-1
intracellular adhesion molecule-1
- IP
immunoprecipitation
- NT
NH2-terminus
- SEA
sperm protein, enterokinase, agrin
- TACI
transmembrane activator and CAML interactor
- TR
tandem repeat
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bram RJ, Crabtree GR. Calcium signaling in T cells stimulated by a cyclophilin B binding protein. Nature. 1994;371:355–358. doi: 10.1038/371355a0. [DOI] [PubMed] [Google Scholar]
- Chervenak JL, Illsley NP. Episialin acts as an antiadhesive factor in an in vitro model of human endometrial-blastocyst attachment. Biol Reprod. 2000;63:294–300. doi: 10.1095/biolreprod63.1.294. [DOI] [PubMed] [Google Scholar]
- Feng P, Park J, Lee BS, Lee SH, Bram RJ, Jung JU. Kaposi’s sarcoma-associated herpesvirus mitochondrial K7 protein targets a cellular calcium-modulating cyclophilin ligand to modulate intracellular calcium concentration and inhibit apoptosis. J Virol. 2002;76:11491–11504. doi: 10.1128/JVI.76.22.11491-11504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendler SJ, Lancaster CS, Taylor-Papadimitriou J, Duhig T, Peat N, Burchell J, et al. Molecular cloning and expression of human tumor-associated polymorphic epithelial mucin. J Biol Chem. 1990;265:15286–15293. [PubMed] [Google Scholar]
- Grant JR, Moise AR, Jefferies WA. Identification of a novel immunosubversion mechanism mediated by a virologue of the B-lymphocyte receptor TACI. Clin Vaccine Immunol. 2007;14:907–917. doi: 10.1128/CVI.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Lopez-Ilasaca M, Dzau VJ. Identification of calcium-modulating cyclophilin ligand (CAML) as transducer of angiotensin II-mediated nuclear factor of activated T cells (NFAT) activation. J Biol Chem. 2005;280:12536–12541. doi: 10.1074/jbc.M500296200. [DOI] [PubMed] [Google Scholar]
- Hattrup CL, Gendler SJ. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol. 2008;70:431–457. doi: 10.1146/annurev.physiol.70.113006.100659. [DOI] [PubMed] [Google Scholar]
- Kawano T, Ahmad R, Nogi H, Agata N, Anderson K, Kufe D. MUC1 oncoprotein promotes growth and survival of human multiple myeloma cells. Int J Oncol. 2008;33:153–159. [PMC free article] [PubMed] [Google Scholar]
- Klein B, Tarte K, Jourdan M, Mathouk K, Moreaux J, Jourdan E, et al. Survival and proliferation factors of normal and malignant plasma cells. Int J Hematol. 2003;78:106–113. doi: 10.1007/BF02983377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Kuwahara I, Lillehoj EP, Lu W, Miyata T, Isohama Y, et al. TNF-αinduces MUC1 gene transcription in lung epithelial cells: its signaling pathway and biological implication. Am J Physiol Lung Cell Mol Physiol. 2007;293:L693–L701. doi: 10.1152/ajplung.00491.2006. [DOI] [PubMed] [Google Scholar]
- Krunkosky TM, Fischer BM, Martin LD, Jones N, Akley NJ, Adler KB. Effects of TNF-αon expression of ICAM-1 in human airway epithelial cells in vitro. Signaling pathways controlling surface and gene expression. Am J Respir Cell Mol Biol. 2000;22:685–692. doi: 10.1165/ajrcmb.22.6.3925. [DOI] [PubMed] [Google Scholar]
- Lan MS, Batra SK, Qi WN, Metzgar RS, Hollingsworth MA. Cloning and sequencing of a human pancreatic tumor mucin cDNA. J Biol Chem. 1990;265:15294–15299. [PubMed] [Google Scholar]
- Levitin F, Stern O, Weiss M, Gil-Henn C, Ziv R, Prokocimer Z, et al. The MUC1 SEA module is a self-cleaving domain. J Biol Chem. 2005;280:33374–33386. doi: 10.1074/jbc.M506047200. [DOI] [PubMed] [Google Scholar]
- Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22:6107–6110. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerzaman D, Xing PX, Kim KC. Construction and characterization of a chimeric receptor containing the cytoplasmic domain of MUC1 mucin. Am J Physiol Lung Cell Mol Physiol. 2000;278:L625–L629. doi: 10.1152/ajplung.2000.278.3.L625. [DOI] [PubMed] [Google Scholar]
- Nagano J, Kitamura K, Hujer KM, Ward CJ, Bram RJ, Hopfer U, et al. Fibrocystin interacts with CAML, a protein involved in Ca2+ signaling. Biochem Biophys Res Commun. 2005;338:880–889. doi: 10.1016/j.bbrc.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- Quin RJ, McGuckin MA. Phosphorylation of the cytoplasmic domain of the MUC1 mucin correlates with changes in cell-cell adhesion. Int J Cancer. 2000;87:499–506. doi: 10.1002/1097-0215(20000815)87:4<499::aid-ijc6>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Rahn JJ, Shen Q, Mah BK, Hugh JC. MUC1 initiates a calcium signal after ligation by intercellular adhesion molecule-1. J Biol Chem. 2004;279:29386–29390. doi: 10.1074/jbc.C400010200. [DOI] [PubMed] [Google Scholar]
- Raina D, Kharbanda S, Kufe D. The MUC1 oncoprotein activates the anti-apoptotic phosphoinositide 3-kinase/Akt and Bcl-xL pathways in rat 3Y1 fibroblasts. J Biol Chem. 2004;279:20607–2012. doi: 10.1074/jbc.M310538200. [DOI] [PubMed] [Google Scholar]
- Ren J, Agata N, Chen D, Li Y, Yu WH, Huang L, et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JA, Masri AA, Adriance MC, Tessier JC, Kotlarczyk KL, Thompson MC, et al. MUC1 overexpression results in mammary gland tumorigenesis and prolonged alveolar differentiation. Oncogene. 2004;23:5739–5747. doi: 10.1038/sj.onc.1207713. [DOI] [PubMed] [Google Scholar]
- Suchanek M, Radzikowska A, Thiele C. Photo-leucine and photo-methionine allow identification of protein-protein interactions. Nat Methods. 2005;2:261–267. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- Tovey SC, Bootman MD, Lipp P, Berridge MJ, Bram RJ. Calcium-modulating cyclophilin ligand desensitizes hormone-evoked calcium release. Biochem Biophys Res Commun. 2000;276:97–100. doi: 10.1006/bbrc.2000.3442. [DOI] [PubMed] [Google Scholar]
- Tran DD, Edgar CE, Heckman KL, Sutor SL, Huntoon CJ, van Deursen J, et al. CAML is a p56Lck-interacting protein that is required for thymocyte development. Immunity. 2005;23:139–152. doi: 10.1016/j.immuni.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Tran DD, Russell HR, Sutor SL, van Deursen J, Bram RJ. CAML is required for efficient EGF receptor recycling. Dev Cell. 2003;5:245–256. doi: 10.1016/s1534-5807(03)00207-7. [DOI] [PubMed] [Google Scholar]
- Varthakavi V, Heimann-Nichols E, Smith RM, Sun Y, Bram RJ, Ali S, et al. Identification of calcium-modulating cyclophilin ligand as a human host restriction to HIV-1 release overcome by Vpu. Nat Med. 2008;14:641–647. doi: 10.1038/nm1778. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- von Bülow GU, Bram RJ. NF-AT activation induced by a CAML-interacting member of the tumor necrosis factor receptor superfamily. Science. 1997;278:138–141. doi: 10.1126/science.278.5335.138. [DOI] [PubMed] [Google Scholar]
- Voynow JA, Gendler SJ, Rose MC. Regulation of mucin genes in chronic inflammatory airway diseases. Am J Respir Cell Mol Biol. 2006;34:661–665. doi: 10.1165/rcmb.2006-0035SF. [DOI] [PubMed] [Google Scholar]
- Wang H, Lillehoj EP, Kim KC. Identification of four sites of stimulated tyrosine phosphorylation in the MUC1 cytoplasmic tail. Biochem Biophys Res Commun. 2003;310:341–346. doi: 10.1016/j.bbrc.2003.09.030. [DOI] [PubMed] [Google Scholar]
- Wang H, Lillehoj EP, Kim KC. MUC1 tyrosine phosphorylation activates the extracellular signal-regulated kinase. Biochem Biophys Res Commun. 2004;321:348–354. doi: 10.1016/j.bbrc.2004.06.167. [DOI] [PubMed] [Google Scholar]
- Xia XZ, Treanor J, Senaldi G, Khare SD, Boone T, Kelley M, et al. TACI is a TRAF-interacting receptor for TALL-1, a tumor necrosis factor family member involved in B cell regulation. J Exp Med. 2000;192:137–43. doi: 10.1084/jem.192.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Yao J, Norris D, Tran DD, Bram RJ, Chen G, et al. Calcium-modulating cyclophilin ligand regulates membrane trafficking of postsynaptic GABAA receptors. Mol Cell Neurosci. 2008;38:277–289. doi: 10.1016/j.mcn.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrihan-Licht S, Baruch A, Elroy-Stein O, Keydar I, Wreschner DH. Tyrosine phosphorylation of the MUC1 breast cancer membrane protein. Cytokine receptor-like molecule. FEBS Lett. 1994;356:130–136. doi: 10.1016/0014-5793(94)01251-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.