Summary
How the epidermal growth factor receptor (EGFR) activates is incompletely understood. The intracellular portion of the receptor is intrinsically active in solution, and to study its regulation we measured autophosphorylation as a function of EGFR surface density in cells. Without EGF, intact EGFR escapes inhibition only at high surface densities. While the transmembrane helix and the intracellular module together suffice for constitutive activity even at low densities, the intracellular module is inactivated when tethered on its own to the plasma membrane and fluorescence cross-correlation shows that it fails to dimerize. NMR and functional data indicate that activation requires an N-terminal interaction between the transmembrane helices, which promotes an antiparallel interaction between juxtamembrane segments and release of inhibition by the membrane. We conclude that EGF binding removes steric constraints in the extracellular module, promoting activation through N-terminal association of the transmembrane helices.
Introduction
Receptor tyrosine kinases, such as the epidermal growth factor receptor (EGFR), play critical roles in regulating metabolism, growth and differentiation (Hubbard and Till, 2000; Lemmon and Schlessinger, 2010). A single transmembrane helix in these receptors connects an N-terminal extracellular ligand-binding module to an intracellular tyrosine kinase domain. Ligand binding increases catalytic activity in the kinase domains and leads to phosphorylation of intracellular tyrosine residues. In EGFR, these tyrosines are principally located in a long C-terminal tail.
In this paper, and a companion one (Arkhipov et al.), we examine how ligand binding to the extracellular module of EGFR activates its kinase domains. EGFR was the first growth factor receptor demonstrated to undergo ligand-dependent dimerization (Yarden and Schlessinger, 1987), and crystal structures have shown how ligand binding promotes the dimerization of the extracellular module (Ferguson et al., 2003; Garrett et al., 2002; Ogiso et al., 2002). A critical step in EGFR activation is the formation of an asymmetric dimer of kinase domains (Zhang et al., 2006), in which the C-terminal lobe of one kinase domain (the activator) and the N-terminal lobe of another kinase domain (the receiver) associate, stabilizing an active conformation of the receiver kinase domain (Zhang et al., 2006). Activation through asymmetric homo- or hetero-dimerization underlies the combinatorial activation of EGFR and its close relatives Her2, Her3 and Her4 (Jura et al., 2011; Yarden and Sliwkowski, 2001).
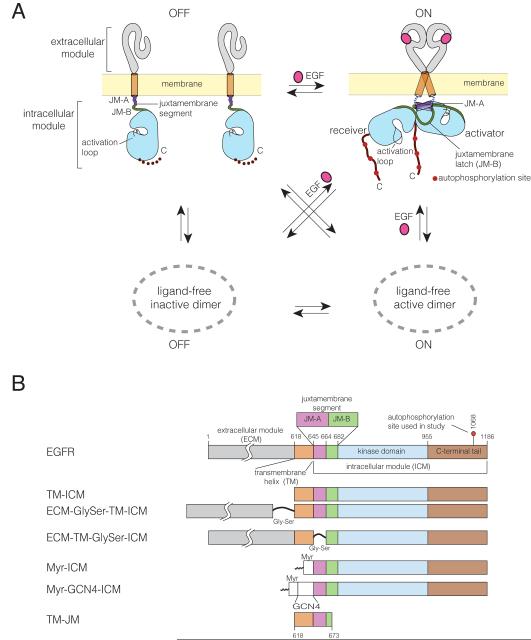
It is natural to think that ligand-driven dimerization of EGFR simply converts inactive monomers into active dimeric receptors, but the mechanism cannot be so simple. The isolated intracellular module of the receptor (consisting of the juxtamembrane segment, kinase domain and C-terminal tail) is active at relatively low concentrations in solution (< 1μM) (Jura et al., 2009; Red Brewer et al., 2009; Thiel and Carpenter, 2007). This is a consequence of the juxtamembrane segments stabilizing the asymmetric dimer necessary for activity (Jura et al., 2009; Red Brewer et al., 2009). The C-terminal portion of the juxtamembrane segment (denoted JM-B) of the receiver kinase latches onto the activator kinase domain (Figure 1A). The N-terminal portion of the juxtamembrane segment (JM-A) is thought to form an antiparallel helical association between subunits, further stabilizing the asymmetric dimer (Jura et al., 2009; Scheck et al., 2012). Clearly, the responsiveness of the receptor to ligand implies that the intrinsic activity of the intracellular module is suppressed in some way when the ligand is not bound.
Figure 1. Model for EGFR Activation and Domain Architecture.
(A) Model for monomer-dimer equilibrium of EGFR in the absence and presence of EGF (Yarden and Schlessinger, 1987).
(B) EGFR constructs used in this study.
EGFR family members are prone to ligand-independent dimerization and activation at high expression levels (Nagy et al., 2010). The coupled equilibria governing EGFR activation, incorporating both ligand-independent and ligand-dependent dimerization, are diagrammed in Figure 1A (Yarden and Schlessinger, 1987). This diagram omits the formation of higher-order oligomers (Clayton et al., 2008) and negative cooperativity in ligand binding (Alvarado et al., 2010; Liu et al., 2012; Macdonald and Pike, 2008), both of which are also likely to be important for EGFR function.
We now present an experimental analysis of EGFR activation aimed at understanding how the conformations of the extracellular and intracellular module are coupled. The companion paper presents the results of molecular dynamics simulations of the receptor in lipid bilayers (Arkhipov et al.), which provided a framework for interpreting some of our experimental results. We begin by using immunofluorescence to measure EGFR autophosphorylation as a function of receptor surface density in cells. Our data lead to the unexpected conclusion that the intrinsic activity of the intracellular module is inhibited when it is tethered to the plasma membrane. We show, using fluorescence cross-correlation spectroscopy (FCCS) that the inhibition of the intracellular module at the membrane is due to a failure to dimerize. These data point to a critical role for the transmembrane helix in dimerizing and activating the intracellular module, but the role of the transmembrane helix in EGFR activation is poorly understood (Lu et al., 2010; Lu et al., 2012; Moriki et al., 2001). We address this by using NMR to analyze the structure of the transmembrane and juxtamembrane segments of EGFR in lipid bilayers.
Results and Discussion
Ligand-independent activation of EGFR depends on the surface density of the receptor and formation of the asymmetric dimer
An essential part of our analysis is to determine the activity of the receptor in cells as a function of its density on the plasma membrane. To do this, we transiently transfected Cos-7 cells with various EGFR constructs fused to mCherry, and monitored phosphorylation at a specific residue in the C-terminal tail (Tyr 1068) using fluorescent phosphospecific antibodies (Figure 2A and Experimental Procedures). Due to the morphology of Cos-7 cells, which resembles that of a fried egg, we could focus our analysis on plasma-membrane localized EGFR by selecting regions at the periphery of the cells (Figure S3). By calibrating our microscope to relate the fluorescence intensity of EGFR in these peripheral regions to its surface density (Galush et al., 2008), we obtain a cell-by-cell quantification of the relative tyrosine phosphorylation level as a function of the surface density of EGFR at the plasma membrane (Figure S1A).
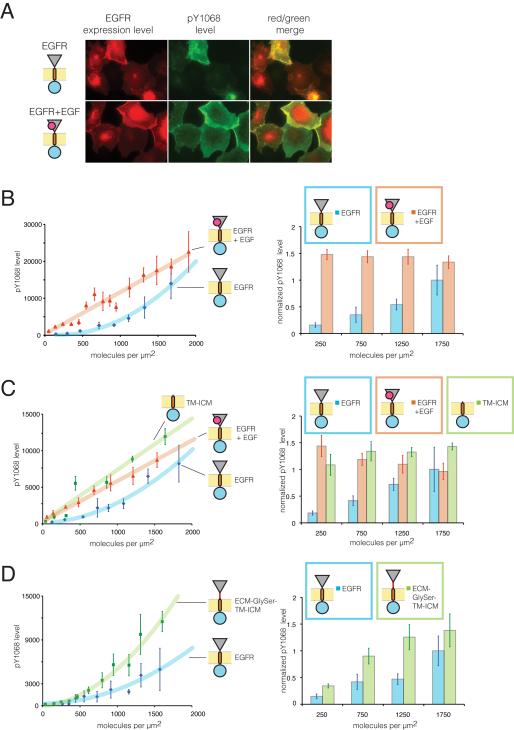
Figure 2.
Surface-Density Dependence of EGFR Activation (See also Figure S1)
(A) Fluorescence microscopy images of EGFR fused to mCherry expressed in Cos-7 cells (left panels, red) compared to phosphorylation level of EGFR at Tyr 1068 (middle panels, green, merged with expression in right panels). In bottom panels, cells were treated with EGF for 3 minutes at 37°C prior to fixation (Experimental Procedures).
(B) Relationship between EGFR surface density and phosphorylation level. In the left panel, individual data points represent the mean surface density and the mean phosphorylation level for selected cells with comparable surface density (within 100 molecules per μm2 of the mean value). Trend lines were calculated using linear and second-order polynomial fits for EGFR with and without ligand, respectively. In the right panel, bars represent the mean ratio of phosphorylation level to surface density for all cells within equal ranges of surface densities (value on x-axis ± 250 molecules per μm2). In these diagrams, as well as all subsequent ones, error bars represent the standard error of the mean.
(C) Surface density-dependent phosphorylation for a construct with extracellular domain deleted (TM-ICM) compared to EGFR ± EGF.
(D) Surface density-dependent phosphorylation levels for a construct with a flexible linker inserted between the extracellular module and the transmembrane (ECM-GlySer-TMICM) compared to EGFR.
The dependence of EGFR activation on surface density, averaged over many cells (~100), is shown in Figure 2B for Tyr 1068. In cells treated with EGF for short times (3-5 minutes) to minimize receptor internalization (Sorkin and Goh, 2009), the relative phosphorylation level per molecule is independent of the receptor surface density over the experimental range considered (50-2000 receptors per μm2) (right panel, Figure 2B). This indicates that EGF binding is sufficient to trigger phosphorylation, even at the lowest surface densities observed in our study.
In the absence of EGF, the relative phosphorylation level per receptor increases with receptor surface density, reaching values at the highest densities that are comparable to those obtained upon EGF stimulation. At a surface density of ~1400 receptors per μm2, the mean phosphorylation level of the unliganded receptor is roughly half of that observed when EGF is added (Figure 2B). This density is equivalent to a local concentration of kinase domains of ~200 μM (Extended Experimental Procedures), ~100-fold higher than that required for robust activation of the intracellular module in solution (Jura et al., 2009). The level of EGF-independent phosphorylation of the receptor is essentially the same as the EGF-induced level when the surface density of the receptor becomes higher than ~2000 receptors per μm2, which corresponds to ~1.3 million receptors per cell (Extended Experimental Procedures). This is comparable to the levels of EGFR expression reported in cancer cells that overexpress EGFR (Haigler et al., 1978).
EGF-independent activation at high expression levels requires formation of the asymmetric dimer. Introduction of the V924R mutation, which disrupts the interface between the activator and receiver kinases in the asymmetric dimer (Zhang et al., 2006), completely suppresses ligand-independent EGFR activity in the range of surface densities examined (Figure S1C). There is a modest increase in phosphorylation level upon EGF treatment in cells expressing the V924R mutant, but this increase is independent of surface density, and is probably due to low levels of endogenous EGFR.
A similar dependence of phosphorylation level on surface density is seen when other tyrosine residues are monitored (such as Tyr 1173, Figure S1B). We observe the same general pattern when we examine these cells using fluorescence-activated cell sorting (FACS), with formation of the asymmetric dimer being required (Figure S1D). Since FACS analysis does not provide an easy way to distinguish between expression of constructs at the cell surface and on internal membranes, we focus primarily on results from immunofluorescence microscopy.
The extracellular module blocks receptor activation in the absence of ligand
Deletion of the extracellular module activates EGFR, although it is not clear if this effect is restricted to high levels of receptor expression (Chantry, 1995; Nishikawa et al., 1994; Zhu et al., 2003). To address this, we measured phosphorylation as a function of surface density for a construct of EGFR in which the extracellular module is deleted, leaving just the transmembrane helix and the intracellular module (TM-ICM, Figure 1B, 2C).
The TM-ICM construct does not localize solely to the plasma membrane (Figure S1E). Nevertheless, by focusing on the peripheral regions in these cells, we find that phosphorylation of TM-ICM at the plasma membrane is much greater than for the unliganded receptor, even at the lowest surface densities studied (Figure 2C). As for the EGF-activated receptor, the normalized phosphorylation level of TM-ICM does not depend on its surface density, consistent with constitutive activation.
To test the importance of the linkage between the extracellular module and the transmembrane helix, we inserted a flexible linker consisting of twenty glycine, serine and threonine residues between them (ECM-GlySer-TM-ICM, Figure 1B). This construct shows a substantial increase in ligand-independent phosphorylation at low receptor densities compared to the wild type receptor (Figure 2D), with similar localization (Figure S1E). This is consistent with previous studies with a similar construct, in cells overexpressing EGFR (Sorokin, 1995).
The intracellular module of EGFR is inhibited at the plasma membrane
To test the role of the transmembrane helix in activation, we generated a construct (Myr-ICM), which has the intracellular module fused to a plasma membrane-targeting motif derived from c-Src (Reuther et al., 2000; Silverman and Resh, 1992). Given the high activity of the intracellular module in solution (Jura et al., 2009), and previous suggestions that the transmembrane helix is likely to play a passive role in coupling to the extracellular module (Lu et al., 2010; Lu et al., 2012), we expected that localization of the intracellular module alone to the plasma membrane would result in robust autophosphorylation, due to its high effective concentrations. Surprisingly, analysis of the surface-density dependence shows that Myr-ICM is substantially inhibited relative to the activated intact receptor (Figure 3A). Confocal images confirm that this inhibition is not due to mislocalization, since Myr-ICM is strongly inhibited on the plasma membrane, relative to the EGF-treated full-length receptor (Figure 3B).
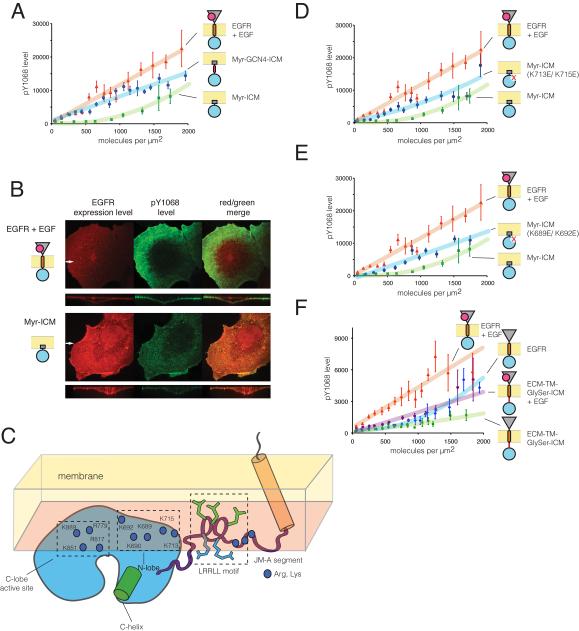
Figure 3. Activity of the Intracellular Module Localized to the Plasma Membrane with the c-Src Motif (See also Figures S2 and S3).
(A) Surface-density dependence of phosphorylation for Myr-ICM compared to EGF-treated EGFR and Myr-GCN4-ICM.
(B) Confocal fluorescence microscopy images of cells expressing EGFR (top panels) or the intracellular module fused to a c-Src membrane localization motif (Myr-ICM, bottom panels), showing expression levels (left panels, red) and antibody detection of phosphorylation at Tyr 1068 (middle panels, green, merge with expression in right panels). Large panels are images in the x-y plane of the basal surface of the cells (closest to the coverslip). Smaller panels are projections in the x-z plane, orthogonal to the basal surface, at the y-coordinate indicated by the white arrow. Note that while expression of Myr-ICM is higher than EGFR, its phosphorylation level is significantly lower.
(C) Schematic model for docking of the EGFR kinase domain against the plasma membrane based on molecular dynamics simulations of unliganded EGFR in lipid bilayers (Arkhipov et al.). In the kinase domain, positively charged residues that interact with negative charged lipids during the simulations are labeled and shown as blue dots. The LRRLL motif in the JM-A segment is shown in stick form, with leucines in green and arginines in blue.
(D) Surface-density dependence of phosphorylation for charge reversal mutations in the N-lobe interaction region of the intracellular domain (Myr-ICM K713E/K715E), compared to Myr-ICM and EGF-treated EGFR.
(E) Surface-density dependence of phosphorylation for another set of charge reversal mutations in the N-lobe (Myr-ICM K689E/K692E), compared to Myr-ICM and EGF-treated EGFR. The data for EGFR and Myr-ICM is same as in Figure 3D, since samples were prepared on the same day.
(F) Surface-density dependence of phosphorylation for ECM-TM-GlySer-ICM and EGFR in the absence and presence of EGF.
The intracellular domain can be activated constitutively at the plasma membrane by the insertion of the coiled-coil segment from the transcription factor GCN4 (O’Shea et al., 1991) between the c-Src membrane-localization motif and the intracellular module (Myr-GCN4-ICM, Figure 1B), which presumably enforces dimerization (Figure 3B). In contrast, a construct in which a flexible linker (GGGTGGGS) is inserted between the c-Src motif and the juxtamembrane segment exhibited low activity (Figure S2A), indicating that direct interference by the c-Src motif is unlikely to be responsible for the inhibition of Myr-ICM.
Molecular dynamics simulations of inactive and unliganded EGFR (Arkhipov et al.) suggest two reasons for inhibition of the intracellular module at the membrane (Figure 3C). First, the JM-A segment interacts tightly with the membrane, with three leucine sidechains (located within an LRRLL motif) that are at the heart of the antiparallel association that stabilizes the asymmetric dimer, buried in the hydrophobic part of the membrane. Second, negatively charged lipids in the membrane interact extensively with positively charged sidechains in the kinase domain and the juxtamembrane segment, lending support to previous speculations that such interactions may inhibit the receptor (McLaughlin et al., 2005).
Many of the membrane-interacting elements are important for activation and substrate binding, so it is difficult to design mutations that do not compromise activation. We focused on four lysine residues in the N-terminal lobe of the kinase domain (residues 689, 692, 713 and 715) that do not appear to be involved in the formation of the asymmetric dimer, but are seen to interact with the membrane in the simulations. Replacement of two of these residues at a time with glutamate results in substantial activation of the Myr-ICM construct in the immunofluorescence microscopy assay (Myr-ICM K713E/K715E and Myr-ICM K689E/K692E, Figures 3D,E), without affecting membrane localization as judged by confocal imaging (Figure S2B). Introduction of the same mutations in the context of the full-length receptor does not result in enhanced phosphorylation (data not shown). Thus, it appears that other inhibitory interactions are dominant in the intact receptor.
The configuration of the EGFR kinase domain is likely to be coupled to the configuration of the JM-A segment. We asked what would happen if the JM-A segment was replaced by a flexible linker. We replaced 12 residues in the JM-A segment by a flexible linker consisting of three repeats of the motif Gly-Gly-Gly-Ser (ECM-TM-GlySer-ICM, Figure 1B). This construct retains the JM-B segment, which stabilizes the active asymmetric dimer by forming the “juxtamembrane latch”. The ECM-TM-GlySer-ICM construct is strongly inhibited both in the absence and presence of EGF at densities as high as 2000 molecules per μM2 (or ~300 μM effective concentration) (see Figure 3F), consistent with the idea that inhibitory interactions between the kinase domains and the plasma membrane play a role in preventing ligand-independent activation.
The intracellular module is predominantly monomeric at the plasma membrane
These results led us to wonder if dimerization of the intracellular module is prevented at the plasma membrane. To assess the level of oligomerization of various EGFR constructs in live cells, we used two-color pulsed-interleaved excitation fluorescence cross-correlation spectroscopy (PIE-FCCS), in which a pair of lasers alternately excites GFP and mCherry with sub-nanosecond pulses (Muller et al., 2005) (Figure 4A). Diffusion of individual molecules in and out of the diffraction-limited laser focus gives rise to fluorescence-intensity fluctuations, from which we calculate a cross-correlation value that is proportional to the fraction of co-diffusing molecules (Larson et al., 2005) (Figure S4A). Correlated diffusion observed with PIE-FCCS is a rigorous indicator of molecular association and is not affected by random, dynamic collisions and crowding effects that complicate FRET-based assays. In contrast to conventional FCCS, PIE-FCCS detects the arrival time of each photon (Becker et al., 2005), enabling unambiguous assignment of each detected photon to an excitation laser source. This ensures that any cross correlation we observe is due to co-diffusion and not spectral cross-talk (Figure S4B, Extended Experimental Procedures) (Muller et al., 2005).
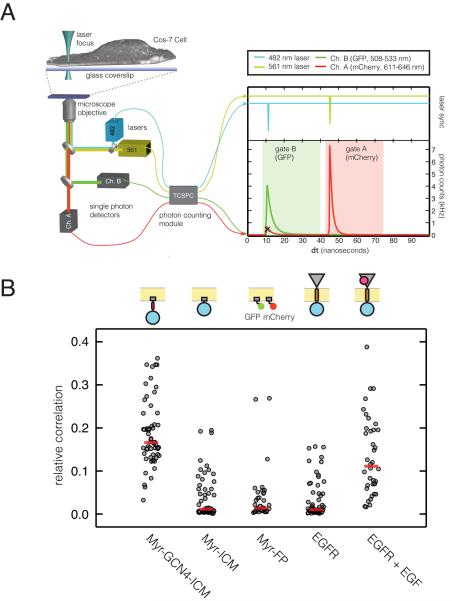
Figure 4. Fluorescence Cross-Correlation Spectroscopy Data for EGFR Constructs on the Plasma Membrane (See also Figure S4).
(A) Schematic of laser excitation and fluorescence detection for two-color pulsed interleaved excitation fluorescence cross-correlation spectroscopy (PIE-FCCS, left). Pulse diagram (right) showing excitation pulses (top panel, with GFP in blue and mCherry in green) and emission (bottom panel, with GFP in green and mCherry in red) is shown. Note that time gating allows us to eliminate mCherry emission when GFP is excited.
(B) Relative cross-correlation values for various EGFR constructs. Myr-FP is a coexpression of GFP and mCherry each fused separately to the c-Src membrane localization motif. Data are represented as a scatter plot, with the red line representing the median value. Surface densities of EGFR constructs ranged from 100 - 1000 molecules per μm2.
To determine the extent to which Myr-ICM dimerized, we compared its cross-correlation values to that of the Myr-GCN4-ICM construct (Figure 4B), which is constitutively active (Figure 3B) and presumably dimerized by the GCN4 coiled-coil. The cross-correlation values for Myr-ICM are comparable to those for a monomeric control, in which GFP and mCherry, each fused to the c-Src localization sequence, are coexpressed (Figure 4B). The cross-correlation values for Myr-GCN4-ICM are significantly higher than for Myr-ICM, and are roughly half that obtained for a fusion of GFP and mCherry in one protein (Figure S4C), consistent with it being a dimer (in a random population of dimers, 50% should be GFP, mCherry pairs). Differences in cross correlation between Myr-ICM and Myr-GCN4-ICM do not change significantly over the range of densities we observe (100-1000 molecules per μm2) (Figure S4D). While the precise oligomeric state cannot be determined from cross-correlation values, our results demonstrate that Myr-ICM oligomerization is inhibited relative to that of Myr-GCN4-ICM on the plasma membrane, and that it is likely to be monomeric over the range of surface densities examined.
The cross-correlation values for unliganded full-length EGFR are similar to those for the monomeric controls, indicating that the intact receptor is also predominantly monomeric in the same range of surface densities for which ligand-independent activation is suppressed (less than 1000 receptors per μm2). Treatment with saturating levels of EGF significantly increases cross-correlation values, indicating that EGF binding increases the oligomeric state of the receptor. This EGF-induced increase in clustering does not depend on the kinase activity of the receptor (Figure S4E). The cross-correlation values for EGF-stimulated EGFR are, on average, less than expected for a constitutive dimer, based on comparison to Myr-GCN4-ICM. This may be due a reduction in the ability of EGF to access EGFR on the basal membrane, which is adhered to the glass surface, as suggested by confocal images in which phosphorylation of Tyr1068 in EGF-treated cells is stronger on the apical membrane (Figure 3A). Our results are consistent with a monomer-to-dimer transition driving EGFR activation at low surface densities.
Structural analysis of the transmembrane and the juxtamembrane segments in lipid bilayers
The transmembrane helix must be critical for activation because its presence converts the inactive intracellular module to an active form at low surface densities. In order to understand the structural basis for this, we used NMR to analyze a fragment of EGFR spanning the transmembrane helix and the first 29 residues of the juxtamembrane segment (TM-JM, residues 618 - 673), reconstituted in lipid bicelles made from 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC). These bicelles have been used previously to analyze the transmembrane helices of EGFR and HER2 (Bocharov et al., 2008; Mineev et al., 2010).
Nearly all (~98%) of the backbone resonances of the TM-JM construct in the membrane were assigned using standard TROSY-based double and triple resonance experiments (Extended Experimental Procedures). The NMR data for the transmembrane helix are consistent with previous reports describing dimeric structures (Bocharov et al., 2008; Mineev et al., 2010) (Figure 5, Figure S5). In addition, the NMR analysis provides evidence for the formation of a helix by the LRRLL motif in the JM-A segment.
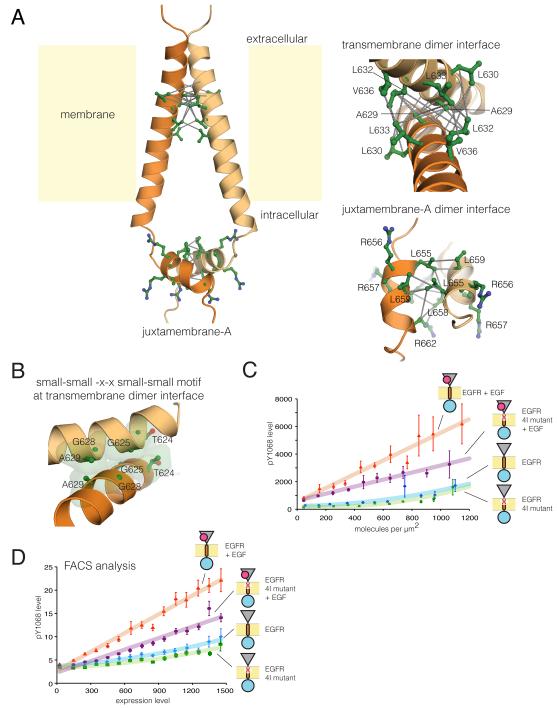
Figure 5. NMR Structure of Transmembrane-Juxtamembrane Dimer in Bicelles and Role of N-terminal Dimer Interface in Receptor Activation (See also Figure S5 and S7).
(A) Structural model of the transmembrane-juxtamembrane segment of EGFR in DMPC/DHPC bicelles as determined from NMR data. Intermolecular NOESY connectivities are shown with grey lines. Dimer interfaces are expanded in the right panels.
(B) Expanded view of the transmembrane dimer interface with residues in the small-small-x-x-small-small motif highlighted.
(C) Surface-density dependence of phosphorylation for EGFR with four residues in the N-terminal interface mutated (4I, T624I/G625I/G628I/A629I). Both wild type and mutant EGFR are compared with or without EGF treatment.
(D) FACS data comparing EGFR expression level (x-axis) to Tyr 1068 phosphorylation level for wild type EGFR and the 4I mutant.
We measured intermolecular NOEs between isotopically labeled and unlabeled proteins incorporated into lipid bicelles in two different sets of experiments (Extended Experimental Procedures) (Tables S1, S2 and Figure S5). The 21 intermolecular NOEs we measured were not enough to determine the configuration of the TM-JM segment unambiguously, so we used observations from molecular dynamics simulations of the TM-JM segment in DMPC lipid bilayers (Arkhipov et al.). These simulations indicated that two of the NOEs we observed are likely to arise from a less populated alternative configuration of the transmembrane helix, and so were removed in our determination of the NMR-based model for the TM-JM segment (Figure 5A, Extended Experimental Procedures, Figure S7, Tables S1 and S2).
The dimerization interface utilizes a classical transmembrane dimerization motif (commonly referred to as a “GxxxG” motif, although the residues represented by G can be any residue with a small sidechain) (Lemmon et al., 1994). The transmembrane helices of the catalytically active members of the EGFR family contain GxxxG motifs at each end of the helix (Fleishman et al., 2002). EGFR has two overlapping N-terminal GxxxG motifs, resulting in a small-small-x-x-small-small motif, with the small sidechains facing the dimer interface in our structure (Figure 5B). This interface is similar to that observed for Her2 (Bocharov et al., 2008), and is consistent with disulfide crosslinking studies of EGFR (Lu et al., 2010).
The right-handed crossing angle of ~ −44 ± 3° in our NMR model results in separation of the C–terminal ends of the transmembrane helices by ~20 Å and provides the appropriate spacing for an antiparallel interaction between the JM-A helices (Figure 5A). As demonstrated by the intermolecular NOE connectivity, this interface is formed by the sidechains of Leu 655, Leu 658 and Leu 659, located within the LRRLL motifs of the two subunits (Figure 5A). These helices interact with each other outside the lipid bilayer, as judged by the NMR-derived water accessibility of the juxtamembrane residues (Figure S5A).
Disruption of the N-terminal dimerization motif of the transmembrane helix inhibits EGFR activity
The activity of EGFR is insensitive to replacement of essentially any residue in the transmembrane segment by other hydrophobic residues (Lu et al., 2010). EGFR activity is not affected significantly, even if two of the interfacial residues in the N-terminal dimerization motif are replaced simultaneously by isoleucine (G625I/A629I, data not shown). In contrast, mutating all four of the small residues in the N-terminal interface to isoluecine (4I, T624I/G625I/G628I/A629I) results in significant inhibition of EGFR, as seen by immunofluorescence microscopy and by FACS analysis (Figure 5 C, D). These results, which are also consistent with observations from molecular dynamics simulations (Arkhipov et al.), provide evidence for the importance of the N-terminal association between transmembrane helices in receptor activation.
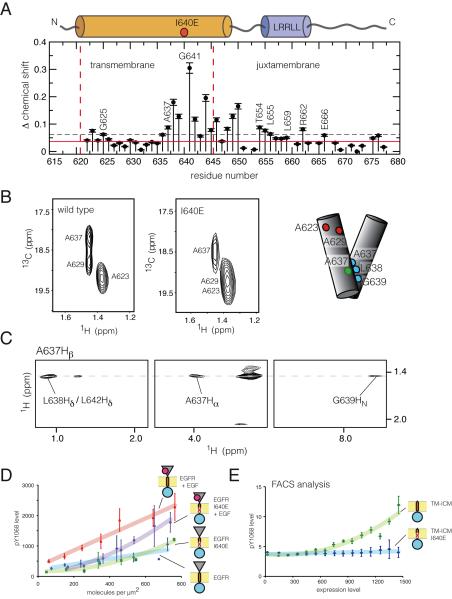
We also sought to disrupt the N-terminal association of the transmembrane helices by stabilizing interactions through the C-terminal dimerization motif, which has been suggested to underlie autoinhibition of the receptor (Fleishman et al., 2002). We mutated Ile 640, located within the C-terminal dimerization motif, to glutamate, which is expected to stabilize its association (Sternberg and Gullick, 1989).
We examined the effect of this mutation on the TM-JM construct in lipid bicelles by NMR. The I640E mutation results in significant chemical shift perturbations throughout the TM-JM construct (Figure 6A and 6B). In particular, chemical shifts for the Cβ atoms of Ala 629 and Ala 637 in the N-terminal and C-terminal dimerization motifs, respectively, are altered (Figure 6B). We also observe chemical shift changes in the LRRLL motif in the JM-A segment. The I640E mutation does not perturb the sequential NOE connectivity (dNN(i,i+1)) or 13Cα secondary-structure induced chemical shifts (⊗Cα) for the transmembrane helix, suggesting that the helical nature of this segment is unaffected (Figure S6).
Figure 6. The Effect of the I640E Mutation on Transmembrane Helix Structure and Receptor Activation (See also Figure S6).
(A) 1H, 15N chemical shift differences between the I640E mutant and the wild type TM-JM segment for each residue. The solid (red) and dashed (black) horizontal lines represent the chemical shift differences expected based on the digital resolution of the spectra and calculated from the average chemical shift, respectively. The vertical red dashed lines represent the predicted membrane-spanning region, based on the sequence analysis.
(B) The Ala Cβ region from the 1H-13C (CT) HSQC spectra of the wild type and I640E TM-JM segments in DMPC/DHPC bicelles, respectively. Schematic representation of the C-terminal dimer is shown at the right, with the uniformly labeled helix on the left and unlabeled one on the right. Residues examined in NMR or cell-based experiments are highlighted.
(C) Representative 2D strip plot showing the Ala 637 Hβ intermolecular NOE cross peak at the 13C frequency of Ala 637 C , from 3D 15 β N-13C F1-filtered/F3-edited NOESY-HSQC spectra.
(D) Surface-density dependence of phosphorylation for the I640E mutation in full-length EGFR. Both wild type and mutant EGFR are compared with or without EGF treatment.
(E) FACS data comparing expression level (x-axis) to Tyr 1068 phosphorylation level for the TM-ICM construct, with or without the I640E mutation.
We measured intermolecular NOEs between labeled and unlabeled TM-JM constructs with the I640E mutation using isotope-filtered NOESY experiments. One of the hallmarks of the N-terminal interface, an NOE between the Hα and Hβ atoms of Ala 629 on different helices (Figure S5G), is missing in the I640E mutant, consistent with disruption of the N-terminal interface. Instead, we observe intermolecular NOEs between the Hα and Hβ atoms of Ala 637, and between the Hβ atom of Ala 637 and the HN atom of Gly 639 across the C-terminal interface (Figure 6C). Thus, dimerization of the transmembrane helices in the I640E mutant brings the residues in the C-terminal dimerization motif into close proximity, although NMR data and molecular dynamics simulations suggest that the dimer formed by the I640E mutant is less stable than the wild type dimer (Figure S6C) (Arkhipov et al.).
The NMR data demonstrate that disruption of the N-terminal interface between the transmembrane helices is correlated with disruption of the antiparallel JM-A interaction. We do not observe any sequential NOE connectivity (dNN(i,i+1)) or intermolecular NOE connectivity for the juxtamembrane segment in the I640E mutant (Figure S6A), indicating a loss of structure in this region. Molecular dynamics simulations of the TM-JM construct in lipid bicelles also indicate that the formation of a C-terminal transmembrane interface is incompatible with formation of the juxtamembrane dimer interface (Arkhipov et al.).
When we introduce the I640E mutation into the intact receptor, EGF-dependent activation is impaired significantly in comparison to that of the wild type EGFR (Figure 6D). Additionally, the constitutive activity of the TM-ICM construct is reduced substantially by introduction of the I640E mutation, as shown by FACS (Figure 6E). These results suggest that interaction of the transmembrane helices through their N-terminal dimerization motif, coupled to formation of an antiparallel interaction between JM-A helices, is essential for activation of the EGFR kinase domain.
Conclusions
The presence of the juxtamembrane segment dimerizes the isolated intracellular module of EGFR in solution, suggesting that the principal function of ligand binding to the intact receptor is to change the structure of the extracellular module such that it does not impede the intrinsic ability of the intracellular module to activate (Jura et al., 2009). Unexpectedly, we find that ligand binding must also play a role in altering the conformation of the transmembrane helix and the JM-A segment to promote receptor activation, because the intracellular module is monomeric and inhibited when it is localized to the plasma membrane on its own.
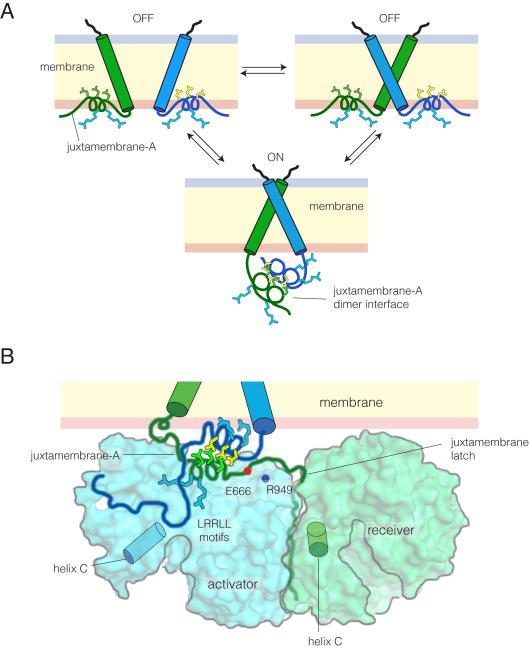
Our NMR analysis shows that the formation of the JM-A helix is coupled to the configuration of the transmembrane helices, and occurs outside of the membrane. Molecular dynamics simulations suggest that when EGFR is in an inactive conformation, the LRRLL motif within the JM-A segment is buried in the membrane (Arkhipov et al.), consistent with NMR data for the isolated juxtamembrane segment of EGFR in detergent micelles (Choowongkomon et al., 2005). These observations suggest that the nature of the interaction between the transmembrane helices toggles the configuration and membrane association of the JM-A portion of the juxtamembrane segments (Figure 7A).
Figure 7. Models for Structural Coupling.
(A) Model for structural coupling between the transmembrane helices and the juxtamembrane segments (JM-A) at the plasma membrane, based on NMR data and molecular dynamics simulations (Arkhipov et al.). The LRRLL motif in JM-A is highlighted, with leucine and arginine sidechains in green and blue, respectively.
(B) Model for asymmetric dimer formation at the plasma membrane. The surface of the kinase domains and the backbone of the juxtamembrane segments are shown. Residues in the LRRLL motif in the JM-A are shown as sticks, with leucine in yellow (activator) or green (receiver) and arginine in blue. Glu 666 of the receiver and Arg 949 of the activator are also shown.
A recent cell-based study clearly links an antiparallel interaction between the JM-A helices to EGFR activation and not just dimerization (Scheck et al., 2012). We propose that the JM-A segments need to be pulled off the membrane in order to promote the asymmetric interaction between kinase domains. The latch formed by the JM-B segment of the receiver on the C-lobe of the activator kinase domain positions the sidechain of Glu 666 of the receiver near Arg 949 of the activator, providing an anchor point for the C-terminal end of the receiver JM-A segment (Red Brewer et al., 2009; Wood et al., 2008). In a crystal structure of the EGFR intracellular module with an intact juxtamembrane segment, the JM-A helix is anchored at this point, but is directed away from the surface of the activator kinase by crystal lattice contacts (Red Brewer et al., 2009). We believe, instead, that the polar face of the JM-A helix of the receiver interacts with the surface of the kinase domain, as modeled earlier (Jura et al., 2009) and seen consistently in the molecular dynamics simulations. If the receiver JM-A helix is docked in this way, then the leucine residues of the LRRLL motif point up from the surface of the kinase domain and are available for interaction with the activator JM-A helix, as shown schematically in Figure 7B. Our NMR data suggest that dimerization of the transmembrane helices through the N-terminal interface facilitates this arrangement.
Molecular dynamics simulations indicate that the extracellular module of EGFR prevents the close approach of the transmembrane helices that would be required for interaction through the N-terminal dimerization motif (Arkhipov et al.), even when the receptor is dimerized in the absence of ligand. Consistent with this idea, ligand-independent activation of the receptor can be increased when flexible linkers are inserted between the extracellular module and the transmembrane helix. Thus, we believe that an essential role for EGF in receptor activation is to cause a specific conformational change in the extracellular modules that allows the transmembrane helices to interact through their N-terminal interface.
A striking feature of the activation mechanism is the strong cooperativity between the external, internal and transmembrane segments of EGFR. We speculate that because the EGFR dimer interface is mediated entirely by the receptor itself, selective pressure for inhibitory mechanisms to prevent ligand-independent activation has been particularly strong. At the same time, because EGFR signaling requires a specific dimeric configuration of kinase domains, there must also be selective pressure to stabilize that configuration. The balance of these competing requirements may have driven the evolution of counter-balanced activating and inhibiting mechanisms that minimize ligand-independent activation and facilitate the formation of appropriate heterodimers in response to external cues.
Experimental Procedures
Mammalian Cell-Based Assays
For immunoflurescence experiments, Cos-7 cells were grown on etched glass coverslips, transfected with Fugene (Promega) and serum starved. When noted, cells were treated with EGF (100 ng/ml, PeproTech) for 3 minutes at 37°C. After fixation in 2% formaldehyde, permeabilization with 0.1% triton X-100, and blocking with 0.2% bovine serum albumin (BSA), cells were stained with Tyr 1068 primary antibody (Cell Signaling) and then anti-rabbit fluorescein-labeled (FITC) antibody (Sigma). Cells were mounted with Thermomount (Thermo Scientific) with 0.2% trans-pyridine-2-azo-p-dimethylaniline (Sigma) for immunofluorescence, or with Prolong Gold (Invitrogen) for confocal imaging. For live and confocal imaging, cells were grown in glass bottom dishes (Matek). Samples for flow cytometry were prepared similarly to immunofluorescence microscopy with an additional step of dissociating cells with trypsin before fixation. See Extended Experimental Procedures for more detailed protocols.
Analysis of phosphorylation as a function of expression level
Cells were imaged with a TE-2000 Nikon fluorescence microscope with a xenon lamp through a 60x TIRF objective (Nikon Instruments Inc.) with standard filters (Extended Experimental Procedures). The microscope was calibrated for mCherry intensity using lipid bilayers containing Texas-Red lipids and mCherry purified from E. coli as described (Galush et al., 2008). For each cell the mean value for both mCherry intensity and FITC intensity were measured in selected peripheral regions and corrected for background using ImageJ (Schneider et al., 2012). Calculation of the phosphorylation level per receptor included correction for background phosphorylation levels in cells expressing low levels of EGFR (less than 100 molecules per μm2), and normalization to EGFR levels for samples fixed on the same day (Extended Experimental Procedures).
Fluorescence correlation spectroscopy
Fluorescence correlation spectroscopy measurements used a custom modified inverted microscope (Nikon Eclipse Ti, Nikon Instruments Inc). A 200 fs pulsed, 560 nm laser beam was synced with a 100 ps pulsed, 482 nm diode laser and delayed by 50 ns. Both lasers were coupled into a single mode optical fiber and then collimated to a 4 mm beam diameter. Samples were excited through a 100x objective (CFI APO 100X Oil TIRF NA 1.49, Nikon Instruments Inc.), with laser powers of ~1 μW measured before the objective. Fluorescence emitted from the sample is directed to a long wave pass dichroic beamsplitter (FF562-Di02-25x36, Semrock Inc.) and on to a pair of bandpass-filtered single photon avalanche diodes (PDM module, Optoelectronic Components). Detector output was measured with a time-correlated single photon counting module (PicoHarp 300, PicoQuant Photonics Inc), with a time resolution of 32 ps (Extended Experimental Procedures). Measurements were made at the cell periphery, and we explicitly avoided large (>1 μm), high intensity features.
Each detected photon is tagged with its absolute arrival time, and the delay time with respect to the laser pulses. Photons are collated into 10 μs time bins after being sorted by detector channel and arrival time. This generates a time-dependent fluorescence signal, free from green to red bleed-through, FRET, and direct mCherry excitation by the 482 nm laser. Fluorescence correlation and cross-correlation spectra are calculated as described in the Extended Experimental Procedures.
NMR Spectroscopy
The transmembrane-juxtamembrane segment (TM-JM, residues 618 – 673) of EGFR was purified from inclusion bodies (Extended Experimental Procedures). The NMR sample was prepared by dissolving the purified protein in deuterated TFE and then mixing with deuterated DMPC and DHPC lipids (Avanti Polar Lipids, Inc). The protein and the lipids were mixed in a ratio of 1:150 and the DMPC to DHPC ratio was set at 0.25. The final NMR sample (at a protein concentration of 0.3 mM) was prepared by resuspending the protein and the lipid mixture in a sample buffer containing 50 mM MES (pH 6.2), 5 mM TCEP, 1 mM EDTA, 0.05 mM ABESF, 7% 2H2O, 0.02% NaN3.
All NMR experiments were performed at 1H frequencies of 600 MHz or 900 MHz on Bruker Avance spectrometers fitted with TCI cryo-probes. The backbone chemical shift assignments were obtained through TROSY-based triple resonance experiments (Kay et al., 1990; Pervushin et al., 1997; Salzmann et al., 1998). Intermolecular NOEs were resolved by 3D 15N-13C F1-filtered/F3-edited NOESY-HSQC and methyl 13C-edited NOESY-HSQC experiments (Breeze, 2000; Stuart et al., 1999). The backbone amide resonances for the I640E mutant were assigned using TROSY HNCA, 13C (CT)-HSQC spectra and 3D NOESY experiments. The structural model for the TM-JM dimer was generated using the simulated annealing protocol of CNS (v1.3) (Brunger, 2007). A detail description of the NMR methods and structure calculation is provided in the Extended Experimental Procedures.
Supplementary Material
Highlights.
Interactions with the membrane help suppress ligand-independent EGFR activation
Dependence of EGFR activation on cell surface density reveals control mechanisms
Intrinsic dimerization of the cytoplasmic module is suppressed at the membrane
Activation requires structural coupling of transmembrane and juxtamembrane segments
Acknowledgements
We thank Jeff Iwig, Brian Kelch, Meg Stratton and Julie Zorn for a careful reading of the manuscript and other Kuriyan lab members for helpful discussions. We thank Eliza Zhang for initial work on the immunofluorescence assay. N.E. was a Leukemia and Lymphoma Society Fellow. R.D. was supported by post-doctoral fellowships from the Natural Sciences and Engineering Research Council of Canada (2009-2011) and the Canadian Institutes of Health Research (2011-2012). Y.H. is a Howard Hughes Medical Institute (HHMI) International Student Research fellow. This work was supported by a grant from the National Cancer Institute (NCI) to JK (2-R01-CA096504-06), and by award U54 CA143836 from NCI to JTG. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI or the NIH. Additional support for the fluorescence spectroscopy system provided by the Director, Office of Science, Office of Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences of the U.S. Department of Energy (DOE) under contract no. DE-AC02-05CH11231. Grants from the National Science Foundation (BBS 87-20134l; 600 MHz NMR), and the National Institute of Health (GM68933; 900 MHz) supported the NMR instrumentation used for this work.
Footnotes
Author Contributions NE, YH, EK and JK developed the cell-based assays for EGFR activity. AS and JTG built and operated the PIE-FCCS microscope. RD conducted the NMR experiments, with guidance from DEW and JP. AA, YS and DES helped couple the interpretation of experimental data and the molecular dynamics simulations. All authors were responsible for editing and writing the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarado D, Klein DE, Lemmon MA. Structural basis for negative cooperativity in growth factor binding to an EGF receptor. Cell. 2010;142:568–579. doi: 10.1016/j.cell.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, Kuriyan J, Shaw DE. Architecture and membrane interactions of the EGF receptor. Cell. doi: 10.1016/j.cell.2012.12.030. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker W, Bergmann A, Geddes CD, Lakowicz JR. Multi-Dimensional Time-Correlated Single Photon Counting. Vol 2005. Springer; US: 2005. [Google Scholar]
- Bocharov EV, Mineev KS, Volynsky PE, Ermolyuk YS, Tkach EN, Sobol AG, Chupin VV, Kirpichnikov MP, Efremov RG, Arseniev AS. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J Biol Chem. 2008;283:6950–6956. doi: 10.1074/jbc.M709202200. [DOI] [PubMed] [Google Scholar]
- Breeze AL. Isotope-filtered NMR methods for the study of biomolecular structure and interactions. Prog Nucl Mag Res Sp. 2000;36:323–372. [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Chantry A. The kinase domain and membrane localization determine intracellular interactions between epidermal growth factor receptors. J Biol Chem. 1995;270:3068–3073. [PubMed] [Google Scholar]
- Choowongkomon K, Carlin CR, Sonnichsen FD. A structural model for the membrane-bound form of the juxtamembrane domain of the epidermal growth factor receptor. J Biol Chem. 2005;280:24043–24052. doi: 10.1074/jbc.M502698200. [DOI] [PubMed] [Google Scholar]
- Chou JJ, Kaufman JD, Stahl SJ, Wingfield PT, Bax A. Micelle-induced curvature in a water-insoluble HIV-1 Env peptide revealed by NMR dipolar coupling measurement in stretched polyacrylamide gel. J Am Chem Soc. 2002;124:2450–2451. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- Clayton AH, Orchard SG, Nice EC, Posner RG, Burgess AW. Predominance of activated EGFR higher-order oligomers on the cell surface. Growth Factors. 2008;26:316–324. doi: 10.1080/08977190802442187. [DOI] [PubMed] [Google Scholar]
- Cohen P. Protein kinases--the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- Fleishman SJ, Schlessinger J, Ben-Tal N. A putative molecular-activation switch in the transmembrane domain of erbB2. Proc Natl Acad Sci U S A. 2002;99:15937–15940. doi: 10.1073/pnas.252640799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galush WJ, Nye JA, Groves JT. Quantitative fluorescence microscopy using supported lipid bilayer standards. Biophys J. 2008;95:2512–2519. doi: 10.1529/biophysj.108.131540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu HJ, Walker F, Frenkel MJ, Hoyne PA, et al. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- Glover KJ, Whiles JA, Wu G, Yu N, Deems R, Struppe JO, Stark RE, Komives EA, Vold RR. Structural evaluation of phospholipid bicelles for solution-state studies of membrane-associated biomolecules. Biophys J. 2001;81:2163–2171. doi: 10.1016/s0006-3495(01)75864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler H, Ash JF, Singer SJ, Cohen S. Visualization by fluorescence of the binding and internalization of epidermal growth factor in human carcinoma cells A-431. Proc Natl Acad Sci U S A. 1978;75:3317–3321. doi: 10.1073/pnas.75.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Till JH. Protein tyrosine kinase structure and function. Annu Rev Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, Wemmer DE, Zhang X, Kuriyan J. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011;42:9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay LE, Ikura M, Tschudin R, Bax A. 3-Dimensional Triple-Resonance Nmr-Spectroscopy of Isotopically Enriched Proteins. J Magn Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Larson DR, Gosse JA, Holowka DA, Baird BA, Webb WW. Temporally resolved interactions between antigen-stimulated IgE receptors and Lyn kinase on living cells. J Cell Biol. 2005;171:527–536. doi: 10.1083/jcb.200503110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Treutlein HR, Adams PD, Brunger AT, Engelman DM. A dimerization motif for transmembrane alpha-helices. Nat Struct Biol. 1994;1:157–163. doi: 10.1038/nsb0394-157. [DOI] [PubMed] [Google Scholar]
- Liu P, Cleveland T.E.t., Bouyain S, Byrne PO, Longo PA, Leahy DJ. A single ligand is sufficient to activate EGFR dimers. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1201114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Mi LZ, Grey MJ, Zhu J, Graef E, Yokoyama S, Springer TA. Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Mol Cell Biol. 2010;30:5432–5443. doi: 10.1128/MCB.00742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Mi LZ, Schurpf T, Walz T, Springer TA. Mechanisms for kinase-mediated dimerization of the EGF receptor. J Biol Chem. 2012 doi: 10.1074/jbc.M112.414391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald JL, Pike LJ. Heterogeneity in EGF-binding affinities arises from negative cooperativity in an aggregating system. Proc Natl Acad Sci U S A. 2008;105:112–117. doi: 10.1073/pnas.0707080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Smith SO, Hayman MJ, Murray D. An electrostatic engine model for autoinhibition and activation of the epidermal growth factor receptor (EGFR/ErbB) family. J Gen Physiol. 2005;126:41–53. doi: 10.1085/jgp.200509274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineev KS, Bocharov EV, Pustovalova YE, Bocharova OV, Chupin VV, Arseniev AS. Spatial structure of the transmembrane domain heterodimer of ErbB1 and ErbB2 receptor tyrosine kinases. J Mol Biol. 2010;400:231–243. doi: 10.1016/j.jmb.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Moriki T, Maruyama H, Maruyama IN. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J Mol Biol. 2001;311:1011–1026. doi: 10.1006/jmbi.2001.4923. [DOI] [PubMed] [Google Scholar]
- Muller BK, Zaychikov E, Brauchle C, Lamb DC. Pulsed interleaved excitation. Biophys J. 2005;89:3508–3522. doi: 10.1529/biophysj.105.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P, Claus J, Jovin TM, Arndt-Jovin DJ. Distribution of resting and ligand-bound ErbB1 and ErbB2 receptor tyrosine kinases in living cells using number and brightness analysis. Proc Natl Acad Sci U S A. 2010;107:16524–16529. doi: 10.1073/pnas.1002642107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red Brewer M, Choi SH, Alvarado D, Moravcevic K, Pozzi A, Lemmon MA, Carpenter G. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell. 2009;34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuther GW, Buss JE, Quilliam LA, Clark GJ, Der CJ. Analysis of function and regulation of proteins that mediate signal transduction by use of lipid-modified plasma membrane-targeting sequences. Methods Enzymol. 2000;327:331–350. doi: 10.1016/s0076-6879(00)27288-1. [DOI] [PubMed] [Google Scholar]
- Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci U S A. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheck RA, Lowder MA, Appelbaum JS, Schepartz A. Bipartite Tetracysteine Display Reveals Allosteric Control of Ligand-Specific EGFR Activation. ACS Chem Biol. 2012 doi: 10.1021/cb300216f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman L, Resh MD. Lysine residues form an integral component of a novel NH2-terminal membrane targeting motif for myristylated pp60v-src. J Cell Biol. 1992;119:415–425. doi: 10.1083/jcb.119.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2009;315:683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Sorokin A. Activation of the EGF receptor by insertional mutations in its juxtamembrane regions. Oncogene. 1995;11:1531–1540. [PubMed] [Google Scholar]
- Sternberg MJ, Gullick WJ. Neu receptor dimerization. Nature. 1989;339:587. doi: 10.1038/339587a0. [DOI] [PubMed] [Google Scholar]
- Stuart AC, Borzilleri KA, Withka JM, Palmer AG. Compensating for variations in H-1-C-13 scalar coupling constants in isotope-filtered NMR experiments. Journal of the American Chemical Society. 1999;121:5346–5347. [Google Scholar]
- Thiel KW, Carpenter G. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc Natl Acad Sci U S A. 2007;104:19238–19243. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood ER, Shewchuk LM, Ellis B, Brignola P, Brashear RL, Caferro TR, Dickerson SH, Dickson HD, Donaldson KH, Gaul M, et al. 6-Ethynylthieno[3,2-d]- and 6-ethynylthieno[2,3-d]pyrimidin-4-anilines as tunable covalent modifiers of ErbB kinases. Proc Natl Acad Sci U S A. 2008;105:2773–2778. doi: 10.1073/pnas.0708281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y, Schlessinger J. Self-phosphorylation of epidermal growth factor receptor: evidence for a model of intermolecular allosteric activation. Biochemistry. 1987;26:1434–1442. doi: 10.1021/bi00379a034. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Zhu HJ, Iaria J, Orchard S, Walker F, Burgess AW. Epidermal growth factor receptor: association of extracellular domain negatively regulates intracellular kinase activation in the absence of ligand. Growth Factors. 2003;21:15–30. doi: 10.1080/0897719031000096424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.