Abstract
S100 proteins are markers for numerous cancers, and in many cases high S100 protein levels are a prognostic indicator for poor survival. One such case is S100B, which is overproduced in a very large percentage of malignant melanoma cases. Elevated S100B protein was more recently validated to have causative effects towards cancer progression via down-regulating the tumor suppressor protein, p53. Towards eliminating this problem in melanoma, targeting S100B with small molecule inhibitors was initiated. This work relies on numerous chemical biology technologies including structural biology, computer-aided drug design, compound screening, and medicinal chemistry approaches. Another important component of drug development is the ability to test compounds and various molecular scaffolds for their efficacy in vivo. This chapter briefly describes the development of S100B inhibitors, termed SBiXs, for melanoma therapy with a focus on the inclusion of in vivo screening at an early stage in the drug discovery process.
Keywords: In vivo screening, Preclinical testing, Intratumoral delivery, Systemic delivery, Pharmacokinetics, Pharmacodynamics, Maximum tolerated dose, Therapeutic window, Genetically modified mouse models, S100 proteins, EF-hand
1. Introduction
In malignant melanoma (MM), the tumor marker S100B binds directly to wild-type p53, dissociates the p53 tetramer, enhances hdm2-dependent ubiquitination of p53, and down-regulates p53-dependent tumor suppression functions (1–3). As a proof of principle for drug design, inhibiting S100B with small interfering antisense RNA (siRNAS100B) or with several S100B inhibitors (SBiXs; X = compound number) was achieved and shown to restore wild-type p53 at the protein level. Inhibiting S100B production was also found to cause an increase in the levels of p53 gene products as necessary to induce cell growth arrest and apoptosis in malignant melanoma (2, 4–7). Also encouraging from a drug development standpoint is that ablation of S100B expression in mice via gene targeting produces unremarkable phenotypes with few, if any, problematic physiological consequences (8–13).
With any drug-design program, there is a need to obtain physiological data at an early stage in the process to help determine whether a compound or a series of compounds induce off-target effects and/or cause other unanticipated toxicities. This is particularly important for S100 inhibitors since there are over 20 structurally similar proteins in the S100 protein family, and they each regulate several physiologically important pathways in a cell-specific manner (14, 15). Thus, an S100 inhibitor could have multiple phenotypes depending on the number of S100 proteins it blocked and the S100 status of the cell-type targeted. In the case of blocking the S100B–p53 protein–protein interaction in malignant melanoma, there exists a separate issue with regard to a feedback loop that is initiated when S100B is inhibited since the gene for S100B itself is up regulated by the tumor suppressor protein, when p53 levels are restored (2). To address these and other issues, this chapter describes the discovery/development of SBiXs for melanoma therapy with a focus on the importance of doing in vivo screening of lead compounds at an early stage in the drug development process. It is likely that such an approach of performing early in vivo screening could benefit many drug discovery programs.
Small molecule inhibitors have been reported for numerous S100 family members, some of which are in clinical trials. Inhibitors of the S100A10–annexin A2 interaction have not been validated in vivo but their predicted clinical uses include angiogenesis and cancer metastasis therapy (16, 17). The anti-allergic drug cromolyn disrupts S100P–RAGE interaction and reduces pancreatic tumor formation in animal models (18). Interestingly, cromolyn also binds other S100 family members (S100A1, S100A12, and S100A13), but its effects on the interaction of these family members with their target proteins have not been fully investigated. Other small molecules that bind S100A1 include pentamidine and propanolol (19). In the case of S100A4, several phenothiazines block S100A4-mediated depolymerization of myosin-IIA filaments (20, 21). S100A4 also binds anti-allergic drugs and a modified version of azaxanthone, which is in clinical trials for treatment of metastatic disease states (21, 22). Two SBiXs developed by our group disrupt S100B–p53 complex formation and prevent unregulated melanoma cell growth and are currently being tested in human and veterinary clinical trials as potential melanoma therapeutics.
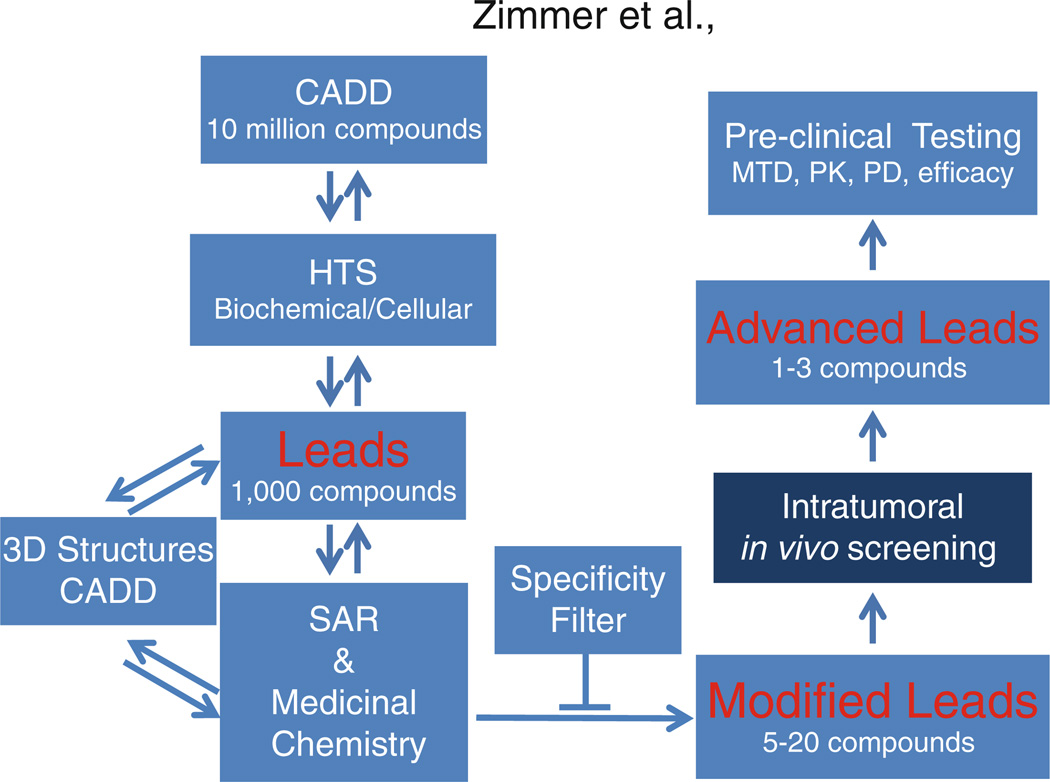
For the development of SBiXs, a combination of computer-aided drug design (CADD), high-throughput screening (HTS), structural biology, medicinal chemistry, and in vivo biology/drug testing approaches is employed. The benefits of a structure-based approach are taken advantage of for developing new compounds as well as addressing issues with regard to target specificity since the 3D structures for several S100 and S100–target complexes are published (4, 15, 19, 23–27). Likewise, structure/activity relationships (SAR) and SAR by NMR approaches are more efficiently directed when the structures are available (28, 29). This is especially important for the difficult hurdle of inhibiting a protein–protein interaction such as the S100B–p53 complex (4, 7, 27, 30). More specifically, modifications on a particular scaffold that depend on the structure of a lead compound bound to S100B are achieved iteratively by using CADD-directed medicinal chemistry; likewise, structural studies can be used to identify multiple sites within the p53-binding cleft on S100B via structural biology approaches (NMR, X-ray), so compounds and combinations of compounds can be rationally linked together synthetically to obtain tight and highly specific S100B binders (i.e., relative to binding S100A1 for example). Such a drug development process is very dependent on the latest new results, so this process occurs in a very iterative manner (Fig. 1). Once promising leads are identified, then it is important that they are screened for their in vivo efficacy and toxicity. Such leads are chosen based on binding affinities (KDs) and/or specificity both in biochemical assays (i.e., S100B thermodynamic/kinetic data versus other S100s) and in cellular assays (i.e., IC50s). For the cellular assays, specificity and other potential off-target effects are evaluated by comparing IC50 values of isogenic cell lines plus/minus S100B present (31, 32). Of the approximately 100–200 lead compounds that undergo extensive in vitro testing each year, eight to ten typically meet the criteria for early-stage in vivo screening.
Figure 1.
Strategy for the discovery and development of SBiXs. An iterative process that includes computer-aided drug design (CADD), high-throughput screening (HTS), structure/activity (SAR) and medicinal chemistry, 3D structure (NMR and X-ray), and a specificity filter is used to engineer modified SBiXs for intratumoral in vivo screening and subsequent advanced leads suitable for preclinical evaluation.
High-throughput biochemical and cellular screens identify two types of lead SBiXs: FDA-approved compounds and new chemical entities. Since there are no effective treatments for melanoma, repurposing of FDA-approved lead SBiXs for melanoma therapy is a high priority. Efficacy evaluation is the primary focus for repurposing approved drugs, since extensive pharmacological/toxicological information is already available for many species, including humans. In fact, phase II human and phase I canine clinical trials for two FDA-approved SBiXs are underway. However, like most drug discovery programs, the vast majority of lead SBiXs are new chemical entities that require extensive development and optimization. In vivo testing in animal models plays a fundamental role in developing new anticancer drugs. Prior to human testing, preclinical tolerability (maximum tolerated dose, MTD), pharmacokinetics (PK), pharmacodynamic (PD), and efficacy assays provide key information that is used to improve advanced leads. For example, a compound may need to be more lipophilic to pass through the cell membrane and reach its target or side groups may need to be added to allow for oral delivery or brain penetration.
In the case of accessible tumors such as melanoma, drugs can be delivered directly to the tumor (intratumoral) without optimization for systemic delivery and/or minimization of toxic effects on normal cells. In addition, intratumoral delivery can achieve significantly higher drug concentrations at the site of action than systemic delivery. In the clinical setting, intratumoral administration has been used to deliver gene therapy constructs, complex biologics, and small molecules to a variety of cancers including adenoviral based p53 genes in head and neck cancer (33); TNFalpha genes in rectal cancer (34); interleukin-2 in melanoma (35); immunostimulant CpG in brain cancers (36); single treatment of a meta-static squamous cell carcinoma lesion (37); BCNU in combination with radiotherapy in glioma (38); and para-toluenesulfonamide in non-small cell lung cancer (39). Intratumoral delivery has been used in the preclinical evaluation of siRNAs (40) and immune modulators (41, 42) as well as to reduce the toxicity of approved agents such as melphalan (43); 2-deoxy- D -glucose alone and in combination with carboplatin (44); and paclitaxel/docetaxel in mammary, bladder, prostate, and head and neck cancers (45–48). Our S100B inhibitor drug development program includes an intratumoral in vivo screen of lead SBiXs before preclinical testing of advanced leads. While in vivo testing prior to medicinal chemistry optimization and ADME testing is atypical, the availability of in vivo screening data early in the drug discovery process focuses resources on SBiXs with the greatest probability of clinical success.
A major consideration for any in vivo trial is the selection of an appropriate animal model. Mouse models are a mainstay in animal testing because it is feasible to perform studies in a short period of time. However, no single mouse model recapitulates the complex genetics and biology of human melanoma (49–52). Therefore, the biological question being asked as well as the advantages and limitations of the various types of models were important criteria in selecting an animal model for intratumoral in vivo screening of SBiXs. Syngeneic transplantation models involve the implantation of well-characterized melanoma cell lines into a syngeneic host and do not recapitulate many aspects of the human disease because characterized mouse lines do not reflect the heterogeneity observed in human tumors. Xenogeneic transplantation models involve the implantation of cell lines or patient-derived cells into an immunocompromised host. Both of these transplantation models can be flank models in which subcutaneous tumors are grown on the backs of mice or orthotopic models in which tumors are grown at the site of origin (i.e., breast cancer cells in the mammary fat pad). Orthotopic models have a better chance to metastasize but are more difficult to monitor. One disadvantage of human xenograft models is the fact that the host is immunocompromised and immunosurvelliance has been shown to play a role in limiting metastasis (53). In the case of SBiXs, S100B’s effects on immune responses in the CNS are well documented although little is known about its role in peripheral immune responses. Nonetheless, human xenograft models are usually considered to be superior to genetically modified mouse models because the composition of the resulting tumor mimics the heterogeneity observed in patients. However, the development of multi-allelic genetically engineered mouse models that mimic spontaneous tumorigenesis and heterogeneity as well as target validation in simulated clinical trials using standard of care chemotherapeutics confirm the utility of genetically modified mouse models in developing cancer therapeutics (54).
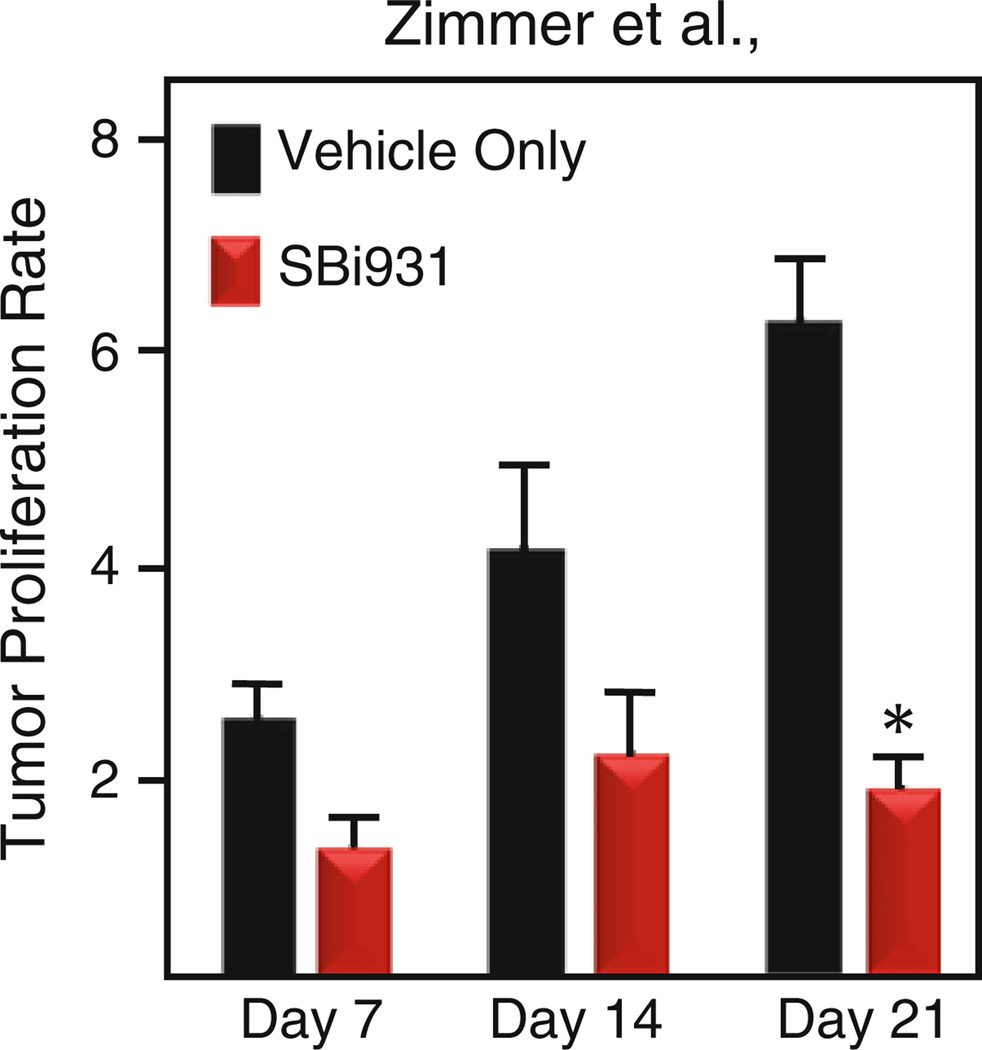
The RAS-induced INK4a/ARF−/− mouse melanoma model (55) was chosen for in vivo screening of lead SBiXs because it has (1) an intact S100B–p53 signaling pathway (elevated S100B and wild-type p53), (2) an intact immune system, (3) tumors which are amenable to intratumoral delivery, and (4) a proven record in developing new melanoma therapies (55, 56). The Tyr::RASG12V / INK4a/ARF−/− line is bigenic and contains two genomic mutations on an FVB background: a mutated H-rasG12V transgene on the Y chromosome and inactivated INK4a/ARF alleles on chromosome 4. This model is not commercially available and we maintain a breeding colony that generates experimental animals as well as breeders for the individual Tyr::RASG12V and INK4a/ ARF−/− lines. Founders for both lines were obtained from the National Cancer Institute Mutant Mouse Resource (Frederick, MD). At 2–3 months of age experimental Tyr::RASG12V /INK4a/ ARF−/− males develop spontaneous cutaneous melanomas in the pinna of the ears (30%), torso (23%), and tail (20%) without distant metastasis (55). Our intratumoral in vivo screening protocol for modified SBiXs is a longitudinal design with a study period of 3–8 weeks and uses the relative tumor proliferation rate (the tumor volume at a particular treatment interval/tumor volume at the time of treatment initiation) as the primary outcome (Fig. 2). Although this trial design is not optimized for garnering tolerability, PK or PD information, the gross/histological pathology, SBiX levels, and p53 pathway reactivation in the tumors are monitored and this information is useful in selecting advanced leads that will proceed to preclinical testing.
Figure 2.
Intratumoral in vivo screening trial. The histogram depicts the mean tumor responsiveness for intratumoral injection of SBi931 (red bars) or vehicle only (black bars) every other day for 21 days. Asterisk denotes p < 0.05 when compared to vehicle only.
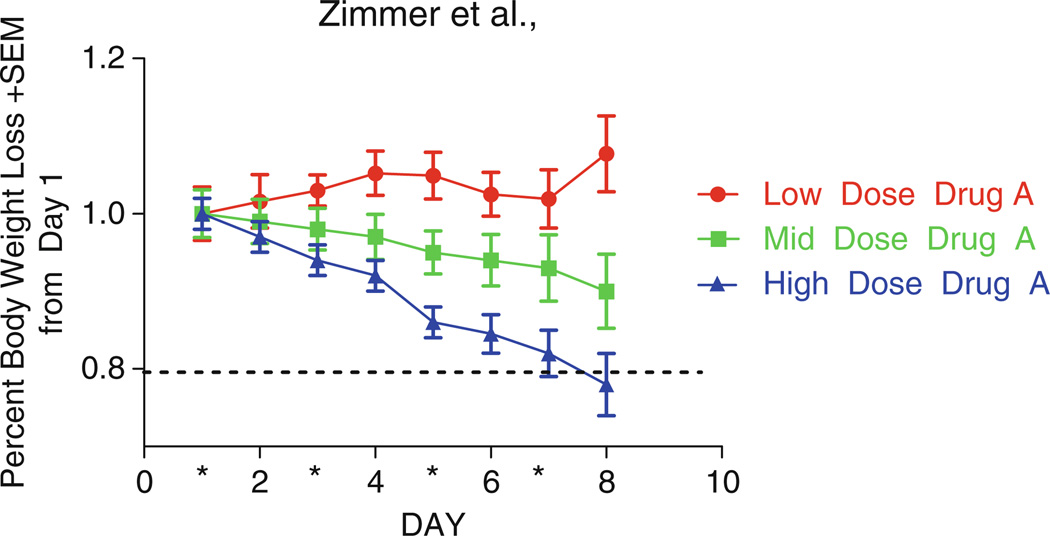
In the case of modified leads with predicted ADME properties favorable for systemic administration, concurrent tolerability (MTD) and pharmacokinetic (PK) assays are also conducted. These trials provide valuable pharmacological information that cannot be garnered from local administration trials including potential effects of the route of administration on tumor responsiveness (57). MTD and PK studies are conducted in the same species/strain and employ the same dosing scheme that will be used in subsequent efficacy studies. For example, if the expected SBiX dosing scheme is 2 weeks of consecutive intraperitoneal (IP) administration in female nu/nu mice, then the MTD experiment should be dosed IP daily × 14 in female nu/nu mice. The route of administration, oral (PO), intraperitoneal (IP), intravenous (IV), or subcutaneous (SC), is determined by the predicted chemical properties of the SBiX. In MTD experiments, animals are monitored post cessation of dosing for delayed toxicity for a minimum of 1 week and optimally 2 weeks. In mouse models, the best indicator for toxicity is weight. MTDs for novel cancer drugs are considered an LD10, the dose at which 10% of mice in one group die or lose 20% of their body weight (58) (Fig. 3). The therapeutic index for cancer drugs is very small in that the MTD can be equivalent to the effective dose. The goal of PK studies is to select a dose and route of administration that allow the plasma/target tissue drug levels to exceed the IC50 value from cellular assays and/or KD from biochemical screens. SBiX levels in the tumor tissue provide useful information about tumor penetrance, half-life in target tissue, and the compound’s ability to leave the bloodstream. Also, SBiX levels in brain tissue assess brain penetration and potential for use in metastatic disease therapy and primary brain cancers. The data from the intratumoral in vivo screening, MTD, and PK trials described below determines if modified leads are suitable for preclinical testing, require additional medicinal chemistry optimization and further testing prior to preclinical testing, or are eliminated.
Figure 3.
Systemic tolerability testing/MTD. The graph depicts the percent body weight loss verses time for groups (n = 3 mice) receiving daily IP doses (mid, low, or high) of a drug for 7 consecutive days. The decrease in mean body weight in the high-dose group was greater than 20% indicating that this dose exceeds the MTD.
2. Materials
SBiX stock: 100 mM in sterile dimethylsulfoxide (DMSO). Dissolve appropriate amount of SBiX in sterile- filtered DMSO to achieve final concentration of 100 mM. Prepare 100 µl aliquots and store at −20°C.
SBiX Injection Solution: For intratumoral in vivo screening, the SBiX injection solution composition is dependent upon the tumor volume and the KD /IC50 of the SBiX, i.e., injection volume ≤ 20% of tumor volume and [SBiX]intratumor = fivefold over KD or IC50, whichever is greater. For systemic tolerability studies, SBiX injection solution composition is dependent upon the dose and appropriate volume range for the route of administration. Dilutions are made in sterile phosphate-buffered saline (PBS).
Vehicle-Only Control: Dilute sterile DMSO into PBS in the same ratios used to prepare the SBiX injection solution in item 2.
Avertin solution: Avertin stock solution (100%) prepared by dissolving tribromoethanol in tertiary amyl alcohol and stored in a brown glass bottle at 4°C. The avertin working solution (2.5%) is prepared fresh weekly by diluting 1.25 ml stock solution (100%) to 48.75 ml sterile filtered PBS. After the pH is adjusted to 7.4, the solution is filtered through a 0.22 micron filter and stored in a brown bottle at 4°C. All containers should be labeled in accordance with institutional and regulatory policies regarding the use of animals.
Syringes: 20 G × 38 mm gastric gavage needles (plastic or metal). 1 cc tuberculin syringes with 25G, 26G, 27G, or 30G needle.
Calipers: TTC Digital Caliper 0–150 mm.
3. Methods
All experiments involving animals described in this section were approved by the institutional IACUC and performed in accordance with the NIH guidelines for the treatment of animals. Appropriate institutional and/or regulatory agency approval is required for these procedures.
3.1. Intratumoral In Vivo Screening
Ten or more experimental Tyr::RASG12V /INK4a/ARF−/− males are obtained from our in-house breeding colony and randomly assigned to a cohort (experimental or control) when tumors ≥ 20 mm3 in size develop.
Assess and record the general health status using the following criteria: body weight, grooming, eating, drinking, alterness/lethargy, mobility, coat (smooth vs. ruffled), and posture (hunch vs. upright) (see Note 1).
Assess and record the gross pathology of the tumor using the following criteria: erosion/invasion, redness, in flammation, and weeping (edema and presence of fluids) (see Note 1).
Restrain the animal and measure the length (l) and width (w) of the tumor (mm) with calipers (see Note 2).
Slowly inject the SBiX injection solution or vehicle-only control directly into the tumor.
Monitor and record the overall health status, gross tumor pathology, and tumor size daily.
Administer SBiX or vehicle-only control every other day. Rotate the injection site to minimize in flammation (see Note 3).
At the conclusion of the study period (3–8 weeks), euthanize animals using anesthetic overdose followed by decapitation. Administer 0.03 ml/g body weight of sterile Avertin (2.5% w/v) by ip injection. Administer additional doses of 0.15 ml as needed to attain a deep plane of anesthesia as determined by pedial and breathing reflexes (see Notes 4–6).
Examine and record the gross pathology of the tumor. Excise and weigh the tumor. Slice the tumor in half along the long axis. Process one-half for histological analysis (formalin fixation and paraffin-embedding) and the other half for biochemical/pharmacological analyses (snap frozen).
Examine internal organs for gross pathology and remove any abnormal organs for processing and histological analysis.
Tumor response is expressed as the mean tumor proliferation rate ± the SEM at 7, 14, and 21 days, i.e., the tumor volume (V = ½ab2) on day 7, 14, or 21 divided by the tumor volume at day 0 (Fig. 2)
3.2. Systemic Tolerability Testing (MTD and PK)
MTD studies are preferably carried out in efficacy species/strain but can be performed in an outbred mouse followed by a small bridging study in efficacy species/strain. Mice should be acclimated for at least 3 days prior to start of experiment. Three mice are the absolute minimum required and more than 5 are utilized if fine-tuning of the dose is required. The dosing schedule and route of administration (PO, IP, SC, IV, or IM) should be chosen based on the desired dosing schedule/route of administration in the proposed efficacy experiment. Mice should be monitored for 1–2 weeks post cessation of dosing to monitor for long-term toxicities.
Divide mice into four groups of three to ten mice. Each group will receive a different dose, i.e., 10×, 5×, 2.5×, and 1× dosing. If the vehicle is known to be nontoxic it may be excluded from the MTD study.
Novel compounds should be dosed IP, IM, SC, or IV using dosing schedule planned for future efficacy experiment. Depending on the route of administration, mice are either restrained by hand (IP, IM, and PO) or immobilized in a mouse restrainer and the tail is warmed by infrared light, hot water, or warm ethanol (IV). Usually mice are not anesthetized for dosing (see Note 7 for dosing volumes and needle size).
Mice should be monitored or observed daily and weighed 3 times per week.
Unacceptable toxicity is the inability of a mouse to ambulate in order to drink and eat in a 24-h period or 20% body weight loss at any time or 15% body weight loss over 72 h as compared to day 1 of the experiment. Animals that meet these criteria should be removed from the study and euthanized (see Note 8) (Fig. 3).
When dosing is complete, mice are monitored for 2 weeks to detect delayed toxicities.
Pharmacokinetic studies are carried out in naïve mice that may be tumor bearing or not. PK experiments are usually carried out with 18 mice (unless the desire is to test more time points).
Mice are divided into six groups of three mice.
Three mice are in the control group (time zero—no drug or vehicle) and 15 mice are in the treated groups (see Note 9) if mice will be implanted with tumor cells.
SBiXs are usually dosed ONCE by the route of administration planned for the efficacy experiment. The dose can range from 10 to 50 mg/kg.
Mice are euthanized (e.g., CO2 inhalation) at specific time points post dosing. For SBiXs, typical time points are 15 min, 30 min, 1 h, 4 h, and 8 h post dosing (see Note 6).
Blood is collected by cardiac puncture (27G needle with 1 ml disposable syringe) and plasma is isolated after centrifugation in BD microtainer heparinized tubes. Mice are placed in left hand and 1 cc syringe with 27 G 1/2 ″ needle is inserted under the sternum towards the heart. The plunger is slowly pulled from the barrel. If no blood is observed, the needle is pulled out and reinserted. The needle is removed from barrel before injecting blood into microtainer tube. The tube is immediately capped and inverted several times to prevent clotting. Samples are centrifuged in a tabletop centrifuge for 10 min at max speed. Plasma is poured off into labeled microfuge tube and stored at −20°C until analysis.
Tumor, brain, or other tissue may be collected at time points noted. Tissue is snap frozen in an ethanol/dry ice bath and stored at −80°C until analysis.
Acknowledgments
These studies were supported by NIH grant CA107331 (DJW) and the Center for Biomolecular Therapeutics (CBT), The University of Maryland School of Medicine, and the Institute for Bioscience and Biotechnology Research (IBBR).
Footnotes
Any animals exhibiting clinically significant health issues should be evaluated for removal from the study.
If a scruff hold blocks tumor access, then isoflurane anesthesia can be used for immobilization.
The trial schedule can be adjusted to match the predicted pharmacokinetics of the SBiX. Trials for SBiXs that are predicted to concentrate in tumor cells have used a 5-day on and 2-day off schedule. Trials for SBiXs that are not predicted to concentrate in tumors have used everyday and every other day schedules.
The trial duration for a typical SBiX in vivo screen is 3 weeks. However, the trial duration can be shortened or lengthened. If tumors progress to complete remission and/or unexpected toxicities are encountered, then trial duration can be shortened. Trial duration for the Tyr::RASG12V /INK4a/ARF−/− model should not exceed 8 weeks because animals will be approaching the age (5.5 months) at which 50% of the mice succumb to tumor growth.
Avertin has been reported to be an irritant and the following steps should be taken to minimize decomposition: (1) store and use under sterile conditions; (2) store in the dark at 4°C; (3) adjust pH of solution to >5.0; (4) discard stock solution after 4 months; (5) discard working solution after 7 days; and (6) discard any solutions that become discolored or have precipitate.
Other approved methods of euthanasia can be used.
| Route | Needle size and volume |
|---|---|
| Oral (PO) | 20G × 38 mm gastric gavage needles (plastic or metal) |
| Dosing volume should be 10 ml/kg and no more than 5 ml/kg | |
| Intraperitoneal (IP) | 26 or 27G needle with 1 ml disposable syringe |
| Dosing volume should be 10 ml/kg and no more than 5 ml/kg | |
| Subcutaneous (SC) | 26 or 27G needle with 1 ml disposable syringe |
| Dosing volume should not exceed 300 ul | |
| Intravenous (IV) | 27G to 30G needle with 1 ml disposable syringe |
| Dosing volume should not exceed 10 ml/kg | |
| Intramuscular (IM) | 27G needle with 1 ml disposable syringe |
| 0.05 ml can be injected into calf muscle | |
If one mouse in a group of three loses 20% body weight loss, this may be past the MTD since the number of animals is very small. The study would have to be repeated using the reported MTD and lower doses.
The cell line of choice will be grown in culture using optimal conditions. Usually, 1 × 106 to 5 × 106 cells are injected with 33% to 50% Matrigel™ (BD Biosciences) on the right fl ank of mice. Cells are injected in 0.15–0.2 ml of volume with a 27G tuberculin syringe. Tumors should be monitored 3 times per week with electronic calipers as described earlier. When tumors reach approximately 400–500 mm3, the mice can be divided into six groups of three mice and the PK study may be initiated.
References
- 1.Lin J, Blake M, Tang C, Zimmer D, Rustandi RR, Weber DJ, Carrier F. Inhibition of p53 transcriptional activity by the S100B calcium- binding protein. J Biol Chem. 2001;276:35037–35041. doi: 10.1074/jbc.M104379200. [DOI] [PubMed] [Google Scholar]
- 2.Lin J, Yang Q, Yan Z, Markowitz J, Wilder PT, Carrier F, Weber DJ. Inhibiting S100B restores p53 levels in primary malignant melanoma cancer cells. J Biol Chem. 2004;279:34071–34077. doi: 10.1074/jbc.M405419200. [DOI] [PubMed] [Google Scholar]
- 3.Lin J, Yang Q, Wilder PT, Carrier F, Weber DJ. The calcium-binding protein S100B down-regulates p53 and apoptosis in malignant melanoma. J Biol Chem. 2010;285:27487–27498. doi: 10.1074/jbc.M110.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markowitz J, Mackerell AD, Jr, Carrier F, Charpentier TH, Weber DJ. Design of Inhibitors for S100B. Current Top Med Chem. 2005;5:1093–1108. doi: 10.2174/156802605774370865. [DOI] [PubMed] [Google Scholar]
- 5.Markowitz J, MacKerell AD, Jr, Weber DJ. A search for inhibitors of S100B, a member of the S100 family of calcium-binding proteins. Mini Rev Med Chem. 2007;7:609–616. doi: 10.2174/138955707780859422. [DOI] [PubMed] [Google Scholar]
- 6.Smith J, Stewart BJ, Glaysher S, Peregrin K, Knight LA, Weber DJ, Cree IA. The effect of pentamidine on melanoma ex vivo. Anticancer Drugs. 2010;21:181–185. doi: 10.1097/CAD.0b013e3283340cee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilder PT, Charpentier TH, Liriano MA, Gianni K, Varney KM, Pozharski E, Coop A, Toth EA, MacKerell AD, Jr, Weber DJ. In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction. Intl J High Throughput Screen. 2010;1:109–126. doi: 10.2147/IJHTS.S8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong Z, O’Hanlon D, Becker LE, Roder J, MacDonald JF, Marks A. Enhanced calcium transients in glial cells in neonatal cerebellar cultures derived from S100B null mice. Exp Cell Res. 2000;257:281–289. doi: 10.1006/excr.2000.4902. [DOI] [PubMed] [Google Scholar]
- 9.Roltsch E, Holcomb L, Young KA, Marks A, Zimmer DB. PSAPP mice exhibit regionally selective reductions in gliosis and plaque deposition in response to S100B ablation. J Neuroinflammation. 2010;7:78. doi: 10.1186/1742-2094-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsoporis JN, Overgaard CB, Izhar S, Parker TG. S100B modulates the hemodynamic response to norepinephrine stimulation. Am J Hypertens. 2009;22:1048–1053. doi: 10.1038/ajh.2009.145. [DOI] [PubMed] [Google Scholar]
- 11.Schulte-Herbruggen O, Hortnagl H, Ponath G, Rothermundt M, Hellweg R. Distinct regulation of brain-derived neurotrophic factor and noradrenaline in S100B knockout mice. Neurosci Lett. 2008;442:100–103. doi: 10.1016/j.neulet.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Kim HS, Seto-Ohshima A, Nishiyama H, Itohara S. Normal delay eyeblink conditioning in mice devoid of astrocytic S100B. Neurosci Lett. 2011;489:148–153. doi: 10.1016/j.neulet.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Dyck RH, Bogoch II, Marks A, Melvin NR, Teskey GC. Enhanced epileptogenesis in S100B knockout mice. Brain Res Mol Brain Res. 2002;106:22–29. doi: 10.1016/s0169-328x(02)00406-0. [DOI] [PubMed] [Google Scholar]
- 14.Leclerc E, Heizmann CW. The importance of Ca2+/Zn2+ signaling S100 proteins and RAGE in translational medicine. Front Biosci (Schol Ed) 2011;3:1232–1262. doi: 10.2741/223. [DOI] [PubMed] [Google Scholar]
- 15.Zimmer DB, Weber DJ. The calcium-dependent interaction of S100B with its protein targets. Cardiovasc Psychiatry Neurol pii. 2010:728052. doi: 10.1155/2010/728052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C, Reddy TRK, Fischer PM, Dekker LV. A Cy5-labeled S100A10 tracer used to identify inhibitors of the protein interaction with annexin A2. Assay Drug Dev Technol. 2010;8:85–95. doi: 10.1089/adt.2009.0218. [DOI] [PubMed] [Google Scholar]
- 17.Reddy TR, Li C, Guo X, Myrvang HK, Fischer PM, Dekker LV. Design, synthesis, and structure-activity relationship exploration of 1-substituted 4-aroyl-3-hydroxy-5-phenyl-1 H-pyrrol-2(5 H)-one analogues as inhibitors of the annexin A2-S100A10 protein interaction. J Med Chem. 2011;54:2080–2094. doi: 10.1021/jm101212e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arumugam T, Ramachandran V, Logsdon CD. Effect of cromolyn on S100P interactions with RAGE and pancreatic cancer growth and invasion in mouse models. J Natl Cancer Inst. 2006;98:1806–1818. doi: 10.1093/jnci/djj498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright NT, Cannon BR, Zimmer DB, Weber DJ. S100A1: structure, function, and therapeutic potential. Curr Chem Biol. 2009;3:138–145. doi: 10.2174/187231309788166460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malashkevich VN, Dulyaninova NG, Ramagopal UA, Liriano MA, Varney KM, Knight D, Brenowitz M, Weber DJ, Almo SC, Bresnick AR. Phenothiazines inhibit S100A4 function by inducing protein oligomerization. Proc Natl Acad Sci USA. 2010;107:8605–8610. doi: 10.1073/pnas.0913660107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.House RP, Garrett SC, Bresnick AR. Moving aggressively: S100A4 and tumor invasion. In: Fatatis A, editor. Signaling pathways and molecular mediators in mestastasis. Chapter 4. New York: Springer; 2012. [Google Scholar]
- 22.Mack GS, Marshall A. Lost in migration. Nat Biotechnol. 2010;28:214–229. doi: 10.1038/nbt0310-214. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharya S, Bunick CG, Chazin WJ. Target selectivity in EF-hand calcium binding proteins. Biochim Biophys Acta. 2004;1742:69–79. doi: 10.1016/j.bbamcr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Fritz G, Botelho HM, Morozova-Roche LA, Gomes CM. Natural and amyloid self-assembly of S100 proteins: structural basis of functional diversity. FEBS J. 2010;277:4578–4590. doi: 10.1111/j.1742-4658.2010.07887.x. [DOI] [PubMed] [Google Scholar]
- 25.Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J. 2006;396:201–214. doi: 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber DJ, Rustandi R, Carrier F, Zimmer DB. Interaction of dimeric S100B(bb) with the tumor suppressor protein p53: A model for Ca2+ dependent S100-target protein interactions. In: Pochet R, editor. CALCIUM: the molecular basis of calcium action in biology and medicine. Dordrecht, The Netherlands: Kluwer Academic; 2000. pp. 521–539. [Google Scholar]
- 27.Charpentier TH, Wilder PT, Liriano MA, Varney KM, Zhong S, Coop A, Pozharski E, MacKerell AD, Jr, Toth EA, Weber DJ. Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography. Biochemistry. 2009;48:6202–6212. doi: 10.1021/bi9005754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 29.Huang Q, Petros AM, Virgin HW, Fesik SW, Olejniczak ET. Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus. Proc Natl Acad Sci USA. 2002;99:3428–3433. doi: 10.1073/pnas.062525799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilder PT, Lin J, Bair CL, Charpentier TH, Yang D, Liriano M, Varney KM, Lee A, Oppenheim AB, Adhya S, Carrier F, Weber DJ. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochium Biophys Acta. 2006;1763:1284–1297. doi: 10.1016/j.bbamcr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 31.Vitolo MI, Weiss MB, Szmacinski M, Tahir K, Waldman T, Park BH, Martin SS, Weber DJ, Bachman KE. Deletion of PTEN promotes tumorigenic signaling, resistance to anoikis, and altered response to chemotherapeutic agents in human mammary epithelial cells. Cancer Res. 2009;69:8275–8283. doi: 10.1158/0008-5472.CAN-09-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss MB, Vitolo MI, Mohseni M, Rosen DM, Denmeade SR, Park BH, Weber DJ, Bachman KE. Deletion of p53 in human mammary epithelial cells causes chromosomal instability and altered therapeutic response. Oncogene. 2010;29:4715–4724. doi: 10.1038/onc.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemunaitis J. Head and neck cancer: response to p53-based therapeutics. Head Neck. 2011;33:131–134. doi: 10.1002/hed.21364. [DOI] [PubMed] [Google Scholar]
- 34.Citrin D, Camphausen K, Wood BJ, Quezado M, Denobile J, Pingpank JF, Royal RE, Alexander HR, Seidel G, Steinberg SM, Shuttack Y, Libutti SK. A pilot feasibility study of TNFerade biologic with capecitabine and radiation therapy followed by surgical resection for the treatment of rectal cancer. Oncology. 2010;79:382–388. doi: 10.1159/000323488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weide B, Eigentler TK, Pflugfelder A, Leiter U, Meier F, Bauer J, Schmidt D, Radny P, Pfohler C, Garbe C. Survival after intratumoral interleukin-2 treatment of 72 melanoma patients and response upon the first chemotherapy during follow-up. Cancer Immunol Immunother. 2011;60:487–493. doi: 10.1007/s00262-010-0957-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carpentier A, Metellus P, Ursu R, Zohar S, Lafitte F, Barrie M, Meng Y, Richard M, Parizot C, Laigle-Donadey F, Gorochov G, Psimaras D, Sanson M, Tibi A, Chinot O, Carpentier AF. Intracerebral administration of CpG oligonucleotide for patients with recurrent glioblastoma: a phase II study. Neuro Oncol. 2010;12:401–408. doi: 10.1093/neuonc/nop047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akeda T, Yamanaka K, Kitagawa H, Kawabata E, Tsuda K, Kakeda M, Omoto Y, Habe K, Isoda K, Kurokawa I, Mizutani H. Intratumoral injection of OK-432 suppresses metastatic squamous cell carcinoma lesion inducing interferon-gamma and tumour necrosis factor-alpha. Clin Exp Dermatol. 2011;37:193–194. doi: 10.1111/j.1365-2230.2011.04151.x. [DOI] [PubMed] [Google Scholar]
- 38.Jenkinson MD, Smith TS, Haylock B, Husband D, Shenoy A, Vinjamuri S, Walker C, Pietronigro D, Warnke PC. Phase II trial of intratumoral BCNU injection and radiotherapy on untreated adult malignant glioma. J Neurooncol. 2010;99:103–113. doi: 10.1007/s11060-010-0113-0. [DOI] [PubMed] [Google Scholar]
- 39.He J, Ying W, Yang H, Xu X, Shao W, Guan Y, Jiang M, Wu Y, Zhong B, Wang D, Tucker S, Zhong N. Gemcitabine plus cisplatin chemotherapy with concurrent para-toluene-sulfonamide local injection therapy for peripherally advanced nonsmall cell lung cancer larger than 3 cm in the greatest dimension. Anticancer Drugs. 2009;20:838–844. doi: 10.1097/CAD.0b013e32832fe48f. [DOI] [PubMed] [Google Scholar]
- 40.Dharmapuri S, Peruzzi D, Marra E, Palombo F, Bett AJ, Bartz SR, Yong M, Ciliberto G, La Monica N, Buser CA, Toniatti C, Aurisicchio L. Intratumor RNA interference of cell cycle genes slows down tumor progression. Gene Ther. 2011;18:727–733. doi: 10.1038/gt.2011.27. [DOI] [PubMed] [Google Scholar]
- 41.Moyer JS, Li J, Wei S, Teitz-Tennenbaum S, Chang AE. Intratumoral dendritic cells and chemoradiation for the treatment of murine squamous cell carcinoma. J Immunother. 2008;31:885–895. doi: 10.1097/CJI.0b013e3181880f1e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Houot R, Levy R. T-cell modulation combined with intratumoral CpG cures lymphoma in a mouse model without the need for chemotherapy. Blood. 2009;113:3546–3552. doi: 10.1182/blood-2008-07-170274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuo M, Yamaguchi K, Feril LB, Jr, Endo H, Ogawa K, Tachibana K, Nakayama J. Synergistic inhibition of malignant melanoma proliferation by melphalan combined with ultrasound and microbubbles. Ultrason Sonochem. 2011;18:1218–1224. doi: 10.1016/j.ultsonch.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 44.Pina Y, Houston SK, Murray TG, Boutrid H, Celdran M, Feuer W, Shi W, Hernandez E, Lampidis TJ. Focal, periocular delivery of 2-deoxy-D-glucose as adjuvant to chemotherapy for treatment of advanced retinoblastoma. Invest Ophthalmol Vis Sci. 2010;51:6149–6156. doi: 10.1167/iovs.09-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Ghananeem AM, Malkawi AH, Muammer YM, Balko JM, Black EP, Mourad W, Romond E. Intratumoral delivery of Paclitaxel in solid tumor from biodegradable hyaluronan nanoparticle formulations. AAPS PharmSciTech. 2009;10:410–417. doi: 10.1208/s12249-009-9222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shikanov A, Shikanov S, Vaisman B, Golenser J, Domb AJ. Paclitaxel tumor biodistribution and efficacy after intratumoral injection of a biodegradable extended release implant. Int J Pharm. 2008;358:114–120. doi: 10.1016/j.ijpharm.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 47.Yoo GH, Subramanian G, Piechocki MP, Ensley JF, Kucuk O, Tulunay OE, Lonardo F, Kim H, Won J, Stevens T, Lin HS. Effect of docetaxel on the surgical tumor microenvironment of head and neck cancer in murine models, Archives Otolaryngology. Arch Otolaryngol Head Neck Surg. 2008;134:735–742. doi: 10.1001/archotol.134.7.735. [DOI] [PubMed] [Google Scholar]
- 48.Shikanov S, Shikanov A, Gofrit O, Nyska A, Corn B, Domb AJ. Intratumoral delivery of paclitaxel for treatment of orthotopic prostate cancer. J Pharm Sci. 2009;98:1005–1014. doi: 10.1002/jps.21492. [DOI] [PubMed] [Google Scholar]
- 49.Sausville EA, Burger AM. Contributions of human tumor xenografts to anticancer drug development. Cancer Res. 2006;66:3351–3354. doi: 10.1158/0008-5472.CAN-05-3627. discussion 3354. [DOI] [PubMed] [Google Scholar]
- 50.Becker JC, Houben R, Schrama D, Voigt H, Ugurel S, Reisfeld RA. Mouse models for melanoma: a personal perspective. Exp Dermatol. 2010;19:157–164. doi: 10.1111/j.1600-0625.2009.00986.x. [DOI] [PubMed] [Google Scholar]
- 51.Heyer J, Kwong LN, Lowe SW, Chin L. Non-germline genetically engineered mouse models for translational cancer research. Nature Rev Cancer. 2010;10:470–480. doi: 10.1038/nrc2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Jong M, Maina T. Of mice and humans: are they the same?—implications in cancer translational research. J Nuc Med. 2010;51:501–504. doi: 10.2967/jnumed.109.065706. [DOI] [PubMed] [Google Scholar]
- 53.Eyles J, Puaux AL, Wang X, Toh B, Prakash C, Hong M, Tan TG, Zheng L, Ong LC, Jin Y, Kato M, Prevost-Blondel A, Chow P, Yang H, Abastado JP. Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. J Clin Invest. 2010;120:2030–2039. doi: 10.1172/JCI42002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh M, Lima A, Molina R, Hamilton P, Clermont AC, Devasthali V, Thompson JD, Cheng JH, Bou Reslan H, Ho CCK, Cao TC, Lee CV, Nannini MA, Fuh G, Carano RAD, Koeppen H, Yu RX, Forrest WF, Plowman GD, Johnson L. Assessing therapeutic responses in Kras mutant cancers using genetically engineered mouse models. Nat Biotechnol. 2010;28:585–593. doi: 10.1038/nbt.1640. [DOI] [PubMed] [Google Scholar]
- 55.Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, Horner JW, 2nd, DePinho RA. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larue L, Beermann F. Cutaneous melanoma in genetically modified animals. Pigment Cell Res. 2007;20:485–497. doi: 10.1111/j.1600-0749.2007.00411.x. [DOI] [PubMed] [Google Scholar]
- 57.Atkinson JM, Shelat AA, Carcaboso AM, Kranenburg TA, Arnold LA, Boulos N, Wright K, Johnson RA, Poppleton H, Mohankumar KM, Feau C, Phoenix T, Gibson P, Zhu L, Tong Y, Eden C, Ellison DW, Priebe W, Koul D, Yung WK, Gajjar A, Stewart CF, Guy RK, Gilbertson RJ. An integrated in vitro and in vivo high-throughput screen identifies treatment leads for ependymoma. Cancer Cell. 2011;20:384–399. doi: 10.1016/j.ccr.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ, Double JA, Everitt J, Farningham DA, Glennie MJ, Kelland LR, Robinson V, Stratford IJ, Tozer GM, Watson S, Wedge SR, Eccles SA. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102:1555–1577. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]