Abstract
We have previously identified the novel Cancer/Testis antigen PASD1 by immunoscreening a testis library with pooled acute myeloid leukemia (AML) patient sera. To develop a cytotoxic T lymphocyte (CTL)-inducing vaccine, we have now investigated the carboxy-terminal region, known to contain serological determinants, for MHC class I (HLA-A⋆0201)-binding peptides. Algorithm-selected natural peptides failed to show detectable HLA-A⋆0201 binding in T2 assays. However, anchor-modified analogue peptides showed enhanced binding, with decreased off-rates. Analogue peptide-loaded antigen-presenting cells (APCs) induced IFN-γ production by T cells from normal donors and patients. In addition, peptide-specific T cells could be expanded from cancer patients by stimulation with the PASD1 analogue peptide Pa14. For clinical application, a DNA fusion gene vaccine encoding Pa14 was designed and tested in “humanized” mice. Splenocytes from vaccinated mice showed in vitro cytotoxicity against tumour cells, either exogenously loaded with the corresponding wild-type peptide (Pw8) or expressing endogenously processed PASD1 protein. We show for the first time that a DNA vaccine encoding an altered PASD1 epitope can induce CTLs to target the natural peptide expressed by human tumour cells.
Keywords: PASD1, immunotherapy, acute myeloid leukemia, DNA vaccine, analogue peptide
Introduction
Cancer/Testis (CT) antigens and, in particular, testis-restricted CT antigens, provide attractive targets for cancer immunotherapy since their expression in normal tissue is highly restricted, being confined to germline tissues such as the testis (reviewed in (1, 2)). However, many CT genes characterized in solid tumours were found to have infrequent expression in myeloid leukemias (3–5).
We used SEREX (reviewed in (6)) with minor modifications (7, 8) to immunoscreen a normal donor testis cDNA library with the aim of expanding the limited number of CT antigens known in acute myeloid leukemia (AML) at that time. We isolated PASD1 (9), which had been similarly identified through the immunoscreening of a testis library with sera from patients with diffuse large B cell lymphoma (DLBCL) (10). PASD1 has two known splice variants, PASD1_v1 and the longer PASD1_v2 (11). Our immunoscreen identified a cDNA which encompassed a.a.263–773, a region unique to the longer PASD1_v2 variant, as well as the region common to both PASD1_v1 and PASD_v2 (a.a.269–639). This cDNA, initially designated as GKT-ATA20, was recognized through immunoscreening by 35% of presentation AML, 6% of chronic myeloid leukemia (CML), and 10% of DLBCL patient sera, but not by 18 normal donor sera (9).
Analysis of PASD1 mRNA and protein expression in tumour cells has confirmed its potential as a target for cancer immune therapy in AML (9), lymphoma (12), and multiple myeloma (13). Ait-Tahar et al. (14) demonstrated that cytotoxic T lymphocyte (CTL) cell lines from DLBCL patients could kill PASD1-positive tumour cells, providing further evidence of the immunogenicity of PASD1. Analysis of the common region of PASD1 by RT-PCR indicates that PASD1 is one of the most frequently expressed CT antigens in presentation AML (33%) (9) when compared to other CT antigens known to be expressed in AML, such as HAGE (23%) (5), BAGE (27%) (15), and RAGE-1 (21%) (16).
We have developed DNA fusion gene vaccines encoding tumour antigens linked to the Fragment C (FrC) sequence of tetanus toxin (TT) (17). The aim of the design is to activate CD4+ T cell help from the anti-TT repertoire to activate immunity against weak tumour antigens and to overcome potential tolerance (18). For induction of CD8+ T cells, a p.DOM-epitope DNA fusion gene vaccine has been designed with the FrC sequence reduced to a single domain (DOM), thereby decreasing the potential for peptide competition but retaining the MHC class II-restricted peptide p30 (19). The target epitope-specific sequence is then inserted at the C-terminus of FrC to aid processing and presentation. In multiple models (reviewed in (17)), this p.DOM-epitope design has been shown to induce high levels of epitope-specific CD8+ T cells. For patients with relapsed prostate cancer, a p.DOM-epitope design incorporating a peptide sequence from prostate-specific membrane antigen has induced significant levels of epitope-specific IFN-γ-producing CD8+ T cell responses in an ongoing clinical trial (20). Based on these clinical results, we wanted to explore the effectiveness of the p.DOM-epitope design for the treatment of myeloid malignancies and to test the capacity of the newly described PASD1 CT antigen as a target for CD8+ T cell attack.
Results
Testing of natural and anchor-modified PASD1-derived peptides for binding to MHC class I molecules
We focused our studies on the region of PASD1 (a.a. 269–773) which was recognized by AML patient sera and previously denoted as GKT-ATA20 (9) (Figure 1A). We identified seven peptides (Pw4–10) with HLA-A⋆0201 binding motifs that were specific to PASD1 and no other known eukaryotic proteins (as determined by BLAST searches) (Table 1). However, the SYFPEITHI binding scores were low and none of the wild-type (wt) peptides showed detectable binding to HLA-A2 molecules in T2 assays. Substitution of a single anchor residue within the Pw4, Pw8, and Pw9 wt peptides was found to improve some of the predicted MHC class I binding scores, and these peptide analogues (Pa11–Pa16) were investigated further (Table 1). All of the Pa11–Pa16 peptide analogues showed detectable binding to HLA-A2 molecules in T2 assays and modification of the peptides led to decreased off rates. Of note, the Pa13 and Pa14 peptide analogues, derived from the Pw8 peptide, showed the greatest stabilization of HLA-A2 molecules (Figure 1, B and C).
Figure 1.
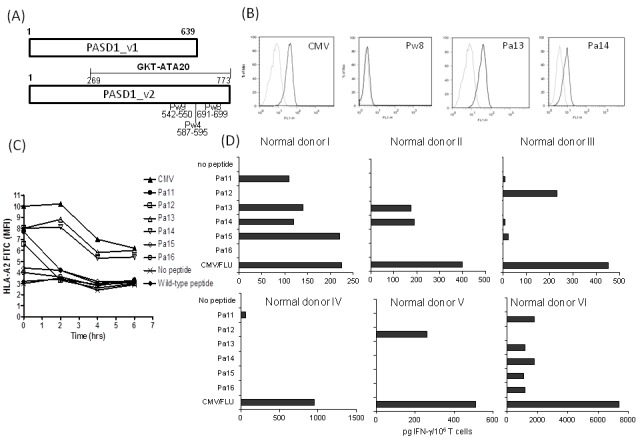
Location and modification of potential target peptides within PASD1. (A) Location of wild peptides (a.a. numbers shown) on variant 1 (_v1) and variant 2 (_v2) of PASD1. GKT-ATA20 is the region of PASD1 identified by AML patient sera (8). (B) Stabilization of HLA-A2 molecules on the surface of T2 cells using wt (Pw8) or analogue peptides (Pa13 and Pa14). A peptide from CMV was used as a positive control. Faint line: FITC-isotype control; bold line: FITC-HLA-A2-specific antibody. (C) Duration of binding of peptides to T2 cells was measured at various time-points after the removal of peptide by FACS analysis of HLA-A2 levels. (D) Purified CD3+ T cells were stimulated with autologous DCs loaded with peptides Pa11–Pa16, CMV, FLU, or no peptide. The positive control (Flu or CMV), which gave optimal results, is shown. Aliquots of culture supernatant were collected on day 7 and analyzed for IFN-γ production levels by ELISA.
Table 1.
Mapping of the PASD1 epitopes, wild-type Pw4-Pw10, and analogues Pa11–Pa16.
| Name | Nonamer sequence | SYFPEITHI score | a.a. location | Wild-type peptide derived from | |
|---|---|---|---|---|---|
| Wild type peptides | Pw4 | LQNPRDVSV | 9 | 587–595 | |
| Pw5 | VVQVNTWSC | 9 | 703–711 | ||
| Pw6 | YQPDQMRSA | 9 | 748–756 | ||
| Pw7 | IVGNERVQI | 16 | 577–585 | ||
| Pw8 | RLWQELSDS | 15 | 691–699 | ||
| Pw9 | KLQERKKWQ | 12 | 542–550 | ||
| Pw10 | QQLREQRKV | 14 | 501–509 | ||
| Peptide analogues | Pa11 | LINPRDVSV | 25 | 587–595 | Pw4 |
| Pa12 | LVNPRDVSV | 25 | 587–595 | Pw4 | |
| Pal3 | RLWQELSDV | 22 | 691–699 | Pw8 | |
| Pa14 | RLWQELSDI | 23 | 691–699 | Pw8 | |
| Pa15 | RLWQELSDL | 27 | 691–699 | Pw8 | |
| Pa16 | KLQERKKWV | 27 | 542–550 | Pw9 |
Modified anchor residue underlined.
Modified PASD1 peptides can stimulate T cells from normal donors
In order to assess whether there is a repertoire of peptide-specific CD8+ T cells capable of being mobilized against target cells, the ability of T cells from normal donors to recognize wt or modified peptides was investigated. The capacity of the wt (Pw4–10) and analogue (Pa11–Pa16) peptides to induce IFN-γ secretion from six normal HLA-A2-positive donor T cell samples were examined. Unlike the wt peptides, which failed to induce significant responses (data not shown), each peptide an-alogue stimulated notable levels of IFN-γ secretion from at least one of the six normal donor T cells, with Pa13 and Pa14 inducing significant levels of IFN-γ secretion in three of six normal donors (normal donors I, II, and VI) (Figure 1D).
Peptide-specific CD8+ cells were detectable in cultures from normal donors and patients after stimulation in vitro with Pa14
Due to the relatively frequent and enhanced ability of Pa14 to induce IFN-γ from normal donors, we selected this analogue for further investigation. In two of the three normal donors which had shown IFN-γ responses against Pa14 (normal donors I and II), it was possible to expand a detectable population of Pa14-specific pentamer-binding T cells after four rounds of stimulation (Figure 2A).
Figure 2.
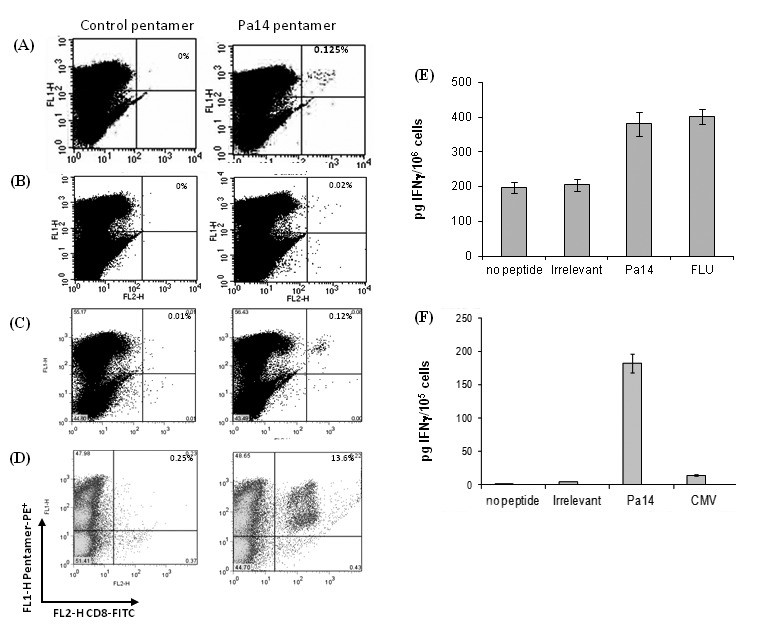
Detection of Pa14-specific T cells from healthy donors and patients. Pa14-specific T cells were detected in peptide-stimulated primary cell cultures from normal donors or from patients. CD3+ cells from (A) normal donor I and (B) AML patient I after 4 (normal donor) or 2 stimulations (AML patient). CD3+ cells from a colon cancer patient after (C) 3 weeks of stimulation and (D) 4 weeks of stimulation in culture. Pentamer-PE (FL2-H)-staining CD8+ T cells are shown as a percentage of the CD8+ T cells; the amount of IFN-γ secretion shown as (E) pg/106 CD8+ T cells from the Pa14-specific T cells from AML patients compared to the response to FLU peptide and (F) pg/105 CD8+ T cells from the Pa14-specific T cells from the colon patient compared to the response to CMV peptide, both measured on day 10 of culture by ELISA.
The capacity of T cells from patients with AML to recognize the selected Pa14 peptide was then investigated. In two of the AML patient cultures (Patients I and II) of the three who expressed PASD1 transcripts, small expansions of pentamer-binding specific T cells were observed after two stimulations with Pa14 (Figure 2A), after which time the T cells died. Pa14-responsive T cells from these AML patients were shown to produce IFN-γ in response to Pa14 10 days and 14 days into the culture period as measured by ELISA (Figure 2B). In contrast, T cells from a HLA-A2-positive colon cancer patient showed a significant increase in Pa14-specific T cells after 3 weeks (Figure 2C) and 4 weeks (Figure 2D). IFN-γ analysis of the culture media indicated that significant IFN-γ was produced by T cells responding to Pa14 but not irrelevant, Pa15 or CMV epitopes (Figure 2, E and F).
DNA vaccination with the PASD1-derived analogue peptide (Pa14) sequence induces responses against Pa14 which cross-reacts with the wt (Pw8) peptide
The previous experiments suggested that there was an available T cell repertoire against the Pa14 peptide and we therefore developed a vaccine strategy for patients. A p.DOM-epitope vaccine was prepared containing the Pw8 wt or Pa14 analogue peptide (Figure 3A). HHD mice were injected with either the p.DOM-Pw8 or p.DOM-Pa14 DNA vaccine and T cell responses examined at day 14. Using an ELISPOT assay for IFN-γ production, T cell responses against the vaccine-encoded peptide or an irrelevant control peptide were measured. We found that in all mice tested, the p.DOM-Pw8 vaccine failed to induce detectable CD8+ T cell responses against Pw8, as measured by IFN-γ production in ELISPOT assays (Figure 3B). In contrast, vaccination with p.DOM-Pa14 alone produced strong T cell responses against Pa14 as measured in vitro by IFN-γ production in ELISPOT assays (Figure 3B).
Figure 3.
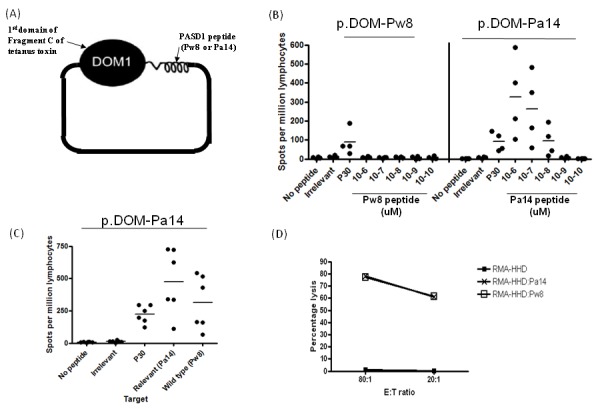
Design and operation of p.DOM-epitope vaccines. Vaccination of HHD mice with p.DOM-Pa14 induced T cell responses against Pa14 which recognized the wt Pw8 peptide. (A) p.DOM-epitope vaccine contains the first domain of tetanus toxin attached by a natural linker to the Pw8 or Pa14 epitopes. (B) In priming experiments, mice were immunized with either p.DOM-Pw8 (n = 5) or p.DOM-Pa14 (n = 6). On day 14, ELISPOT assays for IFN-γ were performed using individual mice. Responses shown are to the immunizing peptide and demonstrate that, unlike p.DOM-Pa14, priming with p.DOM-Pw8 is insufficient to generate an IFN-γ response, although a response to the p30 peptide is induced. Lymphocytes from immunized mice were incubated with a range of peptide concentrations (10−6 – 10−10 uM). (C) In priming experiments, mice were immunized with p.DOM-Pa14 (n = 6) and, 14 days later, ELISPOT assays for IFN-γ were performed. Responses against both Pa14 and the wt Pw8 peptide were detected. Group means are represented by a horizontal bar and data is representative of at least three independent experiments. (D) Splenocytes from mice immunized with p.DOM-Pa14 were stimulated for one week in vitro before assessing cytotoxicity by 51Cr-release assay. Cytotoxicity was assessed against mouse RMA tumour cells stably transduced with the chimeric humanized MHC class I molecule HHD. RMA-HHD cells were pulsed with either Pw8 or Pa14 peptide or no peptide at all. Data is representative of at least three independent experiments.
Crucially, T cells from mice primed with p.DOM-Pa14 could also respond to the wt Pw8 peptide in vitro (Figure 3C). All vaccinated mice also generated a p30 response confirming the operational integrity of the vaccines. Control mice injected with p.DOM vaccine alone failed to induce a response to either Pa14 or Pw8 peptide, as expected (data not shown).
T cells expanded with Pa14 peptide were also able to lyse RMA-HHD target cells loaded with either the Pa14 analogue or wt peptide (Pw8) at comparable levels (Figure 3D). Lysis was not observed against target cells pulsed with an irrelevant peptide (data not shown).
It appears that the wt peptide Pw8 was incapable of inducing an effective CD8+ T cell response under these conditions. However, the vaccine encoding the analogue peptide (p.DOMPa14) induced a high level response against the analogue peptide (Figure 3B) which cross-reacted with the wt Pw8 peptide (Figure 3, C and D).
CTL lines induced by vaccination with p.DOM-Pa14 lysed human target cells naturally expressing full length PASD1
Immunolabeling showed that a proportion of K562 cells expressed PASD1 (Figure 4A), while SW480 cells has been previously shown to express PASD1 (9). K562 is MHC class I-negative, while SW480 expresses HLA-A2 (Figure 4B). Following transduction of the K562 cell line with the HHD-containing retrovirus, the ability of Pa14-specific CTL lines expanded from vaccinated mice to kill the human cell lines were investigated. Mice were primed and boosted with p.DOM-Pa14, and splenocytes from each mouse were stimulated ex vivo with Pa14.
Figure 4.
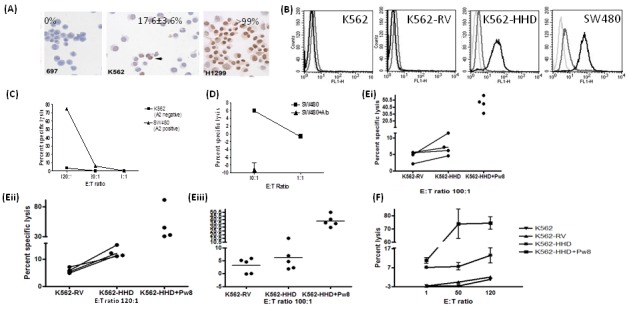
Lysis of HHD-transduced or HLA-A2-positive human cancer cells by CTL lines from p.DOM-Pa14 immunized mice. Following the vaccination of HHD mice with p.DOM-Pa14, splenocytes were stimulated in vitro with 1 μM of Pa14 peptide on a weekly basis. (A) Immunolabeling showed PASD1b expression in a proportion of K562 and in H1299 cells but not 697 cells. (B) K562 cells were transduced with either the MSCV retroviral vector alone (K562-RV) or the MSCV-HHD retrovirus (K562-HHD). Single black lines indicate the expression detected by the HLA-A2 antibody; grey line indicates isotype control; and light grey, cells alone. (C) Lysis of K562 and SW480 in the absence of retroviral transduction demonstrated that CTLs could kill the SW480 cell line, which is HLA-A2-positive, and PASD1 expressing with high efficiency at an E:T ratio of 120:1. (D) Lysis of SW480 cells were reproducibly inhibited by the HLA-A2 binding antibody W6/32, suggesting that lysis was MHC class I-mediated. (E and F) Independent experiments showed that CTL lines could kill Pw8-loaded K562-HHD cells, albeit at high E:T ratios. In addition, CTL lines could lyse endogenously processed Pw8 peptide from PASD1 within the K562-HHD cells. Data is representative of a number of independent experiments.
We showed that killing of the HLA-A2+ SW480 was far more effective than that of the HLA-A2− K562 cells (Figure 4C). SW480 (Figure 4D) cell lysis was reproducibly inhibited by the HLA-A2 binding antibody W6/32 suggesting that lysis was MHC class I-mediated. We showed in multiple experiments that CTL lines could kill Pw8-loaded K562-HHD cells, albeit at high levels which do not indicate whether physiological levels would be recognized (Figure 4, E and F), but do show the capacity of these lines to kill cells presenting wt peptide.
In addition, CTL lines showed reproducible but low levels of killing of K562-HHD cells in the absence of exogenous peptide loading (Figure 4, E and F) when compared to parental (K562) and retroviral vector containing control cells (K562-RV), reflecting the heterogeneous expression of PASD1 in K562 cells. This suggests that the native Pw8 peptide was processed and presented from endogenously produced PASD1_v2.
Discussion
We have developed a p.DOM-epitope vaccine encoding an analogue from the novel AML-associated CT antigen PASD1. When human T cells showed poor responses against the wt PASD1 peptides, we modified single anchor residues to improve MHC class I binding and extended the time period available for T cells to recognize the presented peptide. Success of this strategy is dependent on the TCR binding portion of the peptide not being altered significantly by anchor residue substitution (21). Similar modifications have led to peptide analogues which are effective at inducing immune responses against a range of tumour types including leukemias and solid tumours (22–27). Despite inherent problems demonstrating the ability of modified peptides to recognize and kill tumour cells, some heteroclitic epitopes have now shown promise in phase I clinical trials (28–30). We found that all of the anchor residue substitutions increased the stabilization of the derivative Pa11–Pa16 epitopes which then bound effectively to HLA-A2. In addition, the peptide analogues showed an increased efficacy in inducing IFN-γ secretion from normal and patient T cells and demonstrated that there is a potential Pa14 repertoire available in humans. We focused on one of the most effective peptides (Pa14).
We were unable to expand Pa14-specific T cells from two of the four HLA-A2-positive AML patient samples examined. Of these two, one was PASD1-negative (Patient III) and the other had PASD1-positive AML cells, but had received a single course of chemotherapy treatment. In two AML patients (Patients I and II), a small but clinically relevant number of Pa14-specific T cells were expanded after two rounds of Pa14 stimulation. In both samples, these T cells produced IFN-γ in response to Pa14-loaded targets. Unlike normal donors, further rounds of Pa14-stimulation failed to further expand the Pa14-specific population in AML patients and similar results have been described by others when examining PRAME-specific T cells from leukemia patients (30–32). Using T cells from a patient with colon cancer, we were able to demonstrate the successful expansion of Pa14-specific T cells from 0.01% of the CD3+ T cell population to 0.12% after 3 weeks of stimulation. A further expansion to 13.6% of the population after 4 weeks of stimulation was achieved, suggesting that the prior exposure or the presence of myeloid suppressor cells (33), suppressive factors secreted by myeloid leukemia cells (34), and/or inherent defects in T cell populations from myeloid leukemia patients (35) may be limiting the expansion of T cells from AML patients in our assays.
We wished to examine the efficacy of the PASD1 epitopes in a clinically applicable vaccine strategy. We chose the p.DOM-peptide DNA vaccine design due to its ability to provide CD4+ T cell help and allow effective CD8+ responses against tumour cells, while utilizing a simple and inexpensive production system (17). We have previously shown that the CD4+ T helper cells expanded by the p.DOM-epitope vaccine are essential for effective priming of CD8+ T cells which respond specifically to the linked epitope (18–20, 36–38). The p.DOM-epitope vaccine has also been shown to induce effective and long-term in vivo responses against a number of epitopes, including the myeloid leukemia-relevant WT1 antigen (36)More recently, the p.DOM-epitope vaccine has been used in clinical trials (20), allowing the improvement in its delivery for human therapy (17). In mice, a relatively large intramuscular volume is believed to play a role in the induction of an effective T cell response when administering the p.DOM-epitope DNA vaccine. However, to achieve something similar in humans, an increased transfection rate and inflammation at the injection site would be required. Electroporation has been shown to improve responses dramatically in mice, particularly at boosting (36) and is now being used in clinical trials at our institution with positive long-term CD8+ responses evident (20).
HHD mice provide a humanized model of HLA-A2 in which to examine vaccine efficacy. Using HHD mice, we were able to demonstrate that the Pa14 clinically relevant HLA-A⋆0201-restricted epitope could induce functional T cells in vivo. We demonstrated that the Pa14 modification was essential for an effective immune response and that the p.DOM-Pw8 vaccine, in the absence of a modified anchor residue, was unable to induce T cell responses. To enhance the specificity of the Pa14-induced T cells for the endogenously processed Pw8 epitope, we expanded CTL lines with Pa14 peptide ex vivo. We showed that the CTLs generated could lyse human leukemia cells expressing PASD1 from an endogenous source in a HLA-A2-dependent manner. CTL killing was reproducible but low, reflecting the heterogenous expression of PASD1 found in the target cells. Similarly, heterogenous expression of PASD1 has been demonstrated in other cell lines and patient samples (12).
PASD1 has been shown by us and others to be expressed in a range of haematological and solid tumour samples (9–13). The Pa14 DNA vaccine lends itself to clinical applications in which it could be used in conjunction with other vaccines in a range of tumour types. MHC class I and II expression on primary AML cells provides the necessary signaling pathways for CD8+ CTL and CD4+ helper activity, which will be essential for effective AML tumour cell lysis induced by any vaccine strategy. Immunotherapy is most likely to be applied in first remission which can be achieved in most AML patients. It is hoped that the activation of T cells when the immune function in patients is recovering, following chemotherapy and in first remission, has the potential to remove minimal residual disease and delay and perhaps even prevent relapse.
The p.DOM-Pa14 DNA vaccine, incorporating a modified epitope from the novel CT antigen PASD1, provides a new immunotherapeutic treatment for AML, which combines safety with cogent immunological advantages. This data continues our development and characterization of p.DOM-DNA vaccines and provides the first modified CT antigen immunogenic peptide for inclusion in a PASD1-targeting vaccine with a potential use in clinical trials for solid tumour and leukemia patients.
Acknowledgments
We would like to thank Suzanne Brooks, Nicola Weston-Bell, Mark Townsend, Alison Tutt, Angela Hamblin, and Stuart Dunn for technical assistance, and Linda Barber for useful discussions. These studies were supported by Leukaemia and Lymphoma Research.
Abbreviations
- CT
Cancer/Testis;
- AML
acute myeloid leukemia;
- CTL
cytotoxic T lymphocyte;
- DOM
domain;
- APC
antigen-presenting cell
Materials and methods
Prediction of HLA-A⋆0201-binding wild-type and modified peptides
Seven nonamers specific for human PASD1 were identified using SYFPEITHI (39) and BioInformatics and Molecular Analysis Section (BIMAS) (40) algorithms (Table 1). All peptides were located in PASD1_v1468–639 and PASD1_v2468–773(11). Peptide ‘analogues’ of wt peptides 4–10 (Pw4 – Pw10) were generated by single amino acid substitutions (at position 2 or 9) and denoted Pa11, Pa12, Pa13, Pa14, Pa15, and Pa16. Only those wt and analogue peptides with low similarity to known eukaryotic proteins were selected for study. The promiscuous MHC class II-restricted p30 (41) and the HLA-A2-binding Flu M1 (42), CMV pp65 (43) and, in our study, irrelevant WT1.37 peptides were used where indicated. All peptides were synthesized commercially and supplied at > 95% purity (PPR Ltd., Southampton, U.K.).
Patient and normal donor samples
Normal donor lymphocytes were obtained from buffy coats and CD3+ cells were isolated using Negative Isolation Kits (Miltenyi Biotec, Surrey, U.K.) as per manufacturer’s instructions. All patient samples were received following informed consent and local ethical committee approval in accordance with the Declaration of Helsinki. HLA-A2 positivity was determined by FACS analysis, followed by subtyping at the Anthony Nolan Laboratories, Royal Free Hospital, London. Primary AML blasts were obtained from the peripheral blood of adult patients with high-count AML at diagnosis and prior to the initiation of chemotherapy (Table 2), except for one patient who failed to respond to treatment (Patient IV). Peripheral blood mononuclear cells (PBMCs) from AML patients were purified by Histopaque density gradient centrifugation and cryopreserved in X-VIVO 15 (Cambrex Corporation, Berkshire, U.K.), 10% DMSO, and 50% human AB serum (both Sigma-Aldrich, Poole, U.K.). Primary cells were cultured in X-VIVO 15 medium; AML cells were additionally cultured with recombinant human SCF (20 ng/ml) and IL-3 (10 ng/ml) (R&D Systems, Minneapolis, U.K.). CD14+ cells were purified from remission bone marrow using positive selection MACS CD14 beads (Miltenyi Biotec). In some cases, CD3+ cells from non-remission AML or a single colon cancer patient were positively isolated using CD3 MACS microbeads (Miltenyi Biotec) as per manufacturer’s instructions. Expression of one or both PASD1 transcripts in AML blasts was determined by PASD1-specific RT-PCR using primers targeting the common region of the sequence as described previously (9).
Table 2.
Patient characteristics.
| id | Disease type | Disease stage | Age | Sex | FAB subtype: cytogenetics | % blasts in bone marrow | PASD1 status | HLA-A2 status |
|---|---|---|---|---|---|---|---|---|
| I | AML | diagnosis | 58 | F | M2: 47, XX, +8 | 72% | + | + |
| II | AML | diagnosis | 57 | M | M1: complex cytogenetics | 32% | + | + |
| III | AML | remission | 25 | M | M2: 46, XY, t (6,17) (p21,q21) | 50% | − | + |
| IV | AML | diagnosis (post-DA)‡ | 25 | M | M5: complex cytogenetics | 22% | + | + |
- patient received a single course of daunorubicin and cytarabine but did not respond to treatment.
Flow cytometry
For the analysis of cell-surface molecules, cells were incubated for 20 min at 4°C with directly conjugated antibodies or matched isotype controls (all antibodies BD, Oxford, U.K., except anti-HLA-A2-FITC from AbD Serotec, Oxford, U.K.) and analyzed on a FACSCalibur (BD). Peptide-specific T cells were enumerated by pentamer staining. Briefly, 106 cells were incubated with 10 μl of peptide-specific or irrelevant control PE-labeled HLA-A⋆0201 pentamer (ProImmune, Oxford, U.K.) for 10 min at room temperature, and washed and co-stained with CD8-FITC for 20 min. For detection of intracellular IFN-γ, Brefeldin A (1 mg/ml) was added to stimulated T cells 12 hr prior to intracellular staining. Cells were washed with PBS, stained with CD8-PE, and then washed twice with HBSS, 1% fetal calf serum (FCS). Permeabilization medium (Invitrogen) and anti-IFN-γ-FITC were added and incubated for 20 min at room temperature prior to a final wash and analyzed by flow cytometry.
Immunolabeling
Cytocentrifuge preparations of cells were fixed in acetone for 10 min at room temperature, air-dried, wrapped in foil, and stored at −20°C. For use, the cytocentrifuge preparations were allowed to come up to room temperature before being incubated with the monoclonal antibody PASD1-2 (clone 2ALCC128), which is specific for aa 540–773 present only in the longer PASD1_v2 protein (11). After washing in PBS, the slides were stained using the Mach-Three detection kit following manufacturer’s instructions. Antigen/antibody complexes were visualized using diaminobenzidine tetrahydrochloride substrate (Sigma).
Peptide binding assays using T2 cells
The HLA-A2+ T2 cell line (44) was used to assess binding of peptides to HLA-A2. T2 cells were incubated overnight in complete media (RPMI 1640, 1 mM sodium pyruvate, 2 mM L-glutamine, 1% non-essential amino acids, 50 μM 2-mercaptoethanol, 100 U/ml penicillin, 100 μg/ml streptomycin; all Invitrogen) with 10% FCS alone or with peptide (0.05–100 μM) prior to staining with anti-human HLA-A2-FITC antibody and FACS analysis. To determine the longevity of binding, peptide-pulsed T2 cells were washed three times and replated in fresh medium. Aliquots of cells were analyzed at different time points after the removal of peptide by flow cytometry.
Generation of DCs
Monocytes were obtained from normal donor buffy coats by selection with CD14 MACS beads or by plastic adherence as follows: PBMCs were plated at 106/ml in warm X-VIVO medium and 1% human AB serum and incubated at 37°C for 4 hr. Non-adherent cells were removed by gentle washing with HBSS and were cryopreserved for later use as effectors. The remaining monocytes were cultured in IL-4 (1000 IU/ml) and GM-CSF (800 U/ml) for 5 days to induce differentiation to a dendritic cell (DC) phenotype. On day 5, a maturation cocktail of TNF-α (10 ng/ml), IL-6 (1000 U/ml), and IL-1β (10 ng/ml) (all R&D Systems) was added. Twenty-four hours later, DCs were harvested, washed with HBSS, and used as antigen-presenting cells (APCs). Confirmation of a DC phenotype was determined by flow cytometry.
Stimulation of human T cells
CD3+ or CD8+ cells from normal donors were co-cultured with autologous, peptide-pulsed (50 μg/ml, 4 hr) DCs at an effector to stimulator ratio of 10:1. IL-7 (10 U/ml) was added to the cultures on day 3 and IL-2 (10 U/ml) on day 7. Due to the absence of normal monocytes from presentation AML samples, T2 cells were used as APCs. T cells were restimulated by the addition of irradiated peptide-pulsed APCs on day 7 or day 14, together with IL-2 and IL-7 (both 10 U/ml). Supernatants were harvested for IFN-γ detection by ELISA using the Duo set ELISA Development System (R&D Systems). Stimulated effectors were analyzed for the presence of peptide-specific CD8+ T cells by pentamer staining (ProImmune) or IFN-γ ELISPOT (BD ELISPOT kit, BD Biosciences, UK). IFN-γ ELISPOT assays were carried out using stimulator cells (peptide-pulsed T2 at 2×104/well) incubated either alone or with effector cells (2×105 per well) in ELISPOT wells pre-coated with anti-human IFN-γ antibody. Plates were incubated for 24 hr at 37°C, 5% CO2 and spots developed according to manufacturer’s instructions (BD Biosciences).
Construction of DNA vaccines
Construction of the p.DOM plasmid containing the first domain (DOM) of FrC from tetanus toxin (TT865-1120) with a leader sequence derived from the VH of the IgM of the BCL1 tumour at the N-terminus has been previously described (45). DNA vaccines were constructed encoding PASD1691–699 (either p.DOM-Pw8 or p.DOM-Pa14) peptides fused directly 3’ to DOM. All vaccines were constructed by PCR amplification using p.DOM as template with the forward primer:
5′-TTTTAAGCTTGCCGCCACCATGGGTTGGAGC-3′, and reverse primers as follows:
Pw8: 5′-ATATGCGGCCGCTTAGATATCAGACAACTCTTGCCAAAGCCGGTTACCCCAGAAGTCACG-3′;
Pa14: 5′ATATGCGGCCGCTTATGAATCAGACAACTCTTGCCAAAGCCGGTTACCCCAGAAGTCACG-3′
PCR products were gel purified, digested using HindIII and NotI restriction sites, and cloned into the expression vector pcDNA3 (Invitrogen). Restriction sites within primers are shown in bold and PASD1-peptide encoding sequences are italicized. The modified sequence is underlined. Integrity of the inserted sequences were confirmed by DNA sequencing and translated product size was checked in vitro using the TNT T7 coupled reticulocyte lysate system (Promega, Southampton, U.K.).
Vaccination of HHD transgenic mice
Permission to use the HHD mice was kindly provided by Dr. François Lemonnier, Departement d’Immunologie, Institut Pasteur, France. HHD mice express a hybrid human HLA-A2 transgene, comprising the human α1 and α2 domains, fused to the murine H-2Db α3 transmembrane and intracytoplasmic domain, and covalently linked to human β-2 microglobulin. HHD mice lack mouse MHC class I molecules due to targeted disruption of the H-2Db and mouse β2-microglobulin genes (46). HHD mice at 6 to 10 weeks of age were injected intramuscularly (i.m.) into both quadriceps with a total of 50 μg DNA in saline solution on day 0. Where indicated, mice were boosted with the same DNA vaccine delivered with in vivo electroporation on day 28 as described previously (47). Animal experimentation was conducted within local Ethical Committee and UK Coordinating Committee for Cancer Research (London, U.K.) guidelines under Home Office License.
Mouse IFN-γ-ELISPOT
Vaccine-specific IFN-γ secretion by splenocytes from individual mice was assessed directly ex vivo (BD ELISPOT Set, BD) on day 14 or 36, as described previously (18). Splenocytes were incubated in triplicate, alone, with Pw8 or Pa14 peptides, or with irrelevant peptide to assess CD8+ T cell responses, or with p30 peptide to assess CD4+ T cell responses. Data are expressed as the frequency of spot-forming cells (SFCs) per million lymphocytes. For analysis of peptide-specific T cell sensitivity, cells were incubated with a range of peptide concentrations; the number of SFC/million cells at the peptide concentration inducing the greatest response was assigned a value of 100%. For each peptide concentration tested, the percent maximal response was then calculated by the formula: (experimental SFCs per million cells / maximal SFCs per million cells) × 100% for each individual animal.
Expansion of murine effector cells and lytic assays
For the generation and maintenance of CTL lines, splenocytes from individual mice were resuspended in 10–15 ml complete medium with Pw8 or Pa14 (1 μM) peptides. For further cycles of in vitro restimulation, cells were washed and resuspended at 3×105/mL with 2.5×106/mL syngeneic splenocytes (pre-incubated for 1 hr with the relevant peptide at 1 μM, washed 4 times in unsupplemented RPMI 1640, and irradiated at 2,500 rad). Recombinant human IL-2 was added to cultures at 20 IU/ mL (Perkin-Elmer, Foster City, CA) and cells were incubated at 2 mL/well in a 24-well plate. Subsequent cycles of in vitro restimulation were carried out similarly every 7–10 days. Lytic activity was assessed by standard 5 hr 51Cr release assay as previously described (48). Cells used as targets in CTL assays were the human leukemia line K562 (HLA-A⋆0201−, PASD1+) and the human colon cancer cell line SW480 (HLA-A⋆0201+, PASD1+). K562 were transduced with the MSCV retroviral vector alone to produce K562-RV or with the MSCV retroviral vector containing HHD cDNA to produce K562-HHD cell lines. PASD1 protein expression was determined using the PASD1-2 antibody as described previously (12). For some experiments, target cells were incubated with the anti-human HLA-class I monoclonal antibody W6.32 (Diaclone, Besancon, France) at 5 μg/well for 15 min at 4°C before the addition of T cells and for the duration of the assay. Means of triplicate cultures were expressed as (experimental release – spontaneous release) / (total 51Cr incorporated – spontaneous release) × 100%. The average spontaneous release never exceeded 15% of the total incorporated 51Cr.
References
- 1.Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 2004;4:1. [PubMed] [Google Scholar]
- 2.Hofmann O, Caballero OL, Stevenson BJ, Chen YT, Cohen T, Chua R, Maher CA, Panji S, Schaefer U, Kruger A, Lehvaslaiho M, Carninci P, Hayashizaki Y, Jongeneel CV, Simpson AJ, Old LJ, Hide W. Genome-wide analysis of cancer/testis gene expression. Proc Natl Acad Sci U S A. 2008;105:20422–20427. doi: 10.1073/pnas.0810777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambost H, Brasseur F, Coulie P, de Plaen E, Stoppa AM, Baume D, Mannoni P, Boon T, Maraninchi D, Olive D. A tumour-associated antigen expression in human haematological malignancies. Br J Haematol. 1993;84:524–526. doi: 10.1111/j.1365-2141.1993.tb03111.x. [DOI] [PubMed] [Google Scholar]
- 4.Chambost H, Collette Y, Dutartre H, Thuret I, Olive D. Parameters involved in the recognition of fresh human leukemic blasts by tumor-specific cytolytic T cell clones: a model study. Leuk Res. 2000;24:823–830. doi: 10.1016/s0145-2126(00)00053-9. [DOI] [PubMed] [Google Scholar]
- 5.Adams SP, Sahota SS, Mijovic A, Czepulkowski B, Padua RA, Mufti GJ, Guinn B. Frequent expression of HAGE in presentation chronic myeloid leukaemias. Leukaemia. 2002;16:2238–2242. doi: 10.1038/sj.leu.2402732. [DOI] [PubMed] [Google Scholar]
- 6.Preuss KD, Zwick C, Bormann C, Neumann F, Pfreundschuh M. Analysis of the B-cell repertoire against antigens expressed by human neoplasms. Immunol Rev. 2002;188:43–50. doi: 10.1034/j.1600-065x.2002.18805.x. [DOI] [PubMed] [Google Scholar]
- 7.Guinn BA, Collin JF, Li G, Rees RC, Mufti GJ. Optimised SEREX technique for the identification of leukaemia-associated antigens. J Immunol Methods. 2002;264:207–214. doi: 10.1016/s0022-1759(02)00095-9. [DOI] [PubMed] [Google Scholar]
- 8.Liggins AP, Guinn BA, Banham AH. Identification of lymphoma-associated antigens using SEREX. Methods Mol Med. 2005;115:109–128. doi: 10.1385/1-59259-936-2:109. [DOI] [PubMed] [Google Scholar]
- 9.Guinn BA, Bland EA, Lodi U, Liggins AP, Tobal K, Petters S, Wells JW, Banham AH, Mufti GJ. Humoral detection of leukaemia-associated antigens in presentation acute myeloid leukaemia. Biochem Biophys Res Commun. 2005;335:1293–1304. doi: 10.1016/j.bbrc.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 10.Liggins AP, Guinn BA, Hatton CS, Pulford K, Banham AH. Serologic detection of diffuse large B-cell lymphoma-associated antigens. Int J Cancer. 2004;110:563–569. doi: 10.1002/ijc.20170. [DOI] [PubMed] [Google Scholar]
- 11.Liggins AP, Brown PJ, Asker K, Pulford K, Banham AH. A novel diffuse large B-cell lymphoma-associated cancer testis antigen encoding a PAS domain protein. Br J Cancer. 2004;91:141–149. doi: 10.1038/sj.bjc.6601875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper CD, Liggins AP, Ait-Tahar K, Roncador G, Banham AH, Pulford K. PASD1, a DLBCL-associated cancer testis antigen and candidate for lymphoma immunotherapy. Leukaemia. 2006;20:2172–2174. doi: 10.1038/sj.leu.2404424. [DOI] [PubMed] [Google Scholar]
- 13.Sahota SS, Goonewardena CM, Cooper CD, Liggins AP, Ait-Tahar K, Zojer N, Stevenson FK, Banham AH, Pulford K. PASD1 is a potential multiple myeloma-associated antigen. Blood. 2006;108:3953–3955. doi: 10.1182/blood-2006-04-014621. [DOI] [PubMed] [Google Scholar]
- 14.Ait-Tahar K, Liggins AP, Collins GP, Campbell A, Barnardo M, Lawrie C, Moir D, Hatton C, Banham AH, Pulford K. Cytolytic T cell response to the PASD1 cancer testis antigen in patients with diffuse large B-cell lymphoma. Br J Haematol. 2009;146:396–407. doi: 10.1111/j.1365-2141.2009.07761.x. [DOI] [PubMed] [Google Scholar]
- 15.Greiner J, Ringhoffer M, Taniguchi M, Li L, Schmitt A, Shiku H, Döhner H, Schmitt M. mRNA expression of leukaemia-associated antigens in patients with acute myeloid leukaemia for the development of specific immunotherapies. Int J Cancer. 2004;108:704–711. doi: 10.1002/ijc.11623. [DOI] [PubMed] [Google Scholar]
- 16.Guinn BA, Gilkes AF, Woodward E, Westwood NB, Mufti GJ, Linch D, Burnett AK, Mills KI. Microarray analysis of tumour antigen expression in presentation acute myeloid leukaemia. Biochem Biophys Res Commun. 2005;333:703–713. doi: 10.1016/j.bbrc.2005.05.161. [DOI] [PubMed] [Google Scholar]
- 17.Rice J, Ottensmeier CH, Stevenson FK. DNA vaccines: precision tools for activating effective immunity against cancer. Nat Rev Cancer. 2008;8:108–120. doi: 10.1038/nrc2326. [DOI] [PubMed] [Google Scholar]
- 18.Rice J, Buchan S, Dewchand H, Simpson E, Stevenson FK. DNA fusion vaccines induce targeted epitope-specific CTLs against minor histocompatibility antigens from a normal or tolerized repertoire. J Immunol. 2004;173:4492–4499. doi: 10.4049/jimmunol.173.7.4492. [DOI] [PubMed] [Google Scholar]
- 19.Rice J, Buchan S, Stevenson FK. Critical components of a DNA fusion vaccine able to induce protective cytotoxic T cells against a single epitope of a tumor antigen. J Immunol. 2002;169:3908–3913. doi: 10.4049/jimmunol.169.7.3908. [DOI] [PubMed] [Google Scholar]
- 20.Chudley L, McCann K, Mander A, Tjelle T, Campos-Perez J, Godeseth R, Creak A, Dobbyn J, Johnson B, Bass P, Heath C, Kerr P, Mathiesen I, Dearnaley D, Stevenson F, Ottensmeier C. DNA fusion-gene vaccination in patients with prostate cancer induces high-frequency CD8(+) T cell responses and increases PSA doubling time. Cancer Immunol Immunother. 2012;61:2161–2170. doi: 10.1007/s00262-012-1270-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen JL, Stewart-Jones G, Bossi G, Lissin NM, Wooldridge L, Choi EM, Held G, Dunbar PR, Esnouf RM, Sami M, Boulter JM, Rizkallah P, Renner C, Sewell A, van der Merwe PA, Jakobsen BK, Griffiths G, Jones EY, Cerundolo V. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J Exp Med. 2005;201:1243–1255. doi: 10.1084/jem.20042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuboi A, Oka Y, Udaka K, Murakami M, Masuda T, Nakano A, Nakajima H, Yasukawa M, Hiraki A, Oji Y, Kawakami M, Hosen N, Fujioka T, Wu F, Taniguchi Y, Nishida S, Asada M, Ogawa H, Kawase I, Sugiyama H. Enhanced induction of human WT1-specific cytotoxic T lymphocytes with a 9-mer WT1 peptide modified at HLA-A⋆2402-binding residues. Cancer Immunol Immunother. 2002;51:614–620. doi: 10.1007/s00262-002-0328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinilla-Ibarz J, May RJ, Korontsvit T, Gomez M, Kappel B, Zakhaleva V, Zhang RH, Scheinberg DA. Improved human T cell responses against synthetic HLA-0201 analog peptides derived from the WT1 oncoprotein. Leukaemia. 2006;20:2025–2033. doi: 10.1038/sj.leu.2404380. [DOI] [PubMed] [Google Scholar]
- 24.Fourcade J, Kudela P, Andrade Filho PA, Janjic B, Land SR, Sander C, Krieg A, Donnenberg A, Shen H, Kirkwood JM, Zarour HM. Immunization with analog peptide in combination with CpG and montanide expands tumor antigen-specific CD8+ T cells in melanoma patients. J Immunother. 2008;31:781–791. doi: 10.1097/CJI.0b013e318183af0b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinilla-Ibarz J, Korontsvit T, Zakhaleva V, Roberts W, Scheinberg DA. Synthetic peptide analogs derived from bcr/abl fusion proteins and the induction of heteroclitic human T cell responses. Haematologica. 2005;90:1324–1332. [PubMed] [Google Scholar]
- 26.May RJ, Dao T, Pinilla-Ibarz J, Korontsvit T, Zakhaleva V, Zhang RH, Maslak P, Scheinberg DA. Peptide epitopes from the Wilms’ tumor 1 oncoprotein stimulate CD4+ and CD8+ T cells that recognize and kill human malignant mesothelioma tumor cells. Clin Cancer Res. 2007;13:4547–4555. doi: 10.1158/1078-0432.CCR-07-0708. [DOI] [PubMed] [Google Scholar]
- 27.Christensen O, Lupu A, Schmidt S, Condomines M, Belle S, Maier A, Hose D, Neuber B, Moos M, Kleist C, Terness P, Ho AD, Gold-schmidt H, Klein B, Hundemer M. Melan-A/MART1 analog peptide triggers anti-myeloma T cells through crossreactivity with HM1.24. J Immunother. 2009;32:613–621. doi: 10.1097/CJI.0b013e3181a95198. [DOI] [PubMed] [Google Scholar]
- 28.Maslak PG, Dao T, Gomez M, Chanel S, Packin J, Korontsvit T, Zakhaleva V, Pinilla-Ibarz J, Berman E, Scheinberg DA. A pilot vaccination trial of synthetic analog peptides derived from the BCRABL breakpoints in CML patients with minimal disease. Leukaemia. 2008;22:1613–1616. doi: 10.1038/leu.2008.7. [DOI] [PubMed] [Google Scholar]
- 29.Maslak PG, Dao T, Krug LM, Chanel S, Korontsvit T, Zakhaleva V, Zhang R, Wolchok JD, Yuan J, Pinilla-Ibarz J, Berman E, Weiss M, Jurcic J, Frattini MG, Scheinberg DA. Vaccination with synthetic analog peptides derived from WT1 oncoprotein induces T cell responses in patients with complete remission from acute myeloid leukaemia. Blood. 2010;116:171–179. doi: 10.1182/blood-2009-10-250993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rezvani K, Yong AS, Tawab A, Jafarpour B, Eniafe R, Mielke S, Savani BN, Keyvanfar K, Li Y, Kurlander R, Barrett AJ. Ex vivo characterization of polyclonal memory CD8+ T-cell responses to PRAME-specific peptides in patients with acute lymphoblastic leukaemia and acute and chronic myeloid leukaemia. Blood. 2009;113:2245–2255. doi: 10.1182/blood-2008-03-144071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quintarelli C, Dotti G, Hasan ST, De Angelis B, Hoyos V, Errichiello S, Mims M, Luciano L, Shafer J, Leen AM, Heslop HE, Rooney CM, Pane F, Brenner MK, Savoldo B. High-avidity cytotoxic T lymphocytes specific for a new PRAME-derived peptide can target leukemic and leukemic-precursor cells. Blood. 2011;117:3353–3362. doi: 10.1182/blood-2010-08-300376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molldrem JJ, Lee PP, Kant S, Wieder E, Jiang W, Lu S, Wang C, Davis MM. Chronic myelogenous leukaemia shapes host immunity by selective deletion of high-avidity leukaemia-specific T cells. J Clin Invest. 2003;111:639–647. doi: 10.1172/JCI16398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mougiakakos D, Jitschin R, von Bahr L, Poschke I, Gary R, Sundberg B, Gerbitz A, Ljungman P, Le Blanc K. Immunosuppressive CD14+HLA-DRlow/neg IDO+ myeloid cells in patients following allogeneic hematopoietic stem cell transplantation. Leukaemia. 2013;27:377–388. doi: 10.1038/leu.2012.215. [DOI] [PubMed] [Google Scholar]
- 34.Buggins AG, Patten PE, Richards J, Thomas NS, Mufti GJ, Devereux S. Tumor-derived IL-6 may contribute to the immunological defect in CLL. Leukaemia. 2008;22:1084–1087. doi: 10.1038/sj.leu.2405015. [DOI] [PubMed] [Google Scholar]
- 35.Wendelbo Ø, Nesthus I, Sjo M, Paulsen K, Ernst P, Bruserud Ø. Functional characterization of T lymphocytes derived from patients with acute myelogenous leukaemia and chemotherapy-induced leukopenia. Cancer Immunol Immunother. 2004;53:740–747. doi: 10.1007/s00262-004-0505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaise C, Buchan SL, Rice J, Marquet J, Rouard H, Kuentz M, Vittes GE, Molinier-Frenkel V, Farcet JP, Stauss HJ, Delfau-Larue MH, Stevenson FK. DNA vaccination induces WT1-specific T-cell responses with potential clinical relevance. Blood. 2008;112:2956–2964. doi: 10.1182/blood-2008-02-137695. [DOI] [PubMed] [Google Scholar]
- 37.King CA, Spellerberg MB, Zhu D, Rice J, Sahota SS, Thompsett AR, Hamblin TJ, Radl J, Stevenson FK. DNA vaccines with single-chain Fv fused to fragment C of tetanus toxin induce protective immunity against lymphoma and myeloma. Nat Med. 1998;4:1281–1286. doi: 10.1038/3266. [DOI] [PubMed] [Google Scholar]
- 38.Padua RA, Larghero J, Robin M, le Pogam C, Schlageter MH, Muszlak S, Fric J, West R, Rousselot P, Phan TH, Mudde L, Teisserenc H, Carpentier AF, Kogan S, Degos L, Pla M, Bishop JM, Stevenson F, Charron D, Chomienne C. PML-RARA-targeted DNA vaccine induces protective immunity in a mouse model of leukaemia. Nat Med. 2003;9:1413–1417. doi: 10.1038/nm949. [DOI] [PubMed] [Google Scholar]
- 39.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 40.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 41.Demotz S, Matricardi P, Lanzavecchia A, Corradin G. A novel and simple procedure for determining T cell epitopes in protein antigens. J Immunol Methods. 1989;122:67–72. doi: 10.1016/0022-1759(89)90335-9. [DOI] [PubMed] [Google Scholar]
- 42.Gotch F, Rothbard J, Howland K, Townsend A, McMichael A. Cytotoxic T lymphocytes recognize a fragment of influenza virus matrix protein in association with HLA-A2. Nature. 1987;326:881–882. doi: 10.1038/326881a0. [DOI] [PubMed] [Google Scholar]
- 43.Bodinier M, Peyrat MA, Tournay C, Davodeau F, Romagne F, Bonneville M, Lang F. Efficient detection and immunomagnetic sorting of specific T cells using multimers of MHC class I and peptide with reduced CD8 binding. Nat Med. 2000;6:707–710. doi: 10.1038/76292. [DOI] [PubMed] [Google Scholar]
- 44.Hosken NA, Bevan MJ. Defective presentation of endogenous antigen by a cell line expressing class I molecules. Science. 1990;248:367–370. doi: 10.1126/science.2326647. [DOI] [PubMed] [Google Scholar]
- 45.Rice J, Elliott T, Buchan S, Stevenson FK. DNA fusion vaccine designed to induce cytotoxic T cell responses against defined peptide motifs: implications for cancer vaccines. J Immunol. 2001;167:1558–1565. doi: 10.4049/jimmunol.167.3.1558. [DOI] [PubMed] [Google Scholar]
- 46.Pascolo S, Bervas N, Ure JM, Smith AG, Lemonnier FA, Perarnau B. HLA-A2.1-restricted education and cytolytic activity of CD8(+) T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 mono-chain transgenic H-2Db beta2m double knockout mice. J Exp Med. 1997;185:2043–2051. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buchan S, Gronevik E, Mathiesen I, King CA, Stevenson FK, Rice J. Electroporation as a “prime/boost” strategy for naked DNA vaccination against a tumor antigen. J Immunol. 2005;174:6292–6298. doi: 10.4049/jimmunol.174.10.6292. [DOI] [PubMed] [Google Scholar]
- 48.Rice J, King CA, Spellerberg MB, Fairweather N, Stevenson FK. Manipulation of pathogen-derived genes to influence antigen presentation via DNA vaccines. Vaccine. 1999;17:3030–3038. doi: 10.1016/s0264-410x(99)00171-1. [DOI] [PubMed] [Google Scholar]


