Abstract
Background
Carbohydrate structures of surface-expressed and secreted/excreted glycoconjugates of the human blood fluke Schistosoma mansoni are key determinants that mediate host-parasite interactions in both snail and mammalian hosts. Fucose is a major constituent of these immunologically important glycans, and recent studies have sought to characterize fucosylation-associated enzymes, including the Golgi-localized fucosyltransferases that catalyze the transfer of L-fucose from a GDP-L-fucose donor to an oligosaccharide acceptor. Importantly, GDP-L-fucose is the only nucleotide-sugar donor used by fucosyltransferases and its availability represents a bottleneck in fucosyl-glycotope expression.
Methods
A homology-based genome-wide bioinformatics approach was used to identify and molecularly characterize the enzymes that contribute to GDP-L-fucose synthesis and Golgi import in S. mansoni. Putative functions were further investigated through molecular phylogenetic and immunocytochemical analyses.
Results
We identified homologs of GDP-D-mannose-4,6-dehydratase (GMD) and GDP-4-keto-6-deoxy-D-mannose-3,5-epimerase-4-reductase (GMER), which constitute a de novo pathway for GDP-L-fucose synthesis, in addition to a GDP-L-fucose transporter (GFT) that putatively imports cytosolic GDP-L-fucose into the Golgi. In silico primary sequence analyses identified characteristic Rossman loop and short-chain dehydrogenase/reductase motifs in GMD and GMER as well as 10 transmembrane domains in GFT. All genes are alternatively spliced, generating variants of unknown function. Observed quantitative differences in steady-state transcript levels between miracidia and primary sporocysts may contribute to differential glycotope expression in early larval development. Additionally, analyses of protein expression suggest the occurrence of cytosolic GMD and GMER in the ciliated epidermal plates and tegument of miracidia and primary sporocysts, respectively, which is consistent with previous localization of highly fucosylated glycotopes.
Conclusions
This study is the first to identify and characterize three key genes that are putatively involved in the synthesis and Golgi import of GDP-L-fucose in S. mansoni and provides fundamental information regarding their genomic organization, genetic variation, molecular phylogenetics, and developmental expression in intramolluscan larval stages.
Keywords: Schistosoma mansoni, Schistosome, Fucose, Fucosylation, GDP-L-fucose synthesis, GDP-L-fucose transport, Fucosyltransferase, Miracidium, Sporocyst
Background
The deoxyhexose sugar L-fucose is a major constituent of an array of immunologically important carbohydrates that are presented on surface-expressed and secreted/excreted glycoconjugates of the human blood fluke Schistosoma mansoni (reviewed by [1]). Although the schistosome glycome is perhaps the most extensively characterized among invertebrates, relatively little is known about the enzymatic machinery responsible for its expression. Recent studies by Fitzpatrick et al. [2] and Peterson et al.[3] inventoried the schistosome α3- and α6-fucosyltransferases (FucTs), which transfer L-fucose from a GDP-L-fucose nucleotide-sugar donor to an oligosaccharide acceptor to create α3 and α6 linkages, respectively. These studies also demonstrated stage- and gender-specific variations in FucT gene transcription, which may contribute to differential fucosyl-glycotope expression that has been reported among stages of S. mansoni[4-7].
While the population composition and cellular organization of the expressed glycosyltransferases are key determinants affecting carbohydrate structural diversity, other factors are also important, including nucleotide-sugar donor availability, Golgi membrane dynamics, intralumenal pH, and competition for donor/acceptor substrates [8]. In S. mansoni, this means that GDP-L-fucose synthesis and Golgi import, which dictate fucose donor availability in the Golgi, likely contribute to differential fucosyl-glycotope expression. However, to date, no studies have examined these aspects of fucosylation in schistosomes.
In general, GDP-L-fucose synthesis is localized in the cytosol and can occur by two possible metabolic pathways, the de novo and salvage pathways (reviewed by [9]), which constitute approximately 90% and 10%, respectively, of total GDP-L-fucose synthesis in mammalian cells [10]. In de novo synthesis, GDP-D-mannose is converted to GDP-L-fucose in three steps by GDP-D-mannose-4,6-dehydratase (GMD, EC 4.2.1.47) and the bifunctional enzyme GDP-4-keto-6-deoxy-D-mannose-3,5-epimerase-4-reductase (GMER, EC 1.1.1.271; also called GDP-L-fucose synthase). Alternatively, the salvage pathway generates GDP-L-fucose from free cytosolic L-fucose in two steps, which are generally catalyzed by L-fucokinase (Fuk) and L-fucose-1-phosphate guanylyltransferase (FPGT; also called GDP-L-fucose pyrophosphorylase). Both pathways are summarized in Figure 1. GMD and GMER are well conserved across prokaryotic and eukaryotic taxa in terms of both structure and function [11], but the salvage pathway exhibits some variation. While homologs of Fuk and FPGT have been described in several mammalian species [12-15], the salvage pathway in Bacteroides and Arabidopsis comprises a single bifunctional enzyme (Fkp in Bacteroides; FKGP in Arabidopsis) that exhibits both Fuk and FPGT activities [16,17]. Elements of a salvage pathway do not exist in Drosophila[18] and only a Fuk homolog has been identified in C. elegans[11]. How GDP-L-fucose is synthesized in S. mansoni is unknown.
Figure 1.
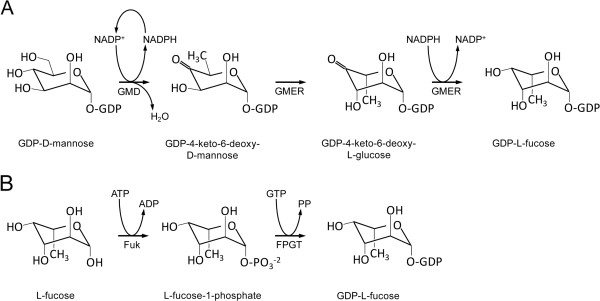
Schematic diagram of GDP-L-fucose synthesis. GDP-L-fucose synthesis occurs by two cytosolic pathways, namely the de novo and salvage pathways. In de novo synthesis (A), GMD with coenzyme NADP+ removes one H2O-equivalent from GDP-D-mannose to form GDP-4-keto-6-deoxy-D-mannose. Then, GMER catalyzes epimerizations at C3 and C5 followed by an NADPH-dependent reduction of C4 to yield GDP-L-fucose. In the salvage pathway (B), Fuk transfers a single phosphate from ATP to free cytosolic L-fucose, yielding L-fucose-1-phosphate and the byproduct ADP. Next, FPGT transfers GMP from GTP to L-fucose-1-phosphate, producing GDP-L-fucose and pyrophosphate. Evidence presented here strongly supports the exclusive use of the de novo synthetic pathway in S. mansoni. GMD, GDP-D-mannose-4,6-dehydratase; GMER, GDP-4-keto-6-deoxy-D-mannose-3,5-epimerase-4-reductase; Fuk, L-fucokinase; FPGT, L-fucose-1-phosphate guanylyltransferase.
In eukaryotes, fucosylation occurs primarily in the Golgi. Consequently, following GDP-L-fucose synthesis in the cytosol, the activated fucose is imported into the Golgi lumen where it can be utilized by Golgi-localized FucTs. This translocation is driven by a GDP-L-fucose transporter (GFT), which couples GDP-L-fucose entry with equimolar exit (i.e., antiportation) of GMP, a downstream byproduct of fucosylation (reviewed by [19]).
Previous studies indicate that GDP-L-fucose synthesis and transport are essential processes in the production of fucosylated glycans. For example, increased expression of GMD, GMER and GFT was linked to higher levels of fucosylation in human hepatocellular carcinoma [20,21] and elevated expression of sialyl Lewis X during inflammation and tumorigenesis [22]. Additionally, Omasa et al.[23] observed decreased fucosylation of recombinant human antithrombin III following RNAi-mediated knockdown of GFT in transfected Chinese hamster ovary cells. The essential role of GFT in proper fucosylation is further evidenced in humans by the rare autosomal recessive syndrome leukocyte adhesion deficiency type II (LADII), which is characterized by severe psychomotor and growth retardation, facial malformation, and persistent and recurrent infections with marked neutrophilia [24]. Red blood cells of LADII patients feature a non-fucosylated variant of the H antigen (called the “Bombay” phenotype), and leukocytes lack the fucosylated Lewis-type blood groups that are requisite for extravasation during immune challenge [25]. Importantly, LADII results from a deficiency in GDP-L-fucose transport, which is attributable to mutations in the GFT gene [26-30]. These observations suggest the possibility that GDP-L-fucose synthesis and Golgi import play key roles in the regulated expression of fucosylated glycotopes in S. mansoni as well.
In the present study, we used a homology-based genome-wide bioinformatics approach to identify and characterize putative GDP-L-fucose synthesis- and transport-associated genes in S. mansoni. This study provides fundamental information about the genomic organization, splicing and molecular phylogenetics of these fucosylation-associated genes as well as important insights regarding their putative roles in glycotope expression in snail-associated larvae, particularly miracidia and primary sporocysts.
Methods
Isolation and cultivation of S. mansoni larvae
Ethics statement: Research protocols involving mice, including routine maintenance and care, have been reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Wisconsin-Madison under assurance number A3368-01. Generation of antibodies against recombinant proteins was performed by GeneTel Laboratories LLC (Madison, WI, USA) in accordance with protocols reviewed and approved by the Office of Laboratory Animal Welfare (OLAW) at the National Institutes of Health under assurance number A4489-01.
Adult and larval S. mansoni (NMRI strain) were collected and cultivated as described by Yoshino and Laursen [31]. Briefly, adults were harvested from infected mice by perforation of the hepatic portal veins, and viable eggs were isolated from liver tissue by homogenization and washed in sterile 0.9% NaCl. Eggs were hatched in artificial pond water [32], and the free-swimming miracidia were either used immediately or transformed to primary sporocysts by cultivation at 26°C in Chernin’s Balanced Salt Solution (CBSS; [33]) containing glucose and trehalose (1 g/L each) as well as penicillin and streptomycin (CBSS+). Transformation of most miracidia was complete within 24 h of culture origination. In this study, primary sporocysts were maintained in CBSS+ for up to 10 days, with refreshment of the culture medium at 2 and 7 days.
GDP-L-fucose synthesis and transport gene identification
The amino acid sequences of previously characterized GDP-L-fucose synthesis- and transport-associated genes, including GMDs, GMERs, GFTs, Fuks, FPGTs, Fkp and FKGP, of Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, and Bacteroides fragilis were downloaded from Reference Sequence (RefSeq) and GenBank online databases at the National Center for Biotechnology Information (NCBI; accession numbers in Tables 1 and 2) and used as queries in a genome-wide tBLASTn [34] screen of genomic scaffolds and predicted genes to identify homologs in the Schistosoma mansoni Database (SchistoDB; [35]).
Table 1.
NCBI accession numbers (number.version) of GDP-L-fucose synthesis-associated genes referenced in this study
| Source organism | Gene identifier a | Nt accession | Prot. accession | Function b | References |
|---|---|---|---|---|---|
|
Homo sapiens |
GMDS |
NM_001500.2 |
NP_001491.1 |
GMD |
[36-39] |
| |
TSTA3/FX |
NM_003313.3 |
NP_003304.1 |
GMER |
[20,37,40,41] |
| |
FUK |
NM_145059.2 |
NP_659496.2 |
FUK |
[14] |
| |
FPGT/GFPP |
NM_003838.3 |
NP_003829.2 |
FPGT |
[13,42,43] |
|
Mus musculus |
Gmds |
NM_146041.2 |
NP_666153.1 |
GMD |
[22] |
| |
Tsta3 |
NM_031201.1 |
NP_112478.1 |
GMER |
[22,44,45] |
| |
Fuk |
NM_172283.2 |
NP_758487.2 |
FUK |
[15] |
| |
Fpgt |
NM_029330.2 |
NP_083606.2 |
FPGT |
" " |
|
Danio rerio |
gmds |
NM_200489.2 |
NP_956783.2 |
GMD |
[46,47] |
|
Drosophila melanogaster |
Gmd |
NM_135044.3 |
NP_608888.2 |
GMD |
[11,18] |
| |
Gmer |
NM_137890.2 |
NP_611734.1 |
GMER |
" " |
|
Caenorhabditis elegans |
gmd-1 |
AM231683.1 |
CAJ77752.1 |
GMD |
[11] |
| |
gmd-2 |
NM_060705.1 |
NP_493106.1 |
" " |
" " |
| |
ger-1 |
NM_066139.3 |
NP_498540.1 |
GMER |
" " |
|
Schistosoma mansoni |
GMD |
GU574757.1 |
ADO17520.1 |
GMD |
Present study |
| |
GMER |
GU574758.1 |
ADO17521.1 |
GMER |
" " |
|
Mortierella alpina |
GMD |
GU299800.1 |
ADC54120.1 |
GMD |
[48] |
| |
GMER |
GU299801.1 |
ADC54121.1 |
GMER |
" " |
|
Arabidopsis thaliana |
GMD1 |
NM_126026.3 |
NP_201429.1 |
GMD |
[11,49] |
| |
MUR1/GMD2 |
NM_114976.3 |
NP_190685.2 |
" " |
[11,49,50] |
| |
GER1 |
NM_105984.3 |
NP_177468.2 |
GMER |
[11,51,52] |
| |
GER2 |
NM_101652.2 |
NP_564040.1 |
" " |
[11] |
| |
FKGP |
NM_100004.3 |
NP_563620.1 |
FUK/FPGT (dual) |
[17] |
|
Bacteroides fragilis |
Gmd |
CR626927.1 |
CAH07586.1 |
GMD |
[16] |
| |
Fcl |
CR626927.1 |
CAH07585.1 |
GMER |
" " |
| Fkp | NC_003228.3 | YP_212230.1 | FUK/FPGT (dual) | [16,53] |
a Official gene names/identifiers are provided. Genes in boldface type were used as query sequences to search for homologs in the SchistoDB [35].
b GMD, GDP-D-mannose-4,6-dehydratase; GMER, GDP-4-keto-6-deoxy-D-mannose-3,5-epimerase-4-reductase; FUK, L-fucose kinase; FPGT, fucose-1-phosphate guanylyltransferase; FUK/FPGT, bifunctional L-fucose kinase/fucose-1-phosphate guanylyltransferase.
Table 2.
NCBI accession numbers (number.version) of nucleotide-sugar transporter genes referenced in this study
| Source organism (tree prefix) a | Gene identifier b | Nt accession | Prot. accession | NST Substrate(s) c | References |
|---|---|---|---|---|---|
|
Homo sapiens (Hs) |
SLC35C1 |
NM_018389.4 |
NP_060859.4 |
GDP-L-Fuc |
[21,27,28,54] |
| |
SLC35B4 |
NM_032826.4 |
NP_116215.1 |
UDP-Xyl, UDP-GlcNAc |
[55] |
| |
SLC35A3/hUGlcNAcT |
NM_012243.1 |
NP_036375.1 |
UDP-GlcNAc |
[56] |
| |
SLC35D2/hUGTrel8/HFRC1 |
NM_007001.2 |
NP_008932.2 |
UDP-Glc, UDP-GlcNAc, GDP-Man |
[57] |
| |
hUGTrel7 |
AB044343.1 |
BAB18586.1 |
UDP-GlcA, UDP-GalNAc |
[58] |
| |
hUGT1 |
D84454.1 |
BAA12673.1 |
UDP-Gal, UDP-GalNAc |
[59-62] |
|
Canis lupus (Cl) |
SLC35A3 |
NM_001003385.1 |
NP_001003385.1 |
UDP-GlcNAc |
[63] |
| |
SLC35A2 |
NM_001003059.2 |
NP_001003059.2 |
UDP-Gal |
[64] |
|
Mus musculus (Mm) |
Slc35c1 |
NM_211358.2 |
NP_997597.1 |
GDP-L-Fuc |
[30,65] |
| |
Slc35b4 |
NM_021435.3 |
NP_067410.1 |
UDP-Xyl, UDP-GlcNAc |
[66] |
| |
mUGT1 |
AB027147.1 |
BAA86885.1 |
UDP-Gal |
[67] |
| |
Slc35a1 |
NM_011895.3 |
NP_036025.2 |
CMP-Sia |
[68] |
|
Cricetulus griseus (Cgr) |
Slc35a1 |
NM_001246755.1 |
NM_001246755.1 |
CMP-Sia |
[69] |
|
Drosophila melanogaster (Dm) |
Gfr |
NM_141525.1 |
NP_649782.1 |
GDP-L-Fuc |
[70,71] |
| |
Efr |
NM_132071.1 |
NP_572299.1 |
" " |
[72] |
| |
Frc |
AB062677.1 |
BAB62105.1 |
UDP-GlcA, UDP-GalNAc, UDP-Gal, UDP-GlcNAc, UDP-Xyl |
[73,74] |
| |
ugt |
AB055493.1 |
BAB62747.1 |
UDP-Gal, UDP-GalNAc |
[62,75] |
|
Caenorhabditis elegans (Ce) |
C50F4.14 |
AF323969.1 |
AAK50396.1 |
GDP-L-Fuc |
[28] |
| |
SQV-7 |
NM_063035.4 |
NP_495436.1 |
UDP-GlcA, UDP-GalNAc, UDP-Gal |
[76] |
|
Schistosoma mansoni (Sm) |
GFT |
GU574756.1 |
ADO17519.1 |
GDP-L-Fuc (putative) |
Present study |
|
Leishmania donovani (Ld) |
LPG2 |
U26175.1 |
AAC46914.1 |
GDP-Man, GDP-Ara, GDP-Fuc |
[77] |
|
Cryptococcus neoformans (Cn) |
GMT1 |
XM_571496.1 |
XP_571496.1 |
GDP-Man |
[78] |
| |
GMT2 |
XM_571874.1 |
XP_571874.1 |
" " |
" " |
|
Saccharomyces cerevisiae (Sc) |
YEA4 |
NM_001178819.1 |
NP_010912.1 |
UDP-GlcNAc |
[79] |
|
Candida albicans (Ca) |
VRG4 |
AF164627.1 |
AAK74075.1 |
GDP-Man |
[80] |
|
Candida glabrata (Cgl) |
Vrg4 |
AF360395.1 |
AAK51897.1 |
GDP-Man |
[81] |
|
Arabidopsis thaliana (At) |
GONST1 |
AJ314836.1 |
CAC69066.1 |
GDP-Man |
[82,83] |
| |
GONST2 |
NM_100603.5 |
NP_172209.4 |
" " |
[83] |
| |
AtUTr1 |
AY115566.1 |
AAM48281.1 |
UDP-Gal, UDP-Glc |
[84] |
| |
NST-K1 |
NM_179196.1 |
NP_849527.1 |
UDP-Gal |
[85] |
| |
udpgalt1 |
AJ633720.1 |
CAG18176.1 |
" " |
[86] |
| udpgalt2 | AJ633721.1 | CAG18177.1 | " " | " " |
a “tree prefix” refers to nomenclature applied in phylogenetic analyses of NSTs (Figure 6, in Additional file 3: Figure S2).
b Official gene names/identifiers are provided. Genes in boldface type were used as query sequences to search for GDP-L-fucose transporter homologs in the SchistoDB [35].
c NST activity has been demonstrated for these substrates. GDP-Fuc, GDP-L-fucose; UDP-Xyl, UDP-D-xylose; UDP-GlcNAc, UDP-D-N-acetylglucosamine; UDP-Glc, UDP-D-glucose; UDP-GlcA, UDP-D-glucuronic acid; UDP-GalNAc, UDP-D-N-acetylgalactosamine; UDP-Gal, UDP-D-galactose; GDP-Man, GDP-D-mannose; CMP-Sia, CMP-sialic acid; GDP-Ara, GDP-D-arabinose.
Primer design
The oligonucleotide primers used in this study were designed using Vector NTI Advance 11.0 software (Invitrogen, Eugene, OR, USA) and the IDT SciTools suite [87] based on available SchistoDB-derived genomic sequence information as well as data obtained by this study, and custom DNA oligonucleotides were purchased from Integrated DNA Technologies (IDT, Coralville, IA, USA). A complete list of primer sequences used in this study is provided in (Additional file 1: Table S1A-E).
Reverse transcriptase-PCR and rapid amplification of cDNA ends for GMD, GMER, and GFT transcript sequencing
Kits and reagents for molecular assays were used according to the manufacturers’ recommendations unless otherwise indicated. Primers used for reverse transcription (RT)-PCR and rapid amplification of cDNA ends (RACE) are provided (see Additional file 1: Table S1A-C). RT-PCR and RACE protocols were performed as detailed in [3] and are summarized as follows: Miracidia, 2-day in vitro-cultivated primary sporocysts and mixed-sex adults (i.e., pooled male and female worms) were washed with artificial pond water (miracidia), CBSS (sporocysts) or mammalian phosphate-buffered saline (8.41 mM Na2HPO4, 1.65 mM NaH2PO4·H2O, 146.4 mM NaCl, pH 7.4; adults), and total (“raw”) RNA was extracted using TRIzol® Reagent (Invitrogen). Genomic contamination was removed with TURBO™ DNase (Applied Biosystems, Foster City, CA, USA), and the resultant DNA-free RNA was converted to RT-PCR-ready cDNA using the SuperScript® III First-Strand Synthesis System (Invitrogen). Reverse transcriptase-PCR reactions were prepared with GoTaq® PCR reagents (Promega, Madison, WI, USA), and amplification products were QIAquick-purified (Qiagen, Germantown, MD, USA), ligated into pCR®4-TOPO® sequencing vector (Invitrogen) and cloned in One Shot® TOP10 Chemically Competent Escherichia coli (Invitrogen). Inserts in QIAprep-isolated plasmids (Qiagen) were sequenced by BigDye Terminator dideoxy PCR sequencing (Applied Biosystems) and, following purification with Agencourt® CleanSEQ® magnetic beads (Beckman Coulter, Brea, CA, USA), reaction products were read by the DNA Sequence Laboratory at the University of Wisconsin Biotechnology Center (Madison, WI, USA). Following RT-PCR confirmation of gene transcription, RACE-ready cDNA was prepared from TRIzol®-derived DNA-free total parasite RNA using a SMART™/SMARTer™ RACE cDNA Amplification Kit (Clontech, Mountain View, CA, USA), and gene-specific cDNA ends were PCR-amplified using an Advantage® 2 PCR Kit (Clontech). Amplification products were isolated, cloned and sequenced as above. Transcript sequences were assembled from the compiled sequence data and edited using Vector NTI Advance 11.0 software. The complete coding sequences (CDSs) were then verified by RT-PCR amplification and sequencing (as above) using primers flanking the open reading frames (ORFs).
Phylogenetic analysis of nucleotide-sugar transporters
Representative amino acid sequences of functionally characterized nucleotide-sugar transporters were compiled from RefSeq and GenBank databases with our data from S. mansoni (Table 2). Sequences were aligned using default settings in MUSCLE v 3.6 [88], with subsequent manual correction in Mesquite [89]. A guide tree was developed for Bayesian phylogenetic inference using neighbor-joining methods in FastTree v 2.0.1 [90] with a Jukes-Cantor + CAT model. Analyses were then performed using mixed amino acid models within MrBayes v 3.1.2 [91] with two parallel runs of four Markov chain Monte Carlo (MCMC) chains, each for five million generations, with subsampling every 100th generation. To ensure the tree search was not trapped at local optima, two independent replicates were conducted [92]. Stationarity of molecular evolutionary parameters was assessed at effective sample sizes >400 in Tracer v1.5 [93]. Additionally, convergence of the MCMC chains was evaluated using the online program AWTY [94]. Trees prior to stationarity were burned-in, and remaining trees were used to assess posterior probabilities for nodal support.
Real-time quantitative PCR analysis of GMD, GMER, and GFT mRNA expression in miracidia and primary sporocysts of S. mansoni
Real-time quantitative (q)PCR protocols used in this study were performed according to the recommendations by Applied Biosystems [95], including strict criteria for qPCR primer design, validation and optimization. Relative transcript abundance in miracidia and primary sporocysts was examined using the comparative CT (ΔΔCT) method. ATP synthase f (herein termed “ATPsf”; SAGE tag 195 corresponding to Smp_140480 in the SchistoDB) and the GroES chaperonin (SAGE tag 132 corresponding to Smp_097380) were selected as endogenous calibrators based on SAGE data [96], which indicate stable expression between miracidia and primary sporocysts. The compatibility of calibrator and gene of interest (GOI) qPCR primers under normal reaction conditions was assessed by plotting ΔCT at 10-fold dilutions of cDNA input and determining the slope of the resultant semi-log regression line; primer efficiencies were deemed compatible if the absolute value of the slope was less than 0.1. Validated calibrator and GOI primer sequences are listed in Additional file 1: Table S1D.
Miracidia and in vitro-cultivated primary sporocysts were washed with artificial pond water and CBSS, respectively, followed by extraction of total RNA and immediate preparation of first-strand cDNA as above. It should be noted that RNA integrity was not routinely assessed prior to cDNA synthesis (as per MIQE guidelines [97]) due to limited raw RNA yields; however, integrity in select samples was visually inspected via electrophoretic fractionation. Also, raw and DNA-free RNA concentrations were estimated using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and only samples exhibiting A260:A280 and A260:A230 ratios >1.8 were processed for inclusion in qPCR analyses. Real-time qPCR reactions (50 μL/rxn) were performed in triplicate using an ABI 7300 Real-Time PCR System (Applied Biosystems), with reaction mixtures comprising 1× SYBR Green PCR Master Mix (Applied Biosystems), 20 ng RNA input-equivalents of parasite cDNA and gene-specific primers (100 nM each forward and reverse for GMD, GMER and GFT; 200 nM each for GroES and ATPsf). Cycling parameters included an initial denaturation at 95°C for 10 min followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Amplification fidelity was confirmed by post-cycling thermal dissociation and agarose gel fractionation of qPCR products. The geometric mean of ATPsf and GroES CT values was used to normalize GOI CT values such that ΔCT = CT-GOI - CT-GeoMean(ATPsf, GroES), and ΔCT values were compared across three independent biological replicates using iterative heteroscedastic two-sample t- and Wilcoxon rank sum tests, with significance set at p≤0.05 and p=0.10, respectively. It should be noted that the nonparametric Wilcoxon rank sum test lacks statistical power when sample size is low (e.g., n=3) and a p-value of 0.10 is acceptable in the current analyses.
Expression and purification of recombinant GMD and GMER for antibody production
Heterologous expression and purification of recombinant GMD and GMER proteins were performed using the GST fusion vector pGEX-6P-1 (GE Healthcare, Piscataway, NJ, USA), which incorporates N-terminal GST and an interceding PreScission™ Protease cleavage site, according to recommendations by Amersham Biosciences (GE Healthcare). The complete CDSs of GMD and GMER were amplified from RT-PCR-ready larval cDNA (generated as above) using 5′-tagged primers designed to incorporate BamHI or EcoRI restriction sites at the amplicon ends (GMD, BamHI-forward and EcoRI-reverse; GMER, BamHI-forward and BamHI-reverse; in Additional file 1: Table S1E). Reverse transcriptase-PCR reactions (25 μL/rxn) comprised 2.5 U GoTaq® Flexi DNA Polymerase, 1× Green GoTaq® Flexi Reaction Buffer, 400 nM each forward and reverse restriction-tagged primers, 1.6 mM dNTP mix (400 μM each), 1.5 mM MgCl2 and ~200 ng RNA input-equivalents of larval cDNA. The thermal profile included initial denaturation at 94°C for 3 min, 40 cycles of 94°C for 15 sec, 58°C for 30 sec and 72°C for 2 min, and final extension at 72°C for 10 min. Following electrophoretic fractionation and ethidium bromide-meditated visualization in 1% agarose gel, the tagged amplification products were isolated by QIAquick gel extraction. To prepare GMD and GMER expression constructs, stock pGEX-6P-1 vector and purified restriction-tagged PCR products were digested with BamHI-HF™ and EcoRI-HF™ (New England BioLabs, Ipswich, MA, USA) (double digest, GMD) or with BamHI-HF™ alone (GMER). Double digests (25 μL/rxn) included 500 U each BamHI-HF™ and EcoRI-HF™, 1× NEBuffer 4 and ~4 μg pGEX-6P-1 stock vector or restriction-tagged GMD amplicon. The protocol for single digests of restriction-tagged GMER and the stock pGEX-6P-1 vector excluded EcoRI-HF™. In both schemes, reactions were incubated at 37°C for 2 h. To prevent self-ligation, 2 U calf intestinal alkaline phosphatase (CIP, New England BioLabs) in 1× NEBuffer 4 was added to the linearized pGEX-6P-1 vector, and reactions (30 μL total volume) were incubated at 37°C for 1 h. Both CIP-treated pGEX-6P-1 vector and restriction-digested GMD/GMER amplicon were purified by electrophoretic fractionation in 1% agarose gel, and the DNA fragments were isolated by QIAquick gel extraction. Next, GMD/GMER amplicon and CIP-treated pGEX-6P-1 vector were combined (5:1 cohesive ends ratio) with 2000 U T4 DNA Ligase and 1× T4 DNA Ligase Reaction Buffer (New England BioLabs), and ligation reactions were incubated at 22°C for 30 min followed by 65°C for 15 min. Recombinant plasmids were cloned in One Shot® BL21 (DE3) Chemically Competent E. coli (Invitrogen), and QIAprep-purified plasmids were sequenced using pGEX 5′ Sequencing Primer 5′-d[GGGCTGGCAAGCCACGTTTGGTG]-3 and pGEX 3′ Sequencing Primer 5′-d[CCGGGAGCTGCATGTGTCAGAGG]-3′ (GE Healthcare).
To express GMD and GMER proteins, plasmid-bearing BL21 cells were grown overnight at 37°C in 2YT medium (1.6% tryptone, 1.0% yeast extract, 0.5% NaCl) containing 100 μg/mL ampicillin (2YTA). Overnight cultures were diluted 1:40 with 2.4 L 2YTA (6*400 mL/flask), and cells were grown at 37°C until A600 reached ~0.6. Then cultures were induced with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (Sigma-Aldrich, St. Louis, MO, USA) and incubated overnight (~16 h) at 26°C with shaking at 200 rpm. Cells were pelleted by centrifugation for 10 min at 7000 g and 4°C, after which pellets were freeze-thawed three times and resuspended in GST-A buffer (20 mM Tris, 1 M NaCl, 0.2 mM EDTA, 1 mM DTT) containing 1× Protease Inhibitor Cocktail Set III (EMD Chemicals, Gibbstown, NJ, USA). Cells were disrupted on ice by four pulses for 25 sec/pulse at output 5 and duty cycle 40% using an S-450A Branson® Sonifier (Branson Ultrasonics Corp., Danbury, CT, USA). Triton X-100 (Sigma-Aldrich) was added to 1.0% final concentration, and homogenates were gently mixed for 30 min at 4°C. After centrifugation for 10 min at 12000 g and 4°C to remove cellular debris, supernatants were filtered with a 0.45 μm Nalgene® syringe filter (Thermo Fisher Scientific), and the GST fusion proteins were affinity-purified on 1 mL GSTrap FF columns (GE Healthcare) using a TRIS™ peristaltic pump (Teledyne Isco, Lincoln, NE, USA). Columns were primed with 10 mL phosphate-buffered saline (PBS: 10 mM Na2HPO4, 1.8 mM KH2PO4, 140 mM NaCl, 2.7 mM KCl, pH 7.3) at a flow rate of 1.0 mL/min, loaded with filtered extract at 0.5 mL/min, washed with 10 mL PBS at 1.0 mL/min, equilibrated with PreScission™ cleavage buffer (PCB: 50 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, pH 7.5) at 1.0 mL/min, incubated 4 h at 4°C with 160 U PreScission™ Protease in 1 mL PCB and eluted from the column with 3 mL PCB. Eluates containing cleaved GMD/GMER were concentrated 10-fold with an Amicon® Ultra-4 30 kDa MWCO centrifugal filter (Millipore, Billerica, MA, USA) and fractionated on a preparative 10% polyacrylamide gel (~1-2 mg protein load). Proteins were visualized with Bio-Safe Coomassie Stain (Bio-Rad Laboratories, Hercules, CA, USA), and GMD and GMER bands were excised and stored at −20°C in PBS. Polyclonal chicken IgY antibodies against gel-isolated GMD and GMER proteins were commercially produced by GeneTel Laboratories LLC (Madison, WI, USA).
Antibody purification using blotted recombinant GMD and GMER proteins
To reduce nonspecific binding and cross-reactivity of GMD and GMER chicken IgY antibodies, 200 μg purified GMD/GMER protein was fractionated in 12.5% polyacrylamide gel and electroblotted for 1.5 h at 100 mA onto 0.2 μm nitrocellulose (Bio-Rad Laboratories) using a TE 77 Semi-Dry Transfer Unit (Hoefer, San Francisco, CA, USA). Following transfer, the membrane-immobilized proteins were visualized with Ponceau S stain (Sigma-Aldrich), and bands were excised by razorblade. After destaining, the protein-bearing membrane strips were blocked overnight at 4°C with 5% nonfat dry milk in tris-buffered saline (TBS: 20 mM Tris, 150 mM NaCl, pH 7.5), rinsed two times with TBS containing 0.05% Tween® 20 (Thermo Fisher Scientific) (TBST), incubated overnight at 4°C with 10 mL crude pre-immune or gene-specific chicken IgY and washed three times with TBST. Finally, bound antibodies were eluted twice by incubation for 10 min in 5 mL 0.1 M Glycine-HCl (pH 2.7), with eluates being immediately neutralized with 400 μL 2 M tris (pH 8.0) followed by dialysis in PBS overnight at 4°C using a 7K MWCO Pierce Slide-A-Lyzer® Dialysis Cassette (Thermo Fisher Scientific). This antibody isolation procedure was repeated twice more using the same antigen-bound strips. Dialyzed eluates were combined and concentrated ~250-fold with a 9K MWCO Pierce Protein Concentrator (Thermo Fisher Scientific), and stored for later use at 4°C in 50% glycerol.
Preparation of cytosolic, membrane/organelle, nuclear and cytoskeletal protein fractions from larval S. mansoni
Subcellular fractionation of miracidia, primary sporocysts and mixed-sex adult worms was performed using a modification of the ProteoExtract® Subcellular Proteome Extraction Kit (EMD Chemicals) protocol, which was originally optimized for use with mammalian cell/tissue samples. Parasites were gently washed four times with artificial pond water (miracidia), CBSS (sporocysts) or mammalian PBS (adults), followed by two washes with Calbiochem® Wash Buffer (kit component). After the final wash, the parasites were pelleted by centrifugation for 1 min at 300 g and 4°C, resuspended in 1.5 mL Extraction Buffer I containing 1× protease inhibitor cocktail (PIC) (kit components), and gently agitated for 10 min at 4°C on a LABQUAKE® Rotatory shaker (Barnstead/Thermolyne, Dubuque, IA, USA). The parasite residua were pelleted by centrifugation for 10 min at 1100 g and 4°C, and the supernatant (cytosolic fraction, F1) was transferred to a clean tube on ice. Residua were then resuspended in 1.5 mL Extraction Buffer II containing 1× PIC (kit components) and incubated 30 min at 4°C on the rotary shaker. Following centrifugation for 10 min at 6500 g and 4°C, the supernatant (membrane/organelle fraction, F2) was placed on ice. Parasite residua were resuspended again in 0.75 mL Extraction Buffer III containing 1× PIC and 562.5 U Benzonase® (kit components), and suspensions were incubated on the rotary shaker for 10 min at 4°C. The insoluble material was pelleted by centrifugation for 10 min at 8200 g and 4°C, and the supernatant (nuclear fraction, F3) was set aside on ice. Finally, the residua were resuspended in 0.75 mL Extraction Buffer IV containing 1× PIC (kit components) and incubated for 30 min at room temperature on the rotary shaker. Insoluble cell debris was pelleted for the last time by centrifugation at 8200 g and room temperature and the final fraction (cytoskeletal fraction, F4) was set on ice. All fractions were then dialyzed in PBS overnight at 4°C using 6-8K MWCO D-Tube™ Dialyzers (EMD Chemicals) and concentrated ~15 fold with a Microcon® YM-10 Centrifugal Filter Device (Millipore).
SDS-PAGE and western blot analyses of schistosome subcellular protein fractions
Subcellular protein extracts (8.5 μg protein/lane) were fractionated in 12.5% polyacrylamide gel, and proteins were electroblotted for 1.5 h at 100 mA onto 0.2 μm nitrocellulose. Membranes were blocked overnight with 5% milk in TBS at 4°C, incubated 2 h at room temperature with membrane-purified chicken IgY diluted 1/20 with 5% milk in TBS, washed three times with TBST (5 min/wash), treated 2 h with alkaline phosphatase-conjugated rabbit α-chicken IgY (GeneTel Laboratories LLC) diluted 1/10000 with 5% milk in TBST, washed three more times with TBST and developed in alkaline phosphatase buffer (100 mM Tris, 100 mM NaCl, 50 mM MgCl2, pH 9.5) containing 5-bromo-4-chloro-3-indoylphosphate p-toluidine salt and nitro-blue tetrazolium chloride (Thermo Fisher Scientific).
Processing of schistosome larvae for confocal laser scanning microscopy
Preparation of parasite larvae for confocal laser scanning microscopy was performed as described by Peterson et al. [7] with modifications. All in-tube washes and treatments were performed at 4°C on a rotary shaker, and parasite larvae were pelleted by centrifugation for 2 min at 300 g between incubations. Briefly, miracidia and 2- and 10-day in vitro-cultivated primary sporocysts were washed five times with artificial pond water (miracidia) or snail PBS (sPBS: 8.41 mM Na2HPO4, 1.65 mM NaH2PO4·H2O, 45.34 mM NaCl, pH 7.4; sporocysts) and transferred to a Sigmacote®-treated (Sigma-Aldrich) microfuge tube. Larvae were simultaneously fixed and permeabilized by overnight incubation in 4% paraformaldehyde and 1% Triton X-100 (Sigma-Aldrich) in sPBS (pH 7.2), washed five times with 2% bovine serum albumin (BSA) and 0.02% azide in sPBS (15 min/wash) and blocked overnight in sPBS containing 5% BSA and 0.02% azide (blocking buffer). Blocked larvae were incubated for 3 days in membrane-purified anti-GMD/GMER antibody concentrates diluted 1/100 in blocking buffer containing 0.1% Tween® 20. Following primary treatment, larvae were washed six times with 1% BSA, 0.02% azide and 0.1% Tween® 20 in sPBS (wash buffer) (20 min/wash) and treated overnight with a mixture of Hoechst 33258 dye (50 μg/mL; Invitrogen), Alexa Fluor®546-conjugated phalloidin (7.5 U/mL; Invitrogen) and Alexa Fluor®488-conjugated goat anti-chicken IgY secondary antibody (4 μg/mL, Invitrogen) in blocking buffer containing 0.1% Tween® 20. Finally, larvae were washed six times with wash buffer (five 20 min washes, one overnight wash), mounted in Vectashield® mounting medium (Vector Laboratories, Burlingame, CA, USA) and imaged at 600× total magnification under oil immersion using an A1R confocal microscope (Nikon Instruments Inc., Melville, NY, USA) equipped with laser lines of 408 nm, 488 nm and 561 nm for the excitation of Hoechst, Alexa Fluor®488 and Alexa Fluor®546 dyes, respectively. Confocal fluorescence images were processed using Adobe Photoshop CS v9.0 (Adobe Systems Inc., San Jose, CA, USA), and antibody reactivities were assessed against secondary-only and membrane-purified preimmune controls.
Results and discussion
Composition, genomic organization, and splicing of schistosome GDP-L-fucose synthesis and transport genes
An exhaustive homology-based search of the Schistosoma mansoni Database (SchistoDB; [35]) using a diversity of previously characterized GDP-L-fucose synthesis- and transport-associated enzymes (see Tables 1–2) identified three homologs in the schistosome genome, herein termed GMD, GMER and GFT (genes and corresponding SchistoDB annotations listed in Table 3). GMD and GMER putatively constitute a complete de novo pathway for GDP-L-fucose synthesis. No homologs of salvage pathway-associated genes (Fuk, FPGT, Fkp, FKGP) were identified, suggesting that GDP-L-fucose synthesis in S. mansoni occurs only by de novo conversion of GDP-D-mannose. Unlike Caenorhabditis and Arabidopsis, which encode multiple paralogs of GMD and GMER [11,49], only one homolog of each gene occurs in S. mansoni. In addition to known Golgi-associated GFTs, search queries included the ER-resident transporter Efr, which imports GDP-L-fucose donor substrates for consumption by ER-associated protein O-FucTs in Drosophila. These searches failed to identify a homologous ER-type GFT in S. mansoni despite the previous finding that schistosomes express two putative ER-resident protein O-fucosyltransferases [3]. Notably, Ishikawa et al. [72] observed that Drosophila Golgi- and ER-resident GFTs (Gfr and Efr, respectively) function redundantly in the O-fucosylation of Notch receptor, suggesting the existence of two pathways for supplying GDP-L-fucose to ER-resident protein O-FucTs. Therefore, a second, ER-type GFT may not be necessary for O-fucosylation in S. mansoni.
Table 3.
Genomic organization of GDP-D-mannose-4,6-dehydratase (GMD), GDP-4-keto-6-deoxy-D-mannose-3,5-epimerase-4-reductase (GMER) and a GDP-L-fucose transporter (GFT) in Schistosoma mansoni
| Gene | Gene ID a | Scaffold ID a | Approx. size (bp) | No. of exons | ORF length (nt) b | Prot. length (aa) b |
|---|---|---|---|---|---|---|
|
GMD |
Smp_153490 |
Smp_scaff000159 |
>4,911 |
10 |
1,089 |
363 |
|
GMER |
Smp_104720 |
Smp_scaff001995 |
≥7,696 |
7 |
954 |
318 |
| GFT | Smp_155830 | Smp_scaff000188 | >13,167 | 11 | 1,149 | 383 |
a Smp gene and scaffold IDs refer to nomenclature in the SchistoDB [35].
b ORF and protein sizes are provided for the main/major transcripts. Alternative splicing may alter ORF length and protein coding.
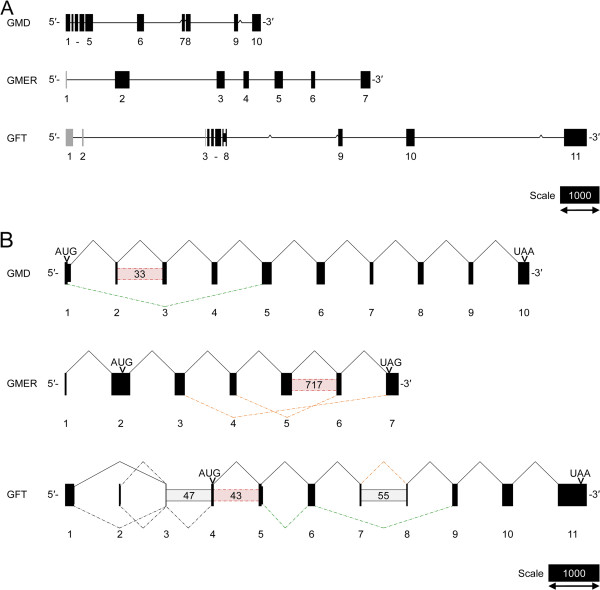
To confirm mRNA expression of GMD, GMER and GFT in S. mansoni and obtain full-length CDSs, transcript sequences were RT-PCR and RACE-amplified from miracidial, primary sporocyst and adult worm cDNAs. Complete nucleotide sequences were submitted to GenBank at NCBI (accession numbers in Tables 1–2). While GMD and GMER sequence data generally validate the corresponding SchistoDB predictions, the data indicate that annotation Smp_155830 erroneously combines a portion of the GFT CDS with an upsteam gag-pol polyprotein-coding gene, which comprises ~65% of the predicted GFT CDS. Mapping sequence data onto the corresponding SchistoDB-derived genomic scaffolds demonstrated that schistosome GMD, GMER and GFT are all multiexonic, with CDSs spanning 10, 6 and 8 exons, respectively (Table 3, Figure 2A).
Figure 2.
Genomic organization and splicing of GDP-L-fucose synthesis- and transport-associated genes in Schistosoma mansoni. The mRNA transcript sequences of GMD, GMER and GFT were mapped onto genomic scaffolds in the SchistoDB [35](A). Exons (boxes, numbered below) and introns (connecting lines) are drawn to scale (bar = 1000 nt) with GMD/GMER/GFT-coding elements, including exons and a subset of retained introns, depicted as black boxes and non-coding exons depicted as gray boxes. Caret marks indicate gaps in the genomic sequence. Alternative splicing, including exon skipping, intron retention, mutual exclusivity and use of alternate splice donor sites, was observed during transcript sequencing (B). Bent connectors indicate splicing between exons (boxes, numbered below), with solid lines representing splicing in the main/major full-length GMD/GMER/GFT-coding transcripts and dotted lines representing alternative splice events. In this panel, exons are drawn to scale (bar = 1000 nt) and spacing of exons is arbitrary. Interexonic boxes represent retained introns (estimated lengths in parentheses) with solid outlines signifying retention in the main/major transcript and dotted lines indicating retention in other isoforms. The positions of the prototypical start and stop codons (AUG and UAA/UAG, respectively) are shown. Colors convey the in silico consequences of splicing: black, conservation of the prototypical ORF; red, introduction of a PTC; orange, induction of a downstream frameshift; green, in-frame deletion/addition.
Alternative splicing was observed for all schistosome GDP-L-fucose synthesis- and transport-associated genes (Figure 2B). Because many of these observations were based on data obtained by RT-PCR and RACE, which target specific sections of each transcript rather than complete CDSs, the relationships among alternative splice events (i.e., whether splice events occur co-dependently in the formation of particular isoforms) are largely unknown. Most modes of alternative splicing were observed, including exon skipping (GMD, GMER and GFT), intron retention (GMD, GMER and GFT), mutual exclusion (e.g., exons 1 and 2 of GFT) and use of alternative splice donor sites (GMD and GMER). An in silico analysis to determine the consequences of alternative splicing revealed that many of these events altered protein coding by introducing a premature termination codon (PTC), forcing a downstream frameshift, or effecting an in-frame deletion or addition. However, additional studies are required to determine the true biochemical effects of these variations.
In eukaryotes, alternative splicing is often an important source of phenotypic complexity, which is driven by splice-mediated expansion of the proteome, posttranscriptional gene regulation (e.g., introduction of a PTC that leads to nonsense-mediated decay) and alteration of cis-regulatory elements that control mRNA translation efficiency, stability and localization (reviewed by [98]). Additionally, in many biological systems, alternative splicing is an important mechanism of modulating physiological activity during development, differentiation and stress responses, and such developmentally regulated alternative splicing has been well documented in S. mansoni (e.g., [99-101]). While a comprehensive investigation of splice variation in the context of parasite development was beyond the scope of the present study, the data feature multiple examples of variant splice events that potentially modulate GMD, GMER and GFT expression. For instance, the observed splice-mediated introduction of PTCs and frameshifts could target the affected GMD, GMER and GFT transcripts for nonsense-mediated decay, and developmental regulation of these processes could yield stage- and/or tissue-specific GDP-L-fucose synthesis and transport activities. Moreover, this could affect FucT activity in the Golgi and ultimately determine the developmental expression of fucosylated glycotopes.
In silico characterization of schistosome GMD, GMER, and GFT
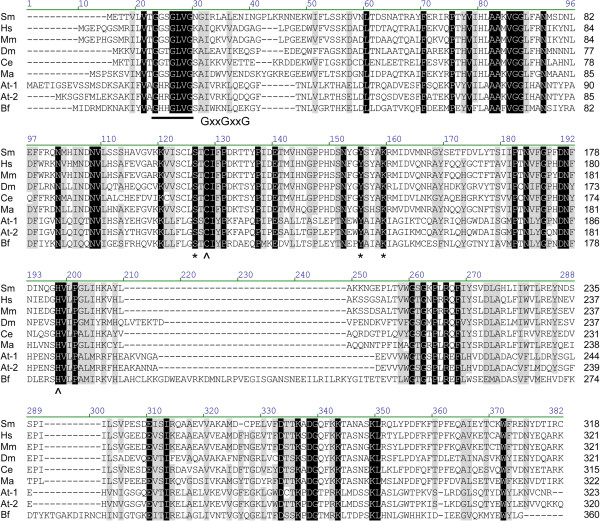
To provide support for their putative roles in GDP-L-fucose synthesis and transport, the predicted amino acid sequences of schistosome GMD, GMER and GFT were compared against previously characterized homologs of other organisms, and proteins were examined for the presence of key primary sequence elements. GMDs and GMERs of other organisms are cytosolic soluble enzymes of the short-chain dehydrogenase/reductase (SDR) gene family and feature a Rossman dinucleotide-binding domain (reviewed by [9]; also see references in Table 1). Amino acid alignment of schistosome GMD and GMER to functionally characterized homologs of humans, Mus, Danio, Drosophila, Caenorhabditis, Mortierella, Arabidopsis and Bacteroides (listed in Table 1) demonstrated that both genes are well conserved across taxa (30.2% overall identity for GMDs, 13.6% for GMERs) (Figures 3 and 4). In pairwise comparisons, schistosome GMD shares ~53-61% of its primary sequence with homologs (61.2% identical to Bacteroides Gmd), while schistosome GMER is ~25-62% identical to its homologs (61.7% identical to human FX). Both GMD and GMER alignments demonstrated the presence of a well-conserved glycine-rich phosphate-binding loop (GxxGxxG; alignment positions 57–63 in GMD; positions 23–29 in GMER), which is key to water-mediated hydrogen bonding between the Rossman fold of redox-associated enzymes and the pyrophosphate of dinucleotide enzyme cofactors (e.g., NAD+/NADP+) [102], and both enzymes feature the catalytically important SDR-associated [S/T]-Y-K triad (alignment positions 187, 211 and 215 in GMD; positions 126, 155 and 159 in GMER) [103-105]. Additionally, schistosome GMER features conserved cysteine and histidine residues (C-128, H-198) that are thought to be involved in proton exchange between GMER and its epimerization reaction intermediates [105]. Finally, analyses using the Simple Modular Architecture Research Tool (SMART; [106]) and Phobius transmembrane topology and signal peptide prediction server [107] demonstrated that schistosome GMD and GMER lack either a transmembrane domain (TMD) or signal sequence, indicating that both proteins are likely soluble and cytosolic.
Figure 3.
Amino acid alignment of GDP-D-mannose-4,6-dehydratases. The predicted amino acid sequence of schistosome GMD (Sm) is compared to GMDs of humans (Hs), Mus musculus (Mm), Danio rerio (Dr), Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce-1, Ce-2), Mortierella alpina (Ma), Arabidopsis thaliana (At-1, At-2) and Bacteroides fragilis (Bf) (accession numbers in Table 1). Alignment position is indicated above each block, and sequence length is reported to the right of each line. Positions exhibiting greater than 80% conservation are highlighted in gray, and identities are identified in black. A well-conserved glycine-rich phosphate-binding loop (GxxGxxG), which is key to water-mediated hydrogen bonding between the Rossman folds of redox-associated enzymes and the pyrophosphates of dinucleotide enzyme cofactors (e.g., NAD+/NADP+) [102], is underlined. Also, the catalytically important [S/T]-Y-K triad common to members of the SDR family of enzymes is indicated by asterisks [103,104]. Vector NTI Advance 11.0 software alignment settings: BLOSUM45 matrix, gap opening penalty = 12, gap extension penalty = 0.1, gap separation penalty range = 0, no residue-specific or hydrophobic residue gaps.
Figure 4.
Amino acid alignment of GDP-4-keto-6-deoxy-D-mannose-3,5-epimerase-4-reductases. The predicted amino acid sequence of schistosome GMER (Sm) is compared to GMERs of humans (Hs), Mus musculus (Mm), Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), Mortierella alpina (Ma), Arabidopsis thaliana (At-1, At-2) and Bacteroides fragilis (Bf) (accession numbers in Table 1). Alignment position is indicated above each block, and sequence length is reported to the right of each line. Positions of identity are indicated in black, and gray-highlighted positions are greater than 80% conserved among the sampled sequences. A well-conserved glycine-rich phosphate-binding loop (GxxGxxG), which mediates the binding of dinucleotide cofactors (e.g., NAD+/NADP+) by redox-associated enzymes [102], is underlined. Additionally, the catalytically important [S/T]-Y-K triad of the short-chain dehydrogenase/reductase-type enzymes is indicated by asterisks (*), and residues thought to be involved in proton exchange between GMER and its epimerization reaction intermediates are marked with carets (^) [105]. Vector NTI Advance 11.0 software alignment settings: BLOSUM45 matrix, gap opening penalty = 12, gap extension penalty = 0.1, gap separation penalty range = 0, no residue-specific or hydrophobic residue gaps.
Previous studies have demonstrated that GFTs are generally Golgi-resident multispan transmembrane proteins with 10 TMDs [27,28,30,70]. Moreover, these genes feature a high degree of conservation across invertebrate and vertebrate taxa. Protein alignments of schistosome GFT with functionally characterized orthologs from Caenorhabditis, Drosophila, Mus and humans (see Table 2) revealed 25.2% overall identity, with pairwise comparisons indicating that schistosome GFT shares ~37-41% identity with orthologous GFTs (Figure 5). A unique feature of the schistosome protein is its conspicuously long C-terminal tail; however, the significance of this extension remains unknown. An analysis of membrane topology using the Phobius server suggested the presence of 10 tightly spaced TMDs, with both N- and C-terminal tails oriented into the cytoplasm (in Additional file 2: Figure S1). For comparison, the positions of the 10 TMDs in known GFTs were also determined using the Phobius server, and alignment with schistosome GFT demonstrated that the spacing of TMDs is roughly conserved across taxa (Figure 5). Altogether, these data support a role for schistosome GFT in GDP-L-fucose transport.
Figure 5.
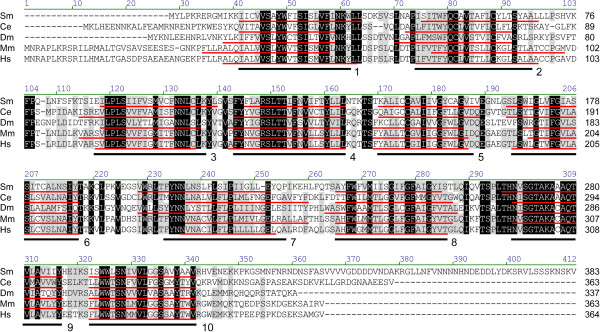
Amino acid alignment of GDP-L-fucose transporters. The predicted amino acid sequence of schistosome GFT (Sm) is compared to GFTs of Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Mus musculus (Mm) and humans (Hs) (accession numbers in Table 2). Alignment position is indicated above each block, and sequence length is reported to the right of each line. Positions of identity and positions exhibiting at least 80% conservation are highlighted in black and gray, respectively. The positions of ten well-conserved TMDs (underlined below sequences and alignment blocks) were determined using the Phobius transmembrane topology and signal peptide prediction server [107]. Vector NTI Advance 11.0 software alignment settings: BLOSUM45 matrix, gap opening penalty = 12, gap extension penalty = 0.1, gap separation penalty range = 0, no residue-specific or hydrophobic residue gaps.
Alternative splice isoforms of GFT that exclude exons 7 and 8 (and the intervening intron) encode a truncated protein featuring 7 TMDs. Importantly, nucleotide-sugar transporters (NSTs), including the GFTs, are part of a diverse drug/metabolite transporter superfamily composed of multispan transmembrane proteins (typically with 4–10 TMDs) that function in drug export, nutrient/metabolite efflux and compartmental metabolite exchange [108,109]. While the observed in-frame deletion of three TMDs likely abolishes GDP-L-fucose transport activity (given it lacks definitive primary sequence characteristics), the truncated GFT could retain its function as an NST (but with altered substrate specificity) or adopt a new class of metabolite transport function altogether. Future studies should assess the biochemical significance of this truncation.
The above topological analyses employed several transmembrane prediction tools (e.g., TMHMM 2.0 and TMpred [110,111]), but Phobius was the only one that predicted all 10 TMDs in most genes. Only TMD 9 of the human GFT was undetected using this method. Lubke et al. [27] reported similar difficulty in demonstrating this same TMD, which they attributed to its unusually high hydrophilicity. In general, in silico predictions of NST membrane topology are inherently difficult because current algorithms do not account for the relative thinness of the Golgi membrane (~20% thinner) and thus fail to recognize the concomitantly short TMDs of Golgi-resident transmembrane proteins such as NSTs [112]. In fact, at a typical length of 17–22 aa, the TMDs of Golgi proteins are on average five aa shorter than those of plasma membrane-associated proteins [113-115].
Phylogenetic analysis of nucleotide-sugar transporters
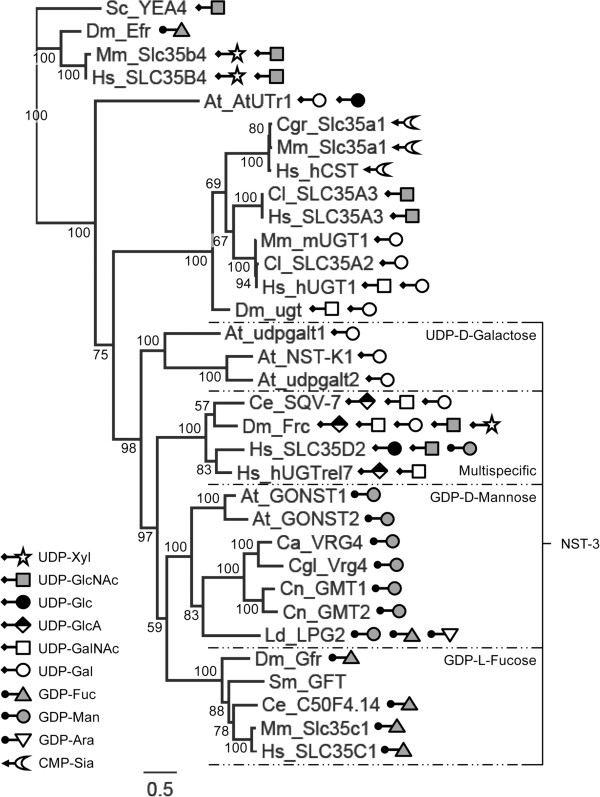
Primary sequence identity alone cannot reliably predict substrate specificity among NST genes [54,112]. NSTs can share as much as 50-60% of their primary sequences and exhibit different substrate specificities while proteins that are only 20% identical can transport the same nucleotide-sugar substrates [63]. However, previous studies have demonstrated that phylogenetic analyses can separate NSTs into functional groups [108,116]. To refine the predicted substrate specificity of schistosome GFT and better understand the structure-function relationship between GFTs and other NSTs, we conducted a phylogenetic analysis of schistosome GFT and a functionally diverse sampling of previously characterized NSTs. The topology of the resultant phylogeny is consistent with observations by Martinez-Duncker et al. [108]: the current repertoire of NSTs can be divided into three main families/groups (NST families 1–3), which form separate monophyletic clades (Figure 6; see in Additional file 3: Figure S2 for a rooted tree demonstrating the three NST families). Consistent with the notion that closely related NSTs can be functionally divergent, all three families include members with aberrant substrate specificities. While structure-function relationships in families 1 and 2 remain somewhat unclear, NST family 3 can be broken down into four daughter clades (corresponding to subfamilies J-M in [108]) that correspond to substrate specificity. Subfamily J includes NSTs exhibiting multispecific UDP-sugar transport activities, while subfamilies K, J and M feature NSTs having relatively narrow substrate specificities (GDP-D-mannose, UDP-D-galactose or GDP-L-fucose, respectively). Of the 18 previously characterized family 3 NSTs examined here, only LPG2 of Leishmania donovani features uncharacteristic activity for its clade, transporting GDP-L-fucose and GDP-D-arabinose in addition to GDP-D-mannose. Schistosome GFT forms a monophyletic clade with known Golgi-resident GFTs, supporting a predicted role in GDP-L-fucose transport. Notably, Drosophila Efr, which delivers GDP-L-fucose to the ER, clusters with NST family 2. This is consistent with other NST family 2 transporters that function in the ER and not the Golgi. Indeed, Martinez-Duncker et al.[108] reported that 54% of NST family 2 members feature a C-terminal di-lysine (KKxx) ER-retention/retrieval signal, and one such signal (KKVE) is present in Drosophila Efr. In contrast, similar ER-retention/retrieval signals do not exist in schistosome GFT or any of the family 3 NSTs examined here.
Figure 6.
Phylogenetic tree of nucleotide-sugar transporters. The amino acid sequences of previously characterized NSTs and putative GFT of Schistosoma mansoni (RefSeq/GenBank accession numbers in Table 2) were included in a phylogeny annotated with substrate specificity data. Posterior probabilities are indicated at each node, and genetic divergence (substitutions per site) is represented by the scale bar. Family 3 NSTs [108], which include transporters of GDP-L-fucose, are labeled on the right. To demonstrate the topology of NST families 1 and 2, these data are also presented as a rooted tree in (Additional file 3: Figure S2). UDP-Xyl, UDP-D-xylose; UDP-GlcNAc, UDP-D-N-acetylglucosamine; UDP-Glc, UDP-D-glucose; UDP-GlcA, UDP-D-glucuronic acid; UDP-GalNAc, UDP-D-N-acetylgalactosamine; UDP-Gal, UDP-D-galactose; GDP-Fuc, GDP-L-fucose; GDP-Man, GDP-D-mannose; GDP-Ara, GDP-D-arabinose; CMP-Sia, CMP-sialic acid.
GMD, GMER and GFT mRNA expression in miracidia and primary sporocysts of S. mansoni
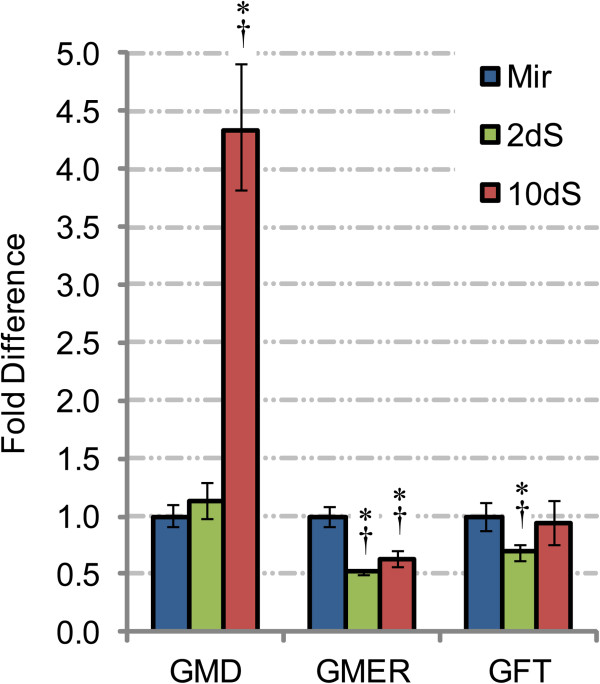
Given recent data demonstrating the abundant expression of fucosylated glycotopes in snail-associated schistosome larvae [7,117] and their predicted immunomodulatory roles in snail hosts, GMD, GMER, and GFT steady-state transcript levels were assayed by qPCR in miracidia and 2- and 10-day in vitro-cultivated primary sporocysts. The results indicate that all three genes are differentially expressed during the miracidium-to-primary sporocyst transformation and subsequent cultivation (Figure 7). In conjunction with larval transformation, GMER and GFT transcript levels declined 48% and 31%, respectively, after two days in culture, while GMD expression remained unchanged. During subsequent in vitro cultivation of primary sporocysts (up to 10 days), GMD transcript abundance climbed ~4-fold while expression of GMER and GFT stayed the same. These results are somewhat confounding since GMD and GMER constitute a single biosynthetic pathway.
Figure 7.
GDP-L-fucose synthesis- and transport-associated gene transcription in larvae of Schistosoma mansoni. Real-time qPCR was used to examine GMD, GMER and GFT transcription in miracidia (Mir) and 2- and 10-day in vitro-cultivated primary sporocysts (2dS and 10dS, respectively). Transcript abundances in primary sporocysts were compared to miracidia (arbitrarily set at 1), and data were analyzed across three biological replicates using heteroscedastic two-sample t- and Wilcoxon rank sum tests, with significance set at p≤0.05 (indicated by *) and p=0.10 (indicated by †), respectively.
For comparison, the S. mansoni Serial Analysis of Gene Expression (SAGE) Database [96] was examined for relevant SAGE tags, and tags 7188 and 10882 corresponding to GMD and GMER, respectively, were identified. Consistent with the present study, the SAGE data indicate that GMD transcript abundance increases ~3-fold from miracidia to 6-day primary sporocysts while the GMER-specific tag 10882 was not detected in either larval stage. Interestingly, both genes exhibited peak expression in 20-day primary sporocysts, suggesting that GDP-L-fucose synthesis potentially increases in older larvae. GFT transcript expression (as indicated by tag 4514) followed a similar profile, with relatively low transcript levels in miracidia and 6-day primary sporocysts and peak expression after 20 days in culture. In the present study, if sporocyst cultivation times had been longer, the expression of all three genes may have peaked similarly in older larvae (i.e., >10 days in culture).
The GMDs in bacteria participate in several overlapping synthetic pathways, with reaction intermediates being converted to GDP-L-fucose, GDP-D-rhamnose or GDP-D-talose by GMER, GDP-6-deoxy-d-lyxo-hexos-4-ulose-4-reductase (RMD) and GDP-6-deoxy-D-talose synthetase (GTS), respectively (reviewed in [118]). Additionally, GMDs of Paramecium bursaria, Chlorella virus 1 and some bacteria (e.g., Pseudomonas aeruginosa) are bifunctional, having the added ability to catalyze the same stereospecific reduction as RMD. A similar, still unknown dual functionality or involvement in other biochemical pathways in schistosomes could explain why the observed GMD and GMER expression profiles vary independently; however, participation of GMD in GDP-D-rhamnose or GDP-D-talose biosynthesis in particular is unlikely because rhamnose and talose, as well as homologs of RMD and GTS, are not observed in S. mansoni.
Recombinant GMD and GMER protein expression, purification, and antibody production
To facilitate analyses of protein expression in larval S. mansoni, GMD and GMER were heterologously expressed and purified, and the recombinant proteins were used to raise GMD- and GMER-specific chicken IgY antibodies (in Additional file 4: Figure S3A-B). To assess specificity, antibodies were tested against blotted crude parasite extracts and pure GMD and GMER recombinant antigens. Initially, immunoblots revealed unacceptable levels of crossreactivity (especially between anti-GMD IgY and recombinant GMER; in Additional file 4: Figure S3C), so antibodies were further purified by membrane adsorption against the purified antigens. Subsequent immunoblots demonstrated that antigen specificities of both IgY preparations were greatly improved, showing essentially monospecific reactivities (in Additional file 4: Figure S3D). Membrane-isolated antibodies were used in downstream immunoblot and microscopic analyses.
Characterization of GMD and GMER protein expression in miracidia and primary sporocysts of S. mansoni
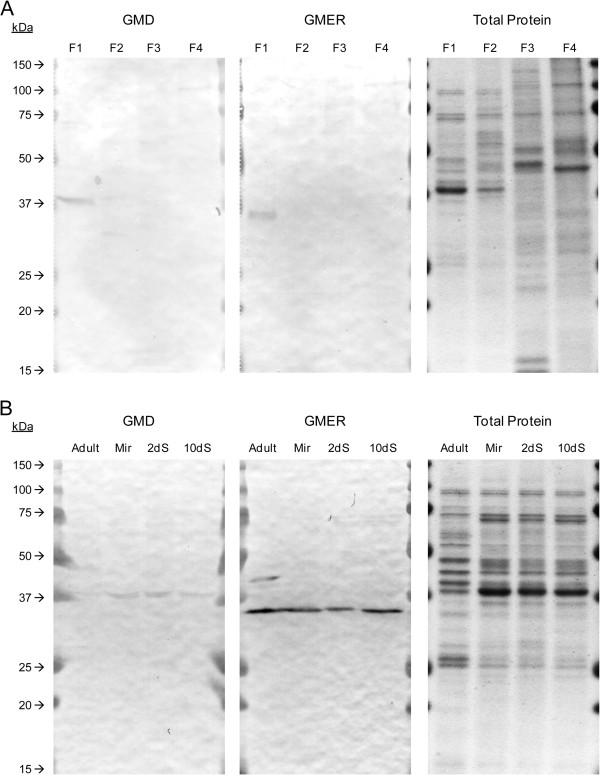
Multiple attempts were made to demonstrate the presence of GMD and GMER in crude adult and larval extracts using western blotting, but only faint bands were produced (Peterson, unpublished data). To enhance detection of GMD and GMER in western blot analyses and concurrently demonstrate their cytosolic localization, 2-day primary sporocysts were serially extracted using a ProteoExtract® Subcellular Proteome Extraction Kit, yielding enriched cytosolic, membrane, nuclear, and cytoskeletal protein fractions. While application of the ProteoExtract® kit for subcellular fractionation of whole schistosome larvae has yet to be experimentally validated regarding the fidelity of differential extraction, Coomassie-stained gels clearly demonstrated compositional differences in the resultant protein fractions and fractionation successfully facilitated detection of GMD and GMER in subsequent immunoblots (Figure 8A). Consistent with their expected roles in cytosolic GDP-L-fucose synthesis, immunoblots revealed the presence of GMD and GMER only in the presumptive cytosolic fraction (bands at 38 and 35 kDa, respectively).
Figure 8.
GMD and GMER protein expression in larval and adult Schistosoma mansoni. Membrane-purified chicken IgY antibodies (see in Additional file 4: Figure S3) were used in western blot analyses of 2-day in vitro-cultivated primary sporocyst subcellular protein extracts (A). Four enriched fractions were examined: (F1) cytosol, (F2) membrane/membrane organelle, (F3) nucleus, and (F4) cytoskeleton. Blots indicate the presence of GMD and GMER only in the cytosolic fraction. Next, cytosolic fractions were used to compare GMD and GMER protein expression among mixed-sex adults, miracidia and 2- and 10-day in vitro-cultivated primary sporocysts (B). In both experiments, total protein was visualized in-gel by Coomassie staining.
In a comparison of cytosolic extracts from miracidia and 2- and 10-day primary sporocysts, GMD and GMER proteins appear to be stably expressed during larval transformation and subsequent in vitro cultivation (Figure 8B). This result seemingly contradicts qPCR and SAGE data described above, which indicate stage-specific differences in GMD and GMER transcript levels among snail-associated larvae. One possible explanation for the apparent discrepancy between transcript and protein abundances is the inability of qPCR and SAGE approaches to adequately differentiate between “functional” GMD/GMER-coding transcripts and variants that are pretranslationally targeted for nonsense-mediated decay or are translated to truncated proteins not detected by the above methods. For example, while GMD gene transcription appears to increase ~4-fold in 10-day in vitro-cultivated primary sporocysts, the absolute abundance of “functional” GMD-coding transcripts might remain unchanged, thus resulting in no detectable alteration in protein expression. Additionally, protein turnover rates may be sufficiently low to permit persistence and stable detection regardless of declining transcript abundance (e.g., GMER). Lastly, it should be noted that colorimetric precipitation-mediated detection of immunoreactive proteins is perhaps inadequate for the demonstration of relatively minor differences in protein abundance and application of more quantitative detection methods (e.g., fluorescence) might have revealed low-level stage-specific variations in GMD and GMER expression that mirror the observed changes in gene transcription.
Immunoblots also examined GMD and GMER expression in mixed-sex adult worms (Figure 8B). Cytosolic extracts were seemingly devoid of immunoreactive GMD, suggesting differential expression between adults and larvae. Additionally, adult extracts featured two anti-GMER IgY-reactive bands, one corresponding to GMER and a second at ~42 kDa. The added band potentially represents the translated product of an adult-specific alternative splice isoform; however, none of the observed variants can account for the increased protein size. Alternatively, the band is an artifact of antibody crossreactivity. That adult worms apparently lack GMD while expressing one or more GMER isoforms is confounding, given their roles in the same biosynthetic pathway. One possible explanation is that GMER or an alternative protein isoform has an unknown role in a separate pathway, which drives its expression independent of GMD.
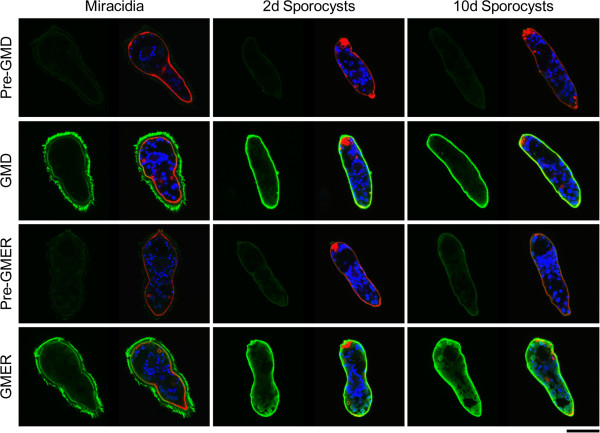
Finally, the membrane-purified antibodies were employed in confocal laser scanning microscopy to demonstrate the tissue localization of GMD and GMER proteins in miracidia and 2- and 10-day primary sporocysts (Figure 9). Both proteins were observed predominantly in the ciliated epidermal plates and tegument of miracidia and sporocysts, respectively, while antibodies exhibited at least minor reactivities in internal somatic tissues. Similar patterns of expression in schistosome larvae were observed for several prominent fucosylated glycotopes, including Fucα1-3GalNAcβ1-4GlcNAc (F-LDN) and Fucα1-3GalNAcβ1-4(Fucα1-3)GlcNAc (F-LDN-F) [7]. Importantly, co-localization of schistosome GMD and GMER implies the presence of a complete de novo pathway for GDP-L-fucose synthesis, and further supports their roles in fucosylation.
Figure 9.
Localization of de novo GDP-L-fucose synthesis in miracidia and primary sporocysts of Schistosoma mansoni. Confocal laser scanning microscopy was used to assess the localization of schistosome GMD and GMER in miracidia and 2- and 10-day in vitro-cultivated primary sporocysts. Fixed and permeabilized larvae were immunostained with membrane-purified chicken IgY raised against schistosome GMD and GMER (rows 2 and 4, respectively), and antibody reactivities were assessed relative to control larvae incubated with membrane-adsorbed pre-immune eluates (rows 1 and 3). Panels include paired micrographs depicting GMD/GMER expression (green) alone and merged with counterstained actin (e.g., muscles, flame cells; red) and DNA (e.g., nuclei; blue). Approximate scale is represented in the lower right corner (bar = 50 μm).
Conclusions
The present study used a genome-wide homology based bioinformatics approach to identify GDP-L-fucose synthesis- and transport-associated genes in the human blood fluke Schistosoma mansoni. The above data indicate that GDP-L-fucose in S. mansoni is generated in the cytosol by a de novo synthetic pathway comprising GMD and GMER enzymes, after which the resulting activated fucose is imported into the Golgi by the multispan transmembrane protein GFT. Importantly, these enzymes represent a bottleneck in the fucosylation process since GDP-L-fucose is the sole nucleotide-sugar donor utilized by Golgi- and ER-resident FucTs. This research has provided a necessary foundation for future investigations that further explore the role of GDP-L-fucose synthesis and transport in schistosome development and immunobiology. Additionally, the genes identified in this study are potential targets for the development of novel anti-schistosomal chemotherapeutics.
Abbreviations
Ara: Arabinose; BSA: Bovine serum albumin; CBSS: Chernin’s Balanced Salt Solution; CDS: Coding sequence; FPGT: L-fucose-1-phosphate guanylyltransferase; Fuc: Fucose; FucT: Fucosyltransferase; Fuk: L-Fucokinase; Gal: Galactose; GalNAc: N-acetylgalactosamine; GFT: GDP-L-fucose transporter; Glc: Glucose; GlcA: Glucuronic acid; GlcNAc: N-acetylglucosamine; GMD: GDP-D-mannose-4,6-dehydratase; GMER: GDP-4-keto-6-deoxy-D-mannose-3,5-epimerase-4-reductase; GOI: Gene of interest; LADII: Leukocyte adhesion deficiency type II; Man: Mannose; NCBI: National Center for Biotechnology Information; NST: Nucleotide-sugar transporter; ORF: Open reading frame; PBS: Phosphate-buffered saline; PCB: PreScission™ cleavage buffer; PIC: Protease inhibitor cocktail; PTC: Premature termination codon; qPCR: Quantitative PCR; RACE: Rapid amplification of cDNA ends; RefSeq: Reference Sequence; RT-PCR: Reverse transcription PCR; SAGE: Serial analysis of gene expression; SchistoDB: Schistosoma mansoni Database; SDR: Short-chain dehydrogenase/reductase; Sia: Sialic acid; sPBS: Snail PBS; TBS: Tris-buffered saline; TBST: Tris-buffered saline with Tween® 20; TMD: Transmembrane domain; Xyl: Xylose.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NAP and TPY designed the above research. NAP and TKA performed experiments and analyzed data. XJW developed the modified subcellular extraction method. NAP wrote the original manuscript, and all authors contributed to its revision and final preparation. All authors have read and approved the manuscript for publication.
Authors’ information
The above investigation was designed and executed by NAP in partial fulfillment of his doctoral degree in the Comparative Biomedical Sciences program at the University of Wisconsin-Madison under the supervision of TPY. TKA and XJW provided valuable technical expertise in phylogenetic analyses and biochemical methods, respectively.
Supplementary Material
This PDF document contains Additional file 1: Table S1, which lists the oligonucleotide primers used for RT-PCR, RACE, qPCR and protein expression in this study.
This TIF document contains Additional file 2: Figure S1, which features the results of an in silico analysis of GFT membrane topology. Transmembrane domains were identified in the schistosome GFT protein using the Phobius transmembrane topology and signal peptide prediction server [107]. The Phobius output suggested 10 TMDs, a number that is consistent with GDP-L-fucose transporters of other organisms [27,28,30,70] (also see Figure 5) (A). A model based on this output was constructed, portraying the arrangement of the 10 TMDs (numbers indicating the amino acid boundaries of each TMD) as well as the most likely orientation for schistosome GMD within the Golgi membrane (B).
This TIF document contains Additional file 3: Figure S2, which features a rooted phylogenetic tree of nucleotide-sugar transporters (see Figure 6 for detailed unrooted tree). The amino acid sequences of NSTs with previously characterized substrate specificities were obtained from RefSeq and GenBank databases at NCBI (accession numbers in Table 2). A tree was constructed using Bayesian methods implemented in MrBayes v3.12 with mixed amino acid evolutionary models. Monophyletic clades representing NST families 1–3 [108] are indicated, and genetic divergence (substitutions per site) is represented by the scale. The tree is rooted on NST family 2.
This TIF document contains Additional file 4: Figure S3, which describes heterologous expression and isolation of recombinant schistosome GMD and GMER proteins and downstream affinity purification of GMD- and GMER-specific polyclonal chicken IgY. GST-GMD and -GMER fusion constructs were created in pGEX-6P-1 vector, and the encoded proteins were expressed in E. coli. Fusion protein expression in induced (Ind) and uninduced (Un) cultures was compared by SDS-PAGE fractionation and Coomassie staining of soluble cellular extracts (A). Fusion protein-containing extracts were passed through a GST-affinity column, and bound GMD and GMER were eluted by PreScission™ Protease-mediated cleavage of the GST fusions. Eluates were then analyzed by SDS-PAGE fractionation and Coomassie staining (B). Polyclonal chicken IgY antibodies were raised against recombinant GMD and GMER proteins, and the resultant antibodies were tested by immunoblotting the pure recombinant antigens (C). Due to crossreactivity among the antibodies and antigens (especially between anti-GMD IgY and recombinant GMER), antibodies were affinity-purified by membrane adsorption using bound GMD and GMER antigen. Following elution, antibody preparations were again tested against blots of pure antigen, demonstrating greatly reduced crossreactivity (D).
Contributor Information
Nathan A Peterson, Email: napeterson@svm.vetmed.wisc.edu.
Tavis K Anderson, Email: tavis.anderson@ars.usda.gov.
Xiao-Jun Wu, Email: wux@svm.vetmed.wisc.edu.
Timothy P Yoshino, Email: yoshinot@svm.vetmed.wisc.edu.
Acknowledgements
This work was supported by NIH grants AI015503 and AI061436 to TPY and NIH-NIAID contract no. HHSN272201000005I (Lewis F, Biomedical Research Institute, Rockville, MD, USA). Additionally, NAP received predoctoral fellowship support through the Cellular and Molecular Parasitology Training Grant NIH T32 AI007414. The authors also thank Laura Gonzalez for her technical assistance.
References
- Hokke CH, Deelder AM, Hoffmann KF, Wuhrer M. Glycomics-driven discoveries in schistosome research. Exp Parasitol. 2007;117:275–283. doi: 10.1016/j.exppara.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JM, Peak E, Perally S, Chalmers IW, Barrett J, Yoshino TP, Ivens AC, Hoffmann KF. Anti-schistosomal intervention targets identified by lifecycle transcriptomic analyses. PLoS Negl Trop Dis. 2009;3:e543. doi: 10.1371/journal.pntd.0000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson NA, Anderson TK, Yoshino TP. In silico analysis of the fucosylation-associated genome of the human blood fluke Schistosoma mansoni: cloning and characterization of the fucosyltransferase multigene family. PLoS One. 2013;8:e63299. doi: 10.1371/journal.pone.0063299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Remoortere A, Hokke CH, van Dam GJ, van Die I, Deelder AM, van den Eijnden DH. Various stages of Schistosoma express LewisX, LacdiNAc, GalNAcβ1-4(Fucα1-3)GlcNAc, and GalNAcβ1-4(Fucα1-2Fucα1-3)GlcNAc carbohydrate epitopes: detection with monoclonal antibodies that are characterized enzymatically synthesized neoglycoproteins. Glycobiology. 2000;10:601–609. doi: 10.1093/glycob/10.6.601. [DOI] [PubMed] [Google Scholar]
- Robijn ML, Wuhrer M, Kornelis D, Deelder AM, Geyer R, Hokke CH. Mapping fucosylated epitopes on glycoproteins and glycolipids of Schistosoma mansoni cercariae, adult worms, and eggs. Parasitology. 2005;130:67–77. doi: 10.1017/S0031182004006390. [DOI] [PubMed] [Google Scholar]
- Wuhrer M, Koeleman CA, Fitzpatrick JM, Hoffmann KF, Deelder AM, Hokke CH. Gender-specific expression of complex-type N-glycans in schistosomes. Glycobiology. 2006;16:991–1006. doi: 10.1093/glycob/cwl020. [DOI] [PubMed] [Google Scholar]
- Peterson NA, Hokke CH, Deelder AM, Yoshino TP. Glycotope analysis in miracidia and primary sporocysts of Schistosoma mansoni: differential expression during the miracidium-to-sporocyst transformation. Int J Parasitol. 2009;39:1331–1344. doi: 10.1016/j.ijpara.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P. Golgi glycosylation. Cold Harbor Springs Perspect Biol. 2011;3:a005199. doi: 10.1101/cshperspect.a005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker DJ, Lowe JB. Fucose: biosynthesis and biological function in mammals. Glycobiology. 2003;13:41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- Yurchenco PD, Atkinson PH. Equilibration of fucosyl glycoprotein pools in Hela cells. Biochemistry. 1977;16:944–953. doi: 10.1021/bi00624a021. [DOI] [PubMed] [Google Scholar]
- Rhomberg S, Fuchsluger C, Rendić D, Paschinger K, Jantsch V, Kosma P, Wilson IB. Reconstitution in vitro of the GDP-fucose biosynthetic pathways of Caenorhabditis elegans and Drosophila melanogaster. FEBS J. 2006;273:2244–2256. doi: 10.1111/j.1742-4658.2006.05239.x. [DOI] [PubMed] [Google Scholar]
- Park SH, Pastuszak I, Drake R, Elbein AD. Purification to apparent homogeneity and properties of pig kidney L-fucose kinase. J Biol Chem. 1998;273:5685–5691. doi: 10.1074/jbc.273.10.5685. [DOI] [PubMed] [Google Scholar]
- Pastuszak I, Ketchum C, Hermanson G, Sjoberg EJ, Drake R, Elbein AD. GDP-L-fucose pyrophosphorylase. Purification, cDNA cloning, and properties of the enzyme. J Biol Chem. 1998;273:30165–30174. doi: 10.1074/jbc.273.46.30165. [DOI] [PubMed] [Google Scholar]
- Hinderlich S, Berger M, Blume A, Chen H, Ghaderi D, Bauer C. Identification of human L-fucose kinase amino acid sequence. Biochem Biophys Res Commun. 2002;294:650–654. doi: 10.1016/S0006-291X(02)00541-7. [DOI] [PubMed] [Google Scholar]
- Niittymäki J, Mattila P, Roos C, Huopaniemi L, Sjöblom S, Renkonen R. Cloning and expression of murine enzymes involved in the salvage pathway of GDP-L-fucose. Eur J Biochem. 2004;271:78–86. doi: 10.1046/j.1432-1033.2003.03904.x. [DOI] [PubMed] [Google Scholar]
- Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- Kotake T, Hojo S, Tajima N, Matsuoka K, Koyama T, Tsumuraya Y. A bifunctional enzyme with L-fucokinase and GDP-L-fucose pyrophosphorylase activities salvages free L-fucose in Arabidopsis. J Biol Chem. 2008;283:8125–8135. doi: 10.1074/jbc.M710078200. [DOI] [PubMed] [Google Scholar]
- Roos C, Kolmer M, Mattila P, Renkonen R. Composition of Drosophila melanogaster proteome involved in fucosylated glycan metabolism. J Biol Chem. 2002;277:3168–3175. doi: 10.1074/jbc.M107927200. [DOI] [PubMed] [Google Scholar]
- Caffaro CE, Hirschberg CB. Nucleotide sugar transporters of the Golgi apparatus: from basic science to diseases. Acc Chem Res. 2006;39:805–812. doi: 10.1021/ar0400239. [DOI] [PubMed] [Google Scholar]
- Noda K, Miyoshi E, Gu J, Gao CX, Nakahara S, Kitada T, Honke K, Suzuki K, Yoshihara H, Yoshikawa K, Kawano K, Tonetti M, Kasahara A, Hori M, Hayashi N, Taniguchi N. Relationship between elevated FX expression and increased production of GDP-L-fucose, a common donor substrate for fucosylation in human hepatocellular carcinoma and hepatoma cell lines. Cancer Res. 2003;63:6282–6289. [PubMed] [Google Scholar]
- Moriwaki K, Noda K, Nakagawa T, Asahi M, Yoshihara H, Taniguchi N, Hayashi N, Miyoshi E. A high expression of GDP-fucose transporter in hepatocellular carcinoma is a key factor for increases in fucosylation. Glycobiology. 2007;17:1311–1320. doi: 10.1093/glycob/cwm094. [DOI] [PubMed] [Google Scholar]
- Niittymäki J, Mattila P, Renkonen R. Differential gene expression of GDP-L-fucose-synthesizing enzymes, GDP-fucose transporter and fucosyltransferase VII. APMIS. 2006;114:539–548. doi: 10.1111/j.1600-0463.2006.apm_461.x. [DOI] [PubMed] [Google Scholar]
- Omasa T, Tanaka R, Doi T, Ando M, Kitamoto Y, Honda K, Kishimoto M, Ohtake H. Decrease in antithrombin III fucosylation by expressing GDP-fucose transporter siRNA in Chinese hamster ovary cells. J Biosci Bioeng. 2008;106:168–173. doi: 10.1263/jbb.106.168. [DOI] [PubMed] [Google Scholar]
- Frydman M, Etzioni A, Eidlitz-Markus T, Avidor I, Varsano I, Shechter Y, Orlin JB, Gershoni-Baruch R. Rambam-Hasharon syndrome of psychomotor retardation, short stature, defective neutrophil motility, and Bombay phenotype. Am J Med Genet. 1992;44:297–302. doi: 10.1002/ajmg.1320440307. [DOI] [PubMed] [Google Scholar]
- Etzioni A, Frydman M, Pollack S, Avidor I, Phillips ML, Paulson JC, Gershoni-Baruch R. Brief report: recurrent severe infections caused by a novel leukocyte adhesion deficiency. N Engl J Med. 1992;327:1789–1792. doi: 10.1056/NEJM199212173272505. [DOI] [PubMed] [Google Scholar]
- Lübke T, Marquardt T, von Figura K, Körner C. A new type of carbohydrate-deficient glycoprotein syndrome due to a decreased import of GDP-fucose into the golgi. J Biol Chem. 1999;274:25986–25989. doi: 10.1074/jbc.274.37.25986. [DOI] [PubMed] [Google Scholar]
- Lübke T, Marquardt T, Etzioni A, Hartmann E, von Figura K, Körner C. Complementation cloning identifies CDG-IIc, a new type of congenital disorders of glycosylation, as a GDP-fucose transporter deficiency. Nat Genet. 2001;28:73–76. doi: 10.1038/ng0501-73. [DOI] [PubMed] [Google Scholar]
- Lühn K, Wild MK, Eckhardt M, Gerardy-Schahn R, Vestweber D. The gene defective in leukocyte adhesion deficiency II encodes a putative GDP-fucose transporter. Nat Genet. 2001;28:69–72. doi: 10.1038/ng0501-69. [DOI] [PubMed] [Google Scholar]
- Helmus Y, Denecke J, Yakubenia S, Robinson P, Lühn K, Watson DL, McGrogan PJ, Vestweber D, Marquardt T, Wild MK. Leukocyte adhesion deficiency II patients with a dual defect of the GDP-fucose transporter. Blood. 2006;107:3959–3966. doi: 10.1182/blood-2005-08-3334. [DOI] [PubMed] [Google Scholar]
- Hellbusch CC, Sperandio M, Frommhold D, Yakubenia S, Wild MK, Popovici D, Vestweber D, Gröne HJ, von Figura K, Lübke T, Körner C. Golgi GDP-fucose transporter-deficient mice mimic congenital disorder of glycosylation IIc/leukocyte adhesion deficiency II. J Biol Chem. 2007;282:10762–10772. doi: 10.1074/jbc.M700314200. [DOI] [PubMed] [Google Scholar]
- Yoshino TP, Laursen JR. Production of Schistosoma mansoni daughter sporocysts from mother sporocysts maintained in synxenic culture with Biomphalaria glabrata embryonic (Bge) cells. J Parasitol. 1995;81:714–722. doi: 10.2307/3283960. [DOI] [PubMed] [Google Scholar]
- Nolan LE, Carriker JP. Observations on the biology of the snail Lymnaea stagnalis appressa during twenty years of laboratory culture. Am Midl Nat. 1946;36:467–493. doi: 10.2307/2421516. [DOI] [Google Scholar]
- Chernin E. Observations on hearts explanted in vitro from the snail Australorbis glabratus. J Parasitol. 1963;49:353–364. doi: 10.2307/3275797. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerlotini A, Heiges M, Wang H, Moraes RL, Dominitini AJ, Ruiz JC, Kissinger JC, Oliveira G. SchistoDB: a Schistosoma mansoni genome resource. Nucleic Acids Res. 2009;37:D579–D582. doi: 10.1093/nar/gkn681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama C, Smith PL, Angata K, Fukuda MN, Lowe JB, Fukuda M. Molecular cloning and expression of GDP-D-mannose-4,6-dehydratase, a key enzyme for fucose metabolism defective in Lec13 cells. J Biol Chem. 1998;273:14582–14587. doi: 10.1074/jbc.273.23.14582. [DOI] [PubMed] [Google Scholar]
- Sullivan FX, Kumar R, Kriz R, Stahl M, Xu GY, Rouse J, Chang XJ, Boodhoo A, Potvin B, Cumming DA. Molecular cloning of human GDP-mannose 4,6-dehydratase and reconstitution of GDP-fucose biosynthesis in vitro. J Biol Chem. 1998;273:8193–8202. doi: 10.1074/jbc.273.14.8193. [DOI] [PubMed] [Google Scholar]
- Bisso A, Sturla L, Zanardi D, De Flora A, Tonetti M. Structural and enzymatic characterization of human recombinant GDP-D-mannose-4,6-dehydratase. FEBS Lett. 1999;456:370–374. doi: 10.1016/S0014-5793(99)00982-5. [DOI] [PubMed] [Google Scholar]
- Imai-Nishiya H, Mori K, Inoue M, Wakitani M, Iida S, Shitara K, Satoh M. Double knockdown of α1,6-fucosyltransferase (FUT8) and GDP-mannose 4,6-dehydratase (GMD) in antibody-producing cells: a new strategy for generating fully non-fucosylated therapeutic antibodies with enhanced ADCC. BMC Biotechnol. 2007;7:84. doi: 10.1186/1472-6750-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonetti M, Sturla L, Bisso A, Benatti U, De Flora A. Synthesis of GDP-L-fucose by the human FX protein. J Biol Chem. 1996;271:27274–27279. doi: 10.1074/jbc.271.44.27274. [DOI] [PubMed] [Google Scholar]
- Zipin A, Israeli-Amit M, Meshel T, Sagi-Assif O, Yron I, Lifshitz V, Bacharach E, Smorodinsky NI, Many A, Czernilofsky PA, Morton DL, Witz IP. Tumor-microenvironment interactions: the fucose-generating FX enzyme controls adhesive properties of colorectal cancer cells. Cancer Res. 2004;64:6571–6578. doi: 10.1158/0008-5472.CAN-03-4038. [DOI] [PubMed] [Google Scholar]
- Quirk S, Seley KL. Substrate discrimination by the human GTP fucose pyrophosphorylase. Biochemistry. 2005;44:10854–10863. doi: 10.1021/bi0503605. [DOI] [PubMed] [Google Scholar]
- Quirk S, Seley KL. Identification of catalytic amino acids in the human GTP fucose pyrophosphorylase active site. Biochemistry. 2005;44:13172–13178. doi: 10.1021/bi051288d. [DOI] [PubMed] [Google Scholar]
- Smith PL, Myers JT, Rogers CE, Zhou L, Petryniak B, Becker DJ, Homeister JW, Lowe JB. Conditional control of selectin ligand expression and global fucosylation events in mice with a targeted mutation at the FX locus. J Cell Biol. 2002;158:801–815. doi: 10.1083/jcb.200203125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker DJ, Myers JT, Ruff MM, Smith PL, Gillespie BW, Ginsburg DW, Lowe JB. Strain-specific modification of lethality in fucose-deficient mice. Mamm Genome. 2003;14:130–139. doi: 10.1007/s00335-002-2212-5. [DOI] [PubMed] [Google Scholar]
- Ohata S, Kinoshita S, Aoki R, Tanaka H, Wada H, Tsuruoka-Kinoshita S, Tsuboi T, Watabe S, Okamoto H. Neuroepithelial cells require fucosylated glycans to guide the migration of vagus motor neuron progenitors in the developing zebrafish hindbrain. Development. 2009;136:1653–1663. doi: 10.1242/dev.033290. [DOI] [PubMed] [Google Scholar]
- Song Y, Willer JR, Scherer PC, Panzer JA, Kugath A, Skordalakes E, Gregg RG, Willer GB, Balice-Gordon RJ. Neural and synaptic defects in slytherin, a zebrafish model for human congenital disorders of glycosylation. PLoS One. 2010;5:e13743. doi: 10.1371/journal.pone.0013743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Perepelov AV, Wang H, Zhang H, Knirel YA, Wang L, Chen W. Biochemical characterization of GDP-L-fucose de novo synthesis pathway in fungus Mortierella alpina. Biochem Biophys Res Commun. 2010;391:1663–1669. doi: 10.1016/j.bbrc.2009.12.116. [DOI] [PubMed] [Google Scholar]
- Bonin CP, Freshour G, Hahn MG, Vanzin GF, Reiter WD. The GMD1 and GMD2 genes of Arabidopsis encode isoforms of GDP-D-mannose 4,6-dehydratase with cell type-specific expression patterns. Plant Physiol. 2003;132:883–892. doi: 10.1104/pp.103.022368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin CP, Potter I, Vanzin GF, Reiter WD. The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc Natl Acad Sci USA. 1997;94:2085–2090. doi: 10.1073/pnas.94.5.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin CP, Reiter WD. A bifunctional epimerase-reductase acts downstream of the MUR1 gene product and completes the de novo synthesis of GDP-L-fucose in Arabidopsis. Plant J. 2000;21:445–454. doi: 10.1046/j.1365-313x.2000.00698.x. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Maeda Y, Jigami Y. Interaction of GDP-4-keto-6-deoxymannose-3,5-epimerase-4-reductase with GDP-mannose-4,6-dehydratase stabilizes the enzyme activity for formation of GDP-fucose from GDP-mannose. Glycobiology. 2003;13:673–680. doi: 10.1093/glycob/cwg099. [DOI] [PubMed] [Google Scholar]
- Wang W, Hu T, Frantom PA, Zheng T, Gerwe B, Del Amo DS, Garret S, Seidel RD 3rd, Wu P. Chemoenzymatic synthesis of GDP-L-fucose and the Lewis X glycan derivatives. Proc Natl Acad Sci USA. 2009;106:16096–16101. doi: 10.1073/pnas.0908248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg CB. Golgi nucleotide sugar transport and leukocyte adhesion deficiency II. J Clin Invest. 2001;108:3–6. doi: 10.1172/JCI13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikov A, Routier F, Fuhlrott J, Helmus Y, Wild M, Gerardy-Schahn R, Bakker H. The human solute carrier gene SLC35B4 encodes a bifunctional nucleotide sugar transporter with specificity for UDP-xylose and UDP-N-acetylglucosamine. J Biol Chem. 2005;280:27230–27235. doi: 10.1074/jbc.M504783200. [DOI] [PubMed] [Google Scholar]
- Ishida N, Yoshioka S, Chiba Y, Takeuchi M, Kawakita M. Molecular cloning and functional expression of the human Golgi UDP-N-acetylglucosamine transporter. J Biochem. 1999;126:68–77. doi: 10.1093/oxfordjournals.jbchem.a022437. [DOI] [PubMed] [Google Scholar]
- Suda T, Kamiyama S, Suzuki M, Kikuchi N, Nakayama K, Narimatsu H, Jigami Y, Aoki T, Nishihara S. Molecular cloning and characterization of a human multisubstrate specific nucleotide-sugar transporter homologous to Drosophila fringe connection. J Biol Chem. 2004;279:26469–26474. doi: 10.1074/jbc.M311353200. [DOI] [PubMed] [Google Scholar]
- Muraoka M, Kawakita M, Ishida N. Molecular characterization of human UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, a novel nucleotide sugar transporter with dual substrate specificity. FEBS Lett. 2001;495:87–93. doi: 10.1016/S0014-5793(01)02358-4. [DOI] [PubMed] [Google Scholar]
- Miura N, Ishida N, Hoshino M, Yamauchi M, Hara T, Ayusawa D, Kawakita M. Human UDP-galactose translocator: molecular cloning of a complementary DNA that complements the genetic defect of a mutant cell line deficient in UDP-galactose translocator. J Biochem. 1996;120:236–241. doi: 10.1093/oxfordjournals.jbchem.a021404. [DOI] [PubMed] [Google Scholar]
- Aoki K, Ishida N, Kawakita M. Substrate recognition by UDP-galactose and CMP-sialic acid transporters. Different sets of transmembrane helices are utilized for the specific recognition of UDP-galactose and CMP-sialic acid. J Biol Chem. 2001;276:21555–21561. doi: 10.1074/jbc.M101462200. [DOI] [PubMed] [Google Scholar]
- Aoki K, Ishida N, Kawakita M. Substrate recognition by nucleotide sugar transporters: further characterization of substrate recognition regions by analyses of UDP-galactose/CMP-sialic acid transporter chimeras and biochemical analysis of the substrate specificity of parental and chimeric transporters. J Biol Chem. 2003;278:22887–22893. doi: 10.1074/jbc.M302620200. [DOI] [PubMed] [Google Scholar]
- Segawa H, Kawakita M, Ishida N. Human and Drosophila UDP-galactose transporters transportUDP-N-acetylgalactosamine in addition to UDP-galactose. Eur J Biochem. 2002;269:128–138. doi: 10.1046/j.0014-2956.2001.02632.x. [DOI] [PubMed] [Google Scholar]
- Guillen E, Abeijon C, Hirschberg CB. Mammalian Golgi apparatus UDP-N-acetylglucosamine transporter: molecular cloning by phenotypic correction of a yeast mutant. Proc Natl Acad Sci USA. 1998;95:7888–7892. doi: 10.1073/pnas.95.14.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olczak M, Guillen E. Characterization of a mutation and an alternative splicing of UDP-galactose transporter in MDCK-RCAr cell line. Biochim Biophys Acta. 2006;1763:82–92. doi: 10.1016/j.bbamcr.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Yakubenia S, Frommhold D, Schölch D, Hellbusch CC, Körner C, Petri B, Jones C, Ipe U, Bixel MG, Krempien R, Sperandio M, Wild MK. Leukocyte trafficking in a mouse model for leukocyte adhesion deficiency II/congenital disorder of glycosylation IIc. Blood. 2008;112:1472–1481. doi: 10.1182/blood-2008-01-132035. [DOI] [PubMed] [Google Scholar]
- Yazbek SN, Buchner DA, Geisinger JM, Burrage LC, Spiezio SH, Zentner GE, Hsieh CW, Scacheri PC, Croniger CM, Nadeau JH. Deep congenic analysis identifies many strong, context-dependent QTLs, one of which, Slc35b4, regulates obesity and glucose homeostasis. Genome Res. 2011;21:1065–1073. doi: 10.1101/gr.120741.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida N, Yoshioka S, Iida M, Sudo K, Miura N, Aoki K, Kawakita M. Indispensability of transmembrane domains of Golgi UDP-galactose transporter as revealed by analysis of genetic defects in UDP-galactose transporter-deficient murine had-1 mutant cell lines and construction of deletion mutants. J Biochem. 1999;126:1107–1117. doi: 10.1093/oxfordjournals.jbchem.a022556. [DOI] [PubMed] [Google Scholar]
- Maggioni A, von Itzstein M, Gerardy-Schahn R, Tiralongo J. Targeting the expression of functional murine CMP-sialic acid transporter to the E. coli inner membrane. Biochem Biophys Res Commun. 2007;362:779–784. doi: 10.1016/j.bbrc.2007.08.070. [DOI] [PubMed] [Google Scholar]
- Eckhardt M, Gerardy-Schahn R. Molecular cloning of the hamster CMP-sialic acid transporter. Eur J Biochem. 1997;248:187–192. doi: 10.1111/j.1432-1033.1997.00187.x. [DOI] [PubMed] [Google Scholar]
- Lühn K, Laskowska A, Pielage J, Klämbt C, Ipe U, Vestweber D, Wild MK. Identification and molecular cloning of a functional GDP-fucose transporter in Drosophila melanogaster. Exp Cell Res. 2004;301:242–250. doi: 10.1016/j.yexcr.2004.08.043. [DOI] [PubMed] [Google Scholar]
- Ishikawa HO, Higashi S, Ayukawa T, Sasamura T, Kitagawa M, Harigaya K, Aoki K, Ishida N, Sanai Y, Matsuno K. Notch deficiency implicated in the pathogenesis of congenital disorder of glycosylation IIc. Proc Natl Acad Sci USA. 2005;102:18532–18537. doi: 10.1073/pnas.0504115102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa HO, Ayukawa T, Nakayama M, Higashi S, Kamiyama S, Nishihara S, Aoki K, Ishida N, Sanai Y, Matsuno K. Two pathways for importing GDP-fucose into the endoplasmic reticulum lumen function redundantly in the O-fucosylation of Notch in Drosophila. J Biol Chem. 2010;285:122–4129. doi: 10.1074/jbc.M109.016964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S, Taniguchi M, Muraoka M, Toyoda H, Sado Y, Kawakita M, Hayashi S. UDP-sugar transporter implicated in glycosylation and processing of Notch. Nat Cell Biol. 2001;3:816–822. doi: 10.1038/ncb0901-816. [DOI] [PubMed] [Google Scholar]
- Selva EM, Hong K, Baeg GH, Beverley SM, Turco SJ, Perrimon N, Häcker U. Dual role of the fringe connection gene in both heparan sulphate and fringe-dependent signalling events. Nat Cell Biol. 2001;3:809–815. doi: 10.1038/ncb0901-809. [DOI] [PubMed] [Google Scholar]
- Aumiller JJ, Jarvis DL. Expression and functional characterization of a nucleotide sugar transporter from Drosophila melanogaster: relevance to protein glycosylation in insect cell expression systems. Protein Expr Purif. 2002;26:438–448. doi: 10.1016/S1046-5928(02)00550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninsone P, Hwang HY, Zemtseva I, Horvitz HR, Hirschberg CB. SQV-7, a protein involved in Caenorhabditis elegans epithelial invagination and early embryogenesis, transports UDP-glucuronic acid, UDP-N-acetylgalactosamine, and UDP-galactose. Proc Natl Acad Sci USA. 2001;98:3738–3743. doi: 10.1073/pnas.061593098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Ma D, Beverley SM, Turco SJ. The Leishmania GDP-mannose transporter is an autonomous, multi-specific, hexameric complex of LPG2 subunits. Biochemistry. 2000;39:2013–2022. doi: 10.1021/bi992363l. [DOI] [PubMed] [Google Scholar]
- Cottrell TR, Griffith CL, Liu H, Nenninger AA, Doering TL. The pathogenic fungus Cryptococcus neoformans expresses two functional GDP-mannose transporters with distinct expression patterns and roles in capsule synthesis. Eukaryot Cell. 2007;6:776–785. doi: 10.1128/EC.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SK, Chiba Y, Takeuchi M, Jigami Y. Characterization of Yeast Yea4p, a uridine diphosphate-N-acetylglucosamine transporter localized in the endoplasmic reticulum and required for chitin synthesis. J Biol Chem. 2000;275:13580–13587. doi: 10.1074/jbc.275.18.13580. [DOI] [PubMed] [Google Scholar]
- Nishikawa A, Poster JB, Jigami Y, Dean N. Molecular and phenotypic analysis of CaVRG4, encoding an essential Golgi apparatus GDP-mannose transporter. J Bacteriol. 2002;184:29–42. doi: 10.1128/JB.184.1.29-42.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa A, Mendez B, Jigami Y, Dean N. Identification of a Candida glabrata homologue of the S. cerevisiae VRG4 gene, encoding the Golgi GDP-mannose transporter. Yeast. 2002;19:691–698. doi: 10.1002/yea.854. [DOI] [PubMed] [Google Scholar]
- Baldwin TC, Handford MG, Yuseff MI, Orellana A, Dupree P. Identification and characterization of GONST1, a golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell. 2001;13:2283–2295. doi: 10.1105/tpc.010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handford MG, Sicilia F, Brandizzi F, Chung JH, Dupree P. Arabidopsis thaliana expresses multiple Golgi-localised nucleotide-sugar transporters related to GONST1. Mol Genet Genomics. 2004;272:397–410. doi: 10.1007/s00438-004-1071-z. [DOI] [PubMed] [Google Scholar]
- Norambuena L, Marchant L, Berninsone P, Hirschberg CB, Silva H, Orellana A. Transport of UDP-galactose in plants. Identification and functional characterization of AtUTr1, an Arabidopsis thaliana UDP-galactos/UDP-glucose transporter. J Biol Chem. 2002;277:32923–32929. doi: 10.1074/jbc.M204081200. [DOI] [PubMed] [Google Scholar]
- Rollwitz I, Santaella M, Hille D, Flügge UI, Fischer K. Characterization of AtNST-KT1, a novel UDP-galactosetransporter from Arabidopsis thaliana. FEBS Lett. 2006;580:4246–4251. doi: 10.1016/j.febslet.2006.06.082. [DOI] [PubMed] [Google Scholar]
- Bakker H, Routier F, Oelmann S, Jordi W, Lommen A, Gerardy-Schahn R, Bosch D. Molecular cloning of two Arabidopsis UDP-galactose transporters by complementation of a deficient Chinese hamster ovary cell line. Glycobiology. 2005;15:193–201. doi: 10.1093/glycob/cwh159. [DOI] [PubMed] [Google Scholar]
- Owczarzy R, Tataurov AV, Wu Y, Manthey JA, McQuisten KA, Almabrazi HG, Pedersen KF, Lin Y, Garretson J, McEntaggart NO, Sailor CA, Dawson RB, Peek AS. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36:W163–W169. doi: 10.1093/nar/gkn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.72. http://mesquiteproject.org.
- Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Bollback JP. Empirical and hierarchical Bayesian estimation of ancestral states. Syst Biol. 2001;50:351–366. [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- Applied Biosystems. Guide to performing relative quantitation of gene expression using real-time quantitative PCR. Foster City, CA; 2008. [Google Scholar]
- Williams DL, Sayed AA, Bernier J, Birkeland SR, Cipriano MJ, Papa AR, McArthur AG, Taft A, Vermeire JJ, Yoshino TP. Profiling Schistosoma mansoni development using serial analysis of gene expression (SAGE) Exp Parasitol. 2007;117:246–258. doi: 10.1016/j.exppara.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantner F, Ziv E, Ram D, Schechter I. Different forms of the mRNA encoding the heat-shock transcription factor are expressed during the life cycle of the parasitic helminth Schistosoma mansoni. Eur J Biochem. 1998;253:390–398. doi: 10.1046/j.1432-1327.1998.2530390.x. [DOI] [PubMed] [Google Scholar]
- Ram D, Ziv E, Lantner F, Lardans V, Schechter I. Stage-specific alternative splicing of the heat-shock transcription factor during the life-cycle of Schistosoma mansoni. Parasitology. 2004;129:587–596. doi: 10.1017/S003118200400602X. [DOI] [PubMed] [Google Scholar]
- DeMarco R, Oliveira KC, Venancio TM, Verjovski-Almeida S. Gender biased differential alternative splicing patterns of the transcriptional cofactor CA150 gene in Schistosoma mansoni. Mol Biochem Parasitol. 2007;150:123–131. doi: 10.1016/j.molbiopara.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Bottoms CA, Smith PE, Tanner JJ. A structurally conserved water molecule in Rossman dinucleotide-binding domains. Protein Sci. 2002;11:2125–2137. doi: 10.1110/ps.0213502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somoza JR, Menon S, Schmidt H, Joseph-McCarthy D, Dessen A, Stahl ML, Somers WS, Sullivan FX. Structural and kinetic analysis of Escherichia coli GDP-mannose 4,6 dehydratase provides insights into the enzyme's catalytic mechanism and regulation by GDP-fucose. Structure. 2000;8:123–135. doi: 10.1016/S0969-2126(00)00088-5. [DOI] [PubMed] [Google Scholar]
- Mulichak AM, Bonin CP, Reiter WD, Garavito RM. Structure of the MUR1 GDP-mannose 4,6-dehydratase from Arabidopsis thaliana: implications for ligand binding and specificity. Biochemistry. 2002;41:15578–15589. doi: 10.1021/bi0266683. [DOI] [PubMed] [Google Scholar]
- Rosano C, Bisso A, Izzo G, Tonetti M, Sturla L, De Flora A, Bolognesi M. Probing the catalytic mechanism of GDP-4-keto-6-deoxy-d-mannose Epimerase/Reductase by kinetic and crystallographic characterization of site-specific mutants. J Mol Biol. 2000;303:77–91. doi: 10.1006/jmbi.2000.4106. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Duncker I, Mollicone R, Codogno P, Oriol R. The nucleotide-sugar transporter family: a phylogenetic approach. Biochimie. 2003;85:245–260. doi: 10.1016/S0300-9084(03)00046-4. [DOI] [PubMed] [Google Scholar]
- Jack DL, Yang NM, Saier MH Jr. The drug/metabolite transporter superfamily. Eur J Biochem. 2001;268:3620–3639. doi: 10.1046/j.1432-1327.2001.02265.x. [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, von Heijne G, Krogh A. In: Procedings of the Sixth International Conference on Intelligent Systems for Molecular Biology: 28 June-1 July, 1998; Montréal, Québec, Canada. Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editor. Menlo Park, CA: Association for the Advancement of Artificial Intelligence Press; 1998. A hidden Markov model for predicting transmembrane helices in protein sequences; pp. 175–182. [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol Chem Hoppe Seyler. 1993;347:166. [Google Scholar]
- Berninsone PM, Hirschberg CB. Nucleotide sugar transporters of the Golgi apparatus. Curr Opin Struct Biol. 2000;10:542–547. doi: 10.1016/S0959-440X(00)00128-7. [DOI] [PubMed] [Google Scholar]
- Munro S. A comparison of the transmembrane domains of Golgi and plasma membrane proteins. Biochem Soc Trans. 1995;23:527–530. doi: 10.1042/bst0230527. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Teasdale RD. Prediction of Golgi Type II membrane proteins based on their transmembrane domains. Bioinformatics. 2002;18:1109–1115. doi: 10.1093/bioinformatics/18.8.1109. [DOI] [PubMed] [Google Scholar]
- Sharpe HJ, Stevens TJ, Munro S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handford M, Rodriguez-Furlán C, Orellana A. Nucleotide-sugar transporters: structure, function and roles in vivo. Braz J Med Biol Res. 2006;39:1149–1158. doi: 10.1590/S0100-879X2006000900002. [DOI] [PubMed] [Google Scholar]
- Lehr T, Beuerlein K, Doenhoff MJ, Grevelding CG, Geyer R. Localization of carbohydrate determinants common to Biomphalaria glabrata as well as to sporocysts and miracidia of Schistosoma mansoni. Parasitology. 2008;135:931–942. doi: 10.1017/S0031182008004514. [DOI] [PubMed] [Google Scholar]
- Mäki M, Renkonen R. Biosynthesis of 6-deoxyhexose glycans in bacteria. Glycobiology. 2004;14:1R–15R. doi: 10.1093/glycob/cwh054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This PDF document contains Additional file 1: Table S1, which lists the oligonucleotide primers used for RT-PCR, RACE, qPCR and protein expression in this study.
This TIF document contains Additional file 2: Figure S1, which features the results of an in silico analysis of GFT membrane topology. Transmembrane domains were identified in the schistosome GFT protein using the Phobius transmembrane topology and signal peptide prediction server [107]. The Phobius output suggested 10 TMDs, a number that is consistent with GDP-L-fucose transporters of other organisms [27,28,30,70] (also see Figure 5) (A). A model based on this output was constructed, portraying the arrangement of the 10 TMDs (numbers indicating the amino acid boundaries of each TMD) as well as the most likely orientation for schistosome GMD within the Golgi membrane (B).
This TIF document contains Additional file 3: Figure S2, which features a rooted phylogenetic tree of nucleotide-sugar transporters (see Figure 6 for detailed unrooted tree). The amino acid sequences of NSTs with previously characterized substrate specificities were obtained from RefSeq and GenBank databases at NCBI (accession numbers in Table 2). A tree was constructed using Bayesian methods implemented in MrBayes v3.12 with mixed amino acid evolutionary models. Monophyletic clades representing NST families 1–3 [108] are indicated, and genetic divergence (substitutions per site) is represented by the scale. The tree is rooted on NST family 2.
This TIF document contains Additional file 4: Figure S3, which describes heterologous expression and isolation of recombinant schistosome GMD and GMER proteins and downstream affinity purification of GMD- and GMER-specific polyclonal chicken IgY. GST-GMD and -GMER fusion constructs were created in pGEX-6P-1 vector, and the encoded proteins were expressed in E. coli. Fusion protein expression in induced (Ind) and uninduced (Un) cultures was compared by SDS-PAGE fractionation and Coomassie staining of soluble cellular extracts (A). Fusion protein-containing extracts were passed through a GST-affinity column, and bound GMD and GMER were eluted by PreScission™ Protease-mediated cleavage of the GST fusions. Eluates were then analyzed by SDS-PAGE fractionation and Coomassie staining (B). Polyclonal chicken IgY antibodies were raised against recombinant GMD and GMER proteins, and the resultant antibodies were tested by immunoblotting the pure recombinant antigens (C). Due to crossreactivity among the antibodies and antigens (especially between anti-GMD IgY and recombinant GMER), antibodies were affinity-purified by membrane adsorption using bound GMD and GMER antigen. Following elution, antibody preparations were again tested against blots of pure antigen, demonstrating greatly reduced crossreactivity (D).