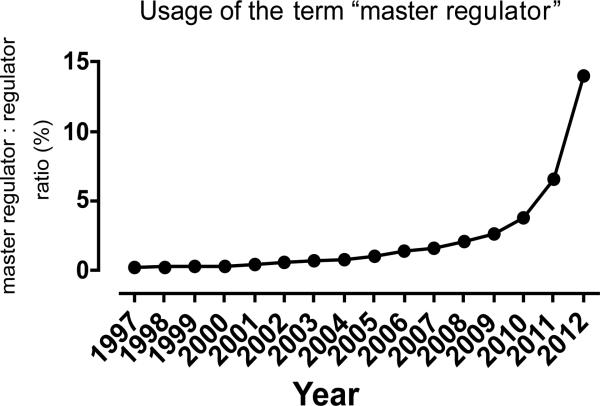
The term “master regulator” or “master regulatory gene” was first coined by Susumu Ohno over 30 years ago for a “gene that occupies the very top of a regulatory hierarchy,” which, “by its very definition should not be under the regulatory influence of any other gene” [1]. While this term was originally proposed to hypothesize a sex determination mechanism, it was subsequently extended to other disciplines to describe the hierarchy of cell specification in yeasts [2,3], Drosophila [4,5] and plants [6]. Although sex determination could conceivably be imagined to be independent of prior regulatory influences, cell type specification clearly requires some sort of prior regulatory influence, and although “other genes” per se might not have been imagined upstream in the earliest usages of the term, as mechanisms of transcriptional control of development were revealed, this original concept of a gene with nothing upstream was forgotten. However the term “master regulator” stayed attached to these earliest developmental specification factors. The meaning evolved to connote governance over a developmental lineage, but without a proper definition as to what sort of governance specifically was referred to. If any gene that is necessary for the establishment of a given lineage is a master regulator, then the genome might have more masters than servants. On the other hand, if the term master can only be applied when a single unique factor governs a specific lineage, then almost no lineages might qualify to have a master regulator. Use of the term is increasing: Google Scholar identifies 139 publications containing the phrase “master regulator” from 1997 and slightly over 7,000 from 2012. Tracking the presence of the term “master regulator” within all publications that mention the term “regulator” over the past decade and a half demonstrates further how popular the term has become (Figure 1), perhaps to the detriment of scientific clarity. Given that the term is not likely to go away, it would be valuable to agree on parameters of definition that would incorporate the current general sense of meaning, but also provide specificity.
Figure 1. Frequency of the term “master regulator” within publications that contain the term “regulator”.
The number of publications mentioning the term “master regulator” is increasing exponentially, but the exponential increase in the total number of publications makes this true of most words. However, by normalizing to the number of publications that mention the term “regulator”, a trend to increased usage of “master regulator” becomes apparent. Data source: Google Scholar.
The current vogue for reprogramming, i.e. converting one lineage in development into another or one cell type into another, by overexpression of a transcription factor, provides a useful metric. We propose that the best interpretation of the term master regulator is a gene that is expressed at the inception of a developmental lineage or cell type, participates in the specification of that lineage by regulating multiple downstream genes either directly or through a cascade of gene expression changes, and critically, when misexpressed, has the ability to respecify the fate of cells destined to form other lineages. This may be most easily tested by ectopic overexpression during embryonic development. The concept of respecification is critical as it provides a distinction between a master and a mere administrator. A master can force cells doing something else to drop what they are doing and attend to a new task, whereas an administrator has little authority outside of their own department.
The classic example of cellular reprogramming by a master regulator is that accomplished by the myogenic transcription factor MyoD [7]. In fact, MyoD was initially identified by the discovery that it converted fibroblasts into myoblasts [8,9]. The extraordinary reprogramming ability of MyoD was further demonstrated in subsequent reports, which showed that it was able to reprogram cell types derived from other non-mesoderm germ layers such as neural and liver cells to acquire a myogenic phenotype [10]. Another example of a master regulator is the hematopoietic transcription factor SCL. Not only does it play a critical early role in the specification of the hematopoietic lineage [11,12], but ectopic expression of SCL in zebrafish embryos can convert non-hematopoietic tissues into hematopoietic precursors [13].
It is important to emphasize that the absence of a particular lineage resulting from a knockout does not entitle the factor in question to be a master regulator under this definition. A prime example is the transcription factor Mesp1. Mesp1 is expressed in nascent mesoderm early during embryo development, and fate-mapping studies showed that Mesp1-expressing progenitors establish all cardiac lineages [14]. The knockout of Mesp1 caused lethal cardiac defects in embryos [15] and Mesp1;Mesp2-null cells had no contribution to cardiac tissues in a chimeric mouse analysis [16]. These observations, together with the report that Mesp1 induced the expression of many cardiogenic transcription factors in vitro, encouraged the notion that Mesp1 is the master regulator of cardiac specification [17]. However, overexpression of Mesp1 alone is insufficient to reprogram fibroblasts into cardiomyocytes [18], and Mesp1 is dispensable in many such cellular reprogramming cocktails [18-20]. It is thus difficult to reconcile these results with the expected properties of a master regulator. We have recently demonstrated that, instead of solely regulating cardiac specification, Mesp1 actually patterns mesoderm into multiple lineages in a context-dependent manner [21]. In early mesoderm, Mesp1 induction leads to a hematopoietic fate, while slightly later, depending on the signaling environment, a cardiac or skeletal myogenic phenotype will be acquired. Furthermore, Mesp1-expressing progenitors mark functional adult hematopoietic and muscle stem cells, and Mesp1-null embryos have defects in yolk sac hematopoiesis [21]. These observations thus strongly argue against the concept of Mesp1 as the master regulator of the cardiac lineage. Nonetheless, it remains possible that Mesp1 is a general mesoderm master regulator.
Recent years have witnessed tremendous excitement in the field of cellular reprogramming, with many investigators pursuing the goal of generating scarce cell types for potential clinical applications. While identifying the master regulators for cell specification is of paramount interest, the ambiguity of the usage of this terminology has provoked confusion, and in some cases hampered progress. For these reasons, it would be valuable to come to a consensus on appropriate usage, and we propose that the definition suggested herein both encompasses the general sense of current common usage of the term and provides meaningful limitations on its application.
Acknowledgement
MK and SSC are supported by NIH grant U01 HL100407.
Footnotes
Publisher's Disclaimer: This article was originally published in a journal by OMICS Publishing Group, and the attached copy is provided by OMICS Publishing Group for the author's benefit and for the benefit of the author's institution, for commercial/research/educational use including without limitation use in instruction at your institution, sending it to specific colleagues that you know, and providing a copy to your institution's administrator. All other uses, reproduction and distribution, including without limitation commercial reprints, selling or licensing copies or access, or posting on open internet sites, your personal or institution's website or repository, are requested to cite properly.
Citation: Chan SSK, Kyba M (2013) What is a Master Regulator? J Stem Cell Res Ther 3: e114. doi:10.4172/2157-7633.1000e114
References
- 1.Ohno S. Major sex-determining genes. Springer-Verlag; Berlin, Germany: 1979. [Google Scholar]
- 2.Nasmyth KA, Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980;19:753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- 3.Herskowitz I. A regulatory hierarchy for cell specialization in yeast. Nature. 1989;342:749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- 4.Lewis EB. Regulation of the genes of the bithorax complex in Drosophila. Cold Spring Harb Symp Quant Biol. 1985;50:155–164. doi: 10.1101/sqb.1985.050.01.021. [DOI] [PubMed] [Google Scholar]
- 5.Lewis EB. The 1991 Albert Lasker Medical Awards. Clusters of master control genes regulate the development of higher organisms. JAMA. 1992;267:1524–1531. [PubMed] [Google Scholar]
- 6.Hamdi S, Teller G, Louis JP. Master regulatory genes, auxin levels, and sexual organogenesis in the dioecious plant Mercurialisannua. Plant Physiol. 1987;85:393–399. doi: 10.1104/pp.85.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, et al. MyoD1: a nuclear phosphoprotein requiring a myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- 8.Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1987;47:649–656. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- 9.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 10.Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci USA. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 12.Robb L, Elwood NJ, Elefanty AG, Kontgen F, Li R, et al. The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J. 1996;15:4123–4129. [PMC free article] [PubMed] [Google Scholar]
- 13.Gering M, Rodaway ARF, Gottgens B, Patient RK, Green AR. The SCL gene specifies haemangioblast development from early mesoderm. EMBO J. 1998;17:4029–4045. doi: 10.1093/emboj/17.14.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saga Y, Hata N, Kobayashi S, Magnuson T, Selden MF, et al. MesP1: a novel basic helix-loop-helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development. 1996;122:2769–2778. doi: 10.1242/dev.122.9.2769. [DOI] [PubMed] [Google Scholar]
- 15.Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, et al. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 16.Kitajima S, Takagi A, Inoue T, Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127:3215–3226. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- 17.Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, et al. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:374–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song K, Nam YJ, Luo X, Qi X, Tan W, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nam YJ, Song K, Luo X, Daniel E, Lambeth K, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan SSK, Shi X, Toyama A, Arpke RW, Dandapat A, et al. Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell. 2013;12:587–601. doi: 10.1016/j.stem.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]