Abstract
Many bacteria swim in liquid or swarm over solid surfaces by synthesizing rotary flagella. The same bacteria that are motile also commonly form non-motile multicellular aggregates held together by an extracellular matrix called biofilms. Biofilms are an important part of the lifestyle of pathogenic bacteria and it is assumed that there is a motility-to-biofilm transition wherein the inhibition of motility promotes biofilm formation. The transition is largely inferred from regulatory mutants that reveal the opposite regulation of the two phenotypes. Here we review the regulation of motility during biofilm formation in Bacillus, Pseudomonas, Vibrio, and Escherichia, and we conclude that the motility-to-biofilm transition, if necessary, likely involves two steps. In the short term, flagella are functionally regulated to either inhibit rotation or modulate the basal flagellar reversal frequency. Over the long term, flagellar gene transcription is inhibited and in the absence of de novo synthesis, flagella are likely diluted to extinction through growth. Both short term and long term control is likely important to the motility-to-biofilm transition to stabilize aggregates and optimize resource investment. We emphasize the newly discovered classes of flagellar functional regulators and speculate that others await discovery in the context of biofilm formation.
Keywords: EpsE, YcgR, c-di-GMP, brake, clutch, motor
Introduction
Bacteria move by a variety of mechanisms but the most studied form of bacterial motility involves the assembly and rotation of propeller-like flagella (Jarrell & McBride, 2008). Each flagellum is assembled from the inside-out starting with a “basal body” inserted into the cytoplasmic membrane (Fig. 1) (Macnab, 2003). Within the basal body, a type III secretion apparatus secretes subunits of the axle-like rod that extends through the peptidoglycan and outer membrane (if present). Next, a short curved hook is polymerized from approximately 200 subunits of the hook structural protein. Finally, the long helical filament is polymerized from approximately 20,000 units of a structural protein called flagellin. Flagella are spontaneously assembled from over thirty proteins, flagellar biosynthesis is thought to consume 2% of a cell’s metabolic resources, and high synthesis cost may have selected for favoring the most economical amino acids in the most abundant structural components during evolution (Macnab, 1996; Smith & Chapman, 2010).
Figure 1. Functional regulation of the flagellum.
Schematic of the Gram-positive and Gram-negative flagellar structures with the filament (green), hook (light blue), basal body (pink) and motor (tan) indicated. The site of clutch or brake motility inhibition is indicated in red. Micrographs of EpsE-GFP and YcgR-GFP localizing as puncta at the flagellum are reprinted from Blair et al., 2008 and Boehm et al., 2010 respectively. Scale bars are 2 μm.
In addition to synthesis cost, bacteria must make further investment by consumption of ion motive forces to power flagellar rotation (Macnab, 1996). Connected to each basal body is a motor complex composed of a rotor and a stator that interact to turn the flagellum (Berg & Anderson, 1973; Silverman & Simon, 1974; Macnab, 1977). The rotor is a cytoplasmic circular gear connected to the basal body and polymerized from monomers of a protein called FliG (Lowder et al., 2005; Sowa et al., 2005; Thomas et al., 2006) (Fig. 1). Anchored around the basal body are stator complexes made of MotB, a membrane protein that is anchored to the peptidoglycan, and MotA, a membrane protein that interacts with FliG (Blair & Berg, 1990; Zhou et al., 1998). MotA and MotB combine to create an ion channel where ion flow through the channel induces a conformational change in MotA that interacts with FliG to generate torque (Garza et al., 1995; Kojima & Blair, 2001; Gabel & Berg, 2003; Berg, 2003). When rotating, the bacterial flagellum is one of the most powerful motors on earth, revolving at 100-1500 Hz and generating 400-3800 pN-nm of torque depending on the species (Magariyama et al., 1994; Sowa et al., 2003; Li & Tang, 2006; Reid et al., 2006; Darnton et al., 2007). The torque generated by the filament powers swimming through liquid and swarming over solid surfaces (Kearns, 2010). Although advantageous under certain conditions, both the high energy investment of synthesis and output power of flagella may account for the observation that motility is extensively regulated at multiple levels.
One way in which flagella are regulated is at the level of gene expression. Flagella genes are often organized into regulatory hierarchies called classes I, II, and III (and in some cases class IV) depending on the organism in question (Kutsukake et al., 1990; Macnab, 1992). Class I genes encode master transcriptional regulators which activate a regulon of genes including the Class II genes. Class II genes typically encode flagellar structural components that form the basal body, the secretion machinery, and the hook. Class III genes typically encode flagellar structural components that form the filament and the motor proteins. Regulation typically occurs at the junctions between the different classes. For example, class I master regulators can be controlled as to whether or not they activate class II gene expression, and regulatory modules that sense class II flagellar assembly states either permit or deny class III gene expression accordingly (Hughes et al., 1993; Kutsukake, 1994; Mukherjee et al., 2011; Patrick & Kearns, 2012). Also commonly encoded among the class III genes are proteins that form the chemoreceptors and the signal transducers for bacterial chemotaxis.
A second way in which flagella are regulated is at the functional level where motile behavior is controlled by chemotaxis (for review see Wadhams & Armitage, 2004; Sourjik & Wingreen, 2011). Chemotaxis is the directed movement up a chemical gradient, and in bacteria it is mediated by a biased random walk controlled by the frequency with which the flagellum changes the direction of rotation (Berg & Brown, 1972). Flagellar motors typically rotate both clockwise and counterclockwise, where rotation in one direction generates straight swimming of the bacterium and the other direction causes cells to acquire a new orientation (Larsen et al., 1974). Gradients of chemoattractants are sensed by binding to chemoreceptors and are transduced by a cytoplasmic two-component regulatory system to bias the motor towards either the clockwise or the counterclockwise state. In this way, cells reinforce smooth swimming when attractants are increasing and encourage new direction sampling when attractants are decreasing. Although metabolism of effectors is unnecessary for chemotaxis, attractants tend to resemble food and energy sources, and repellents tend to resemble toxins (Adler, 1966; Adler, 1969).
Recently, a third form of flagellar regulation has been discovered that operates at the functional level of the motor to either stop or slow rotation. Traditionally, mutants that disrupt motility have been organized into three phenotypic classes called “fla”, “che”, and “mot” (Macnab, 1992). Mutants with a fla phenotype were aflagellate and disrupted either flagellar structural components or regulators required for structural component expression. Mutants with a che phenotype had functional flagella that were defective in chemotaxis and disrupted either the cytoplasmic signal transduction system or the chemoreceptors. Mutants with a mot phenotype were rare mutants and disrupted either MotA or MotB of the stator complex or were point mutants in the rotor FliG. Recently, there has been an increase in the discovery of mutants that have a mot phenotype outside of the structural components of the motor (Ko & Park, 2000; Blair et al., 2008; Pilizota et al., 2009; Boehm et al., 2010). Some of the new mot phenotypes are related to biofilms suggesting that impedance of flagellar rotation is important for biofilm formation.
Biofilms are multicellular aggregates of bacteria bound by a matrix of extracellular polymers (O’Toole et al., 2000; Kolter & Greenberg, 2006; Monds & O’Toole, 2009). The matrix often consists of a complex mixture of extracellular polysaccharide, protein, and DNA, and not only allows the bacteria to cohere to one another but also adhere to solid surfaces (Flemming & Windengender, 2010). The multicellular group of aggregated bacteria may enjoy advantages that their planktonic (free-floating) counterparts do not. Biofilms have been reported to experience increased tolerance to a variety of environmental parameters including antibiotic challenge and grazing by unicellular eukaryotes (Hall-Stoodley et al., 2004; Matz & Kjelleberg, 2005). As we learn more about biofilms it is becoming clear that biofilms are predominantly beneficial in the environment but can be particularly problematic when associated with biofouling and pathogenesis (Hall-Stoodley et al., 2004; Shi & Zhu, 2009; Simões et al., 2010). Importantly, bacteria in biofilms are non-motile (Yildiz et al., 2001; Sauer et al., 2002; Yildiz et al., 2004; Blair et al., 2008; Verstraeten et al., 2008). Thus, it is implicitly assumed that motile bacteria must become non-motile when transitioning to the biofilm state. In support of the notion that the motility-to-biofilm transition is important, mutations in many regulatory genes have diametrically opposed effects on biofilm formation and motility.
A key regulator of the motility-to-biofilm transition is the small cytoplasmic signaling molecule cyclic-di-GMP (c-di-GMP) (Römling et al., 2005; Römling & Amikam, 2006; Wolfe & Visick, 2008; Hengge, 2009). C-di-GMP is the product of the condensation and cyclization of two molecules of GTP catalyzed by diguanylate synthase enzymes containing a GGDEF amino acid motif (Paul et al., 2004; Simm et al., 2004; Ryjenkov et al., 2005). Elevated levels of c-di-GMP have been associated with inhibition of motility and activation of biofilm formation. C-di-GMP is hydrolyzed to two molecules of GMP by the action of phosphodiesterase enzymes containing an EAL or HD-GYP motif of amino acids (Simm et al., 2004; Christen et al., 2005; Ryan et al., 2006). Diminished levels of c-di-GMP have been associated with activation of motility and inhibition of biofilm formation. Organisms can encode dozens of different GGDEF and EAL domain containing proteins and the environmental inputs that govern c-di-GMP levels are poorly understood. C-di-GMP output can be exerted by binding to RNA riboswitches or by binding allosterically to proteins containing PilZ, HD-GYP, or degenerate GGDEF/EAL domains to regulate physiological targets by protein-protein interaction (Amikam & Galperin, 2006; Benach et al., 2007; Christen et al., 2007; Hickman & Harwood, 2008; Sundarsan et al., 2008). How one pool of c-di-GMP controls many different phenotypes, and why different mutants that disrupt c-di-GMP do not always conform to the general phenotypic consequences described above remain poorly understood.
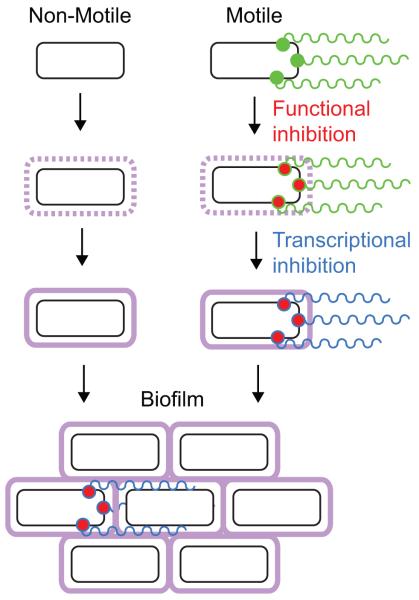
Here we draw upon four model systems, Bacillus, Vibrio, Pseudomonas, and Escherichia, and review what is known about the regulation of flagellar-mediated motility during biofilm formation. We arrive at the conclusion that the inhibition of motility is likely advantageous in the environment but may be either dispensable or easily bypassed in the laboratory. Nonetheless, motility is regulated during the transition to the biofilm and appears to happen in two stages: a fast inhibition at the level of flagellar function and a slow inhibition at the level of inactivation of flagellar gene expression (Fig. 2).
Figure 2. General steps of the motility to biofilm transition.
We propose a model for biofilm formation that incorporates two cell types: those that are non-motile and those that are initially motile. Flagella that are rotating are labeled in green, functional inhibition of the flagellum is labeled in red, decreased transcription of flagella genes is denoted by labeling the flagellum blue, and the extracellular matrix is shown in purple. Non-motile cells increase transcription of and synthesize the extracellular matrix components (purple), while motile cells first inhibit motility functionally at the flagella basal body (red), then decrease transcription of flagella genes (blue) while increasing expression of the extracellular matrix components (purple). Finally, both types of cells form a biofilm together.
Bacillus subtilis
Bacillus subtilis is a Gram-positive bacterium that is motile by assembling many peritrichous flagella arranged along the body of the cell. While the structure of Gram-positive flagella is far less understood than that of Gram-negative flagella, genes homologous to Gram-negative flagellar genes are encoded in the genome, with many concentrated in the long 31 gene fla/che operon (Márquez-Magaña & Chamberlin, 1994). Only a subpopulation of cells synthesize flagella due to a failure to express and activate the Class II encoded sigma factor, σD, that directs the expression of Class III genes encoding, among other things, flagellin (Kearns & Losick, 2005; Cozy & Kearns, 2010). The relative subpopulation that is motile is genetically determined and varies from strain to strain (Kearns & Losick, 2005). B. subtilis laboratory strains have the capacity to swim in liquid media but the ancestral strain also has the ability to swarm over solid surfaces (Kearns & Losick, 2003; Patrick & Kearns, 2009). Ancestral strains swarm because they encode functional alleles of two proteins that domesticated strains lack: Sfp that promotes synthesis of a surfactant and SwrA that activates the fla/che operon (Kearns et al., 2004; Kearns and Losick, 2005; Patrick & Kearns, 2009). The ancestral strain also has the ability to form non-motile biofilms (Branda et al., 2001; McLoon et al., 2011).
B. subtilis forms biofilms either as floating pellicles when grown in liquid, or as colonies with complex architecture when grown on a solid surface (Branda et al., 2001). The biofilm extracellular matrix is complex and composed of many proteins and polysaccharides (Marvasi et al., 2010). One secreted protein found in the matrix is BslA (formerly YuaB) that confers hydrophobicity to the biofilm aggregate (Verhamme et al., 2009; Ostrowski et al., 2011; Kobayashi & Iwano, 2012). The tapA-sipW-tasA operon (tasA operon) encodes other protein components of the matrix including the protein TasA that forms amyloid fibers and is anchored to the cell surface by the protein TapA (Branda et al., 2006; Chu et al., 2006; Romero et al., 2010; Romero et al., 2011). Together TapA and TasA tether and/or organize another major component of the matrix: extracellular polysaccharide (EPS). The EPS is of unknown chemical structure and is synthesized by the proteins encoded within the long 15 gene eps operon (Branda et al., 2001; Kearns et al., 2005). The eps operon contains a pseudoknot RNA-structure called EAR (eps-associated RNA) that acts to promote processive anti-termination of RNA polymerase and ensure complete operon transcription (Irnov & Winkler, 2010). Also encoded within the eps operon is the bifunctional protein EpsE that inhibits flagellar motility (Fig 3; Table 1).
Figure 3. Network of B. subtilis biofilm-related motility regulation.

Transcriptional regulation is labeled in blue, functional regulation labeled in red, and the biofilm matrix products in purple. Green circle highlights the flagellum. Solid lines indicate a confirmed direct interaction while dashed lines indicate that the interaction is poorly understood and could be indirect or direct.
Table 1. Biofilm-related proteins that regulate motility.
| Protein | Function | Annotation | References |
|---|---|---|---|
|
Bacillus subtilis functional regulators
| |||
| EpsE | Flagellar clutch, inhibits FliG | Glycosyltransferase |
Blair et al., 2008
Guttenplan et al., 2010 |
| RemA | Activator of EpsE | No homology/domains | Winkelman et al., 2009 |
| RemB | Activator of EpsE | No homology/domains | Winkelman et al., 2009 |
| YpfA | Inhibits MotA | YcgR homolog/PilZ-domain | Chen et al., 2012 |
|
| |||
|
Bacillus subtilis transcriptional regulators
| |||
| SinI | Antagonist of SinR | Small protein |
Bai et al., 1993
Scott et al., 1999 |
| SinR | Repressor of EpsE and co-inhibitor of fla/che with SlrR |
DNA binding protein |
Kearns et al., 2005
Chai et al., 2010 Cozy et al., 2012 |
| SlrA | Antagonist of SinR | Small protein |
Kobayashi, 2008
Chai et al., 2009 Cozy et al., 2012 |
| SlrR | Co-inhibitor of fla/che with SinR | DNA binding protein |
Chu et al., 2008
Chai et al., 2010 Cozy et al., 2012 |
| SwrA | Activator of fla/che operon | No homology/domains |
Kearns et al., 2004
Kearns and Losick, 2005 Patrick and Kearns, 2009 |
|
| |||
|
Pseudomonas aeruginosa functional regulators
| |||
| BifA | Increase flagella reversals | c-di-GMP hydrolase | Kuchma et al., 2007 |
| FliD | Binds mucin, tethers flagella | Cap of flagellar filament |
Arora et al., 1998
Landry et al., 2006 |
| SadC | Decrease flagella reversals | c-di-GMP synthase |
Caiazza and O’Toole, 2004
Merritt et al., 2007 |
|
| |||
|
Pseudomonas aeruginosa transcriptional regulators
| |||
| ArmZ | Repress fleQ transcription | DNA binding protein |
Garrett et al., 1999
Tart et al., 2005 Tart et al., 2006 |
| FleQ | Activator of flagellar genes/ Regulator of pel transcription |
DNA binding protein |
Arora et al., 1997
Hickman & Harwood, 2008 Baraquet et al., 2012 |
|
| |||
|
Vibrio sp. transcriptional regulators
| |||
| FlrA | Activates flagellar genes | NtrC homolog |
Prouty et al., 2001
Correa & Klose, 2005 |
| HapR | Activates motility | LuxR homolog |
Yildiz et al., 2004
Waters et al., 2008 |
| OpaR | Activates motility | HapR homolog | McCarter, 1998 |
| ScrC | Activates motility | c-di-GMP synthase/hydrolase |
Boles and McCarter, 2002
Ferreira et al., 2008 Trimble and McCarter, 2011 |
| ScrG | Activates motility | c-di-GMP hydrolase | Kim and McCarter, 2007 |
| VpsT | Repressor of flagellar gene expression. |
DNA binding protein |
Casper-Lindley and Yildiz, 2004
Krasteva et al., 2010 |
|
| |||
|
Escherichia coli functional regulator
| |||
| YcgR | Slows rotation and changes reversal frequency at MotA/FliG/FliM |
c-di-GMP binding protein |
Boehm et al., 2010
Fang & Gomelsky, 2010 Paul et al., 2010 |
|
| |||
|
Escherichia coli transcriptional regulators
| |||
| CsrA | Stabilizes flhDC mRNA | RNA binding protein | Wei et al., 2001 |
| OmpR | Represses flhDC expression | DNA binding protein |
Shin and Park, 1995
Prigent-Combaret et al., 1999 |
| RcsBA | Represses flhDC expression | DNA binding complex |
Gottesman et al., 1985
Francez-Charlot et al., 2003 Fredericks et al., 2006 |
EpsE inhibits motility by acting as a clutch on the flagellar rotor (Blair et al., 2008; Guttenplan et al., 2010). When EpsE is expressed, either alone from an artificial promoter or expressed together with the rest of the eps operon during biofilm formation, flagellar rotation comes to a halt. EpsE is thought to inhibit the flagella rotor FliG by direct protein-protein interaction because an EpsE-GFP fusion localizes as puncta along the cell membrane, reminiscent of peritrichously arranged flagellar basal bodies (Fig. 1). Futhermore, EpsE puncta co-localize with flagellar basal bodies and are dependent upon critical amino acids on an exposed surface of FliG (Blair et al., 2008; Guttenplan et al., 2010; Guttenplan et al., 2013). EpsE could have inhibited flagellar rotation either like a brake that caused rotation to lock up, or like a clutch that resulted in unpowered flagella that could be externally rotated. A clutch-like mechanism was supported because when cells were tethered by one flagellum and EpsE was expressed, the cell bodies rotated from the external torque of Brownian motion as freely as one would expect by diffusion. Thus EpsE acts like a clutch by separating the flagellar drive train (the rotor FliG) from the power source (the proton channel MotA/MotB) thereby disabling rotation. EpsE was the first flagellar clutch protein identified and it was speculated that the clutch might be required for a rapid loss of motility during early biofilm formation.
Mutation of EpsE resulted in a severe defect in biofilm formation but attributing the defect to the clutch activity was complicated by the fact that EpsE is homologous to members of the group II glycosyltransferases. Glycosyltransferase enzymes are commonly encoded within operons that synthesize extracellular polysaccharide, and EpsE contains the highly conserved glycosyltransferase active site residues (Guttenplan et al., 2010). Mutation of the active site residues abolish synthesis of the biofilm EPS and biofilm formation but do not abolish flagellar clutch function (Blair et al., 2008; Guttenplan et al., 2010). Clutch activity, rather, requires a series of residues predicted to occupy a common surface of an exposed alpha helix that may represent the contact site with FliG, proving that EpsE is bifunctional (Guttenplan et al., 2010). Interestingly, the residues in EpsE necessary for clutch function are more highly conserved among related glycosyltransferases (even those from non-motile organisms) than the residues in FliG found to be necessary for clutch contact. Thus, under selection for motility inhibition, FliG may have evolved to interact with EpsE, an enzyme that happened to be present during biofilm formation. Finally, both the EPS biosynthetic and clutch functions of EpsE contribute to biofilm formation as mutation of both activities synergize to create a greater defect than either alone (Guttenplan et al., 2010).
What is the function of the clutch during biofilm formation? First, functional inhibition of the flagellum is a rapid mechanism to inhibit motility and may either act early in biofilm formation and/or at intermediate stages to stabilize cell aggregates under stress. Second, if the metaphor of a clutch in an automobile is accurate, EpsE may be reversible if biofilm formation is prematurely aborted. Third, EpsE is the last protein encoded in the eps operon upstream of a series of internal terminators that are counteracted by the EAR anti-termination cis-element (Irnov and Winkler, 2010). If EAR-dependent anti-termination is regulated, there may be conditions under which EpsE inhibits motility without cells progressing immediately to biofilm formation due to termination prior to transcription of the rest of the operon. Finally, the function of the clutch may have a long-term advantage in preserving flagellar function by relieving selective pressure against the enrichment of motility mutants. For example, a strain that is clutch-deficient forms biofilms, but these biofilms accumulate mutations in genes required for flagellar motility (Guttenplan et al., 2010). Thus, in the absence of the clutch the selection for biofilm formation comes at the genetic cost of heritable flagellar defects such that the motility of the biofilm descendants is compromised. The very fact that selective pressure acts against motility in the biofilm provides direct evidence that motility inhibition is indeed an important step (Guttenplan et al., 2010).
A second functional regulator of flagella has recently been discovered in B. subtilis. YpfA is a protein that contains a PilZ-domain attributed to c-di-GMP binding (Chen et al., 2012a) (Table 1). Although c-di-GMP has only been directly observed in Gram-positives once, in Clostridium difficile, indirect reporters suggest it may also be present in B. subtilis (Sudarsan et al., 2008; Bordeluau et al., 2011; Purcell et al., 2012; Chen et al., 2012a). Mutation of a phosphodiesterase homolog called YuxH abolished motility, and motility was restored to the YuxH mutant by mutation of YpfA (Chen et al., 2012a). Interestingly, YpfA interacted with MotA in bacterial two-hybrid analysis and both MotA interaction and motility inhibition was abolished by mutations in the c-di-GMP-binding PilZ domain (Fig 3). Whether c-di-GMP is actually present in B. subtilis has yet to be demonstrated and whether either YuxH or YpfA are related to biofilms is unclear. Mutation of either YuxH or YpfA had little to no detectable effect on biofilm formation but we note that biofilm inhibition could have been masked by the rapid selection of non-motile suppressor mutations, as was found with EpsE clutch activity. Whether YpfA acts as a clutch, brake, or other mechanism remains to be elucidated.
Motility in B. subtilis is also inhibited at the level of transcription during biofilm formation but when and where transcriptional regulation occurs is a complex issue. Expression of flagellar basal body genes decreases as biofilm formation progresses and flagellar filament gene expression is repressed in the mature biofilm (Kobayashi, 2007a; Kobayashi, 2007b; Vlamakis et al., 2008). Furthermore, the control of flagellar gene expression within the biofilm is spatially partitioned. By fluorescently labeling subpopulations with different fluorophores, Vlamakis et al. showed that in a cross-section of a biofilm, cells that express the flagella filament are localized to the periphery and the region directly adjacent to the substrate (Vlamakis et al., 2008). As cells increased their distance from the substrate in three dimensions, flagellar filament expression decreased and gave rise to the expression of the biofilm matrix genes. There appears to be a tight and inverse coupling of the transcriptional regulation of motility and biofilm matrix genes that occurs in both space and time.
Multiple regulators converge to spatially regulate motility and biofilm gene expression. The master regulator of biofilm formation, SinR, represses expression of both the eps and tasA operons (Kearns et al., 2005, Branda et al., 2006; Chu et al., 2006). SinR is a phage-like repressor protein that dimerizes and binds to a consensus sequence (GTTCTCT) to control a relatively small set of genes mostly dedicated to biofilm formation (Gaur et al., 1991; Lewis et al., 1998; Kearns et al., 2005) (Table 1). Encoded upstream of sinR is SinI, a small protein that binds to SinR and inhibits SinR dimerization (Bai et al., 1993; Lewis et al., 1998; Scott et al., 1999) (Table 1). In response to biofilm initiating signals, the response regulator Spo0A activates the expression of SinI, SinR is antagonized, and the biofilm genes are derepressed (Gaur et al., 1988; Branda et al., 2001; Hamon & Lazazzera, 2001; Chai et al., 2011) (Fig. 3). Furthermore, Spo0A activation is heterogeneous in the population and heterogeneous activation likely contributes to the three-dimensional spatial patterning of biofilm gene expression (Vlamakis et al., 2008; Chai et al., 2008).
The genes expressed by SinI antagonism of SinR not only include the matrix synthesizing operons but also include SlrR, a paralog of SinR. Indeed, SinR and SlrR are sufficiently similar that they cross-react with SinR-specific polyclonal antibodies (Chu et al., 2008). SlrR activates expression of the tasA operon and redirects SinR function by forming a SinR/SlrR heterodimer that co-represses expression of the autolysin genes and the fla/che operon that codes for the flagellar basal body proteins (Chu et al., 2008; Chai et al., 2010; Cozy et al., 2012) (Fig. 3; Table 1). Inhibition of the fla/che operon arrests de novo flagellar synthesis. A third protein, YmdB, is also involved in the regulation of flagellar gene expression through its activation of SlrR (Diethmaier et al., 2011). Therefore, the repression of motility gene expression is concomitant with activation of extracellular matrix genes and in the absence of new transcription; the flagella are presumably diluted to extinction through cell growth within the biofilm.
There may be additional mechanisms and pathways to inhibit motility in the biofilm. Other regulators are involved in joint biofilm and motility regulation, but the mechanism of activation or repression is less understood (Fig. 3). SlrA is a paralog of the SinI protein and can antagonize SinR to derepress biofilm genes and inhibit fla/che operon transcript levels (Kobayashi, 2008; Chai et al., 2009; Cozy et al., 2012) (Table 1). RemA and RemB are two other transcriptional activators of biofilm formation that act in parallel to SinR, but the stimulus input and mechanisms of regulatory output of either protein is unknown (Winkelman et al., 2009) (Table 1). DegU is a response regulator that regulates both motility and biofilm formation in a complex manner (Kobayashi, 2007b; Verhamme et al., 2007). When the concentration of phosphorylated DegU is low in the cell, swarming motility and flagellar basal body gene expression is activated (Kobayashi, 2007b; Verhamme et al., 2007; Tsukahara & Ogura, 2008). However, as the amount of phosphorylated DegU increases, DegU-P represses expression of flagellar basal body genes and activates the anti-sigma factor FlgM, which in turn antagonizes the σD sigma factor that expresses the flagellar filament protein (Amati et al., 2004; Hsueh et al., 2011). Additionally, phophorylated DegU activates genes required for biofilm formation (Verhamme et al., 2009; Ostrowski et al., 2011). DegU is phosphorylated by a cytoplasmic signal kinase DegS but the stimulus that controls the amount of phosphorylated DegU remains poorly understood (Mukai et al., 1990).
Motility inhibition during B. subtilis biofilm formation is presumed to be initiated by extracellular signals that are transduced to activate SinI (Kearns et al., 2005; López et al., 2009; Shemesh et al., 2010; López et al., 2010; Chen et al., 2012b) (Fig. 3). SinI antagonizes SinR to derepress the eps and tasA operons that encode proteins that begin assembling the extracellular matrix and the gene that encodes SlrR. As SlrR accumulates, it redirects SinR to repress the fla/che operon transcript abundance thereby inhibiting flagellar gene expression. Flagella are numerous and stable and thus inhibiting transcript is insufficient to inhibit motility. EpsE is also made and inhibits flagellar rotation in the short term while de novo flagellar synthesis is inhibited at the transcriptional level over the long term. Finally, because flagellar gene expression in B. subtilis is heterogeneous, there is a pre-existent non-motile subpopulation that need not inhibit motility and may proceed to biofilm formation directly (Kearns & Losick, 2005; Chai et al., 2008; López et al., 2009; Chai et al., 2010).
Pseudomonas aeruginosa
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that causes dangerous infections of the lungs in patients with cystic fibrosis (CF). P. aeruginosa synthesizes a single polar flagellum to swim in liquid media and also swarms over solid surfaces. During swarming, either the number of flagella at the pole increases and/or the single polar flagellum is augmented by an alternative swarming-specific stator complex (Rashid & Kornberg, 2000; Köhler et al., 2000; Doyle et al., 2004; Toutain et al., 2005). Flagellar biosynthesis is organized in a Class I-IV hierarchy where Class I is represented by the master regulator protein FleQ, classes II and III encode the hook-basal body genes, and Class IV encodes flagellin and the chemotaxis proteins (Arora et al., 1997; Jyot et al., 2002; Dasgupta et al., 2003). P. aeruginosa also uses type IV pilus-based twitching motility to move over solid surfaces but this form of motility will not be discussed further here (for review see Burrows, 2012).
P. aeruginosa forms pellicle biofilms at air-liquid interfaces as well as liquid-solid submerged biofilms attached to glass or other substrates. The primary structural component of the P. aeruginosa biofilm is extracellular polysaccharides (EPS) that depend not only on the strain but also on the conditions under which the biofilm forms. EPS biosynthesis proteins are encoded by genes within either the pel and/or the psl operons depending on the strain considered (Friedman & Kolter, 2004a; Friedman & Kolter, 2004b; Matsukawa & Greenberg, 2004; Jackson et al., 2004). The chemical structure of the PSL polysaccharide has recently been elucidated as a repeating pentasaccharide of D-mannose, D-glucose, and L-rhamnose, but the chemical structure of PEL is unknown (Ma et al., 2007; Byrd et al., 2009). Finally, some clinical isolates of P. aeruginosa isolated directly from the lung of a CF patient synthesize the abundant EPS alginate using enzymes encoded within the alg operons (Govan & Deretic, 1996; Wozniak et al., 2003b; Ramsey & Wozniak, 2005). The chemical structure of alginate an unbranched heteropolymer of O-acetylated β-1,4-linked D-mannuronic and L-guluronic acids (Linker & Jones, 1996; Ramsey & Wozniak, 2005). Other components of the extracellular matrix include DNA and proteins, but the exact composition and contribution of these structures is unknown (Whitchurch et al., 2002; Matsukawa & Greenberg, 2004, Borlee et al., 2010).
P. aeruginosa requires cell motility to form a biofilm, but unregulated motility during biofilm formation destabilizes the biofilm structure (O’Toole & Kolter, 1998; Vallet et al., 2001; Whitely et al., 2001; Sauer et al., 2002; Klausen et al., 2003). Cells that enhanced for swarming motility form a flat and featureless biofilm, while cells that are deficient for swarming motility form a more structured biofilm with large cellular aggregates (Shrout et al., 2006). Thus, a decrease in motility during biofilm formation may promote and stabilize three-dimensional architectural development. Consistent with motility antagonizing biofilms, P. aeruginosa colony morphology variants that inherently form more robust biofilms have a decrease in motility compared to their parental strains (Kirisits et al., 2005). Thus, the proper timing of motility control is required for forming wild type biofilm architecture but motility per se can be bypassed entirely when cells are genetically locked in a biofilm-proficient state.
In P. aeruginosa, flagella are functionally regulated at the level of flagellar reversal frequency (Fig. 4). Strains mutated for the protein SadC were “surface attachment defective”, pellicle-deficient, and had increased motility in swarming assays (Merritt et al., 2007). Increased swarming migration in the sadC mutant was correlated with an increase in basal flagella reversal frequency in a high viscosity medium (Caiazza et al., 2007; Merritt et al., 2007). SadC is a diguanylate cyclase that synthesizes c-di-GMP and sadC mutants showed a reduction in the cytoplasmic c-di-GMP levels (Merritt et al., 2007) (Table 1). Conversely mutants defective in the phosphodiesterase BifA exhibited decreased swarming motility that was correlated with a decrease in basal flagella reversal frequency and an increase in c-di-GMP levels (Kuchma et al., 2007) (Table 1). The output of elevated c-di-GMP levels that controls the flagellar reversal frequency is unknown but the inputs that elevate c-di-GMP seem to be related to the type IV pili that mediate twitching motility and the EPS matrix itself (Kuchma et al., 2010; Kuchma et al., 2012; Irie et al., 2012). The role of controlling flagellar reversal frequency early in biofilm formation is also unclear but maintenance of a critical frequency of basal reversals has been correlated with a cell’s ability to navigate pores in soft agar (Wolfe & Berg, 1989). Thus, reversal frequency control may be important to facilitate spread through a matrix such as the mucus of the CF lung even in the absence of chemotactic signals.
Figure 4. Network of P. aeruginosa biofilm-related motility regulation.

Transcriptional regulation is labeled in blue, functional regulation labeled in red, and the biofilm matrix products in purple. Green circle highlights the flagellum. C-di-GMP in superscript indicates that the protein needs to be bound to c-di-GMP to carry out the indicated activity. Solid lines indicate a confirmed direct interaction while dashed lines indicate that the interaction is poorly understood and could be indirect or direct.
In isolates from CF lungs, motility of P. aeruginosa is mechanically inhibited by the interaction with host mucin, the protein component of mucus found abundantly in the airway of CF patients. When P. aeruginosa is inoculated onto a glass surface coated with mucin, surface motility is inhibited and biofilm formation is enhanced (Landry et al., 2006). One reason for motility inhibition is that the FliD flagellar cap protein found at the tip of the filament binds to mucin to tether the cells and constrain rotation (Arora et al., 1998; Landry et al., 2006) (Table 1). The FliD-mucin interaction may be a mechanism for the cell to abolish motility and promote adherence in a single step (Fig. 4). That FliD also acts as a mucin receptor may contribute to the persistence of biofilms in CF patients’ lungs due to a reduced ability of the cells to disperse.
Once biofilm formation has been initiated, flagellar gene expression decreases as a biofilm matures (O’Toole & Kolter, 1998; Vallet et al., 2001; Whitely et al., 2001; Sauer et al., 2002; Klausen et al., 2003; Wolfgang et al., 2004). One way in which cells coordinate flagellar and biofilm gene expression is by controlling the transcriptional regulator FleQ. FleQ activates flagellar genes and represses pel operon expression (Arora et al., 1997; Hickman & Harwood, 2008) (Fig. 4). However, when c-di-GMP levels rise in the cell, FleQ binds to c-di-GMP and activates rather than represses pel transcription (Hickman & Harwood, 2008; Baraquet et al., 2012). The mechanism of the FleQ functional switch is unclear, but it is likely due to conformational changes caused by the interactions with both c-di-GMP and FleN, the antagonist of FleQ (Baraquet et al., 2012). Binding of FleQ to c-di-GMP does not inhibit flagellar gene expression significantly and presumably, other mechanisms are involved in the decrease in flagellar gene expression during biofilm formation.
Most patients are infected with motile strains of P. aeruginosa but bacteria re-isolated from infected CF lungs are typically non-motile (Luzar et al., 1985; Burke et al., 1991; Mahenthiralingam et al., 1994). Loss of motility is genetic, as selective pressure enriches for a frameshift mutation in a gene mucA that reduces flagellar gene expression and enhances biofilm formation (Martin et al., 1993). The mucA gene encodes the protein MucA, the anti-sigma factor of an alternative sigma factor called AlgT (Govan & Deretic, 1996; Tart et al., 2005). Unchecked, AlgT directs RNA polymerase to express the genes for alginate biosynthesis and the gene armZ (formerly algZ), encoding the transcription factor ArmZ (Wozniak et al., 2003a; Tart et al., 2006). ArmZ in turn represses expression of FleQ, thereby inactivating expression of flagellar biosynthesis genes and inhibiting motility (Garrett et al., 1999; Tart et al., 2005; Tart et al., 2006) (Fig. 4; Table 1). The mutation in mucA occurs at high frequency in a homopolymeric tract and therefore can be reselected for gain-of-motility reversion by restoring the wild type frame (Martin et al., 1993; Henderson et al., 1999). High frequency, reversible frameshifting may reduce selective pressure on the mutation of the flagellar genes themselves and ensures that if the cells are reselected to resume flagellum biosynthesis; all of the required genes are intact.
Late in biofilm maturation, when motility is re-activated to permit cell dispersal under conditions of flow, cells can be seen swimming away from the biofilm (Sauer et al., 2002; Davies & Marques, 2009). Biofilm dispersion is triggered by the cellular release of a small chemical signal, cis-2-decenoic acid (Davies & Marques, 2009). The precise cellular target of cis-2-decenoic acid remains unknown but one protein that has been identified as being involved in dispersion is BdlA (Morgan et al., 2006). In bdlA mutants, biofilms do not disperse upon nutrient starvation conditions, and bdlA mutants have a higher intracellular concentration of c-di-GMP compared to wild type (Morgan et al., 2006). The mechanism of BdlA involvement in dispersion is presumably indirect since the protein does not contain a GGDEF, EAL, or known c-di-GMP binding domain. Other compounds have been discovered to disperse biofilms but if and how these compounds activate motility is unknown (Chen et al., 2000; Banin et al., 2006; Barraud et al., 2006; Kolodkin-Gal et al., 2010; Hibbing & Fuqua, 2011).
The motility regulation in P. aeruginosa biofilms is controlled at multiple levels (Fig. 4). Perhaps, early in an infection, extracellular stimuli signal for an increase in the cellular levels of c-di-GMP, which decreases the flagellar reversal rate. Conversely, an increase in flagellar reversal rate may facilitate navigation of the mucus matrix of the lung. Binding of the flagellar cap to mucin may constrain flagellar rotation and tether the bacteria as c-di-GMP levels continue to rise in the cytoplasm. High levels of c-di-GMP inhibit flagellar gene expression while activating EPS biosynthesis to construct the biofilm matrix. At some point in the process selective pressure may select for MucA mutants to genetically trap a subpopulation of bacteria in a non-motile, biofilm-producing state. Late in biofilm formation the synthesis of cis-2-decenoic acid may reactivate motility and stimulate dispersal of the biofilm.
Vibrio species
Vibrio cholerae is a Gram-negative bacterium that is the causative agent of the severe gastrointestinal infection cholera. V. cholerae swims in liquid by synthesizing a single polar flagellum that is powered by the sodium motive force (Kojima et al., 1999). Flagellar biosynthesis genes are organized in a four level hierarchy (Class I-IV) (Prouty et al., 2001). The master regulator of flagellar biosynthesis is the NtrC-like FlrA protein that activates σ54 RNA polymerase to express class II basal body and secretion apparatus genes. FlrA also activates both a second NtrC-like regulator FlrC that redirects RNA polymerase to express Class III rod and hook genes, and an alternative sigma factor σ 28 that directs RNA polymerase to express the Class IV filament genes (Prouty et al., 2001; Correa & Klose, 2005). Unlike many other bacteria, the flagellum is sheathed by an extension of the outer membrane that surrounds the filament, but the function of the sheath is unknown (Sjoblad et al., 1983). Virulence factor adhesins are co-activated with the flagellar biosynthesis genes and motility per se is required for V. cholerae virulence (Lee et al., 2001; Butler & Camilli, 2005; Morris et al., 2008; Syed et al., 2009).
V. cholerae forms biofilms that manifest as pellicles at air-liquid interfaces, as surface-associated submerged biofilms that attach to glass or other substrates, and finally as rugose colonies of complex architecture (Yildiz & Visick, 2009). The biofilm extracellular matrix is complex and is composed of Vibrio polysaccharide (VPS) and proteins. Two of the proteins, RbmC and Bap1, are seemingly redundant paralogs that coat substrate surfaces and promotes cell adhesion, while another protein RbmA coats cell surfaces and promotes cell-cell cohesion (Fong et al., 2006; Fong et al., 2007; Absalon et al., 2011; Berk et al., 2012). VPS is synthesized by proteins encoded in 18 genes divided between two operons called vps-I and vps-II (Yildiz & Schoolnik, 1999). While the precise structure of the VPS is unknown, sugar composition analysis suggests that it is composed primarily of glucose and galactose with some xylose, mannose, and N-acetyl glucosamine (Yildiz & Schoolnik, 1999). Mutations that disrupt either VPS or matrix protein synthesis disrupt biofilm formation. Extracellular DNA is also a component of the biofilm matrix, but how it interacts with the known VPS and proteins in the matrix is unknown (Seper et al., 2011). Biofilm formation is thought to not be essential for virulence per se, but is rather thought to be essential for survival of cholera-causing bacteria in the environmental water reservoir (Cowell, 1996; Faruque et al., 2006).
Production of c-di-GMP is correlated with the motility-to-biofilm transition. V. cholerae encodes 53 putative diguanylate cyclase (GGDEF domain-containing) and phosphodiesterase (EAL domain-containing) homologs and at least 5 proteins with PilZ domains that potentially serve as c-di-GMP effector proteins (Lim et al., 2006; Pratt et al., 2007). Mutants of the putative phosphodiesterases CdgC, CdgJ, RocS, and MbaA are less motile compared to wild type, consistent with high levels of c-di-GMP inhibiting motility (Lim et al., 2006; Liu et al., 2010). Conversely, mutants of putative diguanylate cyclases CdgD, CdgH, and VpvC are more motile compared to wild type, consistent with low levels of c-di-GMP promoting motility (Lim et al., 2006, Beyhan & Yildiz, 2007; Beyhan et al., 2008). Finally, mutations in the PilZ domain-containing proteins PlzB and PlzC resulted in a decrease in motility and biofilm formation compared to wild type (Pratt et al., 2007). Perhaps PlzB and PlzC promote early biofilm formation through motility. Taken together, c-di-GMP regulates both motility and VPS production, but how the small molecule information is integrated over time remains mysterious.
The study of the transition from motility-to-biofilm formation has been aided by the isolation of spontaneous mutants that produce smooth or rugose colony variants (White, 1938). The rugose variant is enhanced for biofilm formation, exhibits increased VPS synthesis and has reduced motility (Yildiz et al., 2004). By contrast, the smooth colony variant is motile but biofilm deficient (Yildiz et al., 2004). One mutation that mediates the smooth-to-rugose genetic transition is a gain-of-function allele of the VpvC diguanylate cyclase that increases c-di-GMP levels in the cell (Beyhan & Yildiz, 2007) (Fig. 5). Thus, the rugose variants are locked in a non-motile biofilm state because c-di-GMP is constitutively high. Increased c-di-GMP levels decrease motility and increase colony rugosity in part by increasing the levels of two cytoplasmic regulatory proteins VpsR and VpsT (Yildiz et al., 2004; Lim et al., 2006).
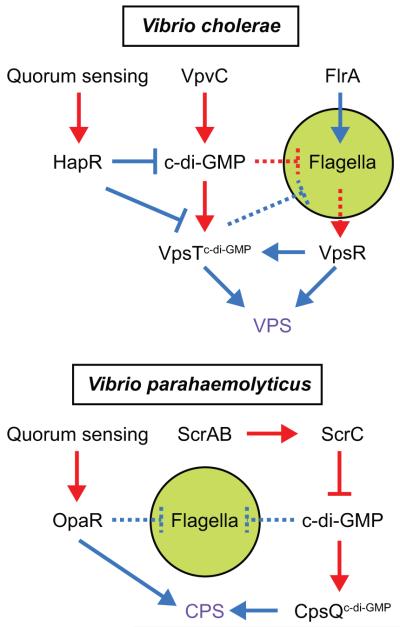
Figure 5. Network of V. cholerae and V. parahaemolyticus biofilm-related motility regulation.
Transcriptional regulation is labeled in blue, functional regulation labeled in red, and the biofilm matrix products in purple. Green circle highlights the flagellum. C-di-GMP in superscript indicates that the protein needs to be bound to c-di-GMP to carry out the indicated activity. Solid lines indicate a confirmed direct interaction while dashed lines indicate that the interaction is poorly understood and could be indirect or direct.
The two-component response regulator proteins VpsR and VpsT are required to activate the expression of VPS biosynthesis genes, and mutation of either gene abolishes biofilm formation (Yildiz et al., 2001; Casper-Lindley & Yildiz, 2004) (Fig. 5). VpsR binds to the vps promoter region when phosphorylated on a conserved aspartate residue but the histidine kinase responsible for phosphorylation is unknown (Lauriano et al., 2004). VpsT on the other hand, only binds to the vps promoter region when c-di-GMP is bound (Krasteva et al., 2010) (Fig 5; Table 1). Although VpsR and VpsT have very similar regulons, VpsR exerts larger effects on gene expression perhaps because VpsR also activates VpsT expression (Beyhan et al., 2007; Srivastava et al., 2011). Mutation of genes required for VPS synthesis in a rugose variant abolishes rugose colony morphology and biofilm formation (Yildiz et al., 2001; Yildiz et al., 2004).
The regulation of motility during biofilm formation in V. cholerae is complex and relatively poorly understood. Flagella are required for early stages of biofilm development and mutants lacking the flagellum have a decrease in biofilm formation (Watnick & Kolter 1999). Flagella function itself may be directly required for vps gene expression, as paralysis of flagellar rotation abolished vps gene expression in a manner that could be bypassed by constitutive expression of VpsR (Lauriano et al., 2004) (Fig. 5). As the biofilm matures, however, motility appears to be lost or at least diminished. One possible explanation for reduced motility is due to flagella tethering by VPS overproduction and indeed, four flagellins comprising the flagellar filament co-purified with the biofilm matrix (Absalon et al., 2011). Simply constraining flagella, however, is likely not sufficient as mutation of genes required for VPS synthesis in a rugose variant abolishes biofilm formation, but did not restore motility (Yildiz et al., 2001; Yildiz et al., 2004). Furthermore, the rugose variant retained polar flagella, despite being non-motile suggesting a level of as-yet-unidentified functional regulation on the flagellar motor (Yildiz et al., 2004). Finally, cells in a mature biofilm decrease expression of the flagella filament, flaA (Watnick et al., 2001; Moorthy & Watnick, 2004). VpsT in its c-di-GMP bound state inhibits flagellar gene expression by an unknown mechanism that could be mediated by direct binding to flagellar promoters or by controlling a secondary regulator (Fig. 5; Lim et al., 2006; Beyhan et al., 2006; Krasteva et al., 2010).
In addition to decreasing motility to promote biofilm formation, V. cholerae may also increase motility late in the cycle to induce dispersion from a mature biofilm. V. cholerae is found in the aquatic environment attached to surfaces as well as in the planktonic form between human outbreaks (Islam et al., 1994). Quorum sensing may be involved in the transition back to the planktonic state since a LuxR homolog, HapR, promotes motility and directly represses expression of VpsT at high cell densities (Yildiz et al., 2004; Waters et al., 2008) (Table 1). Mutation of HapR increases biofilm formation, VPS production, and decreases motility (Zhu et al., 2002; Hammer & Bassler, 2003; Zhu & Mekalanos, 2003). HapR also decreases the amount of c-di-GMP in the cell by regulating expression of putative diguanylate cyclases and phosphodiesterases (Waters et al., 2008; Hammer & Bassler, 2009). Thus, as cell density increases quorum signals activate HapR to mediate the transition from the biofilm state back towards motility.
Flagella and motility are required early in biofilm formation perhaps for locating and interacting with an appropriate surface. Motility appears to be promoted by low levels of c-di-GMP potentiated by PilZ-containing proteins such as PlzB and PlzC. The environmental stimuli to initiate biofilm formation are diverse and include low salt, indole, and polyamines (Karatan et al., 2005; Shikuma & Yildiz, 2009; Mueller et al., 2009; Shikuma et al., 2012). Once biofilm formation is initiated, c-di-GMP levels rise, VpsR and VpsT activate matrix biosynthesis genes, and cells begin to aggregate in the biofilm. Flagella become inhibited at the functional level; perhaps either paralyzed, like B. subtilis, or altered for behavior, like P. aeruginosa. Furthermore, flagella gene expression is inhibited by some mechanism involving, at least in part, c-di-GMP and VpsT. Finally, very late in biofilm formation quorum sensing signals reprogram the regulatory state of the cells, reactivate motility and promote biofilm dispersal. Biofilms and motility are clearly intertwined, and the pathogenesis of V. cholerae may hang on the precise alternation between the biofilms that promote survival in the environment and the planktonic cells that are virulent (Rice et al., 1992; Wai et al., 1998, Yildiz & Schoolnik, 1999; Tamayo et al., 2010).
Another Vibrio species, which was among the earliest studies to investigate the regulation of the motility during biofilm formation, is Vibrio parahaemolyticus. V. parahaemolyticus is motile and synthesizes two entirely separate flagellar systems: a sheathed polar flagellum driven by the sodium motive force and unsheathed lateral flagella driven by the proton motive force (Atsumi et al., 1992; Kim & McCarter, 2000; Stewart & McCarter, 2003). The two flagellar systems have different functions where the polar flagellum powers swimming in liquid media and the lateral flagella powers swarming over solid surfaces (for review see McCarter, 2004). V. parahaemolyticus forms non-motile biofilms that manifest as a pellicle on a liquid interface as well as colonies with complex architecture that grow on solid surfaces (Güvener & McCarter, 2003). The biofilm matrix is composed of extracellular polysaccharide that is referred to as capsule polysaccharide (CPS) due to the tight association with the cell surface (Enos-Berlage & McCarter, 2000). CPS is synthesized from the 11-gene cps operon and is composed of fucose, galactose, glucose, and N-acetylglucosamine (Enos-Berlage & McCarter, 2000; Güvener & McCarter, 2003).
Like V. cholerae, V. parahaemolyticus produces high frequency genetic subpopulations that grow to either translucent or opaque colonies. Opaque colony variants contain mutations in a HapR homolog called OpaR, and opaR mutants confer an opaque phenotype that is enhanced for biofilm formation and reduced for motility (McCarter, 1998) (Table 1). Similarly, mutation of two proteins ScrC and ScrG that contain both GGDEF and EAL domains also result in enhanced biofilm formation and decreased motility (Boles & McCarter, 2002, Kim & McCarter, 2007; Ferreira et al., 2008) (Fig 6; Table 1). Simultaneous mutation of ScrC and ScrG resulted in a synergistically enhanced biofilm formation (Kim & McCarter, 2007). Thus, OpaR, ScrG, and ScrC normally function to promote motility in V. parahaemolyticus.
Figure 6. Network of E. coli biofilm-related motility regulation.

Transcriptional regulation is labeled in blue, functional regulation labeled in red, and the biofilm matrix products in purple. Green circle highlights the flagellum. C-di-GMP in superscript indicates that the protein needs to be bound to c-di-GMP to carry out the indicated activity. Solid lines indicate a confirmed direct interaction while dashed lines indicate that the interaction is poorly understood and could be indirect or direct.
ScrC regulates motility by controlling c-di-GMP in response to quorum sensing signals that accumulate in the environment. ScrC is encoded within a three gene operon scrABC, which also encodes ScrA, a periplasmic putative aminotransferase and ScrB a periplasmic solute binding protein (Boles & McCarter, 2002). ScrC is a transmembrane protein with cytoplasmic GGDEF and EAL domains, and ScrC has the capacity to function as both a diguanylate cyclase and a phosphodiesterase (Ferreira et al., 2008). Regulation of ScrC, however is complex, and is modulated by spent media that accumulates an as-yet-unidentified signal (S signal), the synthesis of which depends of ScrA (Trimble & McCarter, 2011) (Fig 5). Genetic evidence suggests that ScrB is a receptor for the S signal that specifically enhances ScrC phosphodiesterase activity (Trimble & McCarter, 2011). Therefore, at high cell density a pheromone modulates the activity of the bifunctional ScrC to degrade c-di-GMP and enhance motility by an unknown effector. Conversely, at low cell density, c-di-GMP is high and promotes biofilm formation through a recently discovered VpsT homolog, CpsQ that activates CPS gene expression (Ferreira et al., 2011) (Fig 5). V. cholerae does not seem to encode the Scr operon (Ferreira et al., 2008).
V. cholerae and V. parahaemolyticus are two closely related species that both inversely regulate motility and biofilm formation. Both species even seem to have quorum sensing systems that coordinate the lifestyle transition, and yet despite these similarities, the regulatory mechanisms differ. Biofilm formation is better studied in V. cholerae than motility and it will be interesting to understand how motility is inhibited in the biofilm. Motility is better studied in V. parahaemolyticus than in V. cholerae, and it will be interesting to learn how biofilm formation is coordinated between the two different flagellar systems that govern swimming and swarming.
Escherichia coli regulation
Escherichia coli and its close relative Salmonella enterica are the paradigms of bacterial flagellar motility; in these organisms flagellar synthesis, structure, regulation, and chemotaxis have been extensively studied. S. enterica synthesizes between 3-8 flagella per cell organized in a peritrichous arrangement (Iino, 1969; Erhardt & Hughes, 2010). Flagellar number is controlled by the master transcriptional regulator FlhDC which binds to and activates promoters that drive basal body gene expression (Fig. 6; Claret & Hughes, 2000; Claret & Hughes, 2002). Both E. coli and S. enterica swim in liquid media and both are capable of swarming motility under specific permissive conditions (Harshey & Matsuyama, 1994; Kim & Surette; 2005; Girgis et al., 2007).
E. coli biofilms manifest as aggregates at the meniscus liquid-air-surface found on partially submerged coverslips or on the sides of microtiter plate wells. The biofilm extracellular matrix is composed of EPS and proteins but the identity and role of each component differs depending on the strain background. In strain K-12, curli are required for initial biofilm attachment. Curli are proteinaceous amyloid fibers that are nucleated by the outer membrane protein CsgB and assembled by polymerization of CsgA monomers (Hammar et al., 1996; Bian et al., 1997). Following curli formation, an EPS called colanic acid is synthesized by proteins expressed from the 19 gene wca cluster and is required for biofilm maturation (Stevenson et al., 1996; Danse et al., 2000). By contrast, in strain MG1665, the products of the pgaABCD operon synthesize ß-1,6-poly-N-acetylglucosamine (PGA) that is thought to act as an adhesin during early attachment and subsequent biofilm maturation (Wang et al., 2004). The cells within an E. coli biofilm are non-motile.
Some E. coli strains require motility early in biofilm formation. By comparing multiple strains of E. coli in flow cells, motility and biofilm formation were found to be correlated such that strains with the most robust biofilm formation also displayed the most vigorous motility as planktonic cells (Wood et al., 2006). At the molecular level, mutations in flhD, which encodes the FlhD subunit of the master regulator for flagella gene transcription, and mutations in fliC, which encodes the flagella filament protein flagellin, result in a severe biofilm defect (Pratt & Kolter, 1998). Likewise, mutations that paralyzed the flagellar motor but left the structure intact also crippled biofilm formation (Pratt & Kolter, 1998; Vallet et al., 2001). Thus, it appears that motility is important, at least at the early stages, of initiating biofilm formation.
At some point in biofilm formation, it is inferred that flagella become functionally inhibited by the protein YcgR. YcgR was first shown to inhibit motility in cells mutated for the histone-like DNA binding protein H-NS and later in cells mutated for YhjH, an EAL-domain containing c-di-GMP phosphodiesterase (Ko & Park, 2000; Simm et al., 2004; Ryjenkov et al., 2006). In the absence of YhjH, c-di-GMP levels rise, YcgR binds to c-di-GMP via a PilZ-domain and inhibits motility by interacting with the flagellar basal body (Amikam & Galperin, 2006; Ryjenkov et al., 2006) (Fig. 6; Table 1). YcgR has been reported to make direct contact with components of the flagellar motor including the FliG rotor, the MotA stator protein, and the FliM protein that couples the chemotaxis machinery to the direction of flagellar rotation (Boehm et al., 2010; Fang & Gomelsky, 2010; Paul et al., 2010). Supporting these interactions, translational fusions between YcgR and a fluorescent fluorophore localize as puncta along the cell membrane in a manner dependent upon FliG, FliM, and MotA respectively (Fig. 1; Boehm et al., 2010; Paul et al., 2010). The complex interaction between YcgR and multiple pieces of the flagellum perhaps accounts for the observation that YcgR both reduces swimming speed (invoking FliG and MotA), and impairs chemotaxis by biasing flagellar rotation in favor of the counterclockwise direction (invoking FliG and FliM) (Boehm et al., 2010; Fang & Gomelsky, 2010; Paul et al., 2010). In sum, YcgR is thought to functionally inhibit motility in the presence of c-di-GMP at a time when cells are presumably preparing to form a biofilm. The role of YcgR-dependent inhibition of motility on biofilm formation has not been directly investigated.
As shown in other systems, transcriptional regulation of motility also occurs in the transition from motile to sessile cells in a biofilm. Prigent-Combaret et al. found that cells in a biofilm decrease expression of the flagella filament, fliC (Prigent-Combaret et al., 1999). One potential candidate for the inhibition of motility during biofilm forming conditions is the Rcs phosphorelay system. The Rcs system consists of a transmembrane kinase RscC, a membrane bound phosphotransfer protein RcsD, and a response regulator transcription factor RcsB (Majdalani & Gottesman, 2006). RcsB along with the accessory protein RcsA activates expression of colanic acid capsule polysaccharide synthesis genes and inhibits motility by directly repressing expression of the flhDC promoter (Gottesman et al., 1985; Francez-Charlot et al., 2003; Fredericks et al., 2006) (Fig 6; Table 1). Furthermore, mutation of Rcs components impairs biofilm formation and enhances motility (Ferrières & Clarke, 2003; Girgis et al., 2007). Thus, the Rcs signal transduction system appears to coordinate both phenotypes in the motility-to-biofilm transition at the level of gene transcription.
Inverse regulation of the flagella and EPS can also occur at the post-transcriptional level. CsrA is a small RNA-binding protein that acts as a global regulator in many bacteria by post-transcriptional regulation (Romeo, 1998) (Fig 6; Table 1). In E. coli, CsrA increases transcription of flhDC by directly binding the mRNA to stabilize the transcript (Wei et al., 2001). Conversely, CsrA decreases expression of the pgaABCD PGA polysaccharide synthesis operon by binding to the mRNA and occluding the ribosome binding site (Wang et al., 2005). Consistent with the reciprocal regulation, mutants of CsrA are enhanced for biofilm formation and impaired for motility (Wei et al., 2001; Jackson et al., 2002; Jonas et al., 2008). Biologically, CsrA may be important in biofilm dispersion since induction of CsrA in a mature biofilm causes disaggregation within hours (Jackson et al., 2002). In support of the artificial induction results, CsrA levels gradually increase during the course of biofilm development (Jackson et al., 2002). The signal for the increase in CsrA levels is unknown.
Many commonly studied laboratory strains of E. coli do not form biofilms until they acquire a gain-of-function point mutation in the gene ompR that codes for an enhanced allele of the OmpR transcriptional regulator (Vidal et al., 1998). Enhanced OmpR has increased binding to the promoter of csgD, encoding the CsgD master transcriptional activator of CsgA and curli synthesis (Hammar et al., 1995; Prigent-Combaret et al., 2001). In addition, enhanced OmpR inhibits expression of the flagellar filament over the course of biofilm formation as it binds to and directly represses expression of the promoter for flhDC (Shin & Park, 1995; Prigent-Combaret et al., 1999) (Fig. 6; Table 1). Consequently, the enhanced OmpR strains over-produce curli and are defective in flagellar motility, yet form biofilms that are motility-independent (Prigent-Combaret et al., 2000). It appears that E. coli strains that over-produce curli during biofilm formation either undergo a different developmental pathway or the excess curli override the importance of motility in biofilm formation.
Curli and flagella inhibit each other in a reciprocal fashion. Motile cells co-express the protein FliZ with other flagellar genes. FliZ is a DNA binding protein that inhibits curli production by decreasing expression of CsgD and CsgD-activators (Pesavento et al., 2008; Pesavento & Hengge, 2012). FliZ also serves as a negative feedback mechanism that inhibits FlhDC at multiple levels (Wada et al., 2011; Pesavento & Hengge, 2012). As cells transition into the biofilm forming state, c-di-GMP levels rise, activating CsgD, which in turn activates curli gene expression (Pesavento et al., 2008). How c-di-GMP activates CsgD expression is unknown, but once activated, CsgD also directly represses flagellar basal body gene operons to decrease flagellar synthesis (Ogasawara et al., 2011). Finally, a small RNA called McaS inhibits curli by repression of CsgD and activates motility by activating flhDC expression (Thomason et al., 2012). The inverse regulation of flagella genes and curli is similar to the inverse regulation of flagella and EPS in other bacteria and perhaps ensures that the cell does not waste energy on two opposing behaviors.
While we must draw on information from multiple E. coli strains and even extrapolate from S. enterica, the inhibition of motility during biofilm formation may proceed as follows. Motility is required early perhaps to locate a suitable environment or surface on which to form a biofilm. Once biofilm formation is initiated, c-di-GMP levels rise, perhaps by the inhibition of phosphodiesterases like YhjH. Elevated c-di-GMP levels activate YcgR that interacts with the flagellar rotor to decrease rotation speed and increase the frequency of direction reversals. In parallel, c-di-GMP activates CsgD, which in turn activates curli biosynthesis and inhibits flagellar gene expression. As the biofilm matures, post-transcriptional regulators like CsrA may accumulate to reactivate motility and antagonize matrix expression and thereby promote cellular dispersal for a new round of biofilm formation.
Concluding Remarks
It is important to acknowledge that the notion of the motility-to-biofilm transition is largely theoretical. Biofilm formation takes place over prolonged periods of time during which the event of a cell or cells transiting between physiological states is difficult to capture. In addition, many bacteria are non-motile, and some motile bacteria generate non-motile subpopulations that may proceed directly to biofilm formation without further motility regulation (Kearns & Losick, 2005; Cozy & Kearns, 2010). Despite these complications, the fact that some bacterial species can exist in two diametrically distinct modes of growth, either as motile individual cells or as non-motile aggregates of a biofilm, prompts the inference that the bacteria must switch physiologies at some point. The inference of a transition is supported by mutations that enhance biofilm formation and reduce motility thereby revealing the reciprocal regulation of the two phenotypes (other examples Wei & Bauer, 1999; Cano et al., 2002; Yao et al., 2004; Bahlawane et al., 2008). Thus, it has become clear that motility and biofilms are oppositely controlled but precisely when, where, and how the transition takes place during biofilm formation is unclear.
The inhibition of motility is often thought to be an essential event in biofilm formation perhaps because motility is sometimes required early, but inhibited late in mature biofilms. Furthermore, some mutants with enhanced motility are associated with decreased biofilm formation suggesting that the maintenance of motility may destabilize nascent multicellular aggregates. Empirically proving that motility inhibition is required for biofilm formation, however, is challenged by the fact that in many cases the mechanisms of motility inhibition are unknown, flagellar regulation is complex, and matrix biosynthesis gene expression is often co-regulated with motility regulons. Even in instances where motility inhibition can be genetically separated from matrix synthesis, biofilm inhibition may appear transient due to rapidly selected suppressors that abolish motility through independent pathways (Guttenplan et al., 2010). In fact, the arrival and dominance of non-motile suppressors may be the best evidence that motility inhibition is important because they suggest that, under biofilm forming conditions, the selective pressure to inhibit motility is strong (Tart et al., 2005; Tart et al., 2006; Guttenplan et al., 2010).
The concept of the motility-to-biofilm transition, regardless of biological importance, has proven powerful for the study of each individual phenotype. Understanding that the regulation of motility and biofilms is diametrically opposed has been critical in revealing the widespread importance of the secondary-messenger molecule c-di-GMP. Bacteria encode dozens of enzymes homologous to c-di-GMP synthases and hydrolases that control the cytoplasmic accumulation of c-di-GMP, which in turn promotes a biofilm-like state where motility is inhibited at multiple levels. The inputs that control c-di-GMP pools and the reason that mutation of various enzymes acting on a common pool gives rise to a spectrum of diverse phenotypes are unclear. Many output effectors of c-di-GMP remain to be discovered (Roelofs et al., 2011).
The relationship of motility to biofilms is complicated and likely invokes other regulatory systems like quorum sensing. Quorum sensing is a general term describing the process by which genes are regulated at high cell density in response to a secreted pheromone and is relevant as high cell density is achieved within a biofilm. In some systems, quorum sensing is necessary for EPS synthesis and formation of a biofilm whereas in other systems quorum sensing induces motility and biofilm dispersal (Atkinson et al., 1999; Quiñones et al., 2005; Hoang et al., 2008; Waters et al., 2008). The differences in the way that quorum sensing regulates biofilms may be due to ecology. Inducing dispersion in V. cholerae may be advantageous in water for colonizing new regions and in scavenging more nutrients (Yildiz et al., 2004; Waters et al., 2008). In P. syringae and S. meliloti, biofilms are formed in nutrient rich environments of plants, colonizing new areas to scavenge nutrients may be unnecessary (Quiñones et al., 2005; Hoang et al., 2008). Thus, the relationship of motility and biofilms may be further complicated by the density of cells being considered.
Most regulation of the flagellum has historically focused on mechanisms that govern the expression of flagellar genes to ensure the ordered assembly of the machine. Thus, regulation has essentially focused on the non-motile-to-motile transition, and biofilms introduce a new regulatory problem: the inhibition of motility. Motility inhibition by transcriptional repression is slow and may be important for the long-term stability of the biofilm. Flagella, however, are complex, stable structures integrated into the fabric of the cell envelope and abolishing de novo synthesis does not necessarily inhibit motile behavior in the short term. Thus the pre-existent flagella must either be segregated to extinction by growth or there must be a parallel level of control that alters flagellar function in the short term. The context of biofilm formation has enabled the discovery and interpretation of regulators of flagellar function.
Functional regulators of the flagellum are still relatively rare in the literature and are poorly understood (Table 1). EpsE of B. subtilis acts like a clutch likely by binding to the inside surface of FliG, bending the rotor, and disengaging it from the stator (Fig. 1; Blair et al., 2008; Guttenplan et al., 2010). YcgR of E. coli reduces rotation speed and changes basal reversal frequency likely by binding to the outside surface of FliG and deforming the rotor (Fig. 1; Boehm et al., 2010; Fang & Gomelsky, 2010; Paul et al., 2010). The YcgR homolog YpfA in B. subtilis inhibits motility by interacting with the MotA stator protein (Chen et al., 2012a). In Caulobacter crescentus, DgrA is a c-di-GMP binding protein that inhibits flagellar rotation perhaps through another regulator of flagella function, FliL (Jenal et al., 1994; Christen et al., 2007). Fumarate reductase and the F1F0 ATPase of E. coli interacts with FliG to increase the basal tumbling frequency of motile behavior, and CheY locks the flagellar rotor like a brake by interacting with FliM in Rhodobacter sphaeroides (Cohen-Ben-Lulu et al., 2008; Pilizota et al., 2009; Zarbiv et al., 2012). Finally, the mutant phenotypes of cells lacking SadC in P. aeruginosa and VpsR/VpsT in rugose V. cholerae suggest as-yet-undisovered regulators that act during biofilm formation to inhibit motility (Yildiz et al., 2001; Yildiz et al., 2004; Caiazza & O’Toole, 2004; Merritt et al., 2007). Functional regulation of the flagellum is most likely present in many more bacteria and the relationship of this regulatory strategy to biofilm formation remains to be clarified (Wolfe & Visick, 2008).
How motility is regulated during biofilm formation is under studied for a variety of reasons. Research that explores the motility-to-biofilm transition generally favors regulatory mechanisms governing biofilm matrix synthesis. Studying two complex processes simultaneously is difficult and it is typically easier to document the induction of a phenotype than its disappearance. Furthermore, flagellar biosynthesis and regulation is so extensively understood in a relatively narrow group of organisms, E. coli and S. enterica specifically, that the flagella and motile behavior of other model organisms may be taken for granted and understudied in their own right. While it is true that flagellar biosynthesis genes are highly conserved, individual bacteria encode novel elements particularly with respect to the regulators of flagellar gene expression and function. Finally, besides analogies to automobile mechanics there are few conceptual models for how the functional inhibition of motility is mediated. Inhibition is a new frontier of motility regulation and biofilms have already proven to be a powerful context for the discovery of new regulatory mechanisms. Flagella are classic models of bacterial machines and by understanding their regulation we might discover new paradigms for the regulation of other machines like secretion apparati and polymerases.
ACKNOWLEDGEMENTS
We would like to thank Urs Jenal, George O’Toole, and Fitnat Yildiz for thoughtful discussions and material support. We would like to thank all labs whose work addresses the challenging issue of the motility-to-biofilm transition. Work in the Kearns lab is supported by NIH grant GM093030.
References
- Absalon C, Van Dellen K, Watnick PI. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Path. 2011;7(8):e1002210. doi: 10.1371/journal.ppat.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler J. Chemotaxis in bacteria. Science. 1966;153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- Adler J. Chemoreceptors in bacteria. Science. 1969;166:1588–1597. doi: 10.1126/science.166.3913.1588. [DOI] [PubMed] [Google Scholar]
- Amati G, Bisicchia P, Galizzi A. 2004) DegU-P repression expression of the motility fla-che operon in Bacillus subtilis. J Bacteriol. 186:6003–6014. doi: 10.1128/JB.186.18.6003-6014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R. A transcriptional activator, FleQ, regulates mucin adhersion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect Immun. 1998;66:1000–1007. doi: 10.1128/iai.66.3.1000-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson S, Chang C-Y, Sockett RE, Cámara M, Williams P. Quorum sensing in Yersinia enterocolitica controls swimming and swarming motility. J Bacteriol. 2006;188:1451–1461. doi: 10.1128/JB.188.4.1451-1461.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi T, McCarter L, Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature. 1992;355:182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- Bahlawane C, McIntosh M, Krol E, Becker A. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol Plant-Microbe Inter. 2008;21:1498–1509. doi: 10.1094/MPMI-21-11-1498. [DOI] [PubMed] [Google Scholar]
- Bai U, Mandic-Mulec I, Smith I. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 1993;7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- Banin E, Brady KM, Greenberg EP. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl Environ Microbiol. 2006;72:2064–2069. doi: 10.1128/AEM.72.3.2064-2069.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraquet C, Murakami K, Parsek MR, Harwood CS. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Research. 2012;40:7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benach J, Swaminathan SS, Tamayo R, et al. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC, Brown DA. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- Berg HC, Anderson RA. Bacteria swim by rotating their flagellar filaments. Nature. 1973;245:380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- Berk V, Fong JC, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhan S, Tischler AD, Camilli A, Yildiz FH. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J Bacteriol. 2006;188:3600–3613. doi: 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhan S, Yildiz FH. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signaling pathway. Mol Microbiol. 2007;63:995–1007. doi: 10.1111/j.1365-2958.2006.05568.x. [DOI] [PubMed] [Google Scholar]
- Beyhan S, Bilecen K, Salama SR, Casper-Lindley C, Yildiz FH. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J Bacteriol. 2007;189:388–402. doi: 10.1128/JB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhan S, Odell LS, Yildiz FH. Identification and Characterization of Cyclic Diguanylate signaling systems controlling rugosity in Vibrio cholerae. J Bacteriol. 2008;190:7392–7405. doi: 10.1128/JB.00564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Z, Normark S. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 1997;16:5827–5836. doi: 10.1093/emboj/16.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair DF, Berg HC. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Boles BR, McCarter LL. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J Bacteriol. 2002;184:5946–5954. doi: 10.1128/JB.184.21.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau E, Fortier L-C, Malouin F, Burrus V. C-di-GMP turn-over in Clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLoS Genet. 2011;7:e1002039. doi: 10.1371/journal.pgen.1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. Pseudomonas aeruginosa uses a cyclic-di-GMP regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol. 2010;75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. P Natl Acad Sci USA. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- Burke V, Robinson JO, Richardson CJL, Bundell CS. Longitudinal studies of virulence factors of Pseudomonas aeruginosa in cystic fibrosis. Pathology. 1991;23:145–148. doi: 10.3109/00313029109060814. [DOI] [PubMed] [Google Scholar]
- Burrows LL. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol. 2012;66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- Butler SM, Camilli A. Going against the grain: chemotaxis and infection in Vibrio cholerae. Nat Rev Microbiol. 2005;3:611–620. doi: 10.1038/nrmicro1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd MS, Sadovskaya I, Vinogradov E, et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol. 2009;73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza NC, O’Toole GA. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J Bacteriol. 2004;186:4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza NC, Merritt JH, Brothers KM, O’Toole GA. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:3603–3612. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano DA, Dominguez-Bernal G, Tierrez A, Portillo FG, Casadesus J. Regulation of capsule synthesis and cell motility in Salmonella enterica by the essential gene igaA. Genetics. 2002;162:1513–1523. doi: 10.1093/genetics/162.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper-Lindley C, Yildiz FH. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J Bacteriol. 2004;186:1574–1578. doi: 10.1128/JB.186.5.1574-1578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Kolter R, Losick R. Paralogous antirepressors acting on the master regulator for biofilm formation in Bacillus subtilis. Mol Microbiol. 2009;74:876–887. doi: 10.1111/j.1365-2958.2009.06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Norman T, Kolter R, Losick R. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev. 2010;24:754–765. doi: 10.1101/gad.1915010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Norman T, Kolter R, Losick R. Evidence that metabolism and chromosome copy number control mutually exclusive cell fates in Bacillus subtilis. EMBO J. 2011;30:1402–1413. doi: 10.1038/emboj.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Stewart PS. Biofilm removal caused by chemical treatments. Water Res. 2000;34:4229–4233. [Google Scholar]
- Chen Y, Chai Y, Guo J, Losick R. Evidence for Cyclic Di-GMP-mediated signaling in Bacillus subtilis. J Bacteriol. 2012a;194(18):5080. doi: 10.1128/JB.01092-12. DOI: 10.1128/JB.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cao S, Chai Y, Clardy J, Kolter R, Guo JH, Losick R. A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol Microbiol. 2012b;85:418–430. doi: 10.1111/j.1365-2958.2012.08109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a Cyclic di-GMP specific phosphodiesterase and its allosteric control by GTP. J Biol Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- Christen M, Christen B, Allan MG, Folcher M, Jenö P, Grzesiek S, Jenal U. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. P Natl Acad Sci USA. 2007;104:4112–4117. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006;59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- Chu F, Kearns DB, McLoon A, Chai Y, Kolter R, Losick R. A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;68:1117–1127. doi: 10.1111/j.1365-2958.2008.06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret L, Hughes C. Functions of the subunits in the FlhD2C2 transcriptional master regulator of bacterial flagellum biogenesis and swarming. J Mol Biol. 2000;303:467–478. doi: 10.1006/jmbi.2000.4149. [DOI] [PubMed] [Google Scholar]
- Claret L, Hughes C. Interaction of the atypical prokaryotic transcription activator FlhD2C2 with early promoters of the flagellar gene hierarchy. J Mol Biol. 2002;321:185–189. doi: 10.1016/s0022-2836(02)00600-9. [DOI] [PubMed] [Google Scholar]
- Cohen-Ben-Lulu GN, Francis NR, Shimoni E, Noy D, Davidov Y, Prasad K, Sagi Y, Cecchini G, Johnstone RM, Eisenbach M. The bacterial flagellar switch compleix is getting more complex. EMBO J. 2008;27:1134–1144. doi: 10.1038/emboj.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa NE, Klose KE. Characterization of enhancer binding by the Vibrio cholerae flagellar regulatory protein FlrC. J Bacteriol. 2005;187:3158–3170. doi: 10.1128/JB.187.9.3158-3170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell R. Global climate and infectious disease: The Cholera paradigm. Science. 1996;274:2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- Cozy LC, Kearns DB. Gene position in long operon governs motility development in Bacillus subtilis. Mol Microbiol. 2010;76:273–285. doi: 10.1111/j.1365-2958.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozy LM, Phillips AM, Calvo RA, Bate AR, Hsueh Y, Bonneau R, Eichenberger P, Kearns DB. SlrA/SinR/SlrR inhibits motility gene expression upstream of a hypersensitive and hysteretic switch at the level of σD in Bacillus subtilis. Mol Microbiol. 2012;83:1210–1228. doi: 10.1111/j.1365-2958.2012.08003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese PN, Pratt LA, Kolter R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol. 2000;182:3593–3596. doi: 10.1128/jb.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnton NC, Turner L, Rojevsky S, Berg HC. On torque and tumbling in swimming Escherichia coli. J Bacteriol. 2007;189:1756–1764. doi: 10.1128/JB.01501-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003;50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- Davies DG, Marques CNH. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diethmaier C, Pietack N, Gunka K, Wrede C, Lehnik-Habrink M, Herzberg C, Hübner S, Stülke J. A novel factor controlling bistability in Bacillus subtilis: the YmdB protein affects flagellin expression and biofilm formation. J Bacteriol. 2011;193:5997–6007. doi: 10.1128/JB.05360-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle TB, Hawkins AC, McCarter LL. The complex flagellar torque generator of Pseudomonas aeruginosa. J Bacteriol. 2004;186:6341–6350. doi: 10.1128/JB.186.19.6341-6350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enos-Berlage JL, McCarter LL. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J Bacteriol. 2000;182:5513–5520. doi: 10.1128/jb.182.19.5513-5520.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Hughes KT. C-ring requirement in flagellar type III secretion is bypassed by FlhDC upregulation. Mol Microbiol. 2010;75:376–393. doi: 10.1111/j.1365-2958.2009.06973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque SM, Biswas K, Udden SMN, Ahmad QS, Sack DA, Nair GB, Mekalanos JJ. Transmissibility of cholera: In vivo-formed biofilms and their relationship to infectivity and persistence in the environment. P Natl Acad Sci USA. 2006;103:6350–6355. doi: 10.1073/pnas.0601277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Gomelsky M. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol Microbiol. 2010;76:1295–1305. doi: 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- Ferreira RBR, Antunes LC, Greenberg EP, McCarter LL. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J Bacteriol. 2008;190:851–860. doi: 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RBR, Chodur DM, Antunes LC, Trimble MJ, McCarter LL. Output targets and transcriptional regulation by a cyclic dimeric GMP-responsive circuit in the Vibrio parahaemolyticus Scr network. J Bacteriol. 2011;194:914–924. doi: 10.1128/JB.05807-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrières L, Clarke DJ. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. 2003;50:1665–1682. doi: 10.1046/j.1365-2958.2003.03815.x. [DOI] [PubMed] [Google Scholar]
- Flemming H-C, Wingender The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Fong JCN, Karplus K, Schoolnik GK, Yildiz FH. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J Bacteriol. 2006;188:1049–1059. doi: 10.1128/JB.188.3.1049-1059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JCN, Yildiz FH. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol. 2007;189:2319–2330. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francez-Charlot A, Laugel B, Van Gemert A, Dubarry N, Wiorowski F, Castanié-Cornet M, Gutierrex C, Cam K. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol. 2003;49:823–832. doi: 10.1046/j.1365-2958.2003.03601.x. [DOI] [PubMed] [Google Scholar]
- Fredericks CE, Shibata S, Aizawa S, Reimann SA, Wolfe AJ. Acetyl phosphate-sensitive regulation of flagellar biosynthesis and capsular biosynthesis depends on the Rcs phosphorelay. Mol Microbiol. 2006;61:734–747. doi: 10.1111/j.1365-2958.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004a;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 2004b;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel CV, Berg HC. The speed of the flagellar rotary motor of Escherichia coli varies linearly with proton motive force. P Natl Acad Sci USA. 2003;100:8748–8751. doi: 10.1073/pnas.1533395100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett ES, Perlegas D, Wozniak DJ. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma facter AlgT (AlgU) J Bacteriol. 1999;181:7401–7404. doi: 10.1128/jb.181.23.7401-7404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza AG, Harris-Haller LW, Stoebner RA, Manson MD. Motility protein interactions in the bacterial flagellar motor. P Natl Acad Sci USA. 1995;92:1970–1974. doi: 10.1073/pnas.92.6.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur NK, Cabane K, Smith I. Structure and expression of the Bacillus subtilis sin operon. J Bacteriol. 1988;170:1046–1053. doi: 10.1128/jb.170.3.1046-1053.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur NK, Oppenheim J, Smith I. The Bacillus subtilis sin gene a regulator of alternate developmental processes, codes for a DNA-binding protein. J Bacteriol. 1991;173:678–686. doi: 10.1128/jb.173.2.678-686.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgis HS, Liu Y, Ryu WS, Tavazoie S. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 2007;9:e154. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Trisler P, Torres-Cabassa A. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J Bacteriol. 1985;162:1111–1119. doi: 10.1128/jb.162.3.1111-1119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan JRW, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenplan SB, Blair KM, Kearns DB. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 2010;6(12):e1001243. doi: 10.1371/journal.pgen.1001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenplan SB, Shaw S, Kearns DB. The cell biology of peritrichous flagella in Bacillus subtilis. Mol Microbiol. 2013;87:211–229. doi: 10.1111/mmi.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güvener ZT, McCarter LL. Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus. J Bacteriol. 2003;185:5431–5441. doi: 10.1128/JB.185.18.5431-5441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious disease. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive organelles in Escherichia coli. P Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–114. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Distinct sensory pathways in Vibrio cholerae El Tor and Classical biotypes modulate cyclic dimeric GMP levels to control biofilm formation. J Bacteriol. 2009;191:169–177. doi: 10.1128/JB.01307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon MA, Lazazzera BA. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol. 2001;42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- Harshey RM, Matsuyama T. Dimophic transition in Escherichia coli and Salmonella typhimurium: surface-inducted differentiation into hyperflagellate swarmer cells. P Natl Acad Sci USA. 1994;91:8631–8635. doi: 10.1073/pnas.91.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Owen P, Nataro JP. Molecular switches - the ON and OFF of bacterial phase variation. Mol Microbiol. 1999;33:919–932. doi: 10.1046/j.1365-2958.1999.01555.x. [DOI] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C. Inhibition and dispersal of Agrobacterium tumefaciens biofilms by a small diffusible Pseudomonas aeruginosa exoproduct(s) Arch Microbiol. 2012;194:391–403. doi: 10.1007/s00203-011-0767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang HH, Gurich N, Gonzalez JE. Regulation of motility by the ExpR/Sin quorum-sensing system in Sinorhizobium meliloti. J Bacteriol. 2008;190:861–871. doi: 10.1128/JB.01310-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh Y-H, Cozy LM, Sham L-T, Calvo RA, Gutu AD, Winkler ME, Kearns DB. DegU-phosphate activates expression of the anti-sigma factor FlgM in Bacillus subtilis. Mol Microbiol. 2011;81:1092–1108. doi: 10.1111/j.1365-2958.2011.07755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- Iino T. Polarity of flagellar growth in Salmonella. J Gen Microbiol. 1969;56:227–239. doi: 10.1099/00221287-56-2-227. [DOI] [PubMed] [Google Scholar]
- Irie Y, Borley BR, O’Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2012;109:20632–20636. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irnov I, Winkler WC. A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales. Mol Microbiol. 2010;76:559–575. doi: 10.1111/j.1365-2958.2010.07131.x. [DOI] [PubMed] [Google Scholar]
- Islam MS, Drasar BS, Sack RB. The aquatic flora and fauna as reservoirs of Vibrio cholerae: a review. J Diarrhoeal Dis Res. 1994;12:87–96. [PubMed] [Google Scholar]
- Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J Bacteriol. 2002;184:290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell KF, McBride MJ. The surprisingly diverse ways that bacteria move. Nat Rev Microbiol. 2008;6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- Jenal U, White J, Shapiro L. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J Mol Biol. 1994;243:227–244. doi: 10.1006/jmbi.1994.1650. [DOI] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Simm R, Romeo T, Römling U, Melefors O. The RNA binding protein CsrA controls cyclic-di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol. 2008;70:236–257. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyot J, Dasgupta N, Ramphal R. FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J Bacteriol. 2002;184:5251–5260. doi: 10.1128/JB.184.19.5251-5260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatan E, Duncan TR, Watnick PI. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J Bacteriol. 2005;187:7434–7443. doi: 10.1128/JB.187.21.7434-7443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol. 2003;49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Rudner R, Losick R. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol Microbiol. 2004;52:357–369. doi: 10.1111/j.1365-2958.2004.03996.x. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 2005;19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Surrette MG. Prevalence of surface swarming behavior in Salmonella. J Bacteriol. 2005;187:6580–6583. doi: 10.1128/JB.187.18.6580-6583.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-K, McCarter LL. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J Bacteriol. 2000;182:3693–3704. doi: 10.1128/jb.182.13.3693-3704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, McCarter LL. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J Bacteriol. 2007;189:4094–4107. doi: 10.1128/JB.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisits MJ, Prost L, Starkey M, Parsek MR. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. App Env Micro. 2005;71:4809–4821. doi: 10.1128/AEM.71.8.4809-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella, and type IV pili mutants. Mol Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- Ko M, Park C. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol. 2000;303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- Kojima S, Blair DF. Conformational change in the stator of the bacterial flagellar motor. Biochem. 2001;40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- Kobayashi K. Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J Bacteriol. 2007a;189:4920–4931. doi: 10.1128/JB.00157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol. 2007b;66:395–409. doi: 10.1111/j.1365-2958.2007.05923.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi K. SlrR/SlrA controls the initiation of biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;69:1399–1410. doi: 10.1111/j.1365-2958.2008.06369.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Iwano M. BslA (YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol. 2012;85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- Köhler T, Curty LK, Barja F, van Delden C, Pechere J-C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol. 2000;182:5990–5996. doi: 10.1128/jb.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Yamamoto K, Kawagishi I, Homma M. The polar flagellar motor of Vibrio cholerae is driven by an Na+ motive force. J Bacteriol. 1999;181:1927–1930. doi: 10.1128/jb.181.6.1927-1930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. D-amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R, Greenberg EP. Microbial sciences: The superficial life of microbes. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MVAS, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O’Toole GA. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formatioon and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchma SL, Ballok AE, Merrit JH, Hammond JH, Lu W, Rabinowitz JD, O’Toole GA. Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: the pilY1 gene and its impact on surface-associated behaviors. J Bacteriol. 2010;192:2950–2964. doi: 10.1128/JB.01642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchma SL, Griffin EF, O’Toole GA. Minor pilins of the type IV pilus system participate in the negative regulation of swarming motility. J Bacteriol. 2012;194:5388–5403. doi: 10.1128/JB.00899-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K, Ohya Y, Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet. 1994;243:605–612. doi: 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- Landry RM, An D, Hupp JT, Singh PK, Parsek MR. Mucin-Pseudomonas aeruginosa interactions promote biofilm formation and antibiotic resistance. Mol Microbiol. 2006;59:142–151. doi: 10.1111/j.1365-2958.2005.04941.x. [DOI] [PubMed] [Google Scholar]
- Larsen SH, Reader RW, Kort EN, Tso W-W, Adler J. Change in the direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature. 1974;249:74–77. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- Lauriano CM, Ghosh C, Correa NE, Klose KE. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J Bacteriol. 2004;186:4864–4874. doi: 10.1128/JB.186.15.4864-4874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Butler SM, Camilli A. Selection for in vivo regulators of bacterial virulence. P Natl Acad Sci USA. 2001;98:6889–6894. doi: 10.1073/pnas.111581598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RJ, Brannigan JA, Offen WA, Smith I, Wilkinson AJ. An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex. J Mol Biol. 1998;283:907–912. doi: 10.1006/jmbi.1998.2163. [DOI] [PubMed] [Google Scholar]
- Li G, Tang JX. Low flagellar motor torque and high swimming efficiency of Caulobacter crescentus swarmer cells. Biophys J. 2006;91:2726–2734. doi: 10.1529/biophysj.106.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Beyhan S, Meir J, Yildiz FH. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol Microbiol. 2006;60:331–348. doi: 10.1111/j.1365-2958.2006.05106.x. [DOI] [PubMed] [Google Scholar]
- Linker A, Jones RS. A new polysaccharide resembling alginic acid isolated from Pseudomonads. J Biol Chem. 1966;241:3845–3851. [PubMed] [Google Scholar]
- Liu X, Beyhan S, Lim B, Linington RG, Yildiz FH. Identification and characterization of a phosphodiesterase that inversely regulates motility and biofilm formation in Vibrio cholerae. J Bacteriol. 2010;192:4541–4552. doi: 10.1128/JB.00209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Losick R, Kolter R. Paracrine signaling in a bacterium. Genes Dev. 2009;23:1631–1638. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Gontang EA, Kolter R. Potassium sensing histidine kinase in Bacillus subtilis. Meth Enzymol. 2010;471:229–251. doi: 10.1016/S0076-6879(10)71013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder BJ, Duyvesteyn MD, Blair DF. FliG subunit arrangement in the flagellar rotor probed by targeted cross-linking. J Bacteriol. 2005;187:5640–5647. doi: 10.1128/JB.187.16.5640-5647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzar MA, Thomassen MJ, Montie TC. Flagella and motility alterations in Pseudomonas aeruginosa strains from patients with cystic fibrosis: relationship to patient clinical condition. Infect Immun. 1985;50:577–582. doi: 10.1128/iai.50.2.577-582.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Lu H, Sprinkle A, Parsek MR, Wozniak DJ. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J Bacteriol. 2007;189:8353–8356. doi: 10.1128/JB.00620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM. Bacteria flagella rotating in bundles: a study in helical geometry. P Natl Acad Sci USA. 1977;74:221–225. doi: 10.1073/pnas.74.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM. Flagella and motility. In: Neidhart FC, editor. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2nd edition American Society of Microbiology; Washington, DC: 1996. pp. 123–145. [Google Scholar]
- Macnab RM. How bacteria assemble flagella. Ann Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- Magariyama Y, Sugiyama S, Muramoto K, Maekawa Y, Kawagishi I, Imae Y, Kudo S. Very fast flagellar rotation. Nature. 1994;371:752. doi: 10.1038/371752b0. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolate from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol. 2006;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- Márquez-Magaña LM, Chamberlin MJ. Characterization of the sigD transcriptional unit of Bacillus subtilis. J Bacteriol. 1994;176:2427–2434. doi: 10.1128/jb.176.8.2427-2434.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DW, Schurr MJ, Mudd MH, Govan JRW, Holloway BW, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. P Natl Acad Sci USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvasi M, Visscher PT, Martinez LC. Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis. FEMS Microbiol Lett. 2010;313:1–9. doi: 10.1111/j.1574-6968.2010.02085.x. [DOI] [PubMed] [Google Scholar]
- Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:4449–4456. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C, Kjelleberg S. Off the hook-how bacteria survive protozoan grazing. Trends in Microbiol. 2005;13:302–307. doi: 10.1016/j.tim.2005.05.009. [DOI] [PubMed] [Google Scholar]
- McCarter LL. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J Bacteriol. 1998;180:3166–3173. doi: 10.1128/jb.180.12.3166-3173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter LL. Dual flagellar systems enable motility under different circumstances. J Molec Microbiol Biotechnol. 2004;7:18–29. doi: 10.1159/000077866. [DOI] [PubMed] [Google Scholar]
- McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol. 2011;193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt JH, Brothers KM, Kuchma SL, O’Toole GA. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol. 2007;189:8154–8164. doi: 10.1128/JB.00585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monds RD, O’Toole GA. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends in Microbiol. 2009;17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Moorthy S, Watnick PI. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol Microbiol. 2004;52:573–587. doi: 10.1111/j.1365-2958.2004.04000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R, Kohn S, Hwang S, Hassett DJ, Sauer K. BdlA, a Chemotaxis Regulator Essential for Biofilm Dispersion in Pseudomonas aeruginosa. J Bacteriol. 2006;188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DC, Peng F, Barker JR, Klose KE. Lipidation of an FlrC-dependent protein is required for enhanced intestinal colonization by Vibrio cholerae. J Bacteriol. 2008;190:231–239. doi: 10.1128/JB.00924-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RS, Beyhan S, Saini SG, Yildiz FH, Bartlett DH. Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J Bacteriol. 2009;191:3504–3516. doi: 10.1128/JB.01240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai K, Kawata M, Tanaka T. Isolation and phosphorylation of the Bacillus subtilis degS and degU gene products. J Biol Chem. 1990;265:20000–20006. [PubMed] [Google Scholar]
- Mukherjee S, Yakhnin H, Kysela D, Sokoloski J, Babitzke P, Kearns DB. CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol Microbiol. 2011;82:447–461. doi: 10.1111/j.1365-2958.2011.07822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara H, Yamamoto K, Ishihama A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagella synthesis. J Bacteriol. 2011;193:2587–2597. doi: 10.1128/JB.01468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski A, Mehert A, Prescott A, Kiley TB, Stanley-Wall NR. YuaB functions synergistically with the exopolysaccharide and TasA amyloid fibers to allow biofilm formation by Bacillus subtilis. J Bacteriol. 2011;193:4821–4831. doi: 10.1128/JB.00223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Kaplan HB, Kolter R. Biofilm formation as microbial development. Ann Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Patrick JE, Kearns DB. Laboratory strains of Bacillus subtilis do not exhibit swarming motility. J Bacteriol. 2009;191:7129–7133. doi: 10.1128/JB.00905-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick JE, Kearns DB. Swarming motility and the control of master regulators of flagellar biosynthesis. Mol Microbiol. 2012;83:14–23. doi: 10.1111/j.1365-2958.2011.07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Gene Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “Backstop Brake” mechanism. Mol Cell. 2010;38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Gene Dev. 2008;22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento C, Hengge R. The global repressor FliZ antagonizes gene expression by σS-containing RNA polymerase due to overlapping DNA binding specificity. Nucleic Acids Res. 2012;40:4783–4793. doi: 10.1093/nar/gks055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilizota T, Brown MT, Leake MC, Yakushi T, Homma M, Ishijima A, Berry RM. A molecular brake, not a clutch, stops the Rhodobacter sphaeroides flagellar motor. P Natl Acad Sci USA. 2009;106:11582–1158. doi: 10.1073/pnas.0813164106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- Pratt JT, Tamayo R, Tischler AD, Camilli A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem. 2007;282:12860–12870. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol. 1999;181:5993–6002. doi: 10.1128/jb.181.19.5993-6002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret C, Prensier G, Thi TTL, Vidal O, Lejeune P, Dorel C. Developmental pathway for biofilm formation in curli-rpducing Escherichia coli strains: role of flagella, curli, and colanic acid. Env Microbiol. 2000;2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, Brombacher E, Bidal O, Ambert A, Lejeune P, Landini P, Dorel C. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J Bacteriol. 2001;183:7213–7223. doi: 10.1128/JB.183.24.7213-7223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty MG, Correa NE, Klose KE. The novel σ54- and σ 28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol Microbiol. 2001;39:1595–1609. doi: 10.1046/j.1365-2958.2001.02348.x. [DOI] [PubMed] [Google Scholar]
- Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R. Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J Bacteriol. 2012;194:3307–3316. doi: 10.1128/JB.00100-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones B, Dulla G, Lindow SE. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol Plant-Microbe Inter. 2005;18:682–693. doi: 10.1094/MPMI-18-0682. [DOI] [PubMed] [Google Scholar]
- Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infection in cystic fibrosis. Mol Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- Rashid MH, Kornberg Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. P Natl Acad Sci USA. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. P Natl Acad Sci USA. 2006;103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice EW, Johnson CJ, Clark RM, Fox KR, Reasoner DJ, Dunnigan ME, Panigrahi P, Johnson JA, Morris JG., Jr. Chlorine and survival of ‘rugose’ Vibrio cholerae. Lancet. 1992;340:740. doi: 10.1016/0140-6736(92)92289-r. [DOI] [PubMed] [Google Scholar]
- Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. P Natl Acad Sci USA. 2011;108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. P Natl Acad Sci USA. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D, Vlamakis H, Losick R, Kolter R. An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol Microbiol. 2011;80:1155–1168. doi: 10.1111/j.1365-2958.2011.07653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signaling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Römling U, Amikam D. Cyclic di-GMP as a second messenger. Curr Opin Microbiol. 2006;9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Lucey JF, et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. P Natl Acad Sci USA. 2006;103:6702–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryjenkov DA, Simm R, Römling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP. J Biol Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DJ, Leejeerajumnean S, Brannigan JA, Lewis RJ, Wilkinson AJ, Hoggett JG. Quaternary re-arrangement analysed by spectral enhancement: the interaction of a sporulation repressor with its antagonist. J Mol Biol. 1999;293:997–1004. doi: 10.1006/jmbi.1999.3221. [DOI] [PubMed] [Google Scholar]
- Seper A, Fengler W, Roier S, Wolinski H, Kohlwein SD, Bishop AL, Camilli A, Reidl J, Schild S. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol Microbiol. 2011;82:1015–1037. doi: 10.1111/j.1365-2958.2011.07867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh M, Kolter R, Losick R. The biocide chlorine dioxide stimulates biofilm formation in Bacillus subtilis by activation of the histidine kinase KinC. J Bacteriol. 2010;192:6352–6356. doi: 10.1128/JB.01025-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Zhu X. Biofilm formation and food safety in food industries. Trends Food Sci Tech. 2009;20:407–413. [Google Scholar]
- Shikuma NJ, Yildiz FH. Identification and characterization of OscR, a transcriptional regulator involved in osmolarity adaptation in Vibrio cholerae. J Bacteriol. 2009;191:4082–4096. doi: 10.1128/JB.01540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikuma NJ, Davis KR, Fong JNC, Yildiz FH. The transcriptional regulator, CosR, controls compatible solute biosynthesis and transport, motility, and biofilm formation in Vibrio cholerae. Mol Microbiol. 2012 doi: 10.1111/j.1462-2920.2012.02805.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Park C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol. 1995;177:4696–4702. doi: 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek M. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- Silverman M, Simon M. Flagellar rotation and the mechanism of bacterial motility. Nature. 1974;249:73–74. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Römling U. GGDEF and EAL domains inversely regulate cyclic-di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Simões M, Simões LC, Vieira MJ. A review of current and emergent biofilm control strategies. LWT-Food Sci Tech. 2010;43:573–583. [Google Scholar]
- Sjoblad RD, Emala CW, Doetsch RN. Bacterial flagellar sheaths: structures in search of a function. Cell Motil. 1983;3:93–103. doi: 10.1002/cm.970030108. [DOI] [PubMed] [Google Scholar]
- Smith DR, Chapman MR. Economical evolution: microbes reduce the synthetic cost of extracellular proteins. mBio. 2010;1:e00131–10. doi: 10.1128/mBio.00131-10. doi: 10.1128/mBio.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Wingreen NS. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol. 2011;24:1–7. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa Y, Hotta H, Homma M, Ishijima A. Torque-speed relationship of the Na+-driven flagellar motor of Vibrio alginolyticus. J Mol Biol. 2003;327:1043–1051. doi: 10.1016/s0022-2836(03)00176-1. [DOI] [PubMed] [Google Scholar]
- Sowa Y, Rowe AD, Leake MC, Yakushi T, Homma M, Ishijima A, Berry RM. Direct observation of steps in rotation of the bacterial flagellar motor. Nature. 2005;437:916–919. doi: 10.1038/nature04003. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Harris RC, Waters CM. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol. 2011;193:6331–6341. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart BJ, McCarter LL. Lateral flagellar gene system of Vibrio parahaemolyticus. J Bacteriol. 2003;185:4508–4518. doi: 10.1128/JB.185.15.4508-4518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed KA, Beyhan S, Correa N, Queen J, Liu J, Peng F, Stachell KJF, Yildiz F, Klose KE. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J Bacteriol. 2009;191:6555–6570. doi: 10.1128/JB.00949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Patimalla B, Camilli A. Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect Immun. 2010;78:3560–3569. doi: 10.1128/IAI.00048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tart AH, Wolfgang MC, Wozniak DJ. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J Bacteriol. 2005;187:7955–7962. doi: 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tart AH, Blanks MJ, Wozniak DJ. The AlgT-dependent transcriptional regulator ArmZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J Bacteriol. 2006;188:6483–6489. doi: 10.1128/JB.00636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DR, Francis NR, Xu C, DeRosier DJ. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar typhimurium. J Bacteriol. 2006;188:7039–7048. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason MK, Fontaine F, De Lay N, Storz G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol. 2012;84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toutain CM, Zegans ME, O’Toole GA. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J Bacteriol. 2005;187:771–777. doi: 10.1128/JB.187.2.771-777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble MJ, McCarter LL. Bis-(3′-5′)-cyclic dimeric GMP-linked quorum sensing controls swarming in Vibrio parahaemolyticus. P Natl Acad Sci USA. 2011;108:18079–18084. doi: 10.1073/pnas.1113790108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara K, Ogura M. Promoter selectivity of the Bacillus subtilis response regulator DegU, a positive regulator of the fla/che operon and sacB. BMC Microbiol. 2008;8:8. doi: 10.1186/1471-2180-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. The chaperone/usher pathways of Pseudomonas aeruginosa: dentification of fimbrial gene clusters (cup) and their involvement in biofilm formation. P Natl Acad Sci USA. 2001;98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhamme DT, Kiley TB, Stanley-Wall NR. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol Microbiol. 2007;65:554–568. doi: 10.1111/j.1365-2958.2007.05810.x. [DOI] [PubMed] [Google Scholar]
- Verhamme DT, Murray EJ, Stanley-Wall NR. DegU and Spo0A Jointly Control Transcription of Two Loci Required for Complex Colony Development by Bacillus subtilis. J Bacteriol. 2009;191:100–108. doi: 10.1128/JB.01236-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten N, Braeken K, Debkumari B, Fauvart M, Fransaer J, Vermant J, Michiels J. Living on a surface: swarming and biofilm formation. Trends Microbiol. 2008;16:496–506. doi: 10.1016/j.tim.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Gene Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tanabe Y, Kutsukake K. FliZ acts as a repressor of the ydiV gene, which encodes an anti-FlhD4C2 factor of the flagellar regulon in Salmonella enterica serovar typhimurium. J Bacteriol. 2011;193:5191–5198. doi: 10.1128/JB.05441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Microbiol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- Wai SN, Mizunoe Y, Takade A, Kawabata SI, Yoshida SI. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl Environ Microbiol. 1998;64:3648–3655. doi: 10.1128/aem.64.10.3648-3655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Prestion JF, III, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol. 2008;190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick PI, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick PI, Lauriano CM, Klose KE, Croal L, Kolter R. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol Microbiol. 2001;39:223–235. doi: 10.1046/j.1365-2958.2001.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Bauer WD. Tn5-Induced and spontaneous switching of Sinorhizobium meliloti to faster-swarming behavior. App Environ Microbiol. 1999;65:1228–1235. doi: 10.1128/aem.65.3.1228-1235.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei BL, Brun-Zinkernagel A, Simecka JW, Prüß BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science Brevia. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- White BP. The rugose variant of Vibrios. J Pathol. 1938;46:1–6. [Google Scholar]
- Whitely M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- Winkelman JT, Blair KM, Kearns DB. RemA (YlzA) and RemB (YaaB) regulate extracellular matrix operon expression and biofilm formation in Bacillus subtilis. J Bacteriol. 2009;191:3981–3991. doi: 10.1128/JB.00278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Berg HC. Migration of bacteria in semisolid agar. P Natl Acad Sci USA. 1989;86:6973–6977. doi: 10.1073/pnas.86.18.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Visick KL. Get the message out: cyclic di-GMP regulates multiple levels of flagellum/based motility. J Bacteriol. 2008;190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang MC, Jyot J, Goodman AL, Ramphal R, Lory S. Pseudomonas aeruginosa regulates flagellin expression as part of a global response to airway fluid from cystic fibrosis patients. P Natl Acad Sci USA. 2004;101:6664–6668. doi: 10.1073/pnas.0307553101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TK, González Barrios AF, Herzberg M, Lee J. Motility influences biofilm architecture in Escherichia coli. App Microbial Cell Physiol. 2006;72:361–367. doi: 10.1007/s00253-005-0263-8. [DOI] [PubMed] [Google Scholar]
- Wozniak DJ, Sprinkle AB, Baynham PJ. Control of Pseudomonas aeruginosa algZ expression by the alternative sigma factor AlgT. J Bacteriol. 2003a;185:7297–7300. doi: 10.1128/JB.185.24.7297-7300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak DJ, Wyckoff TJO, Starkey M, Keyser R, Azadi P, O’Toole GA, Parsek MR. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. P Natl Acad Sci USA. 2003b;100:7907–7912. doi: 10.1073/pnas.1231792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Luo L, Har KJ, Becker A, Ruberg S, Yu G, Zhu J, Cheng H. Sinorhizobium meliloti ExoR and ExoS proteins regulate both succinoglycan and flagellum production. J Bacteriol. 2004;186:6042–6049. doi: 10.1128/JB.186.18.6042-6049.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: Identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. P Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Dolganov NA, Schoolnik GK. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPSETr-associated phenotypes in Vibrio cholerae O1 El Tor. J Bacteriol. 2001;183:1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol. 2004;53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 2009;17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarbiv G, Li H, Wolf A, Cecchini G, Caplan SR, Sourjik V, Eisenbach M. Energy complexes are apparently associated with the switch-motor complex of bacterial flagella. J Mol Biol. 2012;416:192–207. doi: 10.1016/j.jmb.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role of Asp32 of MotB. J Bacteriol. 1998;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell. 2003;5:647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. P Natl Acad Sci USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]