Abstract
The functional importance of gene enhancers in regulated gene expression is well established(1–3). In addition to widespread transcription of long non-coding RNAs (ncRNA) in mammalian cells(4–6), bidirectional ncRNAs referred to as eRNAs are transcribed on enhancers(7–9). However, it has remained unclear whether these eRNAs are functional, or merely a reflection of enhancer activation. Here, we report that 17β-estradiol (E2)-bound estrogen receptor α (ERα) on enhancers causes a global increase in eRNA transcription on enhancers adjacent to E2-upregulated coding genes. These induced eRNAs, as functional transcripts, appear to exert important roles for the observed ligand-dependent induction of target coding genes, causing an increased strength of specific enhancer:promoter looping initiated by ERα binding. Cohesin, present on many ERα-regulated enhancers even prior to ligand treatment, apparently contributes to E2-dependent gene activation, at least in part, by stabilizing E2/ERα/eRNA-induced enhancer:promoter looping. Our data indicate that eRNAs are likely to exert important functions in many regulated programs of gene transcription.
ERα ChIP-seq analysis using 1 hour E2-treated (100nM) MCF7 cells revealed 31,052 ERα binding sites genome-wide, including only 902 on promoters (Fig.S1a), analogous to reported analyses(10–12) and 7,174 ERα-bound potential enhancers based on the presence of H3K4me1(13,14) and H3K27ac(15) (Fig.S1b). GRO-seq analysis of MCF7 cells in similar conditions, identified 1,309 E2-upregulated coding genes, of which 1,145 exhibited an E2/ERα-binding enhancer within 200kb from their transcription start sites (TSS), thus considered as direct estrogen target genes and hereafter referred to as UP-genes (Fig.S1c). Of these, only 112 exhibited ERα binding to their promoters (Fig.S1c), consistent with suggestions(10,11) that ERα occupancy on enhancers is a key strategy underlying E2-induced gene expression. Most E2-regulated enhancers displayed a rapid bidirectional activation of eRNAs, exemplified by the FOXC1 locus (Fig.S1e), which is about ~1.5 kb long by GRO-seq, although ~10% exhibited an apparent unidirectional eRNA transcription (Fig.1a; Fig.S1f,g)(8). These data suggest that eRNA induction in response to ERα binding is a predictive mark of enhancer activity(9). Binding of ERα did not cause clear alterations in enhancer marks on ERα-bound enhancers, such as H3K27ac (Fig.S1h).
Fig1. E2induction of eRNA in breast cancer cells.
(a) Heat map of GRO-seq showing bidirectional eRNA transcription at enhancers induced by E2. (b) UP-enhancers are closer to the UP-gene cohort [median ~52kb] in comparison to enhancers with non-upregulated eRNAs [median ~270kb]. (c) ERα binds more robustly to UP-enhancers than to the enhancers with nonupregulated eRNA. (d) Among the UP-enhancers, those within 200kb from any E2-upregulated gene TSSs exhibit higher ERα binding intensity than the cohort of UP-enhancers located farther. (e) Most of the UP-enhancers are close to E2-upregulated coding genes. (f) ERα binding intensity on UP-enhancers is higher than on 112 promoters of E2-activated genes, which itself is higher than 790 ERα-bound promoters of non-E2-upregulated genes. A log10 scale is used for panels b,c and f. p-values are given using Student's t-test.
In contrast to androgen receptor-regulated genes(9), there was often more than one ERα-bound enhancer adjacent to UP-genes, exemplified by the FOXC1 locus (Fig.S2f). Approximately 83% of enhancers with detectable GRO-seq signals adjacent to UP-genes exhibited E2-induced eRNA upregulation (Fig.1a); for the remaining 17%, the tag count was not sufficient to assign upregulation bioinformatically. E2-induction of eRNA was not observed on non-ERα-bound H3K27ac-marked enhancers (Fig.S1i). The median distance between enhancers exhibiting E2-induced eRNAs (n=1248, referred as UP-enhancers) and their closest UP-genes was ~52kb, compared with a median distance of ~270kb between enhancers exhibiting no E2 induction of eRNAs with UP-genes (Fig.1b). ChIP-seq analysis revealed that UP-enhancers exhibited significantly stronger binding of ERα than enhancers not exhibiting eRNA upregulation (Fig.1c). The proximal (<200kb) UP-enhancers constituted a majority of all UP-enhancers and exhibited a higher affinity for ERα than distal UP-enhancers (Fig.1d,e). The strength of ERα binding is much higher on UP-enhancers than on 112 ERα-bound promoters of coding genes showing E2 induction, while the remaining 790 ERα-bound promoters of genes with no E2-upregulation exhibit the weakest ERα binding (Fig.1f).
Based on GRO-seq analyses, we selected ten highly upregulated transcription units for further experimentation, each associated with adjacent UP-enhancers exhibiting ~2.5-5-fold E2-induction of eRNAs (Fig.S1j). Despite increasing evidence for crucial nuclear functions of lncRNAs(4–6), it remains an unresolved question whether eRNAs are merely a byproduct of enhancer activation or whether they might serve as key regulators of coding gene transcription(7–9). To begin to investigate the potential roles of ligand-induced eRNAs on gene activation events, both specific siRNAs(16) and locked nucleic acid antisense oligonucleotides (ASO-LNAs)(17) directed against each eRNA transcript were designed based on the peaks of eRNA exhibited by GRO-seq. To exclude off-target effects, experiments were performed with two different LNAs or siRNAs targeting each eRNA.
With a high efficiency of transfection (Fig. S2a), both siRNA and LNA-mediated knock-down of the TFF1, FOXC1 or CA12 eRNAs revealed that, for each transcription unit, the induction of both the eRNA and of the adjacent coding gene, as assessed by QPCR and GRO-seq, respectively, was significantly inhibited (Fig.2a,b,e; Fig.S2b,c). In contrast, these siRNAs/LNAs did not affect housekeeping genes tested (e.g. GAPDH, Fig.2f) or E2-regulated or non-E2-regulated transcription units more distal to the regulated enhancers (Fig.2c–d). Ligand-induced increase of ERα binding occurred even after eRNA knock-down (Fig.S2d,e). Similar eRNA requirements for coding gene induction by E2 were observed based on knock-down of eRNAs adjacent to the PGR, SIAH2, KCNK5, P2RY2, SMAD7, GREB1 and NRIP1 genes using either of two siRNAs/LNAs (Fig.S2b; Fig.3g,i). GRO-seq data were consistent with the notion that LNA against eRNA reduces the levels of eRNA transcript post-transcriptionally, but not its nascent transcription (Fig.2e, bar graph). Knock-down of an eRNA on ERα-bound distal enhancer (~222 kb from FOXC1 TSS) (FOXC1-e222) that did not exhibit E2-induced eRNA and with lower ERα binding affinity, did not affect neighboring FOXC1 gene induction (Fig.S2f), further indicating that eRNA induction potentially marks E2-regulated functional enhancers. While GRO-seq results (Fig.2e) already argue against any LNA-mediated transgene silencing, further assays, including methyl miner and enzyme digestion assay (Fig.S3a–c) indicated no evidence of altered enhancer methylation on either of the FOXC1, P2RY2 or NRIP1 enhancers. Additional supporting evidence was provided by using an LNA targeting the sense transcript of GREB1-RR, a regulatory region near GREB1 gene, exhibiting overlapping-bidirectional transcription (Fig.S3d,e), where we observed no significant change in transcript levels from antisense strand by strand-specific QPCR. We also failed to observe any significant LNA effects on levels of total histone H3, H3K9me3 or H3K27me3 silencing marks on several targeted enhancers (Fig.S3g). Together, these data suggest that siRNA/LNA-mediated knock-down of eRNAs do not elicit transgene silencing of the interrogated enhancers.
Fig2. Importance of eRNA for target gene activation.
(a,b) siRNA/LNA knock-down of eRNAs. Efficacy and effects on coding gene transcription assessed by QPCR for the TFF1, FOXC1, and CA12 eRNAs and coding transcription units. (c) QPCR analysis showing no significant change of several E2-target coding genes when FOXC1 eRNA was knocked-down using LNA. (d) Lack of effect of NRIP1, TFF1, or CA12 eRNA knock-down on expression of other coding genes located much more distal, including USP25 (520kb from NRIPe), RSPH1, (120kb from TFF1e) and APHIb (110kb from the CA12e). (e) GRO-seq data from FOXC1e LNA treated cells showing its inhibitory effect on the transcription of FOXC1 coding locus, but not on the targeted enhancer region itself. The bar graph (right) shows that the FOXC1e LNA knocked-down E2-induction of FOXC1 mRNA (tag counts over the whole gene length), but not transcription of the enhancer region. (f) A similar snapshot as in panel e showing the lack of effect from FOXC1e LNA on GAPDH transcription. Data represent mean±SEM (a,b) and mean±SD (c,d). (n=3). *p<0.05, **p<0.01.
Fig3. Ligand-induced eRNA is functionally important.
(a) Schematic diagram of the Box-b-λN tethering system on FOXC1 enhancer which is upstream of a FOXC1 native promoter-linked luciferase cassette. 5XUAS sites are fused downstream to FOXC1 enhancer. GAL4-λN fusion tethers Box-b-FOXC1 eRNA to the 5XUAS sites. Bar graph shows the effects of the FOXC1 eRNA on FOXC1 promoter-driven luciferase activity in the presence of E2 (24hrs). Blue bars: the activating function of the native full length enhancer (Bar:2) over random DNA (Bar:1) is lost when sense eRNA cassette is substituted with 5XUAS site (Bar:3). Orange bars: this loss was largely rescued upon FOXC1 eRNA tethering to sense eRNA deleted enhancer cassette (Bar 6). (b) eRNA function is sequence-specific: FOXC1e sense eRNA but not anti-sense strand (- strand) eRNA could rescue the activity of sense eRNA deleted enhancer, in the Box-b tethering assay. (c) Gel picture showing plasmid-based eRNA expression from full length enhancer but not from sense eRNA deleted enhancer construct. (d) Bar graph showing efficiency of GAL4 tethering on various pGL3b constructs. (e) 3D-DSL data for the P2RY2 locus, revealing enhanced promoter:enhancer interaction over the basal conditions after 1hr E2 treatment. For all 3D-DSL data, the y-axis exhibits the log10 intensities of interaction counts plus 1 or 0.25 for presentation purpose, and the x-axis depicts coordinates from UCSC genome browser. Interaction data are overlaid with positions of enhancer, ERα and HindIII sites on the regions interrogated. The pertinent promoter:enhancer interaction is shown in red and other interactions are shown in blue. (f) 3D-DSL data for the KCNK5 locus after 1hr E2 treatment. (g) LNA knock-down of NRIP1e eRNA effectively reduced the level of both eRNA and associated coding gene transcripts. (h) 3D-DSL data demonstrating significant reduction in promoter:enhancer interaction upon NRIP1e eRNA-specific LNA treatment. (i) GREB1e siRNA knock-down diminished the levels of eRNA and associated coding gene transcript. (j) 3D-DSL data for the GREB1 locus showing significantly reduced enhancer:promoter looping as well as other genomic interaction, after GREB1e specific siRNA treatment. Mean±SD. (n=3). *p<0.05, **p<0.01.
To independently validate that eRNAs per se are important for quantitative increases in target gene expression, we took advantage of a GAL4-Box-b tethering based reporter assay(18). For this we engineered a chimera RNA by fusing FOXC1 sense eRNA to five copies of Box-b RNA, permitting Box-b-FOXC1e RNA to be recruited by the RNA binding domain of λN protein fused with GAL4 DNA binding domain. Thus eRNA can be artificially tethered to 5XUAS sites just downstream of the FOXC1 enhancer in the reporter plasmid where Luc is under control of the native FOXC1 promoter (Fig.S8). We observed that the presence of full-length FOXC1 enhancer increased luciferase expression to ~2.5 fold when compared to random DNA in place of the enhancer (Fig.3a, blue bar). This effect was abolished when the sense eRNA sequence was deleted and substituted with 5XUAS sites, generating a non-functional “mis-sense” eRNA (Fig. 3a, blue bars, Fig.S8). Tethering of Box-b-FOXC1e chimeric RNA, but not Box-b alone, could fully rescue the loss of activity of sense-eRNA deleted enhancer (Fig.3a, orange bars), while the antisense FOXC1 eRNA (-strand) could not (Fig.3b). We confirmed the loss of plasmid-driven native FOXC1 eRNA expression from sense-eRNA deleted reporter construct and that GAL-4 tethering was not altered (Fig.3c,d). These data further support the suggestion that the sequence-specific eRNA transcript per se, rather than merely the process of enhancer transcription, is required for the actions of the eRNA on enhancer-dependent coding gene activation events. This observation is consistent with a study of roles of ncRNAs in p53-dependent gene activation(19) and regulation of the SNAI1 gene(20).
We next investigated whether enhancer:promoter looping events are induced in the E2-activation process(21), employing a strategy very analogous to 5C, the 3D-DSL(22), to study the spatial organization of genomes(23,24). We first examined two E2-regulated transcription units: P2RY2 and KCNK5. For P2RY2, E2 treatment significantly increased the specific promoter:enhancer interaction (Fig.3e), also induced a new E2-dependent interaction between enhancer and gene terminus region. Similarly, for the KCNK5 locus, E2 treatment caused clear increase in promoter:enhancer interactions, as well as loops to the terminator region of the gene and the other that might represent an enhancer-specific loop (Fig.3f). These observations indicate that a major effect of ligand is to enhance specific promoter:enhancer interactions in parallel to induction of eRNA.
We next investigated whether E2-induced enhancer:promoter interactions are affected by eRNAs. The NRIP1 locus exhibited specific enhancer:promoter and promoter:gene terminus loops, whereas treatment with LNA against its eRNA caused a marked inhibition of these interactions (Fig.3h) and E2-activation of the NRIP1 gene (Fig.3g). siRNA-mediated GREB1 eRNA knock-down also coordinately inhibited GREB1 gene induction and the two specific enhancer:promoter interactions induced by E2 and two additional non-enhancer loops (Fig.3i,j). Together these experiments indicate that estrogen causes quantitative, as well as some qualitative, alterations in the interactions between enhancers and coding gene promoters, and eRNAs are apparently of functional importance for robust enhancer:promoter interactions.
To address the possibility that eRNAs might also work in trans, we first estimated the absolute expression levels of the eRNAs, finding that most of the investigated eRNAs were present at levels of <5–15 copies/cell, although several, including TFF1e, present at ~70–95 molecules/cell (Fig.S4a,b), suggesting that these eRNAs were likely to function primarily in cis. Furthermore, we employed the Chromatin Isolation by RNA Purification (ChIRP)(25) to identify potential sites where FOXC1 eRNA localizes in the genome; despite robust detection of FOXC1 eRNA from its transcribing site establishing the efficacy of the biotin-labeled probes used (Fig.S4c), only 15 peaks could be confidently called, in none was the nearest genes an E2-regulated gene (Fig.S4c). In addition, QPCR analysis following knock-down of FOXC1 eRNA revealed no significant effects on E2 activation of NRIP1, TFF1, PGR or KCNK5 genes (Fig.2c). GRO-seq following LNA transfection against FOXC1 eRNA revealed that a large majority (>95%) of the E2-upregulated coding genes continued to exhibit clear E2-dependent upregulation. Therefore, any trans-effects of eRNAs are likely to be relatively infrequent or quantitatively small. Of course, there are inevitably indirect effects observed after knock-down of any eRNA that down-regulates a functional coding gene. However, at least for a few gene areas, there may be effects of enhancer-based long-range interactions. We identified at least one such example, between the NRIP1 and TFF1 loci, separated by ~27 Mb on Chr.21, exhibiting an E2-induced increase of co-localization by FISH (Fig.S5a–c). Surprisingly, knock-down of NRIP1e eRNA by LNA caused a clear decrease in the interactions between these two genomic loci (Fig.S5b,c), suggesting that such E2-induced co-localization was eRNA-dependent.
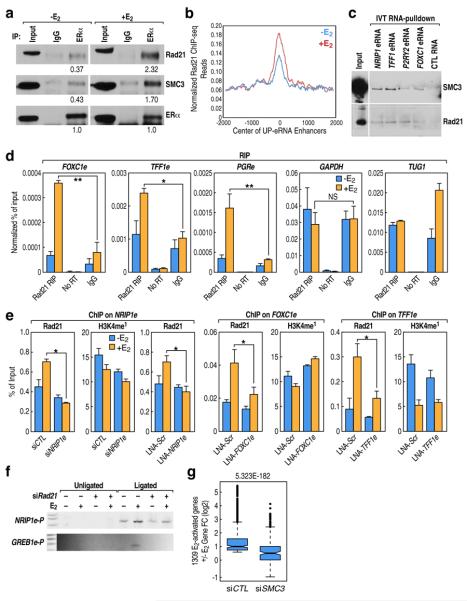
Since several studies have strongly implicated a role for Cohesin in chromosomal interactions and enhancer:promoter looping events(26–28), we investigated whether Cohesin was involved in the observed eRNA functions. First, co-immunoprecipitation showed that ERα can interact with Cohesin subunits (Fig.4a). Rad21 ChIP-seq revealed ~30–40% of its binding sites overlap with putative H3K4me1/H3K27ac-marked enhancers in MCF7 cells (Fig.S7h)(28). After E2 treatment, both ChIP-seq (Fig.4b) and ChIP-QPCR data revealed a reproducible, but modest (50–200%), increased occupancy of the Cohesin subunits Rad21 and SMC3 on the interrogated enhancers, as exemplified by FOXC1e, NRIP1e and TFF1e (Fig.4e;Fig.S6a). By in vitro transcribed (IVT) RNA pull-down, the investigated eRNAs could pull-down SMC3 and Rad21 from MCF7 nuclear extracts (Fig.4c; Fig.S6b). RIP-QPCR confirmed the interaction between Cohesin and several eRNAs, but not with GAPDH or another nuclear RNA, TUG1 (Fig.4d). To test possible direct or indirect involvement of RNAs in Cohesin recruitment to enhancers, we found that RNase treatment caused some decrease of the Cohesin level in chromatin-bound fraction of cells (Fig.S6c). Knock-down of specific eRNAs by LNA or siRNA resulted in a decrease of Cohesin recruitment (Fig.4e), to enhancers in response to E2with essentially no significant alteration of the H3K4me1 mark (Fig.4e), or in ligand-dependent increase of ERα recruitment (Fig.S2d,e). Expression levels of Cohesin subunits and ERα were not affected by knock-down of eRNAs (Fig.S7a). siRNA-mediated depletion of Rad21 caused loss of enhancer:promoter interactions, both basal and E2-induced, when assessed by 3C assay for the NRIP1 and GREB1 gene loci (Fig.4f). When we tested the role of Cohesin in the estrogen transcription program by GRO-seq, we noted that siSMC3 caused a broad inhibition of coding gene activation by E2 (Fig.4g; Fig.S7e–f), with only ~34% of E2-upregulated genes remaining induced (Fig.S7g). Similarly, Rad21 knock-down inhibited E2-induction of genes, as revealed by the five targets evaluated (Fig.S7d). We excluded alterations in levels of ERα as the cause for these dramatic effects of Cohesin depletion (Fig.S7b,c). Based on these results, we are tempted to speculate that many regulatory genomic regions, such as enhancers, harbor the Cohesin complex, that “poises” the enhancer for the stable eRNA-induced looping necessary for gene activation events, but could not exclude the possibility that the role of Cohesin could also reflect non-enhancer-based regulation.
Fig4. Role of eRNA in Cohesin-dependent gene activation.
(a) Co-immunoprecipitation of Rad21 and SMC3 with ERα from E2 or ethanol treated MCF7 whole cell extracts showing physical interaction between ERα and Cohesin subunits, which is further enhanced by E2 treatment. Numbers below blots depict the band density (Image J) relative to that of corresponding density of ERα. (b) Rad21 enrichment centered at UP-enhancers as determined by ChIP-seq, which shows moderate E2-induced increase. (c) In vitro transcribed (IVT) RNA pull-down assay showing the interaction between Cohesin subunits and eRNAs, but not a control RNA (RNA fragment of Xenopus elongation factor α). (d) RIP-QPCR showing binding of Rad21 to selected regulated eRNAs but not to GAPDH or TUG1. (e) ChIP-QPCR analyses represent the inhibitory effect from knock-down of NRIP1e (siRNA and LNA), FOXC1e or TFF1e on E2-indcued Rad21 additional recruitment, but not on H3K4me1. (f) Effect of Rad21 depletion on physical interaction between promoter:enhancer for the GREB1 and NRIP1 genes, assessed by 3C assay. (g) 1,309 E2-induced coding genes were defined by GRO-seq from siCTL (±E2) group and then their fold change (log2 FC) in siCTL versus siSMC3 transfected MCF7 cells was plotted). Mean ± SD. (n=3). *p<0.05, **p<0.01.
Conclusions
Despite the discovery of enhancers more than 35 years ago(1,2), full understanding of the mechanisms by which they regulate gene expression has been a relatively resistant problem. Here, we have provided several lines of evidence that induced eRNA transcripts are functionally important for the actions of estrogen-regulated gene enhancers, at least in part by contributing to the dynamic generation/stabilization of enhancer:promoter looping between the regulated coding transcription units and these ERα-bound enhancers.
Supplementary Full Methods
Antibodies
The antibodies for this study were: anti-ERα (HC-20, Santa Cruz); anti-H3K4me3 (07-473, Santa Cruz); anti-H3K4me1 (ab8899, Abcam); anti-H3K27ac (ab4729, Active motif); anti-Rad21 (ab992, Abcam); anti-SMC3 (ab9263, Abcam); anti-α-tubulin (T5168, Sigma), anti-Gal4 (DBD)(06-262, Millipore) and anti-IgG (I5006, Sigma).
Cell culture
MCF7 obtained from ATCC were cultured in DMEM media supplemented with 10% FBS in a 5% CO2 humidified incubator. They were hormone-stripped for 3 days in phenol-free media with charcoal-stripped FBS before receiving 100nM 17β-estradiol (E2) (Sigma) or ethanol treatment for 1 hour for estrogen signaling induction. MCF10A cells were a kind gift from Dr. Park (The Johns Hopkins University School of Medicine, Baltimore) and they were essentially grown as described in(31). For E2 induction, the culture media was stripped of EGF.
siRNA & Locked Nucleic Acid (LNA) Transfections
LNAs were obtained from Exiqon (Woburn, MA); siRNAs were from Bioneer and Sigma-Aldrich® (Tables 2 and 3). For transfection of both siRNA/LNAs, cells were first hormone-stripped for one day followed by siRNA/LNA (both at 40nM) transfection using Lipofectamine 2000. After 2 days they were then treated with ethanol or E2 for 1hr. For some experiments, transfection were performed twice to achieve higher efficiency. Similarly, LNA transfections were performed 2 days after starvation in stripped media and thus the LNA treatment lasted 24hrs in some experiments.
RT-QPCR
RNA was isolated using Trizol (Invitrogen) or RNeasy column (Qiagen), and total RNA was reverse-transcribed using SuperScript® III Reverse Transcriptase (Invitrogen). Quantitative PCRs were performed mostly with StepOne Plus (Applied Biosystem). For normalization, ΔCt values were calculated relative to the levels of ACTB/GAPDH transcripts.. The results were repeated for at least 3 times and one representative plot is shown in figures; most p-values were obtained using a two-tailed Student's t-test. Primers are listed in Table 4.
ChIP-seq
ChIP was performed as previously described(9). Briefly, approximately 107 cells were cross-linked with 1% formaldehyde at room temperature for 10 min and neutralized with 0.125M glycine. After sonication, ~75μg soluble chromatin was incubated with 1–5μg of antibody at 4°C overnight. Immunoprecipitated complexes were collected using Dynabeads A/G (Invitrogen). Subsequently, immuno-complexes were washed, DNA extracted and purified by QIAquick Spin columns (Qiagen). For ChIP-seq, the extracted DNA was ligated to specific adaptors followed by deep sequencing with the Illumina's HiSeq 2000 system according to the manufacturer's instructions. Usually, the first 48bp for each sequence tag returned by the Illumina Pipeline was aligned to the hg18 assembly using BFAST or Bowtie2. Only uniquely mapped tags were selected for further analysis. The data was visualized by preparing custom tracks on the University of California, Santa Cruz, (UCSC) genome browser using HOMER(32) (http://biowhat.ucsd.edu/homer). The total number of mapable reads were normalized to 107 for each experiment presented in this study.
Identification of ChIP-seq Peaks
The ChIP-seq peaks were identified by HOMER. Given different binding patterns of transcription factors and histones, parameters were optimized for the narrow tag distribution characteristic of transcription factors by searching for high read enrichment regions within a 200bp sliding window. Regions of maximal density exceeding a given threshold were called as peaks, and adjacent peaks were set to be >500bp away to avoid redundant detection. The common artifacts from clonal amplification were circumvented by considering only one tag from each unique genomic position. The threshold was set at a false discovery rate (FDR) of 0.001 determined by peak finding using randomized tag positions in a genome with an effective size of 2 × 109 bp. For ChIP-seq of histone marks, seed regions were initially found using a peak size of 500bp (FDR<0.001) to identify enriched loci. Enriched regions separated by <1kb were merged and considered as blocks of variable lengths. All called peaks were then associated with genes by cross-referencing with the RefSeq TSS database. Peaks from individual experiments were considered overlapping if their peak centers were located within 200bp (for some analysis may extend to 1kb). The peaks within ±1kb apart from RefSeq gene TSS site were considered as promoter-bound
GRO-seq
GRO-seq experiments were performed as previously reported(9,29,33). Briefly, MCF7 cells were swelled in swelling buffer (10mM Tris-Cl pH7.5, 2mM MgCl2, 3mM CaCl2) for 5 min on ice and then lysed in lysis buffer (swelling buffer with 0.5% IGEPAL and 10% glycerol). before being finally re-suspended in 100μl of freezing buffer (50mM Tris-Cl pH8.3, 40% glycerol, 5mM MgCl2, 0.1mM EDTA). For the run-on assay, re-suspended nuclei were mixed with an equal volume of reaction buffer (10mM Tris-Cl pH 8.0, 5mM MgCl2, 1mM DTT, 300mM KCl, 20 units of Superase.In, 1% sarkosyl, 500μM ATP, GTP, and Br-UTP, 2μM CTP) and incubated for 5 min at 30°C. The nuclear-run-on RNA (NRO-RNA) was then extracted with TRIzol LS reagent (Invitogen) following manufacturer's instructions. After base hydrolysis on ice for 40min and followed by treatment with DNase I and antarctic phosphatase, the Br-UTP labeled NRO-RNA was purified by an anti-BrdU argarose beads (Santa Cruz Biotech) in binding buffer (0.5×SSPE, 1mM EDTA, 0.05% tween) for 3hr at 4°C while rotating. Then T4 PNK (NEB) was used to repair the end of NRO-RNA. Subsequently, cDNA synthesis was performed as reported(9,33) with few modifications. The RNA fragments were subjected to poly-A tailing reaction by poly-A polymerase (NEB) for 30 min at 37°C. Reverse transcription was then performed using superscript III (Invitrogen) with oNTI223 primer (for sequence see Table 5). The cDNA products were separated on a 10% polyacrylamide TBE-urea gel with right product (~100–500bp) being excised and recovered by gel extraction. After that, the first-strand cDNA was circularized by CircLigase (Epicentre) and re-linearized by Ape1 (NEB). Re-linearized single strand cDNA were separated by TBE gel and the products of desired size was excised (~120–320bp) for gel extraction. Finally, cDNA template was amplified by PCR using the Phusion High-Fidelity enzyme (NEB) with primers oNTI200 and oNTI201 for deep sequencing (primers listed in Table 5).
Computational analysis of GRO-seq
The sequencing reads were aligned to hg18 using Bowtie2. For analyzing estrogen effects on gene transcriptin, we counted the reads from the first 30kb (assuming a RNA polymerase speed of ~0.5 kb/min during 1hr E2 treatment) of entire gene body, excluding the promoter-proximal region on the sense strand with respect to the gene orientation by using BED Tools or HOMER. EdgeR (http://www.bioconductor.org/) was used to compute the significance of the differential gene expression (FC≥1.5, FDR≤0.01). Additionally, a read density threshold (i.e. GRO-seq normalized read counts/kb) was used in order to exclude lowly expressed genes.
De novo identification of GRO-seq transcripts
GRO-seq read densities were analyzed in a similar manner to ChIP-seq. Provided GRO-seq generates strand-specific data, separate tracks were uploaded onto the UCSC genome browser; tag-enriched sites were identified using a sliding window of 250bp. Transcript initiation sites were identified as regions where the GRO-seq read density increased threefold relative to the preceding 1kb region. Transcript termination sites were defined by either a reduction in reads below 10% as compare to that of TSS or when another transcript's start was identified on the same strand. Individual high-density peaks spanning a region less than 250bp were considered artifacts and removed from the analysis. Transcripts were defined as putative eRNAs if their de novo called start sites was located distal to RefSeq TSS (≥3kb) and were associated with ERα and H3K27ac co-bound regions.
Bioinformatics characterization of ERα enhancers
The ERα-H3K27ac co-bound regions are defined as that the distance from the center of an ERα peak to the H3K27ac peak-occupied region is ≤1kb. Overall, two methods were used to assign the ERα bound enhancers to E2 upregulated genes: 1) identifying the E2-upregulated coding genes from GRO-seq first and coupling each of them to their closest ERα-H3K27ac co-bound enhancer within certain distance (200kb) (a “gene-centric” view); 2) characterizing the ERα-H3K27ac co-bound enhancers first and then coupling each of them to their closest TSS that belongs to 1,309 E2 upregulated coding genes (an “enhancer-centric” view). The comparison of ChIP-seq tag intensity, GRO-seq transcription levels or distances between different categories (Fig.1,4&S1) are presented as boxplots by using either log or normal scales. The p-values were determined by two-tailed Student's t-test.
ChIRP-seq
The ChIRP experiment was performed essentially following the original protocol (25), except for a few modifications. First, we designed antisense DNA probes targeting FOXC1 eRNA (“odd” and “even”) (~40mer) based on high oligo specificity (using the BLAST & BLAT), moderate GC content (40–60%) and a Tm around 65°C, with probes for LacZ RNA as control (all probes listed in Table 8). All DNA probes were biotinylated and purified using the Label IT® Nucleic Acid Labeling Kit (Mirus Bio.). The sequencing reads were aligned to hg18 by Bowtie2 and the peaks were called by HOMER with 3 criteria: a) they are consistently called in both the “odd” and “even” ChIRP-seqs; b) they do not intersect the peaks in lacZ sample; c) they do not intersect the satellite repeats or retrotransposons sequences. The remaining ChIRP peaks were divided into 2 categories: 1) highly confident peaks (peak score > 8), and 2) weak peaks (peak score ≤ 8). The peaks were extended with 1kb for intersection analysis by using BedTools, and the peak annotation was carried out in HOMER.
RNA copy number quantification
To quantify each transcript, the PCR product using the QPCR primers for this transcript was purified and the concentration was measured. The absolute copy numbers of the PCR product were calculated by the following. For example: for GAPDH, the number of ssDNA molecules from 1μl of 17ng/μl PCR product of GAPDH fragment (142bp) with 87,788.56Da molecular weight is about 2*(17*10−9 × 6.023*1023)/87,788.56 = 2.32*1011. The number of ssDNA molecules from 1μl of 16ng/μl PCR product of TFF1e fragment (82bp) with 50,696.92Da molecular weight is about 2*(16*10−9 × 6.023*1023)/50,696.92 = 3.8*1011. Using these PCR products with known molecule copy numbers, standard curves can be generated by QPCR, which will be the basis to quantify the copies of eRNAs from cDNA samples. For cDNA samples, 3μg of total RNA (~according to QIAgen manual, is ~2×105 cells) were converted into 20μl cDNA. During multiple times of QPCR from cells of different batches, the cycle number of target eRNA being amplified will vary within 2–3 cycles(~4–8 folds). The copy number of GAPDH mRNA is largely consistent with previous reports(34). Considering the efficiency of reverse transcriptase on GAPDH mRNA is estimated to be ~50%(35), which might be even lower for eRNAs, the real numbers of eRNA copies could be higher than the estimation.
Immunoprecipitation
Cells were collected with cold PBS and lysed with RIPA buffer (50mM Tris pH7.4, 150mM NaCl, 1mM EDTA, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, 0.5mM DTT, protease inhibitor). The lysate was diluted 2–4 times with dilution buffer (50mM Tris pH 7.4, 100mM NaCl, 1mM EDTA, 0.1% NP-40 and 10% glycerol, protease inhibitor). 2–5μg of antibodies were added into the diluted cell lysate and incubated overnight at 4°C. The next day, the protein complexes were collected by magnetic Dynabeads G for 2hr at 4°C with rotation. The beads-antibody-protein complexes were then washed 4 times with wash buffer (50mM Tris pH 7.4, 125mM NaCl, 1mM EDTA and 0.1% NP-40) and boiled for Western blots analysis.
RIP and in vitro transcribed (IVT) RNA pull down
RIP experiment was done largely following a previous protocol(36). Briefly, cells were crosslinked with 0.3% formaldehyde for 10min at 37°C. 2.5M glycine was added (1/20 of the medium volume) to neurtralize for 10min at room temperature. The cell pellet was re-suspended in 0.6ml of RIPA buffer (50mM Tris pH7.4, 150mM NaCl, 1mM EDTA, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, 0.5mM DTT, protease inhibitor and Superase-In 40u/ml), sonicated once and incubated on ice with frequent vortex for 25mins. Subsequently, the supernatant was diluted with RIP dilution buffer (50mM Tris pH 7.4, 150mM NaCl, 1mM MgCl2, 0.05% NP40) and pre-cleared with ~25ul protein A sepharose slurry for 30mins at 4°C. Antibodies were added and incubated overnight at 4°C with rotation. The next day, collect the RNA-protein complex by pre-washed ~60μl protein A sepharose beads for 1.5–2.5hr at 4°C. After wash in RIPA buffer and RIPA-500 buffer (RIPA with higher salt - 500mM NaCl), then the beads were re-suspended in 150ul of RIPA buffer with proteinase K at 45°C for 45min. RNA were extracted with TRIzol followed by DNaseI digestion. Reverse transcritpion was performed with SuperScript® III RT kit (Invitrogen). For RIP-QPCRs, the amount of pull-downed RNAs was calculated as the percentage of input GAPDH RNA of its respective group. The results were repeated for at least two times but was presented as a representative plot. p-values were obtained using two-tailed Student's t-test.
For IVT RNA pull-down, first, plasmids carrying DNA sequences of investigated eRNA were linearized and in vitro transcribed using MEGAtranscript kit (Ambion) with 25% of UTP being replaced by biotinylated UTP (Ambion). About 10μg of biotinylated RNA was heated to 90°C for 3 minutes, put on ice for 2 min and added into RNA structure buffer (10mM Tris pH7.2, 0.1M KCl, 10mM MgCl2, 1μl Superase.In) for 20min to form structure. The biotinylated RNA was then mixed with pre-washed Streptoavidin magnetic beads and incubated at room temperature for 30min to conjugate RNA with beads, following the manufacturer's protocol. After that, nuclear extract ~10mg in RIP buffer (20mM Tris pH7.4, 1mM EDTA, 150mM NaCl, 0.5mM DTT, 0.1% NP-40, 5% glycerol, protease inhibitor and Superase.In 40u/ml) was then mixed with biotinylated RNA and incubated at 4°C for 4hrs. Following four times of wash with high salt buffer (20mM Tris pH7.4, 500mM NaCl, 0.05% TX-100, 10u/ml Superase.In), the beads were boiled for Western blots.
DNA enzyme digestion methylation assay
The protocol largely follows a previous paper(17). Briefly, genomic DNA was extracted from MCF7 cells (AccuPrep, Bioneer) 24hrs after transfection with LNA/siRNA against eRNAs, and digested with HpaII (NRIP1e) or HhaI (FOXC1e), both from NEB, prior to QPCR amplification using primers (Table 4) that spanning the enzyme digestion sites (Fig.S3a). The relative resistance to restriction digestion was calculated by dividing the amount of DNA remains after digestion by the amount before digestion.
Methyl miner assay
MCF7 Cells were transfected with LNA or siRNA for 24 hrs, after which they were subjected to DNA isolation using QIAgen DNA isolation column. 1μg of each DNA was used for Biotin tagged-MBD peptide pull down as per manufacturer protocol (Invitrogen), after which unmethylated and methylated DNA fractions were collected, purified and subjected to QPCR analysis using primers specified (Table 4).
FISH and imaging
The cells were processed for DNA Immuno-FISH essentially as described in(29), with BAC probes from Empire Genomics (Table 1). MCF7 cells were. Ethanol or E2 treated cells, grown onto acid-washed poly-lysine coated coverslips, were washed with 1XPBS and immediately fixed with freshly made 4% paraformaldehyde/PBS for 10min. Permeabilization was achieved by incubating in PBS containing 0.5% Triton X-100 for 15min. FISH pre-hybridization treatments include incubating the cover-slips in 0.1N HCl for 5min at room temperature, followed by digestion with 0.01N HCl/ 0.002% pepsin for 5min at 37°C, stopped by 50mM MgCl2/PBS and equilibrated in 50% formamide/2XSSC 2hrs prior to hybridization. 5μl of probe/hybridization buffer mix was used per coverslip, with a hybridization program of 76°C for 3mins followed by overnight hybridization at 37°C in a humidified dark chamber. The coverslips were then washed with pre-warmed WS1 (0.4xSSC/0.3% NP-40), WS2 (2xSSC/0.1% NP-40) and PBS before being finally mounted with prolong gold-DAPI anti-fade mounting reagent (Invitrogen).
For FISH Image acquisition and data analysis(37), images were acquired using the Leica SP5 II confocal microscopy (63X objective lens) with a resonance scanner. Z-stack data acquisition was set up across 3.2 μm thickness at 0.4 μm each step (9 steps for each 3D image set). The 3D images were then generated in Volocity (v6.0.1). The FISH-positive gene loci were identified using the “Find Object Using % Intensity” (generally >20%) function in combination with “Exclude Objects by Size” (generally >0.1 μm3). The overlap between two FISH-positive gene loci was calculated by function “Intersect” with size exclusion (> 0.03 μm3). The cells counted (n>100 for each group, Fig.S5) were from 8 images/fields; the percentage of overlapping events from each one was calculated separately, which together generates the mean and SD.
Box-b-λN tethering assay
Similar to the previous method(18), as described in Fig.3a and Fig.S8, the Box-b system utilizes viral RNA-protein interaction where Box-b is a viral RNA that can be recognized and bound by viral anti-terminator protein λN. Fusion of FOXC1 eRNA with Box-b enables the fused Box-b-FOXC1e to be bound by λN. Subsequently λN protein was fused to GAL4-DBD, which then recognizes 5XUAS sites on the reporter plasmid DNA. Using this technique Box-b-eRNA can be tethered to the 5XUAS sites on a reporter plasmid with the help of GAL4-DBD-λN fusion protein(18). Full length FOXC1 eRNA was cloned in pCDNA3.1 downstream to 5 copies of Box-b. This construct was co-transfected along with the reporter plasmids and λN-GAL4-DBD vector (Fig.S8), which is also based on a pCDNA3.1 with CMV promoter.
FOXC1 promoter was cloned in KpnI and BglII sites in pGL3-basic vector, 5XUAS sites were cloned at upstream SalI site in pGL3-basic vector, FOXC1 full-length enhancer (1.2 kb) was placed just upstream to 5XUAS sites at BamHI site. For deletion of sense eRNA, enhancer region was amplified including full antisense transcript, core region and 20 nucleotide from sense eRNA, thus called FOXC1 enh-sense del enhancer, was also cloned at BamHI site upstream to 5XUAS site (Fig.S8).
Luciferase reporter assay
Tethering plasmids alone or in combinations were transfected along with Renilla-TK plasmid into 3 days stripped MCF7 cells. Six hours post-transfections, they were treated with 10nM E2 for 24 hrs further; they were subjected to luciferase assay using Dual-Lucifersae reporter assay kit (Promega); plates were read in Veritas Microplate Luminometer (Turner Biosystems).
Chromatin Conformation Capture (3C)
3C was performed as per ref(22,24). Briefly 25×106 MCF7 cells were fixed by adding 1% formaldehyde at room temperature for 10 minutes, and the reaction was stopped by glycine. Lysis buffer (500μl 10mM Tris-HCl pH8.0, 10mM NaCl, 0.2% Igepal CA630; protease inhibitors was added and cells were incubated on ice. Next, cells were lysed with a Dounce homogenizer, and the suspension was spun down at 5,000 rpm at 4°C. The supernatant was discarded and the pellet was washed twice with 500μl ice-cold 1x NEBuffer 2 (NEB, Ipswich, MA). The pellet was then re-suspended in 1X NEBuffer 2 and split into five separate 50μl aliquots. The extracted chromatin was then digested overnight by 400 Units HindIII (NEB). Each digested chromatin mixture was ligated by T4 DNA Ligase (800U) in 20 times of initial volume for 4hrs at 16°C. The ligase step was omitted in one chromatin aliquot from the five mentioned above as the unligated control. The chromatin was subsequently decrosslinked overnight at 65°C and purified twice with phenol and then with phenol:chloroform:IAA (25:24:1). DNA was precipitated and pellets were air-dried before re-suspending in 250μl 1XTE buffer. To degrade any carryover RNA, 1μl RNAse A (1 mg/ml) was added to each tube and incubated at 37°C for 15 minutes. DNA was further purified using Phenol:Chloroform:IAA and precipitated. The digestion and ligation efficiencies were checked and normalized before 3D-DSL.
Probe design for 3C
Donor and acceptor probes were designed on HindIII sites covering the enhancers and the gene body of the following genes: GREB1, NRIP1, KCNK5 and P2RY2, by using custom Perl scripts (available upon request). The chosen regions for probe design covered the most prominent ERα binding sites as well as enhancers. The uniqueness of the probe sequences was verified by Bowtie alignment to the human genome hg18 assembly. Universal adaptor sequences that are compatible with HiSeq 2000 flow cell design were added to the probe ends for bridge amplification of the ligation products and for direct sequencing. Acceptors were phosphorylated and both acceptors and donors were pooled individually in equimolar amounts for 3D-DSL (Table 6).
3D-DSL
The DSL ligation products were prepared as described in(12,22). 3D-DSL was performed as described in(22). Briefly, after 3C efficiency estimation, equal amount of 3C chromatin was biotinylated using the Photoprobe Kit (Vector Lab). Donor and acceptor probe pools (20fmol per probe) were annealed to the biotinylated 3C samples at 45°C for 2 hours followed by 10min at 95°C. The biotinylated DNA was immunoprecipitated with magnetic beads conjugated to streptavidin, and during this process unbound oligonucleotides were removed by stringent washes. The 5′-phosphate of acceptor probes and 3′-OH of donor probes were ligated using Taq DNA ligase at 45°C for 1hr. These ligated products were washed and eluted from beads and then amplified by PCR using primers A and B-AD (or Primer B-BC1 and -BC2 if bar coding was used) for deep sequencing on the Illumina HiSeq 2000 using Primer A as sequencing primer.
3D-DSL data analysis
After removing the adaptor sequences, the reads are aligned to a custom library that includes all the combinations donors-acceptors. The alignment was performed with Novoalign, and the reads were counted for every possible interaction by using custom Perl scripts (available upon request). The reads that were generated by donor-acceptor ligations on the same restriction site are removed: the remaining number of reads included both intra- and inter-chromosomal interactions. We used the median value (~6 millions) of the all the samples from the same sequencing run for normalization; the reads accounting for ligation products in unligated controls were subtracted. In addition to standard tools such as my5C(38) and HiTC(39) we used an intensity based method to characterize the set of interactions a related method was also in 3C-seq procedures(40). A p-value is assigned to an interaction based on Poisson probability distribution function, p(x) = e−λ * λx/x!, where p(x) is the probability of an interaction, x is the interaction intensity, and λ is the average interaction intensity considering all the potential interactions in the library, i.e. the ratio (total number of usable reads)/(total number of all possible interactions given the set of acceptors and the set of donors). The p-values were corrected for multiple testing by using Bonferroni correction method. In addition, for each interaction, we define supplementary parameters, such as 1) fold enrichment over Poisson's λ, and 2) a fold enrichment over background (where the background represents the average intensity of the ligations between the probes on the neighboring restriction sites). We consider significant for downstream analyses the interactions that meet the following criteria: a) a (corrected) p-value < 0.01, and b) a fold enrichment over background > 2, although for display purposes, in the figure plots, we may also show the weak interactions. To generate 3D-DSL plots, Matlab was used; a 10-kb window was used to bundle the interactions, except for a 20-kb window for NRIP1. The interactions were plotted using a Bezier curve between the two positions with the third point in the middle of the positions with the y-axis corresponding to the log 10 intensity. For example, if the two x-axis positions are 1 and 2, and the intensity is 4, a Bezier curve is drawn between (1,0),(1.5,4), and (2,0). The peak locations were then added on the bottom of the plot as stated in the legend. Interactions at distances generally < 10kb were not plotted for the NRIP1 locus.
Supplementary Material
Acknowledgements
We thank Dr. Kasey Hutt for help with statistical analyses; Dr. Majid Ghassemian from the UCSD Biomolecular/Proteomics Mass Spectrometry Facility for assistance in mass-spectrometry. C. Nelson for cell culture assistance; J. Hightower for assistance with figure and manuscript preparation; We thank Dr. Howard Chang for generously providing the Box-b, λN-GAL4 constructs. We acknowledge the UCSD Cancer Center Specialized Support Grant P30 CA23100 for confocal microscopy. W.L. (BC110381) and D.N. (BC103858) are DoD Postdoctoral Fellows. M.G.R. is an Investigator with the Howard Hughes Medical Institute. This work was supported by grants DK 039949, DK18477, NS034934, HL065445, CA173903 to M.G.R. and from DoD to M.G.R.
Footnotes
Author contributions: M.G.R., W.L., D.N., E.N. and C.K.G. conceived the project. W.L. and D.N. performed most of the experiments reported with important contributions from E.N. and A.Y.C. Q.M. B.T., and D.M. performed the bioinformatic analyses. Additional experiments/methods were contributed by X.S., S.O. and H-S.K. J.Z. and K.O. assisted in deep-seq library preparations and sequencing. W.L., D.N. and M.G.R. wrote the final paper with input from C.K.G.
The authors declare no conflict of interests.
Author information statement: The sequencing datasets are deposited in Gene Expression Omnibus database (GSE45822).
References
- 1.Newman JJ, Young RA. Connecting transcriptional control to chromosome structure and human disease. Cold Spring Harb Symp Quant Biol. 2010;75:227–35. doi: 10.1101/sqb.2010.75.016. Review. [DOI] [PubMed] [Google Scholar]
- 2.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144(3):327–39. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12(4):283–93. doi: 10.1038/nrg2957. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–46. doi: 10.1038/nature10887. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145(4):622–34. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474(7351):390–4. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28(10):1418–28. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–97. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 12.Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, Benner C, Wang D, Ye Z, Bibikova M, Fan JB, Duan L, Glass CK, Rosenfeld MG, Fu XD. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci U S A. 2007;104(12):4852–7. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19(6):541–9. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahlenstiel CL, Lim HG, Cooper DA, Ishida T, Kelleher AD, Suzuki K. Direct evidence of nuclear Argonaute distribution during transcriptional silencing links the actin cytoskeleton to nuclear RNAi machinery in human cells. Nucleic Acids Res. 2012;40(4):1579–95. doi: 10.1093/nar/gkr891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic Transcripts Regulate the Epigenetic State of rRNA Genes. Mol Cell. 2006;5(3):351–61. doi: 10.1016/j.molcel.2006.03.028. 22. [DOI] [PubMed] [Google Scholar]
- 18.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melo CA, et al. eRNAs Are Required for p53-Dependent Enhancer Activity and Gene Transcription. Mol Cell. 2013;49(3):524–35. doi: 10.1016/j.molcel.2012.11.021. 7. [DOI] [PubMed] [Google Scholar]
- 20.Lai F, Orom UA, Matteo Cesaroni O, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494(7438):497–501. doi: 10.1038/nature11884. 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fullwood MJ, Liu MH, Pan YF, Liu J, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462(7269):58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu X-D, Topol EJ, Rosenfeld MG, Frazer KA. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470(7333):264–8. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic Maps of LongNoncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460(7253):410–3. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20(5):578–88. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai S, Kohwi-Shigematsu T. Intranuclear relocalization of matrix binding sites during T cell activation detected by amplified fluorescence in situ hybridization. Methods. 1999;19(3):394–402. doi: 10.1006/meth.1999.0875. [DOI] [PubMed] [Google Scholar]
- 30.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abukhdeir AM, et al. Physiologic estrogen receptor alpha signaling in non-tumorigenic human mammary epithelial cells. Breast Cancer Res Treat. 2006;99:23–33. doi: 10.1007/s10549-006-9177-0. [DOI] [PubMed] [Google Scholar]
- 32.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingolia NT, et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White AK, et al. High-throughput microfluidic single-cell RT-qPCR. Proc Natl Acad Sci U S A. 2011;108:13999–4004. doi: 10.1073/pnas.1019446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong JF, et al. A microfluidic processor for gene expression profiling of single human embryonic stem cells. Lab Chip. 2008;8:68–74. doi: 10.1039/b712116d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai MC, et al. Long non-coding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rueden CT, et al. Visualization approaches for multidimensional biological image data. Biotechniques. 2007;43:31, 33–6. doi: 10.2144/000112511. [DOI] [PubMed] [Google Scholar]
- 38.Lajoie BR, et al. My5C: web tools for chromosome conformation capture studies. Nat Methods. 2009;6(10):690–1. doi: 10.1038/nmeth1009-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Servant N, et al. HiTC: exploration of high-throughput `C' experiments. Bioinformatics. 2012;28(21):2843–4. doi: 10.1093/bioinformatics/bts521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stadhouders R, et al. Dynamic long-range chromatin interactions control Myb proto-oncogene transcription during erythroid development. EMBO J. 2011;31:986–99. doi: 10.1038/emboj.2011.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.