Abstract
The aim of the present study was to diagnose the presence of Giardia cysts and Cryptosporidium oocysts in household animals using nested polymerase chain reaction (PCR) and sequence analysis. One hundred faecal samples obtained from 81 dogs and 19 cats were investigated. The Cryptosporidium genotypes were determined by sequencing a fragment of the small subunit (SSU) rRNA gene, while the Giardia Assemblages were determined through analysis of the glutamate dehydrogenase (GDH) locus. Isolates from five dogs and two cats were positive by PCR for the presence of Giardia, and their sequences matched the zoonotic Assemblage A of Giardia. Cryptosporidium spp. isolated from one dog and one cat were both found to be C. parvum. One dog isolate harboured a mixed infection of C. parvum and Giardia Assemblage A. These findings support the growing evidence that household animals are potential reservoirs of the zoonotic pathogens Giardia spp. and Cryptosporidium spp. for infections in humans.
Keywords: Cryptosporidium parvum, Giardia, Assemblages, genotypes, zoonosis, Germany
Abstract
Le but de cette étude était de diagnostiquer la présence de kystes de Giardia et d’oocystes de Cryptosporidium chez des animaux de compagnie en utilisant la réaction en chaîne de la polymérase (PCR) et l’analyse des séquences. Cent échantillons de fèces obtenus de 81 chiens et 19 chats ont été étudiés. Les génotypes de Cryptosporidium ont été déterminés en séquençant un fragment de la petite unité (SSU) du gène rRNA, et les Assemblages de Giardia ont été déterminés par une analyse du locus de la glutamate déshydrogénase (GDH). Des isolats de cinq chiens et de deux chats ont été positifs par PCR pour la présence de Giardia, et leurs séquences correspondaient à l’Assemblage A de Giardia. Les Cryptosporidium isolées d’un chien et d’un chat appartenaient tous deux à l’espèce C. parvum. Un chien avait une infection mixte par C. parvum et Giardia Assemblage A. Ces résultats confirment que les animaux de compagnie sont des réservoirs potentiels des pathogènes zoonotiques Giardia spp. et Cryptosporidium pour des infections humaines.
Introduction
Cryptosporidium and Giardia are protozoan pathogens that cause diarrhoeal illness when they colonise and reproduce in the intestines of humans or domestic animals, particularly dogs, cats, and livestock respectively. Both Giardia and Cryptosporidium are capable of completing their life cycle within a single host, resulting in cyst or oocyst stages that are excreted in the faeces. Epidemiological studies have focused on the transmission route of Giardia and Cryptosporidium [39, 40] and have sought to determine their zoonotic potential [45, 50]. Giardia intestinalis assemblages have been defined (A-H) by DNA sequence analysis so far, of which assemblages A and B are mainly virulent for humans [38, 49] and are often referred to as “zoonotic” assemblages [12, 27], but are also found in a number of other mammalian hosts [43]. Animals kept as household and/or pets play a significant role in the zoonotic transmission routes of the parasites due to their close association with their owners and the abundance of parasite cysts/oocysts excreted in large quantities [9, 16]. Household animals may also come into contact with free-living and/or domestic animals and can contract infections from them [9]. Animals can harbour infections of either zoonotic or host-specific Giardia Assemblages [14, 47, 48]. In general, cats and dogs may be affected by the host-specific C. felis and C. canis, respectively, and dogs can be infected with C. parvum due to the broader host range of this species [30]. Genotyping of oocysts recovered from the faeces of infected cats has shown that cats can also be infected with C. muris [36, 42]. Giardia host-adapted Assemblages in cats are usually A and F, whereas for dogs, the Assemblages include A, C and D [4, 21, 43].
The objective of the present study was to examine household animals for Cryptosporidium and Giardia infections.
Materials and methods
Sample collection
Faecal samples from domestic dogs and cats with clinical suspicion for giardiasis or cryptosporidiosis (predominantly diarrhoea of variable duration) were submitted by veterinary clinics from Germany and other European countries to a private veterinary laboratory in Germany (Idexx Vet Med Lab, Ludwigsburg, Germany). Fresh faecal samples from 81 dogs and 19 cats were collected during 2007 and were labelled and stored immediately at −20 °C. The samples were shipped to the laboratory at Cologne University for purification and processing and were kept frozen until used.
Sample purification
Purification of the faecal samples and isolation of the oocysts/cysts were performed using the diethyl ether sedimentation technique combined with the saturated salt flotation technique, as described by Joachim et al. [22]. Briefly, each faeces sample was suspended in a 50 mL polypropylene tube with phosphate-buffered saline (PBS, pH 4) and homogenised by shaking vigorously. The excess faecal debris and lipids were removed by mixing the suspension with a one-fourth volume of diethyl ether until the sample became a homogenised emulsion; the suspension was then centrifuged at 2,500 g for 10 min. The supernatant was discarded and the remaining sediment was resuspended in distilled water and centrifuged twice as described above to remove other residues. The resulting pellet was resuspended in 5M cold saturated sodium chloride solution and carefully overlaid with cold distilled water so that a visible gradient was obtained. The samples were centrifuged at 2,300 g for 10 min, and oocysts/cysts were recovered from the interphase were washed twice with distilled water and stored in PBS (pH 7.4) with streptomycin (200 μg/mL) and amphotericin B (5 μg/mL) at 4 °C.
DNA extraction
DNA was extracted from the purified faecal suspension and used for molecular analysis and sequencing. DNA extraction was performed using the modified method described by Karanis et al. [25] followed by the use of the QIAamp Stool Kit (Qiagen GmbH, Hilden, Germany). In brief, the oocysts/cysts were ruptured using 10 freeze-thaw cycles in the presence of lysis buffer in a dry thermo device (DTU-2B, Taitec, Japan) and were further processed according to the Qiagen manufacturer’s instructions. DNA was eluted in 100 μL AE buffer and stored at –20 °C.
Molecular assays for the detection of Giardia and Cryptosporidium in faecal samples
To characterise the Giardia and Cryptosporidium species isolated from each positive sample, the DNA of all samples was extracted from the oocysts/cysts and was consequently tested by nested polymerase chain reaction (PCR). Positive faecal specimens were sequenced to identify the Cryptosporidium species and the Giardia Assemblages of the parasites found in the positive samples of the infected animals.
Giardia spp. A fragment of the glutamate dehydrogenase (GDH) gene of approximately 220 bp in length was amplified using previously published primers and conditions [1]. All PCR amplifications were performed in an ABI 2720 Thermal cycler (Applied Biosystems, Foster, CA) in standard mixtures of 50 μL containing 1× PCR buffer, 200 nmol of each primer, 200 μM of dNTPs, 1.5 mM of MgCl2, 2.5 U of HotStarTaq DNA polymerase (Qiagen GmbH, Hilden, Germany), 2 μL of bovine serum albumin (BSA, acetylated, 10 mg/mL) (Promega, Madison, WI), 2 μL DNA template and distilled water. The PCR program included one incubation at 96 °C for 15 min and 40 amplification cycles (94 °C for 30 s, 55 °C for 30 s and 72 °C for 60 s), followed by one final extension incubation of 7 min at 72 °C. The PCR products were separated on a 1.6% agarose gel, stained with ethidium bromide and visualised on a UV transilluminator. PCR negative-control samples omitted template DNA, which was replaced by distilled water, and PCR positive-control samples, containing DNA extracted from 10 Giardia cysts, were always included for each test.
Cryptosporidium spp. Cryptosporidium DNA was amplified by nested PCR using primers and conditions as described by Nichols et al. [32] and Karanis et al. [25] to produce a DNA fragment of approximately 440 bp in length of the SSU rRNA gene. PCRs were performed in 50 μL volumes containing 200 nmole of each primer, 1× PCR buffer, 200 μM dNTP, 3 mM MgCl2, 2.5 U GoTaq DNA polymerase (Promega, Wisconsin, USA), 2 μL BSA (10 mg/mL), 2 μL DNA template and distilled water. PCR consisted of one initial denaturation cycle at 96 °C for 15 min and 35 cycles (94 °C for 30 s, primary PCR 68 °C and secondary PCR 60 °C for 60 s, 72 °C for 30 s), followed by one final extension at 72 °C for 10 min. The PCR products were analysed as described above. Positive controls containing DNA extracted from 10 C. parvum oocysts and negative controls containing distilled water were included for each test.
PCR product purification and sequencing
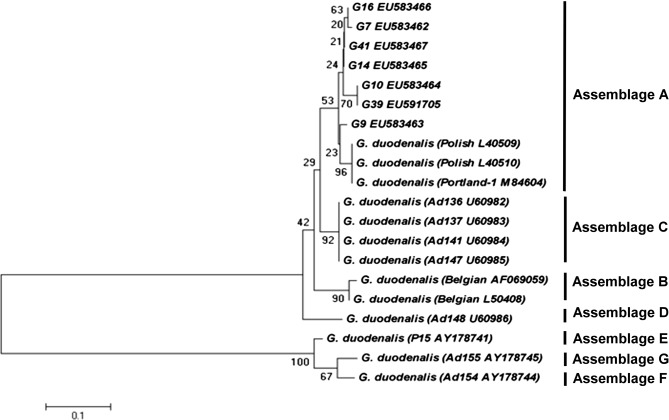
All PCR products from the molecular assays for both Giardia cysts and Cryptosporidium oocysts were cut out of agarose gels and purified with QIAquick Gel Extraction Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. The purified PCR products were directly sequenced in both directions on an ABI Prism 3100 (Applied Biosystems, Japan) Genetic Analyser using a Big Dye Terminator V.3.1 cycle sequencing kit (Applied Biosystems, Japan). The accuracy of the data was confirmed by two directional sequencing of the obtained sequences and by the alignment of the nucleotide sequences of the GDH gene for Giardia cysts and the SSU rRNA for Cryptosporidium oocysts against reference sequences retrieved from GenBank using the program ClustalW. Sequence similarity was also determined using the Basic Local Alignment Search Tool (BLAST) and the phylogenetic and molecular evolutionary analyses were conducted using MEGA version 5. The evolutionary history was inferred using the Neighbor-Joining method [41] and the respective evolutionary analyses were conducted using MEGA 5 [44]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches in Figure 1 [17]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method [26] and are in the units of the number of base substitutions per site.
Figure 1.
Phylogenetic relationship between the 7 Giardia Assemblages (G7, G9, G10, G14, G16, G41 and G39) obtained in this study and other previously published Giardia Assemblages. Nucleotide sequences of the GDH gene obtained in this study were aligned against reference sequences retrieved from GenBank using ClustalW and MEGA 5. The evolutionary history was inferred using the Neighbor-Joining method and the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura two-parameter method with the pairwise deletion option and are in the units of the number of base substitutions per site.
The nucleotide sequences of the positive samples have been submitted to the GenBank and they are available in the GenBank database under the Accession Nos. EU583462–EU583467 and EU591705 (93%–97% homology) for Giardia and EU606193 (94% homology) and EU606194 (99% homology) for Cryptosporidium.
Results and Discussion
A total of 100 faecal samples from household animals, 19 cats and 81 dogs, were investigated by nested PCR for the presence of Cryptosporidium spp. oocysts and Giardia spp. cysts. To test for Giardia, all samples were amplified with the primers GDF3 and GDB5 and the PCR products were sequenced. Five dog and two cat faecal samples were found to be positive for Giardia. The results are presented in Table 1. In all cases, the samples found positive by PCR were confirmed by sequence analysis and no sequences were identified other than those of the typical Giardia Assemblage A in the dog and cat samples. In faecal samples of two dogs (6 year old, mix-breed with diarrhoea and vomiting and 1 year old with persistent diarrhoea), additional laboratory examinations identified the presence of Escherichia coli and Campylobacter jejuni (diagnosed by aerobe cultivation), respectively, but no other infections with yeasts and parasites have been found.
Table 1.
Positive household animals for Giardia infections by PCR of the GDH locus, sequencing analysis results and accession numbers (n = 19 cats and n = 81 dogs).
| Sample name | Species |
Giardia PCR result of the investigation of the GDH locus |
Giardia Assemblage by sequencing analysis |
GenBank Accession No. |
|---|---|---|---|---|
| G7 | Dog | + | A | EU583462 |
| G9§ | Cat | + | A | EU583463 |
| G10 | Dog | + | A | EU583464 |
| G14 | Dog | + | A | EU583465 |
| G16 | Cat | + | A | EU583466 |
| G41 | Dog | + | A | EU583467 |
| G39* | Dog | + | A | EU591705 |
Positive.
Cat from Denmark.
Dog with mixed infection (Giardia and Cryptosporidium).
SSU rDNA PCR and sequencing detected Cryptosporidium in one dog and one cat sample. Interestingly, both samples had high identity values to C. parvum (Table 2). Direct sequencing of these PCR products for both Giardia and Cryptosporidium showed that one dog carried a mixed infection with both C. parvum and Giardia Assemblage A (Tables 1 and 2). According to the study of Nichols et al. [33] outer amplification primers developed by them (N18SF2/R2) showed nonspecific amplification of DNA from the dinoflagellate Gymnodinium, but in both positive cases of our study, the results of nested PCR were confirmed by sequence analysis and no sequences were identified other than those of the typical C. parvum.
Table 2.
Positive household animals for the presence of Cryptosporidium genotypes by PCR of the SSU rRNA and sequencing analysis results (n = 19 cats and n = 81 dogs).
| Sample name | Species |
Cryptosporidium PCR result of the investigation of the SSU rRNA gene |
Cryptosporidium genotype by sequencing analysis |
GenBank Accession No. |
|---|---|---|---|---|
| G39* | Dog | + | C. parvum | EU606193 |
| G37 | Cat | + | C. parvum | EU606194 |
Positive.
Dog with mixed infection (Giardia and Cryptosporidium).
The phylogenetic relation of Giardia species is shown in Figure 1, using sequences of 13 Giardia Assemblages downloaded from GenBank as reference sequences. The phylogenetic tree indicates that the sequences obtained in our study cluster to Giardia Assemblage A. Even though the number of positive samples was relatively low, these results clearly demonstrate the presence of Giardia and Cryptosporidium among household animals from different regions in Germany.
According to a 5-year survey (1993–1997) of dairy herds in five German state veterinary laboratories, Cryptosporidium was diagnosed in 19–34% of the examined faecal samples and 20–36% of the post mortem cases [23] by conventional techniques. The primary cause of diarrhoea was acknowledged to be Cryptosporidium by only 1/5 of the investigators, indicating that the role of Cryptosporidium in Germany was underestimated. The recorded mixed infections were mostly associated with the presence of rotavirus and E. coli.
Only few studies have been carried out to evaluate the occurrence of different Giardia genotypes and Cryptosporidium species/genotypes in household animals in Germany and the actual risk of transmission to their owners. Broglia [11] genotyped Cryptosporidium isolates and defined the subtypes of C. parvum that originated from neonatal calves in Germany. All of the calf isolates in their study were identified as C. parvum. Other reports have also indicated that C. parvum is the most frequently found species of Cryptosporidium in pre-weaned calves [2, 37]. Erinaceus europaeus L. (European hedgehogs) in Germany were found to be naturally infected with Cryptosporidium in 29.8% by coproantigen analysis, while molecular analysis revealed IIa, IIc and VIIa subtype families of C. parvum [15].
The retrospective study of Barutzki and Schaper [5] reported a prevalence rate of 16.6% and 12.6% for Giardia infections in dogs and cats, respectively, in Germany between 1999 and 2002. The same authors reported that Giardia spp. were the most commonly found parasites using coprological examinations (coproantigen ELISA or SAF technique) with prevalence rates of 18.6% and 12.6% in dogs and cats, respectively [6].
Giardia human isolates belong to Assemblages A and B [13, 24]. Each Assemblage consists of two distinct subgroups; Assemblage A can be divided into A-I and A-II and Assemblage B can be divided into subgroups B-III and B-IV [3, 20]. Zoonotic genotypes of Giardia Assemblage A that are specific for hosts other than humans and Assemblages C and D were specifically assigned for dogs [21, 31, 38, 46]. In contrast, Assemblages A and B are not human-specific and infect a wider host range including dogs, cats, livestock and wildlife and are considered to have zoonotic potential. Barutzki et al. [7] investigated two different groups of dogs: one group presented clinical symptoms of gastrointestinal disorders and the other group was randomly selected. They concluded that 7% of the randomly selected Giardia positive dogs carried zoonotic species of Giardia belonging to Assemblage A. Interestingly, Leonhard et al. [29] examined asymptomatic dogs in southern Germany that were kept isolated or in groups and found that Assemblage A was most prevalent. They also identified Assemblages A and C, but Assemblages C and D were very rarely found.
Only one case was recorded as an outbreak of gastroenteritis caused by C. parvum after diagnosis by serological tests, such as ELISA for specific IgG, analysis of stool specimens, genotyping of the isolates and epidemiological analysis in Germany. In an August 2001 field training session, half of the recruits (n = 201) of the German armed forces became ill with acute gastroenteritis [10]. The zoonotic transmission of the disease was excluded after analysis of faecal droppings of the sheep that grazed in the area. Although the investigators were not able to identify the source of infection, the analysis of the risk factors correlated between drinking of tap water during the field exercise or the consumption of various meals at the beginning of the field training and gastroenteritis. Gallas-Lindemann et al. [18] examined a large area of 650 km2 in Lower Rhine, Germany, and provided substantial evidences on the dissemination and circulation of infected with (oo)cysts waste water. Deposition of (oo)cysts in waste water from households and their presence in all investigated water sources demonstrated the risk of waterborne transmission threatening for human health (from waste water through surface and groundwater to drinking water), since both parasites were detected in the whole surface water system [18].
Giardia and Cryptosporidium infections in household and domestic animals have been described also in other European countries highlighting the role of animals, and in particular of cats and dogs, in transmission of the infection to humans [8, 19, 28, 34, 35].
In spite of the small number of the indicated positive samples, the present study enhances the knowledge of the occurrence of Giardia spp. and Cryptosporidium spp. in privately owned household animals and genetically characterises the isolates found in the infected animals.
In summary, both Giardia and Cryptosporidium parasites are capable of infecting a variety of vertebrate species, from humans to animals, demonstrate a worldwide distribution, and are pathogens of veterinary and public health concern because of their ability to cause gastrointestinal disease, their ubiquitous presence in the environment, and the propensity for waterborne and foodborne outbreaks of these parasites. The absence of clinical symptoms results in the true prevalence of these parasites being underestimated, not only in cats and dogs but also in humans, and should be of major concern to veterinarians and physicians alike. Factors that may be affecting the underestimation of the risk could be related to poor and outdated information, improper interpretation of the published data and lack of sensitive methods. Infected household animals may especially pose a risk to immunocompromised people because they can be more susceptible to infection with Giardia and Cryptosporidium.
Acknowledgments
Note by Editor. This manuscript was accepted for publication in March 2012 but for some unknown reason the revised manuscript was not receive before February 2013. The paper was accepted after additional reviewing.
Cite this article as: Sotiriadou I, Pantchev N, Gassmann D & Karanis P: Molecular identification of Giardia and Cryptosporidium from dogs and cats. Parasite, 2013, 20, 8.
References
- 1.Abe N, Kimata I, Iseki M.2003. Identification of genotypes of Giardia intestinalis isolates from dogs in Japan by direct sequencing of the PCR amplified glutamate dehydrogenase gene. Journal of Veterinary Medical Science, 65, 29–33 [DOI] [PubMed] [Google Scholar]
- 2.Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F.2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. Journal of Clinical Microbiology, 41, 2744–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews RH, Monis PT, Ey PL, Mayrhofer G.1998. Comparison of the levels of intra-specific genetic variation within Giardia muris and Giardia intestinalis. International Journal for Parasitology, 28, 1179–1185 [DOI] [PubMed] [Google Scholar]
- 4.Ballweber LR, Xiao L, Bowman DD, Kahn G, Cama VA.2010. Giardiasis in dogs and cats: update on epidemiology and public health significance. Trends in Parasitology, 26, 180–189 [DOI] [PubMed] [Google Scholar]
- 5.Barutzki D, Schaper R.2003. Endoparasites in dogs and cats in Germany 1999–2002. Parasitology Research, 90, S148–S150 [DOI] [PubMed] [Google Scholar]
- 6.Barutzki D, Schaper R.2011. Results of parasitological examinations of faecal samples from cats and dogs in Germany between 2003 and 2010. Parasitology Research, 109, S45–S60 [DOI] [PubMed] [Google Scholar]
- 7.Barutzki D, Thompson R, Wielinga C, Parka U, Schaper R.2007. Observations on Giardia infection in dogs from veterinary clinics in Germany. Parasitology Research, 101, 153–15617216487 [Google Scholar]
- 8.Beck R, Sprong H, Pozio E, Cacciò SM.2012. Genotyping Giardia duodenalis isolates from dogs: lessons from a multilocus sequence typing study. Vector-Borne and Zoonotic Diseases, 12, 206–213 [DOI] [PubMed] [Google Scholar]
- 9.Berrilli F, D’Alfonso R, Giangaspero A, Marangi M, Brandonisio O, Kaboré Y, Glé C, Cianfanelli C, Lauro R, Di Cave D.2012. Giardia duodenalis genotypes and Cryptosporidium species in humans and domestic animals in Cote d’Ivoire: occurrence and evidence for environmental contamination. Transactions of the Royal Society of Tropical Medicine and Hygiene, 106, 191–195 [DOI] [PubMed] [Google Scholar]
- 10.Brockmann SO, Dreweck C, Wagner-Wiening C, Hagen RM, Kimmig P, Petry F, Jakobi V.2008. Serological and epidemiological analysis of an outbreak of gastroenteritis among military recruits in Germany caused by Cryptosporidium parvum. Infection, 36, 450–457 [DOI] [PubMed] [Google Scholar]
- 11.Broglia A, Reckinger S, Cacciò SM, Nockler K.2008. Distribution of Cryptosporidium parvum subtypes in calves in Germany. Veterinary Parasitology, 154, 8–13 [DOI] [PubMed] [Google Scholar]
- 12.Cacciò SM, Ryan U.2008. Molecular epidemiology of giardiasis. Molecular and Biochemical Parasitology, 160, 75–80 [DOI] [PubMed] [Google Scholar]
- 13.Cacciò SM, Thompson RC, McLauchlin J, Smith HV.2005. Unravelling Cryptosporidium and Giardia epidemiology. Trends in Parasitology, 21, 430–437 [DOI] [PubMed] [Google Scholar]
- 14.Covacin C, Aucoin DP, Elliot A, Thompson RC.2011. Genotypic characterisation of Giardia from domestic dogs in the USA. Veterinary Parasitology, 177, 28–32 [DOI] [PubMed] [Google Scholar]
- 15.Dyachenko V, Kuhnert Y, Schmaeschke R, Etzold M, Pantchev N, Daugschies A.2010. Occurrence and molecular characterization of Cryptosporidium spp. genotypes in European hedgehogs (Erinaceus europaeus L.) in Germany. Parasitology, 137, 205–216 [DOI] [PubMed] [Google Scholar]
- 16.Eligio-García L, Cortes-Campos A, Jiménez-Cardoso E.2005. Genotype of Giardia intestinalis isolates from children and dogs and its relationship to host origin. Parasitology Research, 97, 1–6 [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J.1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 39, 783–791 [DOI] [PubMed] [Google Scholar]
- 18.Gallas-Lindemann C, Sotiriadou I, Plutzer J, Karanis P.2013. Prevalence and distribution of Cryptosporidium and Giardia in wastewater and the surface, drinking and ground waters in the Lower Rhine, Germany. Epidemiology and Infection, 141, 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamnes IS, Gjerde BK, Robertson LJ.2007. A longitudinal study on the occurrence of Cryptosporidium and Giardia in dogs during their first year of life. Acta Veterinaria Scandinavica, 49, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homan WL, van Enckevort FH, Limper L, van Eys GJ, Schoone GJ, Kasprzak W, Majewska AC, van Knapen F.1992. Comparison of Giardia isolates from different laboratories by isoenzyme analysis and recombinant DNA probes. Parasitology Research, 78, 316–323 [DOI] [PubMed] [Google Scholar]
- 21.Hopkins RM, Meloni BP, Groth DM, Wetherall JD, Reynoldson JA, Thompson RC.1997. Ribosomal RNA sequencing reveals differences between the genotypes of Giardia isolates recovered from humans and dogs living in the same locality. Journal of Parasitology, 83, 44–51 [PubMed] [Google Scholar]
- 22.Joachim A, Eckert E, Petry F, Bialek R, Daugschies A.2003. Comparison of viability assays for Cryptosporidium parvum oocysts after disinfection. Veterinary Parasitology, 111, 47–57 [DOI] [PubMed] [Google Scholar]
- 23.Joachim A, Krull T, Schwarzkopf J, Daugschies A.2003. Prevalence and control of bovine cryptosporidiosis in German dairy herds. Veterinary Parasitology, 112, 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karanis P, Ey PL.1998. Characterization of axenic isolates of Giardia intestinalis established from humans and animals in Germany. Parasitology Research, 84, 442–449 [DOI] [PubMed] [Google Scholar]
- 25.Karanis P, Plutzer J, Halim NA, Igori K, Nagasawa H, Ongerth J, Liqing M.2007. Molecular characterization of Cryptosporidium from animal sources in Qinghai province of China. Parasitology Research, 101, 1575–1580 [DOI] [PubMed] [Google Scholar]
- 26.Kimura M.1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16, 111–120 [DOI] [PubMed] [Google Scholar]
- 27.Lasek-Nesselquist E, Welch DM, Sogin ML.2010. The identification of a new Giardia duodenalis assemblage in marine vertebrates and a preliminary analysis of G. duodenalis population biology in marine systems. International Journal for Parasitology, 40, 1063–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lebbad M, Petersson I, Karlsson L, Botero-Kleiven S, Andersson JO, Svenungsson B, Svärd SG.2011. Multilocus genotyping of human Giardia isolates suggests limited zoonotic transmission and association between assemblage B and flatulence in children. PLoS Neglected Tropical Diseases, 5, e1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leonhard S, Pfister K, Beelitz P, Wielinga C, Thompson RC.2007. The molecular characterisation of Giardia from dogs in southern Germany. Veterinary Parasitology, 150, 33–38 [DOI] [PubMed] [Google Scholar]
- 30.Lucio-Forster A, Griffiths JK, Cama VA, Xiao L, Bowman DD.2010. Minimal zoonotic risk of cryptosporidiosis from pet dogs and cats. Trends in Parasitology, 26, 174–179 [DOI] [PubMed] [Google Scholar]
- 31.Monis PT, Andrews RH, Mayrhofer G, Mackrill J, Kulda J, Isaac-Renton JL, Ey PL.1998. Novel lineages of Giardia intestinalis identified by genetic analysis of organisms isolated from dogs in Australia. Parasitology, 116, 7–19 [DOI] [PubMed] [Google Scholar]
- 32.Nichols RA, Campbell BM, Smith HV.2003. Identification of Cryptosporidium spp. oocysts in United Kingdom noncarbonated natural mineral waters and drinking waters by using a modified nested PCR-restriction fragment length polymorphism assay. Applied and Environmental Microbiology, 69, 4183–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichols RA, Connelly L, Sullivan CB, Smith HV.2010. Identification of Cryptosporidium species and genotypes in Scottish raw and drinking waters during a one-year monitoring period. Applied and Environmental Microbiology, 76, 5977–5986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overgaauw PA, van Zutphen L, Hoek D, Yaya FO, Roelfsema J, Pinelli E, van Knapen F, Kortbeek LM.2009. Zoonotic parasites in fecal samples and fur from dogs and cats in The Netherlands. Veterinary Parasitology, 163, 115–122 [DOI] [PubMed] [Google Scholar]
- 35.Paoletti B, Otranto D, Weigl S, Giangaspero A, Di Cesare A, Traversa D.2011. Prevalence and genetic characterization of Giardia and Cryptosporidium in cats from Italy. Research in Veterinary Science, 91, 397–399 [DOI] [PubMed] [Google Scholar]
- 36.Pavlasek I, Ryan U.2007. The first finding of a natural infection of Cryptosporidium muris in a cat. Veterinary Parasitology, 144, 349–352 [DOI] [PubMed] [Google Scholar]
- 37.Plutzer J, Karanis P.2007. Genotype and subtype analyses of Cryptosporidium isolates from cattle in Hungary. Veterinary Parasitology, 146, 357–362 [DOI] [PubMed] [Google Scholar]
- 38.Plutzer J, Ongerth J, Karanis P.2010. Giardia taxonomy, phylogeny and epidemiology: Facts and open questions. International Journal of Hygiene and Environmental Health, 213, 321–333 [DOI] [PubMed] [Google Scholar]
- 39.Pozio E.2008. Epidemiology and control prospects of foodborne parasitic zoonoses in the European Union. Parassitologia, 50, 17–24 [PubMed] [Google Scholar]
- 40.Robertson LJ.2009. Giardia and Cryptosporidium infections in sheep and goats: a review of the potential for transmission to humans via environmental contamination. Epidemiology and Infection, 137, 913–921 [DOI] [PubMed] [Google Scholar]
- 41.Saitou N, Nei M.1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 42.Santin M, Trout JM, Vecino JA, Dubey JP, Fayer R.2006. Cryptosporidium, Giardia and Enterocytozoon bieneusi in cats from Bogota (Colombia) and genotyping of isolates. Veterinary Parasitology, 141, 334–339 [DOI] [PubMed] [Google Scholar]
- 43.Sprong H, Cacciò SM, van der Giessen JW.2009. Identification of zoonotic genotypes of Giardia duodenalis. PLoS Neglected Tropical Diseases, 3, e558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S.2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson RC, Colwell D, Shury T, Appelbee A, Read C, Njiru Z, Olson ME.2009. The molecular epidemiology of Cryptosporidium and Giardia infections in coyotes from Alberta, Canada, and observations on some cohabiting parasites. Veterinary Parasitology, 159, 167–170 [DOI] [PubMed] [Google Scholar]
- 46.Thompson RC, Hopkins RM, Homan WL.2000. Nomenclature and genetic groupings of Giardia infecting mammals. Parasitology Today, 16, 210–213 [DOI] [PubMed] [Google Scholar]
- 47.Thompson RC, Palmer CS, O’Handley R.2008. The public health and clinical significance of Giardia and Cryptosporidium in domestic animals. Veterinary Journal, 177, 18–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Traub RJ, Monis PT, Robertson I, Irwin P, Mencke N, Thompson RC.2004. Epidemiological and molecular evidence supports the zoonotic transmission of Giardia among humans and dogs living in the same community. Parasitology, 128, 253–262 [DOI] [PubMed] [Google Scholar]
- 49.Vanni I, Cacciò SM, van Lith L, Lebbad M, Svärd SG, Pozio E, Tosini F.2012. Detection of Giardia duodenalis assemblages A and B in human feces by simple, assemblage-specific PCR assays. PLoS Neglected Tropical Diseases, 6, e1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao L, Fayer R.2008. Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Veterinary Parasitology, 38, 1239–1255 [DOI] [PubMed] [Google Scholar]