Abstract
Background
Smith-Lemli-Opitz syndrome (SLOS) is a multiple malformation, neurodevelopmental disorder of cholesterol metabolism caused by mutations in 7-dehydrocholesterol reductase (DHCR7). Corpus callosum (CC) malformations and developmental delay are common manifestations of this disorder, but the relationship between the two has not been evaluated. We tested the hypothesis that shorter callosal length and smaller area correlates with higher serum 7-dehydrocholesterol (7DHC) and increased severity of neurodevelopmental delay in a large cohort of SLOS patients.
Methods
Thirty-six individuals with SLOS (18M/18F) between 0.20 and 12.5 years (mean = 3.9, SD = 3.6) and 36 typically developing controls (18M/18F) between 0.12 and 12.8 years (mean = 4.0, SD = 3.6) were each imaged one time on a 1.5T MR scanner. One mid-sagittal image per study was selected for manual measurement of CC cross-sectional area and length. Gross motor, fine motor, and language developmental quotients, anatomical severity score, and serum sterol levels were assessed with imaging measurements.
Results
Shorter CC length and smaller area correlated with lower developmental quotient in gross motor and language domains. Furthermore, length and area negatively correlated with a serum 7DHC, 8DHC, sterol ratio, and anatomical severity score, and positively correlated with total cholesterol. The degree of developmental delay ranged from mild to severe, involving all domains.
Conclusions
For individuals with SLOS, smaller callosal area and length are associated with higher serum 7DHC, anatomic severity, and motor and language delay. These findings suggest the relationship between callosal development, biochemistry, and neurodevelopment may lead to finding predictors of outcome in SLOS.
Keywords: corpus callosum, Smith-Lemli-Opitz syndrome, SLOS, developmental delay, development
INTRODUCTION
Smith-Lemli-Opitz syndrome (SLOS) is an autosomal recessive, multiple malformation, neurodevelopmental disorder caused by mutations in the gene encoding 7-dehydrocholesterolreductase (DHCR7) resulting in impaired cholesterol synthesis[1–4]. Decreased DHCR7 activity results in increased blood and tissue levels of 7-dehydrocholesterol (7DHC) and its isomer 8-dehydrocholesterol (8DHC). In the majority of cases, cholesterol levels are low. SLOS is a rare disorder, and the incidence has been estimated to be on the order of 1 in 20,000 to 60,000 live births[5–8]. There are few published studies describing neuroimaging findings in SLOS[9–10]. Dysgenesis of midline structures, including the corpus callosum (CC), is the most common imaging finding.
The SLOS clinical phenotype is highly associated with characteristic dysmorphic features, autistic behavior and intellectual disability[4,11–13]. While the biochemical disturbances are potentially amendable to therapeutic intervention, no established therapies have been developed. Thus, identification of biomarkers of disease severity would be of value when testing therapeutic interventions, and may provide guidance when prognosticating neurodevelopmental outcome. Although developmental delay and callosal malformations are reported with great frequency in SLOS, their relationship has not been studied. The aims of this study are to examine whether mid-sagittal CC length and cross-sectional area, are associated with developmental delay and sterol levels in a large cohort of individuals with SLOS, and we hypothesize that shorter CC length and smaller area are associated with higher 7DHC levels and severity of developmental delay.
MATERIALS & METHODS
Study Population
This study was approved by the Institutional Review Boards of both the Eunice Kennedy Shriver National Institute of Child Health and Human Development in Bethesda, Maryland and the Hugo Moser Research Institute at the Kennedy Krieger Institute in Baltimore, Maryland. Written informed consent was obtained from parents or legal guardians and documented in the medical record. This study included 36 individuals with SLOS between ages 0.20 to 12.5 years, and 36 typically developing control subjects between ages 0.12 to 12.8 years. The ethnicity of SLOS patients included Caucasian (92.5%), Hispanic (5%), and Asian (2.5%). The diagnosis of SLOS was made by biochemical or molecular analysis, and confirmed by an expert evaluator (FDP) at the NIH Clinical Center. Hearing impairment was reported in two subjects with SLOS. There were no subjects with epilepsy, implanted neurosurgical devices, deafness, blindness or clinically significant visual impairment, or ventilator dependence. Inclusion criteria for control subjects were the absence of neurologic disease based on review of medical records including epilepsy, intellectual disability, developmental delay or autism, brain mass or vascular malformation, or neurometabolic abnormality; and the absence of structural or qualitative abnormalities on MRI of the brain as reported by a Johns Hopkins Hospital neuroradiologist.
Image Acquisition and Analysis
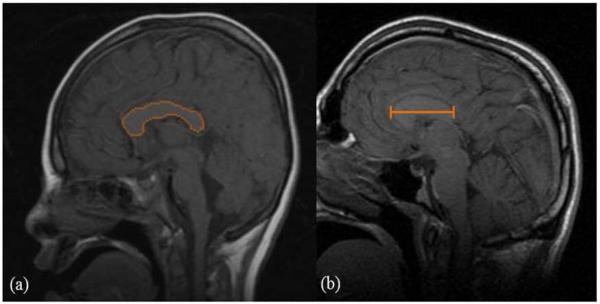
Images of SLOS subjects were acquired on a 1.5 T GE scanner at the NIH Clinical Center, and control images were obtained on a 1.5 T Siemens scanner at the Johns Hopkins Hospital. The MRI examinations for SLOS subjects included sagittal T1-weighted spin echo images (TR 400 ms, TE 90 ms, 240 mm × 240 mm FOV, 8 mm slice thickness, 4 mm interslice gap). Controls were scanned with a sagittal T1-weighted 3D volumetric interpolated breath-hold exam (VIBE) sequence (TR 9.9 ms, TE 4.6 ms, 190 mm × 190 mm FOV, 1 mm slice thickness, 0 mm interslice gap). One mid-sagittal slice per study was selected by a single observer radiologist for measurement of CC area and length. Blinded evaluators (1 radiologist, 1 pediatric neurologist) performed manual CC delineation of each image using DTI Studio and ROIEditor (www.mristudio.org, Johns Hopkins University, Baltimore, MD, USA) to derive the cross-sectional area and anterior-posterior length measurements for each scan (Figure 1)[14].
Figure 1.
Mid-sagittal T1-weighted MR images used for corpus callosum measurements. (a) Example of manual tracing of the CC margin on a mid-sagittal image, used for measurement of the cross-sectional area, (b) Example of CC length measurement performed on a mid-sagittal image.
Developmental and Biochemical Measures
Clinical data from electronic and paper medical records, parent interview, and physical exam were used to determine the SLOS anatomical severity score, sterol levels, and developmental delay. A 93-item questionnaire based on widely-accepted and published age-ranges for developmental milestone acquisition was modified and applied by a single interviewer (neurodevelopmental pediatrician) to determine developmental quotient (DQ) at the time of scan[15–16]. Supplementary developmental data was provided through medical records and clinical exam. Developmental quotient for gross motor (GMDQ), language (LDQ), and fine motor/adaptive skills (FMADQ) domains were calculated as DQ = (developmental age/chronologic age) × 100. The SLOS anatomical severity scale score is a clinical severity score based on organ system dysmorphology[5,17]. Serum 7-dehydrocholesterol (7DHC) (mg/dL), 8-dehydrocholesterol (8DHC) (mg/dL), and total cholesterol (mg/dL) levels were drawn at the time of scan and analyzed by gas chromatography/mass spectrometry (GCMS) at the Clinical Mass Spectrometry Laboratory at Kennedy Krieger Institute. Initial 7DHC and total cholesterol levels at time of diagnosis were available for analysis, but initial 8DHC levels were not available. The sterol ratio was calculated with the following formula: (7DHC + 8DHC) / (7DHC + 8DHC + Total Cholesterol). Statistical analysis was performed using Pearson's correlation (r), Student's T-test (unpaired, two-tailed) and Cohen's Kappa (κ). Significance was determined at the p < 0.05 and the κ > 0.80 level.
RESULTS
Thirty-six individuals with SLOS (18 males, 18 females) between 0.20 and 12.5 years (mean = 3.9, SD = 3.6) and 36 typically developing control subjects (18 males, 18 females) between 0.12 and 12.8 years (mean = 4.0, SD = 3.6) received one MRI scan. There was no significant difference in age or gender between groups. There was no significant difference in CC cross sectional area (Figure 1a) for the SLOS group (mean = 437.2 mm2, SD = 208.9) compared to controls (mean = 498.6 mm2, SD = 149.0) (p= 0.16). Corpus callosum length (Figure 1b) was significantly different between SLOS (mean = 51.2 mm, SD = 10.0) and control groups (mean = 61.5 mm, SD = 8.3) (p < 0.01). Observers performing callosal measurements demonstrated good inter-rater reliability (κ = 0.82). Demographic and comparison of CC measures is provided in Table 1. For the SLOS group, mean language DQ was 36.9 ±22.7; gross motor DQ 40.9 ±28.6; and fine motor/adaptive skills DQ 42.1 ±29.3, indicating a spectrum of developmental delay in the mild to severe range.
Table 1.
Demographics and Callosal Measurements of SLOS (N = 36) and Control (N = 36) Groups.
| SLOS Mean (SD) | Control Mean (SD) | p-value | k | |
|---|---|---|---|---|
| Age at scan, years | 3.9 (3.6) | 4.0 (3.6) | 0.94 | n/a |
| Gender ratio (M/F) | (18/18) | (18/18) | 1.0 | n/a |
| CC Area, mm2 | 437.2 (208.9) | 498.6 (149.0) | 0.16 | 0.82 |
| CC Length, mm | 51.2 (10.0) | 61.5 (8.3) | <0.01 | 0.82 |
N = number of MRI scans. CC = corpus callosum. SD = standard deviation. We considered values of p < 0.05. and Cohen's kappa (k) > 0.80 to be statistically significant.
For individuals with SLOS, shorter CC length correlated with lower developmental quotient in gross motor (r = 0.46; p < 0.01) and language (r = 0.37; p = 0.03), but not with fine motor/adaptive skills (r = 0.18; p = 0.32) (Table 2). Smaller cross-sectional area correlated with gross motor (r = 0.50; p < 0.01) and language (r = 0.37; p = 0.03) developmental quotient, but not with fine motor/adaptive skills (r = 0.17; p = 0.33). Furthermore, CC length and area did not reach statistical significance for an association with the anatomical severity scale score, but did show a trend toward significance. Callosal length (r = −0.52; p < 0.01) and area (r = −0.65; p < 0.01) demonstrated a negative correlation with 7DHC level (Figure 2). Similarly, callosal length (r = −0.52; p < 0.01) and area (r = −0.56; p < 0.01) demonstrated a negative correlation with 8DHC level. Callosal length (r = 0.43; p = 0.01) and area (r = 0.51; p < 0.01) positively correlated with serum total cholesterol level. The sterol ratio was negatively correlated with callosal length (r = −0.45; p = 0.01) and area (r = −0.59; p < 0.01). To examine the question of whether CC measurements are an indication of the baseline status or current state of disease severity, we compared 7DHC at time of diagnosis and 7DHC at time of MRI scan. The results showed corpus callosum measures to be correlated with initial cholesterol and 7DHC levels. A summary of results is provided in Table 2.
Table 2.
Callosal Measures Correlate with Development and Sterols in SLOS (N = 36).
| Length (mm) | Area (mm2) | |||
|---|---|---|---|---|
| r | p-value | r | p-value | |
| GMDQ | 0.46 | <0.01 | 0.50 | <0.01 |
| LDQ | 0.37 | 0.03 | 0.37 | 0.03 |
| FMADQ | 0.18 | 0.32 | 0.17 | 0.33 |
| SS | −0.28 | 0.10 | −0.32 | 0.06 |
| 7DHC (initial) | −0.54 | <0.01 | −0.53 | <0.01 |
| 7DHC (at MRI) | −0.52 | <0.01 | −0.65 | <0.01 |
| 8DHC (at MRI) | −0.52 | <0.01 | −0.56 | <0.01 |
| CHL (initial) | 0.58 | <0.01 | 0.55 | <0.01 |
| CHL (at MRI) | 0.43 | 0.01 | 0.51 | <0.01 |
| Ratio (at MRI) | −0.45 | 0.01 | −0.59 | <0.01 |
Developmental quotients for gross motor (GMDQ), language (LDQ), and fine motor/adaptive skills (FMADQ) domains were calculated as DQ = (developmental age/chronologic age) × 100. SLOS anatomic severity scale score (SS); 7-dehydrocholesterol. mg/dL (7DHC); 8-dehydrocholesterol, mg/dL (8DHC); Total cholesterol, mg/dL (CHL); Ratio = (7DHC + 8DHC) / (7DHC + 8DHC + CHL). Initial sterol level was obtained at time of diagnosis, and sterol level at MRI was obtained at time of scan. We considered p < 0.05 for Pearson's correlation (r) to be significant.
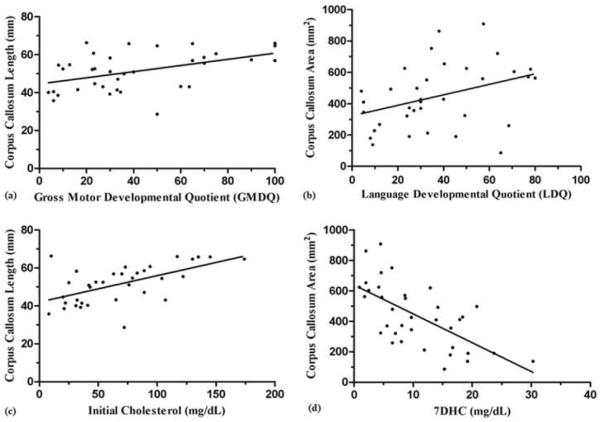
Figure 2.
Sample Correlation Plots between Callosal and Clinical Measures in SLOS (n = 36). (a) Callosal length is positively correlated with gross motor developmental quotient (DQ) (p < 0.01: r = 0.46), (b) Callosal area is positively correlated with language developmental quotient (DQ) (p = 0.03; r = 0.37), (c) Callosal length is positively correlated with serum cholesterol at time of diagnosis (initial) (p < 0.01; r = 0.58), (d) Callosal area is negatively correlated with 7DHC at time of scan (p < 0.01; r = −0.65). We considered p < 0.05 for Pearson's correlation (r) to be significant.
DISCUSSION
For individuals with SLOS, mid-sagittal CC length and area are associated with severity of language and gross motor delay, anatomical severity score, and sterol levels. The degree of developmental disability for our cohort was in the moderate to severe range for each domain. These results support our hypothesis that corpus callosum length and area are associated with developmental delay and biochemistry in individuals with SLOS.
The CC is a midline brain structure comprised of axonal tracts involved in the transfer of information between cortical and subcortical neurons and contralateral brain hemispheric and spinal cord regions. During maturation, axons lengthen and the CC increases in total volume[18]. While studies have mapped anatomic and functional specificity within segments of the CC, our understanding of the role of CC malformation in neurologic outcome is limited[19–20]. One study reports an association between smaller mid-sagittal callosal area and lower intelligence quotient (IQ) in healthy children and adolescents[21]. A few studies have shown neurodevelopmental disorders such as autism and intellectual disability are associated with smaller CC volume[22–26]. Hypotheses suggest impaired neural connectivity and synchronization as causative[27–28]. Furthermore, congenital midline brain malformation syndromes such as Aicardi syndrome, septo-optic dysplasia syndromes, holoprosencephaly, Joubert syndrome, and Chiari malformations are often accompanied by neurodevelopmental disability[29].
Agenesis of the corpus callosum (ACC) can be the result of numerous etiologic factors including gene mutations, infection, metabolic disturbances, and trauma during callosal development[30–32]. While many ACC syndromes are associated with developmental delay, most humans with agenesis or dysgenesis of the CC do not manifest developmental delay. A 10-year longitudinal study of neonates born with ACC reported normal intelligence in 73% of individuals, and borderline intelligence in 27%[33]. However, corpus callosum malformations are increasingly recognized in patients with cognitive and behavioral impairment. Individuals with ACC and normal IQ demonstrate executive function deficits manifested as lower flexibility, inhibition, and inference for contingencies[34–35]. In light of the evidence that populations with neurodevelopmental disability and typically developing children both express the spectrum of CC dysmorphology, further study examining cognitive skills that depend on callosal connectivity are needed.
A potential limitation of this study is that controls were scanned using a different sequence and parameters. The thicker image slices used on the SLOS patients may cause an over-estimation of length and area, because partial volume effects cause them to appear longer and have a larger cross sectional area. The inability to re-orient the thick slices may also lead to over-estimation. Prospective attention to orientation of the images at the time of acquisition would have minimized this effect. However, the errors bias against our findings, thus we do not believe the results were compromised. Furthermore, we are uncertain if the CC measurements reflect the current state of disease or a dynamic state. We attempted to analyze disease state by studying initial sterol levels and sterol level at time of scan. The results showed correlation for callosal measurements for both initial sterol level and level at time of scan, suggesting disease severity and/or sterol levels remain relatively constant over time, or callosal measures reflect both baseline and current state of the disease. Further studies involving longitudinal imaging data are required to address these important questions.
The genetic and cognitive/behavioral phenotype of SLOS has been described, but the mechanisms of neurologic injury in this disease remain largely unknown[4,36]. Cholesterol serves numerous key functions in the developing brain as a co-factor for sonic hedgehog morphogenic signaling, a key component of membrane lipid raft distribution, activity-dependent synaptic plasticity, and neurosteroid formation[37–43]. We hypothesize that multilevel-disturbance of cholesterol-dependent processes during embryologic development are responsible for impaired midline brain formation and developmental delays in SLOS. Further study of in-vivo fiber organization and microstructure with diffusion tensor imaging and tractography represent important tools that will be applied toward increasing our understanding of the relationships reported in this paper[44–45].
In sum, we demonstrate that corpus callosum measurements are associated with developmental delay and biochemical measures of disease severity in SLOS. To date, there are no reliable treatments or markers of neurodevelopmental outcome for this disorder. These novel findings hold promise for future studies that lead to reliable clinical prognosticators of neurodevelopment, with the goal of improving the lives of families and individuals with SLOS.
ACKNOWLEDGEMENTS
This work was supported by the Intramural research program of the Eunice Kennedy Shriver National Institute of Health, the RSH/SLOS Foundation, a Bench to Bedside award from the NIH Clinical Center, the NIH Office of Rare Diseases, and the intramural research program of NICHD. We would also like to acknowledge the MRI technologists (Mary Busse, RT, Lisa Catron, RT, Bonita Damaska, RT, Sandra Hess, RT, Sandra McKee, RT, Hugo Sandoval, RT, James Sedlacko, RT, Maryanne Russell, RT, Ronald White, RT, Betty Wise, RT, and Scott Pryde, RT) and our research coordinators Halima Goodwin, CRNP and Sandra K. Conley, CRNP. The authors would like to acknowledge Richard Kelley and Lisa Kratz for their work on the sterol assays, and Andrea Gropman for her clinical evaluation of patients. The authors would like to express appreciation to the families and patients that participated in these studies.
Footnotes
The authors report no conflicts of interest.
REFERENCES
- 1.Tint GS, Irons M, Elias ER, Batta AK, Frieden R, Chen TS, et al. Defective cholesterol biosynthesis associated with the Smith-Lemli-Opitz syndrome. N Engl J Med. 1994;330(2):107–113. doi: 10.1056/NEJM199401133300205. [DOI] [PubMed] [Google Scholar]
- 2.Irons M, Elias ER, Salen G, Tint GS, Batta AK. Defective cholesterol biosynthesis in Smith-Lemli Opitz syndrome. Lancet. 1993;(8857):341–1414. doi: 10.1016/0140-6736(93)90983-n. [DOI] [PubMed] [Google Scholar]
- 3.Wassif CA, Maslen C, Kachilele-Linjewile S, Lin D, Linck LM, Conner WE, Steiner RD, Porter FD. Mutations in the human sterol delta7-reductase gene at 11q12–13 cause Smith-Lemli-Opitz syndrome. Am J Hum Genet. 1998;63(1):55–62. doi: 10.1086/301936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter FD. RSH/Smith-Lemli-Opitz syndrome: a multiple congenital anomaly/mental retardation syndrome due to an inborn error of cholesterol biosynthesis. Mol Genet Metab. 2000;71(1–2):163–174. doi: 10.1006/mgme.2000.3069. [DOI] [PubMed] [Google Scholar]
- 5.Kelley RI, Hennekam RC. The Smith-Lemli-Optiz Syndrome. J Med Genet. 2000;37(5):321–335. doi: 10.1136/jmg.37.5.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowry RB, Yong SL. Borderline normal intelligence in the Smith-Lemli-Opitz (RSH) syndrome. Am J Med Genet. 1980;5(2):137–143. doi: 10.1002/ajmg.1320050205. [DOI] [PubMed] [Google Scholar]
- 7.Ryan AK, Bartlett K, Clayton P, Eaton S, Mills L, Donnai D, Winter RM, Burn J. Smith-Lemli-Opitz syndrome: a variable clinical and biochemical phenotype. J Med Genet. 1998;35(7):558–565. doi: 10.1136/jmg.35.7.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bzduch V, Behulova D, Skodova J. Incidence of Smith-Lemli-Opitz syndrome in Slovakia. Am J Med Genet. 2000;90(3):260. doi: 10.1002/(sici)1096-8628(20000131)90:3<260::aid-ajmg17>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 9.Caruso PA, Poussaint TY, Tzika AA, Zurakowski D, Astrakas LG, Elias ER, Bay C, Irons MB. MRI and 1H MRS findings in Smith-Lemli-Opitz syndrome. Neuroradiology. 2004;46(1):3–14. doi: 10.1007/s00234-003-1110-1. [DOI] [PubMed] [Google Scholar]
- 10.Weaver DD, Solomon BD, Akin-Samson K, Kelley RI, Muenke M. Cyclopia (synophthalmia) in Smith-Lemli-Opitz syndrome: First reported case and consideration of mechanism. Am J Med Genet C Semin Med Genet. 2010;154C(1):142–145. doi: 10.1002/ajmg.c.30241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tierney E, Nwokoro NA, Porter FD, Freund LS, Ghuman JK, Kelley RI. Behavior phenotype in the RSH/Smith-Lemli-Opitz syndrome. Am J Med Genet. 2001;98(2):191–200. doi: 10.1002/1096-8628(20010115)98:2<191::aid-ajmg1030>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 12.Tierney E, Bukelis I, Thompson RE, Ahmed K, Aneja A, Kratz L, Kelley RI. Abnormalities of cholesterol metabolism in autism spectrum disorders. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(6):666–668. doi: 10.1002/ajmg.b.30368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sikora DM, Pettit-Kekel K, Penfield J, Merkens LS, Steiner RD. The near universal presence of autism spectrum disorders in children with Smith-Lemli-Opitz syndrome. Am J Med Genet A. 2006;140(14):1511–1518. doi: 10.1002/ajmg.a.31294. [DOI] [PubMed] [Google Scholar]
- 14.Jiang H, van Zijl PC, Kim J, Pearlson GD, Mori S. DtiStudio: resonance program for diffusion tensor computation and fiber bundle tracking. Comput Methods Programs Biomed. 2006;81(2):106–116. doi: 10.1016/j.cmpb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Gerber RJ, Wilks T, Erdie-Lalena C. Developmental Milestones: Motor Development. Pediatr Rev. 2010;31(7):267–276. doi: 10.1542/pir.31-7-267. [DOI] [PubMed] [Google Scholar]
- 16.Wilks T, Gerber RJ, Erdie-Lalena C. Developmental Milestones: Cognitive Development. Pediatr Rev. 2010;31(9):364–367. doi: 10.1542/pir.31-9-364. [DOI] [PubMed] [Google Scholar]
- 17.Bialer MG, Penchaszadeh VB, Kahn E, Libes R, Krigsman G, Lesser ML. Female external genitalia and mullerian duct derivatives in a 46,XY infant with the smith-lemli-Opitz syndrome. Am J Med Genet. 1987;28(3):723–731. doi: 10.1002/ajmg.1320280320. [DOI] [PubMed] [Google Scholar]
- 18.Lee BY, Sohn JH, Choi MH, Lee SJ, Kim HS, Yang JW, Choi JS, Kim HS, Yi JH, Tack GR, Chung SC. A volumetric study of the corpus callosum in 20s and 40s Korean people. Brain Struct Funct. 2009;213(4–5):463–467. doi: 10.1007/s00429-009-0209-5. [DOI] [PubMed] [Google Scholar]
- 19.Witelson SF. Hand and sex differences in the isthmus and genu of the human corpus callosum. A postmortem morphological study. Brain. 1989;112(Pt 3):799–835. doi: 10.1093/brain/112.3.799. [DOI] [PubMed] [Google Scholar]
- 20.Hofer S, Frahm J. Topography of the human corpus callosum revisited--comprehensive fiber tractography using diffusion tensor magnetic resonance imaging. Neuroimage. 2006;32(3):989–994. doi: 10.1016/j.neuroimage.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 21.Ganjavi H, Lewis JD, Bellec P, MacDonald PA, Waber DP, Evans AC, Karama S. Negative associations between corpus callosum midsagittal area and IQ in a representative sample of healthy children and adolescents. PLoS One. 2011;6(5):e19698. doi: 10.1371/journal.pone.0019698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anstey KJ, Mack HA, Christensen H, Li SC, Reglade-Meslin C, Maller J, Kumar R, Dear K, Easteal S, Sachdev P. Corpus callosum size, reaction time speed and variability in mild cognitive disorders and in a normative sample. Neuropsychologia. 2007;45(8):1911–1920. doi: 10.1016/j.neuropsychologia.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 23.Frazier TW, Hardan AY. A meta-analysis of the corpus callosum in autism. Biol Psychiatry. 2009;66(10):935–941. doi: 10.1016/j.biopsych.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanfield AC, McIntosh AM, Spencer MD, Philip R, Gaur S, Lawrie SM. Towards a neuroanatomy of autism: a systematic review and meta-analysis of structural magnetic resonance imaging studies. Eur Psychiatry. 2008;23(4):289–299. doi: 10.1016/j.eurpsy.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Hardan AY, Pabalan M, Gupta N, Bansal R, Melhem NM, Fedorov S, Keshavan MS, Minshew NJ. Corpus callosum volume in children with autism. Psychiatry Res. 2009;174(1):57–61. doi: 10.1016/j.pscychresns.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewing-Cobbs L, Prasad MR, Swank P, Kramer L, Mendez D, Treble A, Payne C, Bachevalier J. Social communication in young children with traumatic brain injury: Relations with corpus callosum morphometry. Int J Dev Neurosci. 2012;30(3):247–254. doi: 10.1016/j.ijdevneu.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapleau CA, Larimore JL, Theibert A, Pozzo-Miller L. Modulation of dendritic spine development and plasticity by BDNF and vesicular trafficking: fundamental roles in neurodevelopmental disorders associated with mental retardation and autism. J Neurodev Disord. 2009;1(3):185–96. doi: 10.1007/s11689-009-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsiaras V, Simos PG, Rezaie R, Sheth BR, Garyfallidis E, Castillo EM, Papanicolaou AC. Extracting biomarkers of autism from MEG resting-state functional connectivity networks. Comput Biol Med. 2011;41(12):1166–77. doi: 10.1016/j.compbiomed.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka T, Gleeson JG. Genetics of brain development and malformation syndromes. Curr Opin Pediatr. 2000;12(6):523–528. doi: 10.1097/00008480-200012000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Jeret JS, Serur D, Wisniewski KE, Lubin RA. Clinicopathological findings associated with agenesis of the corpus callosum. Brain Dev. 1987;9(3):255–264. doi: 10.1016/s0387-7604(87)80042-6. [DOI] [PubMed] [Google Scholar]
- 31.Wisniewski KE, Jeret JS. Callosal agenesis: review of pathological and cytogenic features, in clinical description and related disorders. In: Lassonde M, Jeeves MA, editors. Callosal agenesis: A natural split brain. Plenum Press; New York: 1991. pp. 1–6. [Google Scholar]
- 32.Norman MG, McGillivray BC, Kalousek DK, Hill A, Poskitt KJ. Crossing the midline. In: Becker LE, Cochrane DD, Muenke M, editors. Congenital malformations of the brain: pathological, embryological, clinical, radiological and genetic aspects. Oxford University Press; New York: 1995. pp. 309–331. [Google Scholar]
- 33.Moutard ML, Kieffer V, Feingold J, Lewin F, Baron JM, Adamsbaum C, Gélot A, Isapof A, Kieffer F, de Villemeur TB. Isolated corpus callosum agenesis: a ten-year follow-up after prenatal diagnosis (how are the children without corpus callosum at 10 years of age?) Prenat Diagn. 2012;32(3):277–283. doi: 10.1002/pd.3824. [DOI] [PubMed] [Google Scholar]
- 34.Brown WS, Anderson LB, Symington MF, Paul LK. Decision-Making in Individuals with Agenesis of the Corpus Callosum: Expectancy-Valence in the Iowa Gambling Task. Arch Clin Neuropsychol. 2012;27(5):532–544. doi: 10.1093/arclin/acs052. [DOI] [PubMed] [Google Scholar]
- 35.Marco EJ, Harrell KM, Brown WS, Hill SS, Jeremy RJ, Kramer JH, Sherr EH, Paul LK. Processing speed delays contribute to executive function deficits in individuals with agenesis of the corpus callosum. J Int Neuropsychol Soc. 2012;18(3):521–529. doi: 10.1017/S1355617712000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porter FD. Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur J Hum Genet. 2008;16(5):535–541. doi: 10.1038/ejhg.2008.10. [DOI] [PubMed] [Google Scholar]
- 37.Fantini J, Barrantes FJ. Sphingolipid/cholesterol regulation of neurotransmitter receptor conformation and function. BBA Biomemb. 2009;1788(11):2345–2361. doi: 10.1016/j.bbamem.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 38.Korade Z, Kenworthy AK. Lipid rafts, cholesterol, and the brain. Neuropharmacology. 2008;55(8):1265–1273. doi: 10.1016/j.neuropharm.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linetti A, Fratangeli A, Taverna E, Valnegri P, Francolini M, Cappello V, Matteoli M, Passafaro Maria P, Rosa P. Cholesterol reduction impairs exocytosis of synaptic vesicles. Journal of Cell Science. 2010;123(Pt 4):595–605. doi: 10.1242/jcs.060681. [DOI] [PubMed] [Google Scholar]
- 40.Mellon SH, Griffin LD. Neurosteroids: biochemistry and clinical significance. Trends Endocrinol Metab. 2002;13(1):35–43. doi: 10.1016/s1043-2760(01)00503-3. [DOI] [PubMed] [Google Scholar]
- 41.Rakheja D, Boriack RL. Precholesterol sterols accumulate in lipid rafts of patients with Smith-Lemli-Opitz syndrome and X-linked dominant chondroplasia punctata. Pediatr Dev Pathol. 2008;11(2):128–132. doi: 10.2350/06-10-0179.1. [DOI] [PubMed] [Google Scholar]
- 42.Simons K, Ehehalt R. Cholesterol, lipid rafts, and disease. The Journal of Clinical Investigation. 2002;11(5):597–603. doi: 10.1172/JCI16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh P, Paila YD, Chattopadhyay A. Differential effects of cholesterol and 7-dehydrocholesterol on the ligand binding activity of the hippocampal serotonin(1A) receptor: implications in SLOS. Biochem Biophys Res Commun. 2007;358(2):495–499. doi: 10.1016/j.bbrc.2007.04.135. [DOI] [PubMed] [Google Scholar]
- 44.Mori S, Crain BJ, Chacko VP, van Zijl PCM. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol. 1999;45(2):265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 45.Ren T, Anderson A, Shen WB, Huang H, Plachez C, Zhang J, Mori S, Kinsman SL, Richards LJ. Imaging, anatomical, and molecular analysis of callosal formation in the developing human fetal brain. Anat Rec A Discov Mol Cell Evol Biol. 2006;288(2):191–204. doi: 10.1002/ar.a.20282. [DOI] [PubMed] [Google Scholar]