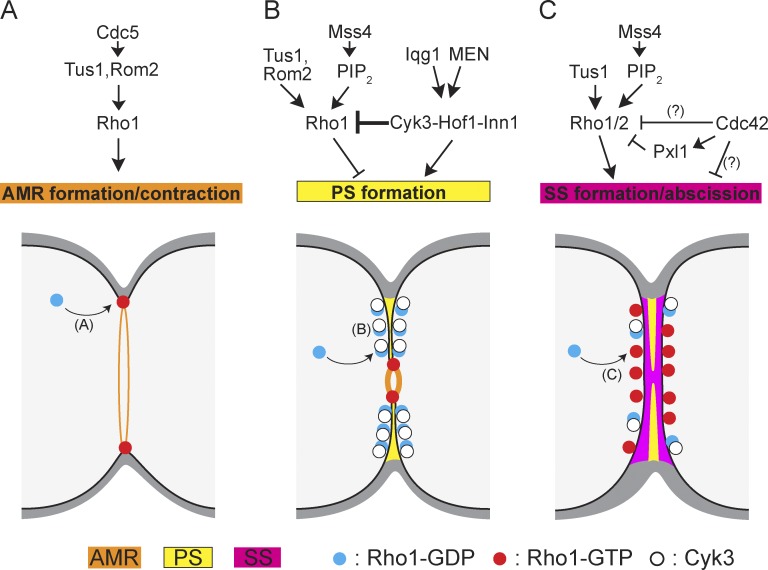
Rho1 inactivation by Cyk3 promotes primary septum formation and cleavage-furrow ingression, whereas Rho1 activation or Cdc42 inactivation promotes secondary septum formation and abscission.
Abstract
In yeast and animal cytokinesis, the small guanosine triphosphatase (GTPase) Rho1/RhoA has an established role in formation of the contractile actomyosin ring, but its role, if any, during cleavage-furrow ingression and abscission is poorly understood. Through genetic screens in yeast, we found that either activation of Rho1 or inactivation of another small GTPase, Cdc42, promoted secondary septum (SS) formation, which appeared to be responsible for abscission. Consistent with this hypothesis, a dominant-negative Rho1 inhibited SS formation but not cleavage-furrow ingression or the concomitant actomyosin ring constriction. Moreover, Rho1 is temporarily inactivated during cleavage-furrow ingression; this inactivation requires the protein Cyk3, which binds Rho1-guanosine diphosphate via its catalytically inactive transglutaminase-like domain. Thus, unlike the active transglutaminases that activate RhoA, the multidomain protein Cyk3 appears to inhibit activation of Rho1 (and thus SS formation), while simultaneously promoting cleavage-furrow ingression through primary septum formation. This work suggests a general role for the catalytically inactive transglutaminases of fungi and animals, some of which have previously been implicated in cytokinesis.
Introduction
Cytokinesis is the postmitotic event that physically separates the cytoplasms of the daughter cells. In both fungal and animal cells, cytokinesis is performed in two steps, cleavage-furrow formation and abscission. Formation and ingression of the cleavage furrow involve many cellular processes such as actomyosin ring (AMR) contraction, targeted membrane deposition, and ECM rearrangements (Balasubramanian et al., 2004; Barr and Gruneberg, 2007; Fededa and Gerlich, 2012), among which the AMR is the most extensively characterized. The majority of AMR components (F-actin, type-II myosin, and other associated proteins) are conserved in fungi, amoebae, and animals (Balasubramanian et al., 2004). Thus, the mechanisms of AMR function are likely to be conserved also in these species, although the degrees to which cleavage-furrow ingression depends on AMR contraction may vary (Balasubramanian et al., 2004; Uyeda and Nagasaki, 2004). Membrane trafficking also appears to be critical for furrow ingression (McKay and Burgess, 2011), and perturbation of various intracellular trafficking systems has a strong impact on this process and/or on subsequent abscission (Albertson et al., 2005). The involvement of ECM rearrangements in cytokinesis is less well understood, but many types of ECM components (proteins, proteoglycans, and polysaccharides) localize to the cleavage furrow, and some of them have been shown to be required for normal cytokinesis (Hwang et al., 2003; Mizuguchi et al., 2003; Ng et al., 2005; Xu and Vogel, 2011).
After cleavage-furrow ingression, the intercellular bridge connecting the two daughter cells is disassembled to irreversibly separate the daughter cells (Byers and Abramson, 1968; Schiel and Prekeris, 2010; Neto and Gould, 2011), a process termed abscission. In mammalian cells, abscission appears to involve anchoring of the plasma membrane to the midbody and subsequent resolution, in which the septins, secretory vesicles, and recycling endosomes are all implicated (Estey et al., 2010; Schiel and Prekeris, 2010; Neto and Gould, 2011). In Drosophila melanogaster S2 cells, F-actin appears to be essential for cleavage-furrow ingression but dispensable for abscission (Echard et al., 2004), suggesting that there are AMR-dependent and -independent events in animal cytokinesis. Among the key questions for a comprehensive understanding of cytokinesis are how these events are organized temporarily and spatially to accomplish successful cytokinesis and whether there is a central regulatory molecule or system that governs them.
The budding yeast Saccharomyces cerevisiae is an ideal model organism for study of the AMR-independent processes involved in cytokinesis because cells lacking the only type II myosin, and hence the AMR, are viable (Bi et al., 1998; Wloka and Bi, 2012). In wild-type cells, contraction of the AMR and cleavage-furrow ingression start concomitantly with synthesis of an extracellular polysaccharide, chitin, between the two layers of plasma membrane in the furrow to form the primary septum (PS; Shaw et al., 1991; Wloka and Bi, 2012). Chitin in the PS is synthesized mainly by the chitin synthase Chs2 (Shaw et al., 1991); the proteins Iqg1, Inn1, Hof1, and Cyk3 appear to be involved in PS formation by directly or indirectly contributing to Chs2 activation (Epp and Chant, 1997; Lippincott and Li, 1998; Korinek et al., 2000; Sanchez-Diaz et al., 2008; Jendretzki et al., 2009; Tully et al., 2009; Nishihama et al., 2009; Meitinger et al., 2010; Palani et al., 2012). Because Chs2 is delivered to the division site by exocytosis (Chuang and Schekman, 1996; VerPlank and Li, 2005), PS formation is a cytokinetic event achieved by the concerted actions of AMR contraction, membrane trafficking, and ECM rearrangement.
Because the transmembrane protein Chs2 and the AMR are both positioned at the leading edge of the cleavage furrow, where membrane fusion should take place during abscission, it is conceivable that furrow ingression and PS formation are inherently incapable of providing a mechanism for abscission because their machineries physically obstruct the process, necessitating another mechanism for abscission. An abscission defect in yeast has been observed as a consequence of perturbed spindle–midzone function (Bouck and Bloom, 2005), and a pathway named NoCut was found (Norden et al., 2006; Mendoza et al., 2009) to be responsible for this checkpoint function, which is thought to transduce a signal from the chromatin and spindle midzone to the cortical membrane. However, the precise mechanisms of abscission in yeast remain obscure.
As PS formation nears completion, the modes of ECM rearrangement and membrane trafficking change, and the secondary septa (SSs), consisting mainly of glucan, begin to be laid down on both mother and daughter sides of the PS (Cabib, 2004). The completed trilamellar septum is then partially degraded by a chitinase and glucanases that are secreted asymmetrically by the daughter cell (Kuranda and Robbins, 1991; Colman-Lerner et al., 2001; Cabib, 2004), thereby concluding cell separation.
In animal cells, the small GTPase RhoA appears to regulate multiple processes in cytokinesis (Piekny et al., 2005; Fededa and Gerlich, 2012). RhoA is bound to the membrane by CAAX prenylation at its C terminus and activated to its GTP-bound state and concentrated at the division site by a guanine nucleotide exchange factor (GEF). RhoA-GTP then binds to various downstream effectors such as the actin-nucleator formin and the Rho-associated protein kinase to regulate AMR assembly and initiation of contraction (Matsumura, 2005). In addition, RhoA may be involved in abscission, at least in some cell types. For example, in HeLa cells, RhoA has been reported to be activated in late telophase (Yoshizaki et al., 2004), localize to the midbody, and promote abscission (together with Citron kinase and anillin; Gai et al., 2011), and expression of a dominant-negative form of the RhoA GEF Ect2 caused a specific defect in abscission (Chalamalasetty et al., 2006).
During S. cerevisiae cytokinesis, the RhoA homologue Rho1 promotes AMR formation through the formin Bni1 (Tolliday et al., 2002). Rho1 activity during this process appears to be controlled mainly by the Polo kinase Cdc5 through two GEFs, Tus1 and Rom2 (Yoshida et al., 2006). Rom2 may also be regulated by phosphatidylinositol 4,5-bisphosphate (PIP2) produced by the Mss4 phosphatidylinositol 5-kinase in the plasma membrane (Audhya and Emr, 2002). PIP2 also contributes to the accumulation of Rho1 at the division site after mitotic exit through interaction with the polybasic sequence at the C terminus of Rho1; this pool of Rho1 can promote cytokinesis in myo1Δ cells via recruitment of the chitin synthase Chs3, without restoring actin ring assembly (Yoshida et al., 2009). In addition, Rho1 is known to use the glucan synthases Fks1/2, the protein kinase Pkc1, and the exocyst subunit Sec3 as effectors to regulate cell wall synthesis, the cell wall integrity MAPK pathway, and exocytosis, respectively, which contribute to a broad range of cellular events such as stress response, bud growth, wound healing, and cell cycle progression (Guo et al., 2001; Levin, 2005; Kono et al., 2008, 2012).
In this study, we investigated the contribution of SS formation to abscission in yeast and identified genes involved in this process by a genetic screen. Further characterization of these genes has suggested a role for Rho1 in SS formation and a novel mechanism for its regulation by the catalytically inactive transglutaminase domain in Cyk3.
Results
Possible contribution of SS formation to abscission in yeast
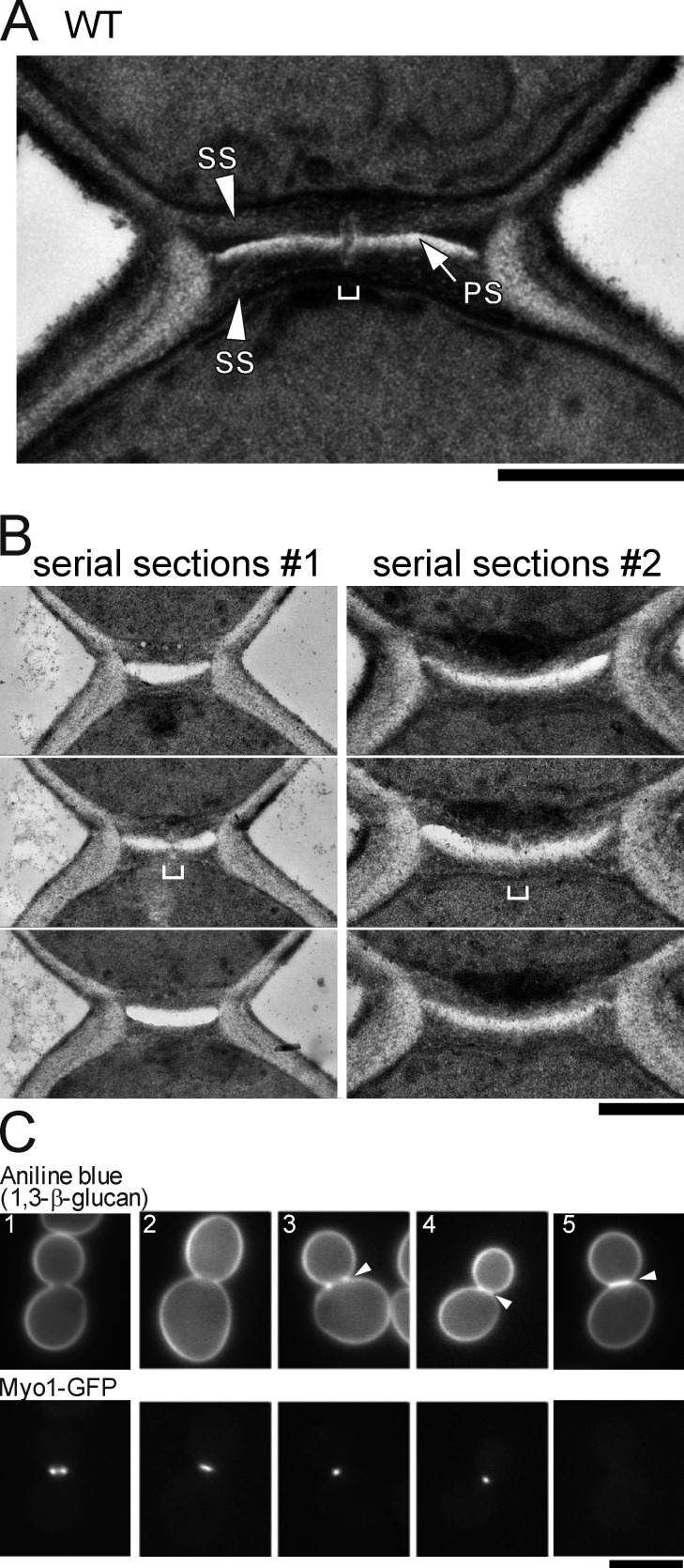
In mutants defective in anaphase-promoting complex function or degradation of Iqg1, such as cdh1Δ, the PS frequently fails to fully span the division plane, and the gap is filled with materials reminiscent of the SS (Tully et al., 2009). Similar discontinuities in the PS, typically ∼40 nm in diameter, were observed also in wild-type cells (Fig. 1 A), although in lower frequency (∼5% vs. the ∼27% observed in cdh1Δ cells). To ask whether such discontinuities were normal structures that had been missed in conventional EM thin sections, we performed serial section EM on wild-type cells. We found such discontinuities in four of the seven cells observed (Fig. 1 B). As the discontinuities are only ∼40 nm and the sections used were ∼80 nm thick, it is likely that some were missed and that most or all wild-type septa have a similar structure. Collectively, these results, those of Tully et al. (2009), and other evidence (see Introduction) suggest that PS formation alone is not sufficient to complete cytokinesis and that abscission is delayed in mutants defective in timely disassembly of the AMR.
Figure 1.
Possible contribution of SS formation to abscission in yeast. (A and B) EM analyses showing discontinuities (brackets) in PSs (electron lucent layer) of wild-type strain YEF473A. (A) A single section. (B) Serial ∼80-nm sections of two cells. Bars, 0.5 µm. (C) Initiation of SS formation before AMR disassembly. Strain RNY1681 (MYO1-GFP) was cultured to exponential phase in SC medium at 24°C, stained in the growth medium with 0.05% aniline blue, and observed using standard YFP and CFP filter sets. Arrowheads show strong staining of the SS. Bar, 5 µm.
Next, we asked whether the process that follows PS formation, SS formation, might contribute to abscission. We stained wild-type cells expressing Myo1-GFP with aniline blue, a dye that binds specifically to 1,3-β-glucan, the main constituent of the SS (Young and Jacobs, 1998). As shown in Fig. 1 C, the dye stained only general cell wall in the majority (∼90%, n = 104) of large-budded cells before (Fig. 1 C, image 1) and during (Fig. 1 C, image 2) AMR constriction. In contrast, in cells in which the AMR had contracted but not disassembled (n = 34), strong staining was usually obvious either at the periphery of the division plane (41%; Fig. 1 C, image 3) or as a continuous line across it (29%; Fig. 1 C, image 4). After AMR disassembly, cells always showed a continuous line of staining across the division plane (100%, n = 37; Fig. 1 C, image 5). Thus, SS formation appears to begin before the plasma membrane is resolved to separate the mother and daughter cytoplasms, suggesting that it might play a role in abscission.
Identification of Rho-related genes as dosage suppressors of PS formation mutants
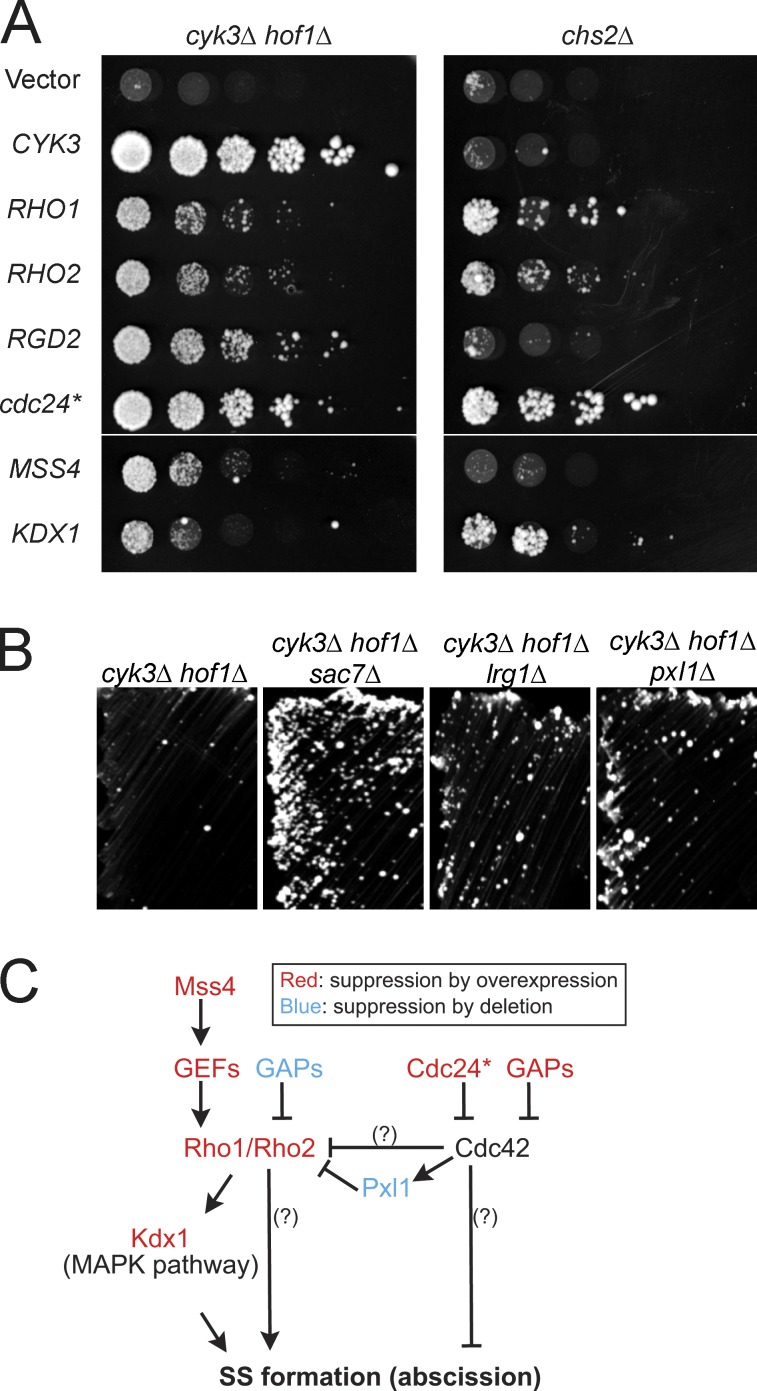
SS formation and its regulation remain poorly understood, mostly because of a lack of mutants or conditions that exhibit specific defects in this process. We reasoned that a dosage-suppressor screen of mutants defective in PS formation might identify genes involved in SS formation. We performed such a screen using a cyk3Δ hof1Δ double mutant (see Materials and methods). This mutant is almost inviable (Fig. 2 A; Korinek et al., 2000), probably as a result of its severe inefficiency in PS formation (Fig. 3, A and E). In addition to the expected CYK3 and HOF1, we isolated nine suppressor genes from two independent genomic libraries (Fig. S1 A). Six of these genes (Fig. 2 A) encode proteins functionally related to two Rho family small GTPases, Rho1 and Cdc42: RHO1 itself and its homologue RHO2, proteins that function upstream and downstream of Rho1 (MSS4 and KDX1, respectively), and direct regulators of Cdc42 (RGD2 and a C-terminally truncated version of CDC24, designated cdc24*; see below). Interestingly, unlike CYK3, which induces Chs2-dependent PS formation (Nishihama et al., 2009), most of these genes were also able to suppress chs2Δ (Fig. 2 A), indicating that the Rho-related genes suppress through a pathway independent of PS formation. Characterization of the other three genes, PSP1, NAB6, and PAP1, will be described elsewhere.
Figure 2.
Suppression of cyk3Δ hof1Δ and chs2Δ growth defects by activation of Rho1 or inactivation of Cdc42. (A) cyk3Δ hof1Δ (MOY68) and chs2Δ (RNY1419) strains carrying a URA3 HOF1 or URA3 CHS2 plasmid were transformed with LEU2-marked high-copy plasmids containing the indicated genes (Table 2). The transformants were cultured overnight in SC-Ura-Leu medium, spotted onto SC-Leu + 5-FOA plates as serial 5× dilutions starting at ∼106 cells per spot, and imaged after 4 d at 24°C. Spotting of equal numbers of cells was confirmed by spotting onto SC-Ura-Leu plates (not depicted). These data are shown again in Fig. S1 A alongside additional strains. (B) Suppression of cyk3Δ hof1Δ by deletion of SAC7, LRG1, or PXL1. Strains of the indicated genotypes (MOY68, MOY438, MOY433, and MOY430) were streaked on a SC + 5-FOA plate to eliminate their URA3 HOF1 plasmids and incubated at 24°C for 4 d. Streaking was used here because the relatively weak suppression was masked in spotting assays by the presence of a few large colonies that presumably represented spontaneous suppressors. (C) Summary of activating and inhibitory interactions inferred from the suppression results. Cdc24* and Pxl1 are discussed in the legends of Fig. S3 and Fig. S4. Question marks show interactions postulated to exist because RHO1 and RHO2 suppress more strongly than does KDX1 and because cdc24* and RGD2 suppress more strongly than does pxl1Δ.
Figure 3.
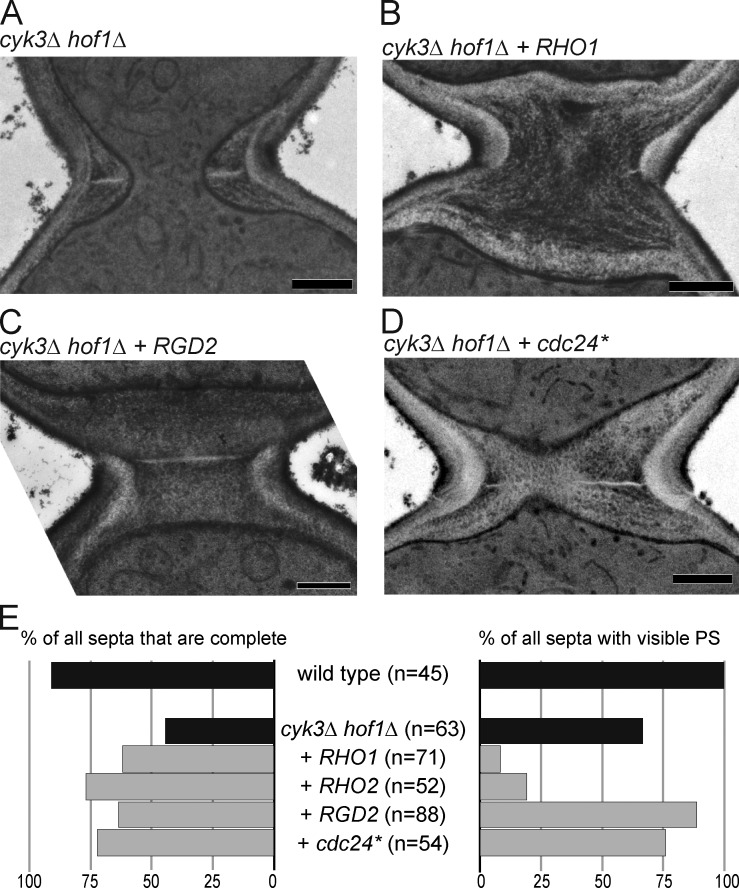
Thickened SS without restoration of PS when Rho-related genes are expressed in cyk3Δ hof1Δ cells. (A–D) cyk3Δ hof1Δ strains overexpressing the indicated genes (Fig. 2 A) were streaked on SC + 5-FOA plates to eliminate the URA3-marked HOF1 plasmid and observed by EM. (A) RNY2127, (B) MOY245, (C) MOY682, and (D) MOY78. Bars, 0.5 µm. (E) For each strain, cells with any visible septal structure were scored for successful bud-neck closure (left) and the presence of PS-like structures (right). Data for RHO2 are from the experiment in Fig. S3 A Black bars show control strains without plasmid. Gray bars show mutant strains transformed with plasmids overexpressing the indicated genes.
Consistent with the independence of suppression from Chs2, little or no improvement in PS formation was observed during suppression by RHO1 or RHO2 (Fig. 3, B and E, right; and Fig. S2 A), RGD2 (Fig. 3, C and E), cdc24* (Fig. 3, D and E), or KDX1 (not depicted). Instead, the suppressed cells had very thick SS-like structures that spanned the bud necks with slightly better efficiency than was seen in control cells (Fig. 3 E, left), probably accounting for the higher growth rates of the suppressed cells. These results suggest that the Rho-related genes hyperactivate SS formation and that the SS can be an alternative to the PS as a driving force for successful (albeit abnormal) cytokinesis. Similar improvements in growth and hyperactivation of SS formation were observed using an iqg1Δ mutant (Fig. S1 A and Fig. S2, B–E), indicating that the suppression mechanisms do not require the AMR, which fails to form in iqg1Δ mutants (Epp and Chant, 1997; Lippincott and Li, 1998). Interestingly, many cyk3Δ hof1Δ cells suppressed by RHO1 or RHO2 had SS without detectable PS (Fig. 3, B and E; and Fig. S2 A), suggesting that these genes (but not cdc24* or RGD2) also have inhibitory effects on PS formation.
Hyperactivation of SS formation by activation of Rho1 or inactivation of Cdc42
Each of the aforementioned suppressors either activates the Rho1 pathway or inactivates the Cdc42 pathway, with the apparent exception of cdc24*. We extended the test to other GEFs and GTPase-activating proteins (GAPs) that regulate these small GTPases. Activation of Rho1 by either overexpression of certain Rho1-GEF genes (TUS1 and ROM1) or deletion of certain Rho1-GAP genes (SAC7 or LRG1) suppressed cyk3Δ hof1Δ (Fig. S1 B and Fig. 2 B, left and middle images). In striking contrast to cdc24*, wild-type CDC24 failed to suppress cyk3Δ hof1Δ (Fig. S1 B). Further analyses (Fig. S3) suggested that the chimeric Cdc24* protein, consisting of portions of Cdc24 and of the tetA tetracycline resistance protein (from the vector backbone), functions as an inhibitor of Cdc42. Consistent with this hypothesis, overexpression of certain Cdc42 GAPs (RGA1 and BEM3) also suppressed the cyk3Δ hof1Δ mutant (Fig. S1 B). Interestingly, RGD1, which encodes a GAP for Rho3 and Rho4 (Prouzet-Mauléon et al., 2008), also suppressed the mutant, suggesting that these proteins may also be involved in cytokinesis. In most cases, suppression of an iqg1Δ mutant paralleled those with the cyk3Δ hof1Δ mutant (Fig. S1, A and B). Collectively, our genetic data support the model that either activation of Rho1 or inactivation of Cdc42 is sufficient to suppress effects in AMR and/or PS formation by inducing SS formation (Fig. 2 C).
We also found that deletion of the paxillin homologue gene PXL1 weakly suppressed both cyk3Δ hof1Δ and hof1Δ (Fig. 2 B and Fig. S4 A). PXL1 was identified as an allele-specific dosage suppressor of cdc42 point mutations, and both genetic and biochemical evidence suggests that Pxl1 is a negative regulator of Rho1 (Gao et al., 2004; Mackin et al., 2004). Because the timing and pattern of Pxl1-GFP localization also coincide with that of SS formation (Fig. S4, B–D), it seems likely that Pxl1 connects Cdc42 and Rho1 during SS formation (Fig. 2 C), serving as a molecular brake for excessive SS formation.
Requirement for Rho1 in SS formation and cell separation
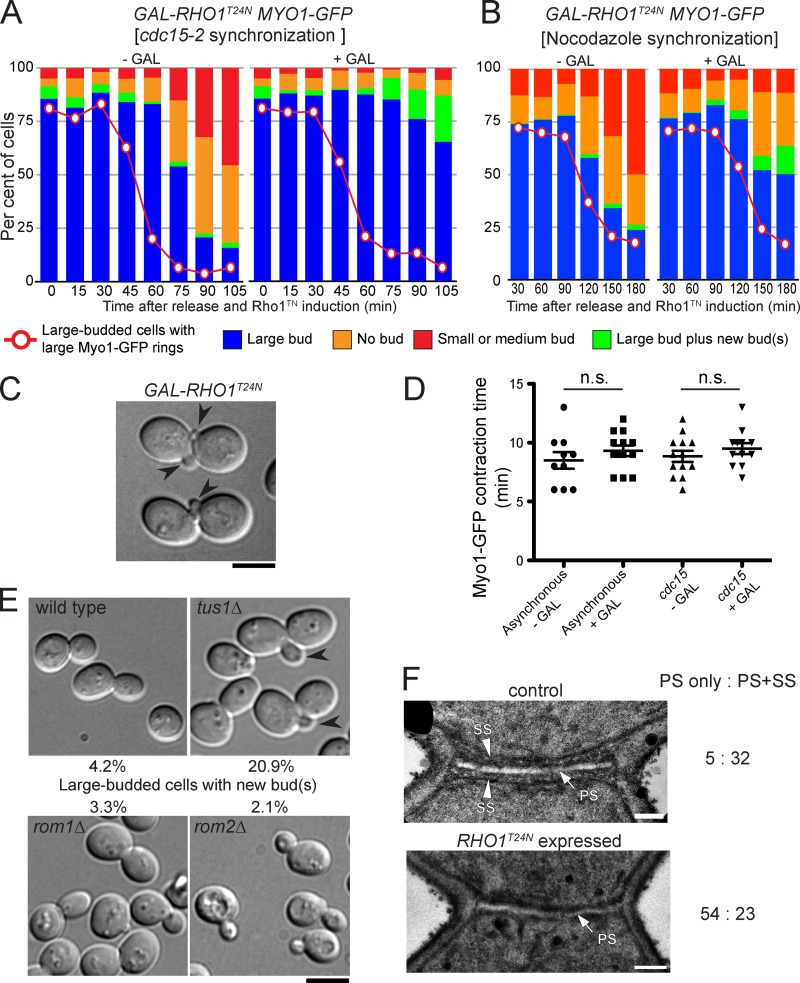
The extensive formation of SS when Rho1 was overexpressed or activated in cytokinesis mutants suggested that it may have a role in SS formation in wild-type cells. To test this hypothesis, we induced the expression of a GDP-locked, dominant-negative mutant of Rho1 (T24N) in cells released from an anaphase block produced by a cdc15-2 mutation. Under these conditions, the AMR was already formed at the time of release (not depicted; Yoshida et al., 2006), and the initiation and rate of furrow ingression were not affected by the expression of Rho1T24N (Fig. 4, A and D). However, the Rho1T24N-expressing cells completely failed to separate even after 105 min (Fig. 4 A) or 12 h (not depicted). Some cells eventually formed tiny buds (Fig. 4, A and C), indicating that the cells did not simply die upon induction of Rho1T24N and thus that the separation defect was triggered specifically by inhibition of a Rho1 pathway. EM analyses revealed that at 75 min, ∼87% of control cells had formed SS (Fig. 4 F, top), whereas only ∼30% of the Rho1T24N-expressing cells had detectable SS even though normal-looking PSs were formed (Fig. 4 F, bottom), indicating that SS formation was blocked or delayed by the dominant-negative Rho1.
Figure 4.
Dependence of SS formation and cell separation, but not cleavage-furrow ingression, on Rho1 activity. (A–D) Effects of dominant-negative Rho1. A cdc15-2 MYO1-GFP (YCp-PGAL-RHO1T24N) strain (MOY542) was synchronized by incubation at 37°C in SC-Leu (2% raffinose) medium (A, C, and D, right) or with nocodazole in YPD (2% raffinose) medium (B; see Materials and methods). Each culture was separated into two and released from the block in the presence (+GAL) or absence (−GAL) of 2% galactose. (A and B) At the indicated times, aliquots of cells were fixed with formaldehyde, sonicated briefly, and scored for the percentages of cells with large (precontraction) Myo1-GFP rings, large buds, large buds plus one or more new buds, no buds, and small or medium buds (n > 200 per sample). Plasmid loss during incubation in the nonselective medium presumably accounts for the reduced efficiency of cell separation blockage in B. (C) Representative cells at 105 min in the +GAL culture of A. Arrowheads show new buds formed before cell separation. (D) Asynchronous cells in SC-Leu (2% raffinose) medium and cdc15-2–synchronized cells were observed by time-lapse microscopy beginning 15 min after addition (or not) of galactose. The time intervals between initiation of Myo1-GFP contraction and Myo1-GFP disappearance were recorded as constriction times. The long and short horizontal bars indicate means and means ± SDs, respectively. Student’s unpaired t tests showed that the differences in mean values were not significant. Data from three separate experiments were combined; n = 10, 12, 13, and 12 (left to right). (E) Cell morphologies of Rho1 GEF mutants. Strains of the indicated genotypes (YEF473A, RNY879, RNY935, and RNY875) were cultured in YM-P medium to exponential phase. Arrowheads show new buds formed before cell separation. The percentages of such cells among total large-budded cells are indicated (n = 300). (F) Effect of Rho1T24N on septum morphology. Control (−GAL) and Rho1T24N-expressing (+GAL) cells from the 75-min sample in A were observed by EM. Counts of septa with PS only or with both PS and SS are shown on the right. Bars: (C and E) 5 µm; (F) 0.2 µm.
Importantly, although Rho1 activity in early stages of cytokinesis is required for AMR formation (and possibly also for its contractility), it appeared to be dispensable for cleavage-furrow ingression, as indicated by nearly normal constriction of Myo1-GFP rings in Rho1T24N-expressing cells that had been synchronized with nocodazole at a stage before actin ring formation (Fig. 4 B and not depicted) as well as in asynchronous culture (Fig. 4 D and not depicted). These results are consistent with the dispensability of the AMR and its contractility for cleavage-furrow formation in yeast (see Introduction).
Examination of Rho1-GEF mutants revealed that tus1Δ cells also exhibited bud emergence before cell separation, whereas rom1Δ and rom2Δ cells did not (Fig. 4 E). Interestingly, of 30 tus1Δ cells examined by EM, 40% had apparently complete PSs without SS, and 50% had SS only on the daughter side of the PS (Fig. S5 A and not depicted). As Tus1 remains at the bud neck after the furrow and AMR have constricted (Yoshida et al., 2006), and overexpression of TUS1 (but not of ROM1 or ROM2) could suppress cyk3Δ hof1Δ, iqg1Δ, and chs2Δ mutations (Fig. S1 B), this GEF may be primarily responsible for Rho1 activation during SS formation. Conversely, hyperactivation of Rho1 either by expression of a constitutively active construct (Rho1Q68L) or by deletion of the GAP gene LRG1 caused formation of abnormally thickened SS (Fig. S5 A). Surprisingly, deletion of another GAP gene, SAC7, did not have a strong impact on SS structure (Fig. S5 A), although this deletion was at least as effective as lrg1Δ in suppressing cyk3Δ hof1Δ (Fig. 2 B). Collectively, these results suggest that Rho1 has a role in SS formation as well as in actin ring formation during normal cytokinesis and that Tus1 and Lrg1 are its major positive and negative regulators for this role.
Temporary inactivation of Rho1 during AMR constriction and PS formation
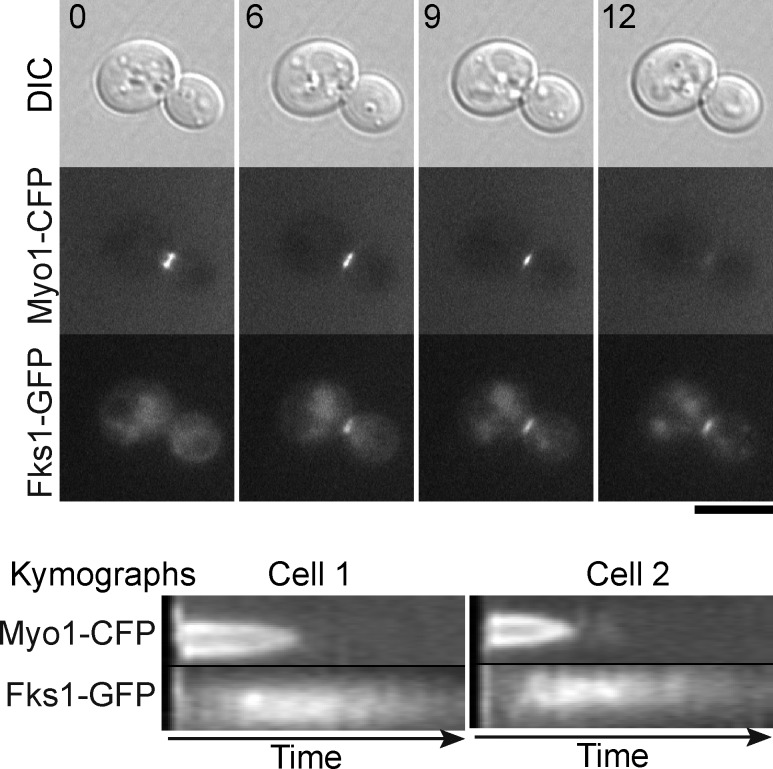
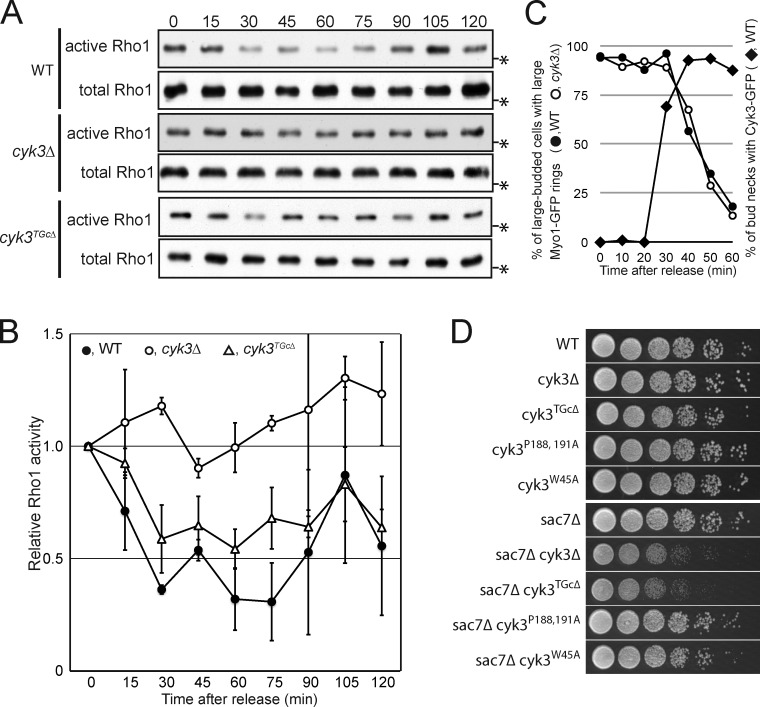
Both Rho1 (Yoshida et al., 2009) and its effector Fks1 (glucan synthase; Fig. 5) arrive at the division site before cleavage-furrow ingression and hence before the initiation of SS formation. These observations suggest that Rho1 activity might be specifically suppressed during PS formation to prevent premature activation of SS formation. Consistent with this hypothesis, expression of the constitutively active Rho1Q68L caused a substantial increase in the number of cell clusters (Fig. S5 B), suggesting that Rho1Q68L interferes with a step in cell division. To explore the hypothesis further, we conducted pull-down assays using cdc15-2–synchronized cells and a GST-fused Rho-binding domain (RBD) from Pkc1, which binds specifically to GTP-bound Rho1 (Kono et al., 2008). As reported previously (Yoshida et al., 2006), Rho1 activity was high in the arrested cells (Fig. 6, A and B, 0 min). It then decreased during the 30 min after release and generally remained low until 75 min, except for a small increase at 45 min that was observed in each of three independent experiments (Fig. 6, A and B). In parallel experiments, furrow ingression started at <40 min after release (Fig. 6 C) and was typically complete within 7–10 min in each cell (not depicted). Because SS formation begins before AMR disassembly (Fig. 1 C), the transient activation of Rho1 coincides with, and is probably responsible for, the initiation of SS formation. The later activation of Rho1 beginning at ∼75–90 min is presumably associated with bud growth in the new cell cycle (Kono et al., 2008).
Figure 5.
Localization of Fks1 to the division site during AMR constriction and before the beginning of SS formation. Strain MOY1303 (FKS1-GFP fks2Δ MYO1-CFP) was observed by time-lapse microscopy in YM-P medium. Whole-cell images of one cell (top) and kymographs of the division planes of two other cells (bottom) are shown. Heights of the kymographs correspond to 2 µm. Numbers at the top are time in minutes after the beginning of observation. DIC, difference interference contrast. Bar, 5 µm.
Figure 6.
Temporary inactivation of Rho1 during PS formation and its apparent regulation by Cyk3. (A and B) Activity of Rho1 during cytokinesis in wild-type, cyk3Δ, and cyk3TGcΔ cells. cdc15-2 3HA-RHO1 (MOY553), cdc15-2 3HA-RHO1 cyk3Δ (MOY552), and cdc15-2 3HA-RHO1 cyk3TGcΔ (MOY973) strains were grown to exponential phase in YM-P medium at 24°C, arrested by incubation at 37°C for 3 h, and then released into mitotic exit by cooling rapidly (∼5 min) to 24°C. Samples were collected at the indicated times (minutes) and subjected to the GST-RBD pull-down assay (see Materials and methods). Numbers of independent experiments: WT, 3; cyk3Δ, 2; and cyk3TGcΔ, 2. (A) Representative Western blots of active and total Rho1. Asterisks show the position of a 25.9-kD molecular mass marker. (B) Quantification of band intensities (means ± SDs) of active Rho1 relative to total Rho1; the values at time 0 were set to 1.0. (C) Normal initiation of Myo1-GFP constriction in cyk3Δ cells and timing of Cyk3 localization to the bud neck. cdc15-2 MYO1-GFP (MOY720), cdc15-2 MYO1-GFP cyk3Δ (MOY721), and cdc15-2 CYK3-GFP (MOY543) strains were synchronized as in A. For Myo1-GFP, large-budded cells were scored for the presence of large (i.e., precontraction) or smaller GFP rings (n = 64–111 per sample). For Cyk3-GFP, large-budded cells were scored for the presence of detectable GFP signal at the neck (n = 124). (D) Genetic interaction between CYK3 and SAC7. Strains of the indicated genotypes were spotted on YPD plates as in Fig. 2 A and incubated at 24°C for 48 h (strains: YEF473B, MOY585, MOY882, MWY636, MWY1412, MOY405, MOY440, MOY967, MOY980, and MOY982). WT, wild type.
Possible regulation of Rho1 inactivation through physical interaction with Cyk3
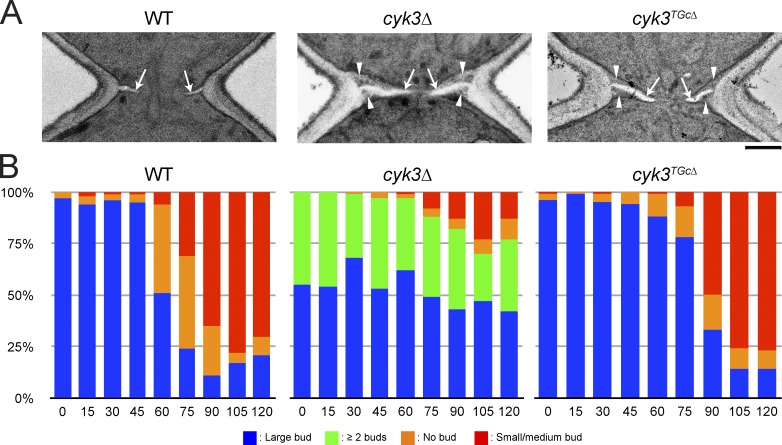
Several observations suggest that Cyk3 might be involved in a Rho1 inactivation mechanism that functions during PS formation. First, in cdc15-2–synchronized cells, Cyk3-GFP localized to the bud neck just before furrow ingression (Fig. 6 C), which coincides with the timing of Rho1 inactivation. Second, in contrast to wild-type cells, cyk3Δ cells formed PS and SS simultaneously (Fig. 7 A, left and center), suggesting that the putative temporary suppression of SS formation by Rho1 inactivation does not function properly in this mutant. Third, overexpression of Cyk3 abolished the simultaneous formation of PS and SS observed in a mitotic exit network mutant that was forced to exit mitosis with the cyclin inhibitor Sic1 (Meitinger et al., 2010). Fourth, the severe clustering phenotype of cyk3Δ cells (Fig. 7 B, left and center) appears to involve activation of Rho1 because deletion of CYK3 did not exacerbate the similar phenotype produced by the constitutively active Rho1Q68L (Fig. S5 B). Fifth, cyk3Δ showed a synergistic growth defect at 24°C with the Rho1-GAP mutation sac7Δ (Fig. 6 D) but not with another Rho1-GAP mutation, lrg1Δ (not depicted). Lrg1, like Cyk3, localizes to the neck (as well as to the bud tip; Watanabe et al., 2001), whereas Sac7 localizes throughout the cell cortex (Fig. 8 D, image 5), and a sac7Δ lrg1Δ mutant is inviable (Lorberg et al., 2001). Thus, collectively, the data suggest that Cyk3, like Lrg1 (Svarovsky and Palecek, 2005), is involved in controlling Rho1 activity at the neck, whereas Sac7 has a more general role, and that the excessive activation of Rho 1 when both systems are inactive is harmful or even lethal. Finally, the temporary inactivation of Rho1 completely disappeared in cyk3Δ cells (Fig. 6, A and B), although Myo1-GFP constriction initiated normally (Fig. 6 C).
Figure 7.
Septum morphology and cell separation in cyk3 mutants. (A) Simultaneous formation of PS and SS in cyk3Δ and cyk3TGcΔ mutants. Wild-type (YEF473A), cyk3Δ (RNY502), and cyk3TGcΔ (MOY882) cells were observed by EM. Arrows show PS; arrowheads show SS. Bar, 0.5 µm. (B) The cdc15-2–synchronized cells from the experiment of Fig. 6 A were scored for bud morphology. n > 200 for each time point. Numbers at the bottom indicate times (in minutes) after release from the cdc15-2 block. WT, wild type.
Figure 8.
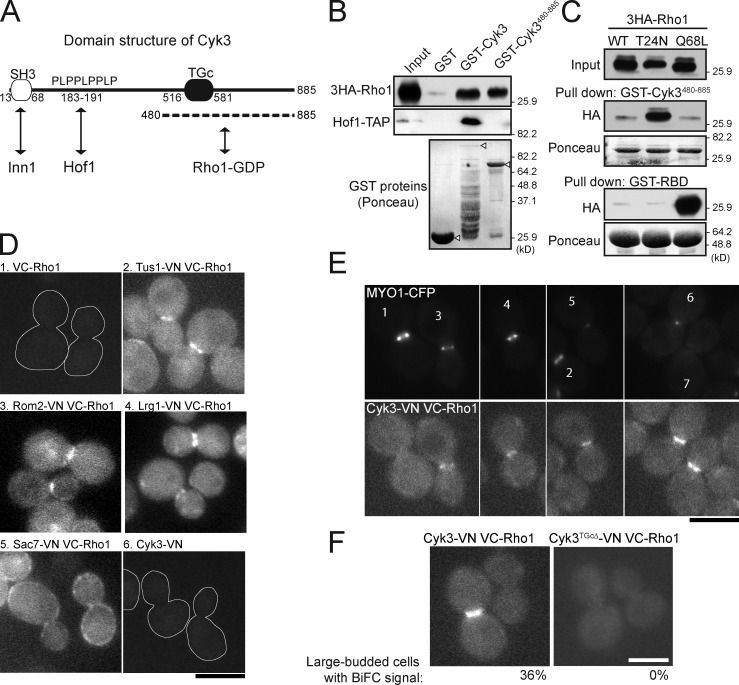
Physical interaction between Cyk3 and Rho1. (A) Domain structure of Cyk3. SH3, src-homology domain; PLPPLPPLP, proline-rich motif; TGc, transglutaminase core domain. The previously known interactions with Inn1 and Hof1 and the interaction with Rho1 identified in this study are indicated. (B) Cells expressing 3HA-tagged Rho1 (MOY522) were grown to exponential phase in YM-P medium at 24°C, and pull-down was performed (top) using bacterially expressed GST, GST-Cyk3, or GST-Cyk3480–885 (see Materials and methods). Hof1-TAP (strain MOY22) was included here as a control (middle) because Hof1 is known to bind to the PLPPLPPLP motif of Cyk3 (unpublished data). (bottom) The GST proteins used were also analyzed by SDS-PAGE; the arrowheads indicate the full-length proteins. (top) Note that there appears to have been relatively little full-length GST-Cyk3 (because of proteolysis or premature translation termination), yet pull-down by this preparation was quite effective. This may mean that binding of Cyk3 to Rho1 normally involves a site in the N-terminal region as well as that in the C-terminal region. The migration positions of molecular mass markers are indicated. (C) Selective affinity of the Cyk3 C terminus for Rho1-GDP. 3HA-tagged wild-type (WT; strain MOY801), dominant-negative (T24N; strain MOY824), and constitutively active (Q68L; strain MOY803) Rho1 proteins were expressed under the control of the GAL1 promoter for 30 min at 24°C. Cell extracts were analyzed as in B using GST-Cyk3480–885 or (as a control) the GST-tagged RBD (Fig. 6). (D–F) BiFC revealing interactions between Rho1 and other proteins. Strains expressing the indicated proteins (MOY1247, MOY1269, MOY1293, MOY1294, MOY1275, MOY1321, MOY1349, MOY1325, and MOY1384) were grown to exponential phase in SC-Leu liquid medium at 24°C and observed by fluorescence microscopy. (D and E) Numbering is for ease of reference in the text. Cell outlines are shown in 1 and 6 where there is no fluorescence signal. (F) The percentage of large-budded cells with detectable BiFC signal at the bud neck is shown for each strain. Bars, 5 µm.
Several lines of evidence indicate that Cyk3 might inactivate or block activation of Rho1 by physically interacting with its inactive form. First, a pull-down assay using bacterially expressed, GST-fused full-length Cyk3 (GST-Cyk3) or its C-terminal half (GST-Cyk3480–885; Fig. 8 A) revealed that Rho1 interacted with the C-terminal half of Cyk3 (Fig. 8 B), in contrast to Hof1, which bound only to full-length GST-Cyk3 in this assay (Fig. 8 B). Second, a similar experiment using wild-type, constitutively active (Q68L), or dominant-negative (T24N) Rho1 revealed that the affinity of Rho1 for the C-terminal half of Cyk3 was significantly increased by the T24N mutation but not by the Q68L mutation (Fig. 8 C). Third, although we have not detected a Cyk3–Rho1 interaction in vivo by coimmunoprecipitation (presumably because of the membrane association of both proteins and/or the transient nature of the interaction), we have detected an interaction using bimolecular fluorescence complementation (BiFC). We constructed a Rho1 BiFC reporter by fusing the C-terminal portion (aa 155–238) of the Venus protein to the N terminus of Rho1 via a short linker (VC155-GGSGGS-Rho1; henceforth simply VC-Rho1) and expressed this construct from a low-copy plasmid in rho1Δ cells. This reporter successfully detected interactions of Rho1 with several GEFs and GAPs (Fig. 8 D, images 1–5). We then tagged the C terminus of Cyk3 with the N-terminal portion of Venus (aa 1–155) containing an amino acid substitution (I153L) to diminish potential self-assembly between the N- and C-terminal portions of Venus (Kodama and Hu, 2010). A CYK3-VN strain showed no detectable fluorescence (Fig. 8 D, image 6), but when VC-Rho1 was also expressed, clear BiFC signal appeared at the bud neck in a temporarily regulated manner (Fig. 8 E). In most cells with large Myo1-CFP rings, signal was not detected (Fig. 8 E, cells 1 and 2), but it did appear in some such cells (cells 3 and 4), and it remained while Myo1-CFP constricted to a dot (Fig. 8 E, cells 5 and 6). After AMR disassembly, BiFC signal remained at the bud neck (Fig. 8 E, cell 7), but this signal persistence may reflect only the tight binding between the N- and C-terminal Venus fragments (Kodama and Hu, 2010) rather than a continuing association of Cyk3 with Rho1 during SS formation.
The N-terminal portion of Cyk3 contains an SH3 domain and a proline-rich region, which bind to Inn1 and Hof1, which are involved in PS formation (Fig. 8 A; our unpublished data; Nishihama et al., 2009; Jendretzki et al., 2009; Meitinger et al., 2011; Labedzka et al., 2012). The C-terminal portion of Cyk3 contains a transglutaminase core (TGc) domain that resembles the protein and peptide cross-linking transglutaminases. Interestingly, several transglutaminases are implicated in the modification and activation of mammalian RhoA (see Discussion). However, the TGc domain in Cyk3 lacks the conserved cysteine that is essential for TGase activity (Makarova et al., 1999; Pollard et al., 2012). We constructed a strain in which the TGc domain (aa 519–580) is deleted (cyk3TGcΔ). Cyk3TGcΔ appears to retain most Cyk3 functionality because a cyk3TGcΔ hof1Δ double mutant (in contrast to a cyk3Δ hof1Δ double mutant: Fig. 2 A) grew as well as a hof1Δ single mutant, Myo1-GFP constricted at a normal rate in cyk3TGcΔ cells, and Cyk3TGcΔ-GFP localized normally to the bud neck (not depicted). However, cyk3TGcΔ sac7Δ cells grew as poorly as cyk3Δ sac7Δ cells (Fig. 6 D). In contrast, mutations in the SH3 domain (cyk3W45A) or the proline-rich region (cyk3P188,191A) did not cause such a synergistic growth defect with sac7Δ (Fig. 6 D). The cyk3TGcΔ strain showed a partial deficiency in Rho1 inactivation as judged by the GST-RBD pull-down assay (Fig. 6, A and B), a moderate premature formation of SS formation (Fig. 7 A, right), and a moderate delay in cell separation (Fig. 7 B, right). Finally, no Cyk3–Rho1 interaction could be detected by the BiFC assay when the TGc domain was deleted (Fig. 8 F). Collectively, these results suggest that the TGc domain of Cyk3 is required for effective blockage of Rho1 activation, that this role of Cyk3 is independent of its binding to the PS formation proteins Inn1 and Hof1, and that the more severe phenotype of cyk3Δ cells (compared with cyk3W45A, cyk3P188,191A, and cyk3TGcΔ cells) may be a synthetic effect of slowed PS formation together with a premature activation of SS formation.
Discussion
SS formation as the mechanism of yeast abscission
In animal cells, it has long been recognized that abscission, the final resolution of the daughter cell membranes, is a step distinct from cleavage-furrow ingression (Schiel and Prekeris, 2010; Neto and Gould, 2011). However, in yeast, this idea has been controversial, in part because conventional EM images have typically shown a PS that appears to span the entire division plane (Weiss, 2012; Wloka and Bi, 2012). Nonetheless, studies using mutants and fluorescent membrane markers have suggested that a distinct abscission step exists and is specifically inhibited in response to spindle midzone and chromosome segregation defects by the NoCut checkpoint pathway (Norden et al., 2006; Mendoza et al., 2009). Some septin mutants also appear to be specifically defective in abscission (Dobbelaere and Barral, 2004; Wloka et al., 2011). Our serial section EM revealed a discontinuity in the PSs of most wild-type cells, supporting the hypothesis that a distinct abscission step is needed to complete cytokinesis. We suggest that the abscission machinery must be distinct from the AMR and PS formation machinery because these elements localize to the leading edge of the cleavage furrow, where they would presumably interfere with the membrane fusion needed for abscission. In support of this idea, the PS discontinuity is enlarged in mutants defective in timely disassembly of the AMR (Tully et al., 2009).
Several lines of evidence suggest that SS formation is involved in abscission. First, 1,3-β-glucan, the major constituent of the SS, began to be deposited at the bud neck before the AMR was disassembled (and thus before abscission). Second, the glucan synthase Fks1 localized on the surface of the cleavage furrow, but in the area trailing the AMR, and so is positioned to function without physically interfering with membrane resolution. Finally, hyperactivation of SS formation restored abscission in mutants defective in PS formation. Mechanisms other than glucan synthesis are probably also involved in SS formation and abscission. In particular, the chitin synthase Chs3 is required for formation of remedial SS in a chs2 mutant (Shaw et al., 1991), as well as in myo1Δ cells overexpressing Rho1 (Yoshida et al., 2009), and so is likely to be involved in normal SS formation as well.
Antagonistic regulation of SS formation by Rho1 and Cdc42
Our suppression data suggest that the small GTPases Rho1 and Cdc42 act antagonistically to regulate SS formation (Fig. 2 C and Fig. 9 C). Rho1 appears to promote SS formation, in that its overexpression or activation hyperactivated SS formation. Promotion of SS formation by Rho1 has also been observed in a myo1Δ mutant (Yoshida et al., 2009), and it has been hypothesized that Rho1 is involved in SS formation in wild-type cells (Weiss, 2012; Wloka and Bi, 2012). We have provided strong support for this hypothesis by showing that expression of a dominant-negative Rho1 in synchronized cells blocks cell division but not cleavage-furrow ingression. During SS formation, Rho1 effectors implicated in cell wall synthesis and stress response are likely to be activated, such as the glucan synthases Fks1 and Fks2 (Mazur and Baginsky, 1996; Qadota et al., 1996), the protein kinase Pkc1 (through activation of the MAPK pathway, in which Kdx1 [also known as Mlp1] is involved; Watanabe et al., 1997; Kim et al., 2008), and Chs3 (Valdivia and Schekman, 2003; Yoshida et al., 2009). In support of this hypothesis, deletions of these genes exhibit synthetic growth defects with myo1Δ (our unpublished data; Rodríguez-Quiñones et al., 2008; Yoshida et al., 2009).
Figure 9.
Model for regulation of Rho1 activity through cytokinesis. (A) In anaphase, Rho1 is recruited to the division site and activated by GEFs, and it promotes actin polymerization through the formin Bni1 to form the AMR (Tolliday et al., 2002). The Polo kinase Cdc5 activates the GEFs by phosphorylation (Yoshida et al., 2006). (B) During PS formation and cleavage-furrow ingression, Cyk3 binds to Rho1 to inhibit its activation (this study), perhaps by competing with the GEFs. Abnormally activated Rho1 at this stage can inhibit PS formation. Localization of Rho1 to the cleavage furrow is mediated through its binding to PIP2 (Yoshida et al., 2009). Cyk3 also interacts with Inn1 and Hof1 to activate PS formation under the control of Iqg1 and the mitotic exit network (see Introduction). (C) During SS formation and abscission, Rho1 (and apparently also Rho2) promotes SS formation through the MAPK pathway, Fks1 activation, and potentially other effectors (this study). Activation of Rho1 is due mostly to the GEF Tus1 and potentially also through binding to PIP2 (Yoshida et al., 2009; this study). Rho1 also appears to be negatively regulated by Cdc42 (through Pxl1 and perhaps also directly), perhaps to avoid hyperactivation. Cdc42 may also inhibit SS formation through Ste20 (unpublished data; Atkins et al., 2013). MEN, mitotic exit network. Question marks show interaction inferred but not demonstrated directly (Fig. 2).
In contrast, Cdc42 appears to inhibit SS formation (Fig. 2 C and Fig. 9 C), as its inactivation by overexpression of certain GAPs or the abnormal Cdc24* suppressed mutants defective in PS formation. The available evidence suggests that Cdc42 functions both by inactivation of Rho1 and by an independent mechanism. Deletion of PXL1, a factor downstream of Cdc42 that inactivates Rho1 (Gao et al., 2004; Mackin et al., 2004), suppressed cyk3Δ hof1Δ, but it did so more weakly than overexpression of either the Cdc42-GAP Rgd2 or Cdc24*. In addition, unlike Rho1, Cdc42 GAPs did not inhibit PS formation while activating SS formation. Although the exact mechanisms remain to be determined, Atkins et al. (in this issue) have shown that the p21-activated kinase Ste20 functions downstream of Cdc42 in cytokinesis, and we have independently confirmed that ste20Δ bypasses the defects in cyk3Δ hof1Δ by hyperactivation of SS formation (unpublished data). The relevant substrate of Ste20 remains to be identified.
The most downstream known components of the NoCut checkpoint pathway are the homologous Boi1 and Boi2 proteins in the cell cortex (Mendoza et al., 2009), which interact both genetically and physically with Cdc42, Rho3, Rho4, and the scaffold protein Bem1 (Bender et al., 1996; Matsui et al., 1996). Thus, it will be interesting to test whether the NoCut pathway acts through these GTPases. Similarly, the ER surveillance pathway delays cytokinesis upon ER stress, and the Pkc1-MAPK cascade downstream of Rho1 becomes essential for cell survival (Babour et al., 2010). An attractive model would be that the ER surveillance pathway delays cytokinesis at abscission, giving the cell time to adjust to the stress, and then hyperactivates SS formation through Rho1 activation to resume cell division.
Dispensability and temporary inactivation of Rho1 during cleavage-furrow ingression
As Rho1 is required for AMR formation in anaphase (Fig. 9 A; Yoshida et al., 2006), our results suggest that Rho1 activity is required at both the beginning and the end of cytokinesis. Because Rho1 localizes to the division site throughout cytokinesis (Yoshida et al., 2009; our unpublished data), it seemed likely that Rho1 simply remained active during the entire process. However, we found that in cdc15-2–synchronized cells, in which AMR formation and cleavage-furrow ingression are temporarily separated, global Rho1 activity decreased substantially before furrow ingression, suggesting that the activity Rho1 might decrease even more dramatically at the division site. Moreover, Rho1T24N did not block AMR constriction or PS formation, suggesting that Rho1 activity is dispensable for these processes. Indeed, overexpression of Rho1 significantly inhibited PS formation in cells (cyk3Δ hof1Δ) in which this process is already inefficient, suggesting that active Rho1 may actually inhibit PS formation. The dispensability of Rho1 for PS formation seems surprising given that proper localization of Chs2 requires the exocyst subunit Sec3 (VerPlank and Li, 2005), which is regulated by Rho1 as an effector (Guo et al., 2001). However, another Rho protein, such as Rho3 or Cdc42 (Wu and Brennwald, 2010), may be primarily responsible for Chs2 delivery during PS formation.
It remains possible that a small quantity of active Rho1 localizes to the leading edge of the cleavage furrow and promotes AMR contractility as seen in many other organisms (Fig. 9 B; Balasubramanian et al., 2004; Piekny et al., 2005; Bement et al., 2006). However, because of the force provided by PS formation, the AMR in S. cerevisiae can constrict at a nearly normal rate even when no force is provided by actomyosin contractility (Lord et al., 2005; Fang et al., 2010), making it difficult to assess intrinsic AMR contractility in this system.
Consistent with our observations in yeast, it appears that in at least some types of mammalian cells, a deficiency in RhoA activity at the division site does not block furrow ingression (O’Connell et al., 1999; Yoshizaki et al., 2004). Thus, once the AMR is formed and its contraction is initiated, RhoA may not be required for further cleavage-furrow ingression, and its role in abscission (Chalamalasetty et al., 2006; Gai et al., 2011) may be more important.
Temporary inactivation of Rho1 by Cyk3 during cleavage-furrow ingression
Cyk3 was originally identified as a dosage suppressor of iqg1Δ (Korinek et al., 2000), and its overexpression has been found to promote PS formation in both wild-type and mutant (hof1Δ, myo1Δ, inn1Δ, and a hypomorphic chs2 mutation) strains (our unpublished data; Nishihama et al., 2009; Oh et al., 2012; Wloka and Bi, 2012). Subsequent studies have shown that binding of the SH3 domain and PXXP motifs of Cyk3 to Inn1 and Hof1, respectively, is important for PS formation (unpublished data; Nishihama et al., 2009; Meitinger et al., 2011). In addition, it has been reported that overexpression of Cyk3 suppressed SS formation in a mitotic exit network mutant forced to exit from mitosis (Meitinger et al., 2010), although the mechanism was not defined. In this study, we found a previously unknown link between Cyk3 and Rho1 that involves the TGc domain of Cyk3. Both cyk3Δ and cyk3TGcΔ mutants showed simultaneous formation of PS and SS, a synthetic growth defect with deletion of the Rho1 GAP gene SAC7, and a complete loss of the temporary inactivation of Rho1 during furrow ingression. Pull-down experiments showed that the C-terminal region of Cyk3 bound preferentially to inactive Rho1 and that this interaction was lost in the cyk3TGcΔ mutant, and BiFC assays reproduced these findings in vivo.
Transglutaminases from bacterial toxins and human cells have been shown to directly deamidate or transglutaminate Gln-63 of human RhoA (Horiguchi et al., 1997; Schmidt et al., 1997; Singh et al., 2001), causing a loss of GTPase activity and thus constitutive activity of the protein. Interestingly, the TGc domain in Cyk3 lacks a Cys (Pro in Cyk3) that is one of the three residues critical for transglutaminase activity (Makarova et al., 1999; Pollard et al., 2012) and so should be enzymatically inactive. Therefore, Cyk3 may function by binding to Rho1 without modifying or activating it. Because the switch II region of RhoA is necessary and sufficient for its recognition by the bacterial toxin transglutaminases CNF1 and DNT (Lerm et al., 1999; Jank et al., 2006) and is also the central recognition site for Rho-GEFs (Hakoshima et al., 2003), the binding of Cyk3 to Rho1-GDP could interfere with its normal activation by Rho-GEFs (Fig. 9 B).
Because Rho1 appears to have an inhibitory effect on PS formation and thus furrow ingression, it is possible that the promotion of PS formation by overexpression of Cyk3 may be through an inhibition of Rho1. However, we consider this to be unlikely because the phenotype of cyk3Δ is more severe than cyk3TGcΔ, and unlike cyk3TGcΔ, mutations in the N-terminal region (cyk3W45A and cyk3P188,191A) did not show genetic interactions with sac7Δ. Both of these results suggest that the N-terminal and C-terminal regions of Cyk3 have genetically separable roles. Therefore, Cyk3 appears to be a bifunctional protein, with its N-terminal region involved in promoting PS formation through interactions with Inn1 and Hof1, and its C-terminal region involved in suppression of SS formation through interaction with Rho1.
It is presently unclear how Rho1 is reactivated for SS formation (Fig. 9 C). Although the BiFC signal between Cyk3 and Rho1 persisted after AMR disassembly, this signal may well be an artifact caused by the inherent irreversibility of binding of the BiFC probes that hinders its use for analysis of dissociation between two molecules (Kodama and Hu, 2010). Thus, it seems more likely that Cyk3 dissociates from Rho1 to allow its reactivation and that the dissociation is regulated by some mechanism such as phosphorylation.
A conserved role for transglutaminase-like domains in cytokinesis?
There appears to exist a subclass of eukaryotic transglutaminase-like proteins that lack one or more of the three residues required for catalytic activity (Makarova et al., 1999), and some of them have been implicated in cytokinesis. For example, a Schizosaccharomyces pombe cyk3Δ mutant shows reduced efficiency of PS formation and premature SS formation in certain mutant backgrounds (Pollard et al., 2012), suggesting that the S. pombe and S. cerevisiae proteins have similar roles despite the apparent differences in cell wall compositions and mechanisms of cytokinesis between these yeasts. Moreover, overexpression of S. pombe Cyk3 induces cell wall damage and morphology defects (Pollard et al., 2012), which could be a consequence of Rho1 inhibition. Drosophila hillarin is concentrated at the cleavage furrow during cell division, and in a septin mutant background, a mutation in hillarin causes increased polyploidy (Ji et al., 2005). Similarly, Caenorhabditis elegans Ltd-1 is expressed during seam cell division and localizes in a pattern reminiscent of actin (Vargas et al., 2002). It will be interesting to determine whether these other transglutaminase-like proteins also function, at least in part, by regulating Rho proteins.
Materials and methods
Strains, plasmids, growth conditions, genetic methods, and reagents
The yeast strains and plasmids used are listed in Table 1 and Table 2. Standard culture media (including the buffered rich medium YM-P) and genetic techniques were used (Lillie and Pringle, 1980; Guthrie and Fink, 1991). The PCR method (Longtine et al., 1998) was used for gene deletion and tagging except where noted. 1 mg/ml 5-fluoroorotic acid (5-FOA; Research Products International) was added where noted. Cell cycle synchronization using cdc15-2 or nocodazole (Toronto Research Chemicals) was performed essentially as described by Futcher (1999). In brief, cdc15-2 cells were cultured at 24°C in either YM-P medium or synthetic complete (SC) medium with appropriate supplements and carbon sources to early log phase (OD600 ≈ 0.2) and then arrested by gradually increasing the temperature to 37°C. After 3 h, the cells were released by rapidly cooling the medium to 24°C on ice. For nocodazole synchronization, cells were cultured in YPD medium at 24°C to early log phase, and nocodazole was added at 15 µg/ml. After 2 h, the cells were collected and washed by filtration and then resuspended in fresh medium. Aniline blue was obtained from EMD Millipore, and a 0.1% Ponceau S solution was obtained from Sigma-Aldrich.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
| YEF473A | MATa his3 leu2 lys2 trp1 ura3 | Bi and Pringle, 1996 |
| YEF473B | MATα his3 leu2 lys2 trp1 ura3 | Bi and Pringle, 1996 |
| KN143 | As YEF473B except LEU2:PGAL1-RHO1Q68L | This studya |
| KO969 | As YEF473A except iqg1Δ::his3MX6 | This study |
| KO1372 | As RNY2242 except (YEp13-RHO1) | This study |
| MWY636 | As YEF473A except cyk3P188,191A | Our laboratory |
| MWY1412 | As YEF473A except cyk3W45A | Our laboratory |
| RNY469 | AsYEF473A except myo1Δ::kanMX6 (pRS316GW-MYO1) | This study |
| RNY502 | As YEF473A except cyk3Δ::TRP1 | This study |
| RNY875 | As YEF473A except rom2Δ::TRP1 | This study |
| RNY879 | As YEF473A except tus1Δ::His3MX6 | This study |
| RNY935 | As YEF473A except rom1Δ::His3MX6 | This study |
| RNY1419 | As YEF473A except chs2Δ::kanMX6 (pJC328) | This study |
| RNY1681 | As YEF473A except MYO1-GFP:kanMX6 | This study |
| RNY2046 | As YEF473B except cdc15-2 MYO1-CFP:kanMX6 | This study |
| RNY2127 | As YEF473A except cyk3Δ::kanMX6 hof1Δ::TRP1 (pRS316GW-HOF1) | This study |
| RNY2150 | As YEF473A except cyk3Δ::URA3-kanMX6 | This study |
| RNY2242 | As YEF473A except iqg1Δ::His3MX6 (pRS316GW-IQG1) | This study |
| RNY2299 | As YEF473A except hof1Δ::kanMX6 | This study |
| MOY22 | As YEF473A except HOF1-TAP:His3MX6 | This study |
| MOY66 | As YEF473B except cyk3Δ::TRP1 hof1Δ::kanMX6 (pRS316GW-HOF1) | This study |
| MOY68 | As YEF473A except cyk3Δ::TRP1 hof1Δ::kanMX6 (pRS316GW-HOF1) | This study |
| MOY78 | As MOY68 except (YEp13-cdc24*) | This study |
| MOY245 | As MOY66 except (YEp13-RHO1) | This study |
| MOY355 | AS RNY2242 except (YEp13-RGD2) | This study |
| MOY403 | As YEF473B except PXL1-GFP:His3MX6 | This study |
| MOY404 | As YEF473B except pxl1Δ::His3MX6 | This study |
| MOY405 | As YEF473B except sac7Δ::His3MX6 | This study |
| MOY407 | As YEF473B except lrg1Δ::His3MX6 | This study |
| MOY428 | As YEF473B except myo1Δ::kanMX6 pxl1Δ::His3MX6 (pRS316GW-MYO1) | This study |
| MOY429 | As YEF473A except cyk3Δ::TRP1 pxl1Δ::His3MX6 | This study |
| MOY430 | As YEF473B except cyk3Δ::TRP1 hof1Δ::kanMX6 pxl1Δ::His3MX6 (pRS316GW-HOF1) | This study |
| MOY433 | As YEF473A except cyk3Δ::TRP1 hof1Δ::kanMX6 lrg1Δ::His3MX6 (pRS316GW-HOF1) | This study |
| MOY438 | As YEF473A except cyk3Δ::TRP1 hof1Δ::kanMX6 sac7Δ::His3MX6 (pRS316GW-HOF1) | This study |
| MOY440 | As YEF473B except cyk3Δ::TRP1 sac7Δ::His3MX6 | This study |
| MOY445 | As YEF473A except hof1Δ::kanMX6 pxl1Δ::His3MX6 | This study |
| MOY522 | As YEF473A except rho1Δ::his3MX6 URA3:3HA-RHO1 | This studyb |
| MOY526 | As YEF473A except MYO1-CFP:kanMX6 PXL1-GFP:His3MX6 | This study |
| MOY542 | As YEF473A except cdc15-2 MYO1-GFP:kanMX6 (YCp-PGAL-RHO1T24N) | This study |
| MOY543 | As YEF473B except cdc15-2 CYK3-GFP:kanMX6 | This study |
| MOY552 | As YEF473B except rho1Δ::his3MX6 URA3:3HA-RHO1 cdc15-2 cyk3Δ::kanMX6 | This study |
| MOY553 | As YEF473B except rho1Δ::his3MX6 URA3:3HA-RHO1 cdc15-2 | This study |
| MOY585 | As YEF473A except cyk3Δ::TRP1 | This study |
| MOY681 | As MOY68 except (pGP564-RHO2) | This study |
| MOY682 | As MOY68 except (YEp13-RGD2) | This study |
| MOY691 | As YEF473B except cyk3Δ::TRP1 LEU2:PGAL1-RHO1Q68L | This study |
| MOY720 | As YEF473A except cdc15-2 MYO1-GFP:kanMX6 | This study |
| MOY721 | As YEF473B except cdc15-2 MYO1-GFP:kanMX6 cyk3Δ::kanMX6 | This study |
| MOY801 | As YEF473A except (YCp-PGAL-3HA-RHO1) | This study |
| MOY803 | As YEF473A except (YCp-PGAL-3HA-RHO1Q68L) | This study |
| MOY824 | As YEF473A except (YCp-PGAL-3HA-RHO1T24N) | This study |
| MOY882 | As YEF473A except cyk3TGcΔ | This studyc |
| MOY967 | As YEF473A except sac7Δ::His3MX6 cyk3TGcΔ | This study |
| MOY973 | As YEF473A except cyk3TGcΔ rho1Δ::His3MX6 URA3:3HA-RHO1 cdc15-2 | This study |
| MOY980 | As YEF473A except sac7Δ::His3MX6 cyk3P188,191A | This study |
| MOY982 | As YEF473A except sac7Δ::His3MX6 cyk3W45A | This study |
| MOY1247 | As YEF473B except rho1Δ::His3MX6 MYO1-CFP:kanMX6 (pRS315-VC-GGS2-RHO1) | This study |
| MOY1269 | As YEF473A except TUS1-VN173:His3MX6 rho1Δ::His3MX6 (pRS315-VC-GGS2-RHO1) | This study |
| MOY1275 | As YEF473A except SAC7-VN173:His3MX6 rho1Δ::His3MX6 (pRS315-VC-GGS2-RHO1) | This study |
| MOY1293 | As YEF473A except ROM2-VN173:TRP1 rho1Δ::His3MX6 [pRS315-VC-GGS2-RHO1] | This study |
| MOY1294 | As YEF473A except LRG1-VN173:TRP1 rho1Δ::His3MX6 (pRS315-VC-GGS2-RHO1) | This study |
| MOY1303 | MATα (ade2 or ADE2) his3 lys2 trp1 ura3 Δfks1::HIS3 ade3::FKS1-GFP::LEU2 MYO1-CFP:kanMX6 Δfks2::LYS2 | This studyd |
| MOY1321 | As YEF473A except CYK3-VN155I152L:TRP1 | This study |
| MOY1325 | As YEF473B except CYK3-VN155I152L:TRP1 rho1Δ::His3MX6 (pRS315-VC-GGS2-RHO1) | This study |
| MOY1349 | As YEF473B except CYK3-VN155I152L:TRP1 rho1Δ::His3MX6 MYO1-CFP:kanMX6 (pRS315-VC-GGS2-RHO1) | This study |
| MOY1384 | As YEF473A except CYK3TGcΔ-VN155I152L:TRP1 rho1Δ::His3MX6 (pRS315-VC-GGS2-RHO1) | This study |
Except where noted, strains were constructed using conventional genetic crosses, the PCR method (Baudin et al., 1993; Longtine et al., 1998) for deletion and tagging of chromosomal genes, and/or plasmid transformations (Table 2). The cdc15-2 marker was introduced as described by Nishihama et al. (2009). In addition to the PCR tagging plasmids described by Longtine et al. (1998), similar plasmids contained CFP sequences (unpublished data), TAP tag sequences (provided by P. Walter, University of California, San Francisco, San Francisco, CA), URA3-kanMX6 (unpublished data), VN173 (Sung and Huh, 2007), or VN155I153L (Table 2).
Constructed by transforming EcoRV-digested YIp128-PGAL1RHO1Q68L into YEF473B.
Constructed by transforming NcoI-digested pRS306-3HA-RHO1 into a YEF473-background rho1Δ::His3MX6/RHO1 heterozygote and selecting Ura + His+ segregants.
Constructed by digesting pRS315-cyk3TGcΔ with SphI (at −384 relative to the CYK3 start codon) and SacII (at 2,935, 464 bp downstream of the stop codon), transforming into strain RNY2150, and selecting Ura− colonies on an SC + 5-FOA plate. Correct integration was confirmed by showing kanamycin sensitivity and PCR amplifying genomic DNA using appropriate primers.
Constructed by crossing YOC2439 (MATa ade2 his3 lys2 trp1 ura3 Δfks1::HIS3 Δfks2::LYS2 ade3::FKS1-GFP::LEU2; a gift from Y. Ohya, University of Tokyo, Tokyo, Japan) and RNY2046.
Table 2.
Plasmids used in this study
| Plasmid | Descriptiona | Reference or source |
| pGEX-2T | For expression of GST in E. coli | GE Healthcare |
| pGEX-RBD (PKC1) | For expression of GST-RBD (Pkc1) in E. coli | Yoshida et al., 2006; Kono et al., 2008 |
| pGEX2T-CYK3 | For expression of GST-Cyk3 in E. coli | See Supplemental material |
| pGEX2T-CYK3480–885 | For expression of GST-Cyk3480-885 in E. coli | See Supplemental material |
| YEp13b | 2µ and LEU2 | Rose and Broach, 1990 |
| pJC328 | CEN, URA3, and CHS2-MYC | Chuang and Schekman, 1996 |
| YCp111-CDC3-CFP | CEN, LEU2, and CDC3-CFP | Nishihama et al., 2009 |
| pRS316GW-IQG1 | CEN, URA3, and IQG1 | Nishihama et al., 2009 |
| pRS316GW-HOF1 | CEN, URA3, and HOF1 | Unpublished data |
| pRS425GW-CYK3b | 2µ, LEU2, and CYK3 | Unpublished data |
| pRS316GW-MYO1 | CEN, URA3, and MYO1 | Unpublished data |
| YEp13-RHO1b | 2µ, LEU2, and RHO1 | This studyc |
| pGP564-RHO2b | 2µ, LEU2, RHO2, and neighboring genesd | This studye |
| YEp13-RGD2b | 2µ, LEU2, and RGD2 | This studyc |
| YEp13-cdc24*b,f | 2µ, LEU2, and cdc24* | This studyc |
| YEp13-CDC24b,f | 2µ, LEU2, and CDC24 | See Supplemental material |
| pSD1b | 2µ, LEU2, and MSS4 | M.N. Hallg |
| YEp13-KDX1b | 2µ, LEU2, and KDX1 | This studyc |
| pGP564-PSP1b | 2µ, LEU2, PSP1, and neighboring genesd | This studye |
| YEp13-NAB6b | 2µ, LEU2, NAB6, and neighboring genesd | This studyc |
| YEp13-PAP1b | 2µ, LEU2, and PAP1 | This studyc |
| pRS425GW-TUS1b | 2µ, LEU2, and TUS1 | See Supplemental material |
| pRS425GW-ROM1b | 2µ, LEU2, and ROM1 | See Supplemental material |
| pRS425GW-ROM2b | 2µ, LEU2, and ROM2 | See Supplemental material |
| pRS425GW-RGD1b | 2µ, LEU2, and RGD1 | See Supplemental material |
| YEp181-RGA1b | 2µ, LEU2, and RGA1 | Caviston et al., 2003 |
| YEp181-RGA2b | 2µ, LEU2, and RGA2 | See Supplemental material |
| YEp181-BEM2b | 2µ, LEU2, and BEM2 | Unpublished data |
| YEp13-BEM3b | 2µ, LEU2, and BEM3 | Bi and Pringle, 1996 |
| pRS425GW-SAC7b | 2µ, LEU2, and SAC7 | See Supplemental material |
| pRS425GW-LRG1b | 2µ, LEU2, and LRG1 | See Supplemental material |
| pGP564-BAG7b | 2µ, LEU2, BAG7, and neighboring genesd | This studye |
| YEp13-cdc24ΔPB1f | 2µ, LEU2, and cdc24ΔPB1 | See Supplemental material |
| YEp13-cdc24*-DH8f | 2µ, LEU2, and cdc24* with N452G and E453G mutations | See Supplemental material |
| pRS425-CDC24-GFPf | 2µ, LEU2, and CDC24-GFP | See Supplemental material |
| YEp13-cdc24*-GFPf | 2µ, LEU2, and cdc24ΔPB1-GFP-tetA | See Supplemental material |
| pRS425-cdc24ΔPB1-GFPf | 2µ, LEU2, and cdc24ΔPB1-GFP | See Supplemental material |
| YCp-PGAL-RHO1T24N | CEN, LEU2, and PGAL-RHO1T24N | See Supplemental material |
| YIp128-PGAL-RHO1Q68L | Integrative, LEU2, and PGAL-RHO1Q68L | See Supplemental material |
| pRS306-3HA-RHO1 | Integrative, URA3, and 3HA-RHO1 | See Supplemental material |
| YCp-PGAL-3HA-RHO1 | CEN, LEU2, and PGAL-3HA-RHO1 | See Supplemental material |
| YCp-PGAL-3HA-RHO1Q68L | CEN, LEU2, and PGAL-3HA-RHO1Q68L | See Supplemental material |
| YCp-PGAL-3HA-RHO1T24N | CEN, LEU2, and PGAL-3HA-RHO1T24N | See Supplemental material |
| pFA6a-VN173:TRP1 | PCR template for chromosomal tagging with VN173 | Sung and Huh, 2007 |
| pFA6a-VN173:His3MX6 | PCR template for chromosomal tagging with VN173 | Sung and Huh, 2007 |
| pFA6a-VC155-TRP1 | PCR template for chromosomal tagging with VC155 | Sung and Huh, 2007 |
| pFA6a-VN155I153L-TRP1 | PCR template for chromosomal tagging with VN155I153L | See Supplemental material |
| pRS315-VC-GGS2-RHO1 | CEN, LEU2, and VC155-GGSGGS-RHO1 | See Supplemental material |
| pRS315-CYK3TGcΔh | CEN, LEU2, and CYK3 lacking bp 1,556–1,739 | See Supplemental material |
CEN indicates a low-copy number plasmid; 2µ indicates a high-copy number plasmid; integrative indicates a plasmid without a yeast replication origin.
Used in Fig. 2 A and/or Fig. S1.
Isolated in the suppressor screen using the YEp13-based library (see Materials and methods) or by subcloning a single-gene fragment from the originally isolated plasmid back into YEp13.
The actual plasmids used in the experiments shown in this paper are listed. The genes responsible for suppression were confirmed by further truncation.
Isolated from a genomic tiling library (Thermo Fisher Scientific).
Used in Fig. S3.
University of Basel, Basel, Switzerland.
Used for construction of strain MOY882.
Plasmid constructions
To construct pGEX2T-CYK3 and pGEX2T-CYK3480–885, the appropriate regions were PCR amplified from yeast genomic DNA with BamHI and HindIII sequences on the 5′ and 3′ ends, respectively, and inserted between the corresponding sites in pGEX-2T. YEp13-CDC24 was constructed by PCR amplifying a DNA fragment containing nucleotides 2,296–2,859 relative to the start site of CDC24 and 1–40 relative to the SphI of YEp13 (incorporated in the primer) using pRS315-CDC24 (provided by E. Bi, University of Pennsylvania, Philadelphia, PA) as a template. This fragment was cloned by gap repair into SphI-digested YEp13-cdc24* (Table 2) by cotransforming into YEF473A.
pRS315GW-HOF1, pRS315GW-MYO1, pRS425GW-CYK3, pRS425GW-TUS1, pRS425GW-ROM1, pRS425GW-ROM2, pRS425GW-RGD1, pRS425GW-SAC7, and pRS425GW-LRG1 were constructed as follows. Genomic DNA fragments containing each gene were amplified by PCR (positions relative to the start sites: HOF1, −1,000 to 2,510; MYO1, −782 to 6,150; CYK3, −900 to 3,248; TUS1, −500 to 4,424; ROM1, −1,000 to 3,968; ROM2, −500 to 4,571; RGD1, −1,000 to 2,501; SAC7, −1,000 to 2,465; and LRG1, −1,000 to 3,554), cloned into the pCR8/GW/TOPO vector (Invitrogen), and transferred into pRS315-attR or pRS425-attR (this study) by Gateway recombination (Invitrogen). YEp181-RGA2 was constructed by ligating a SacI–KpnI fragment from pDLB1981 (Gladfelter et al., 2002) into the corresponding sites of YEplac181 (Gietz and Sugino, 1988).
YEp13-cdc24ΔPB1 was constructed by using a mutagenesis kit (QuikChange; Agilent Technologies) to introduce a stop codon into YEp13-cdc24* immediately before the BamHI site at the junction between the CDC24 and tetA coding sequences. YEp13-cdc24*-DH8 was constructed in two steps. First, a fragment containing nucleotides 764–1,659 relative to the start site of CDC24 and two mutations (N452G and E453G) within the DH (Dbl homology) domain was PCR amplified using plasmid pRS414-3×mycCdc24-DH8 (provided by R. Arkowitz, Université de Nice, Nice, France) as a template. The PCR product was then cloned by gap repair into BbvCI-cut YEp13-cdc24*. To construct pRS425-CDC24-GFP, an XhoI fragment from pRS315-CDC24-GFP (provided by E. Bi) was integrated by gap repair into NotI-digested pRS425 (Christianson et al., 1992). YEp13-cdc24*-GFP was constructed in three steps. First, a DNA fragment containing base pairs 2,261–2,281 of CDC24, the GFP sequence, and base pairs 1–40 relative to the BamHI site of YEp13 (incorporated in the primer) was PCR amplified using pRS315-CDC24-GFP as a template. Second, pRS315-CDC24-GFP was digested by NheI and BglII to generate a DNA fragment containing base pairs −146 to −1 relative to the BamHI site of YEp13 and the CDC24 gene. Finally, these two fragments were integrated into BamHI-digested YEp13 by gap repair. YEp13-cdc24ΔPB1-GFP was constructed in the same way except that a primer generating a stop codon between the GFP sequence and the BamHI site was used in the PCR step.
YCp-PGAL-RHO1T24N was constructed as follows. First, site-directed mutagenesis was performed in pRS316-RHO1 (this study) using the QuikChange kit (primer 5′-CTGTGGTAAGAACTGTTTATTAATC-3′ and its complement). The resulting pRS316-RHO1T24N did not yield any yeast transformants, presumably as a result of dominant lethality of this allele. Next, the RHO1T24N ORF was PCR amplified with DNA fragments homologous to base pairs −40 to −1 and 1–40 relative to the SalI site of YCpIF2 (Foreman and Davis, 1994) at the 5′ and 3′ ends, respectively. Finally, this PCR product was integrated into SalI-digested YCpIF2 by gap repair. YIp128-PGAL-RHO1Q68L was constructed as follows. First, the NdeI–SacI fragment containing RHO1Q68L from pRS304-Pcdc42-12myc-RHO1Q68L was inserted into the corresponding sites of pRS304-PGAL1 (both plasmids provided by D. Lew, Duke University, Durham, NC). From the resulting plasmid, a DNA fragment containing PGAL-RHO1Q68L was released by digestion with EcoRI and SacI and ligated into the corresponding sites of YIp128 (Gietz and Sugino, 1988).
To construct pRS306-3HA-RHO1, plasmid pRS316-RHO1 was digested at the NotI site in the vector, blunt ended, and self-ligated to eliminate the site. A new NotI site was then introduced immediately downstream of the RHO1 start codon by site-directed mutagenesis using the QuikChange kit. A DNA fragment encoding 3HA with NotI site ends was then PCR amplified using YEpHA-BUD8F (Schenkman et al., 2002) as a template and inserted at the new NotI site. Finally, the insert of this plasmid was transferred into pRS306 using the HindIII and SacI sites in both plasmids. YCp-PGAL-3HA-RHO1, YCp-PGAL-3HA-RHO1T24N, and YCp-PGAL-3HA-RHO1Q68L were constructed using the same approach as for YCp-PGAL-RHO1T24N (see preceding paragraph), using pRS316-RHO1, pRS316-RHO1T24N, and pRS304-Pcdc42-12myc-RHO1Q68L (provided by D. Lew) as PCR templates. The PCR products were integrated into SalI–PstI-digested YCpIF2-3HA-BUD8 (this study).
Plasmid pFA6a-VN155I153L-TRP1 was constructed by PCR amplifying a fragment encoding aa 1–155 of Venus using pFA6a-VN173-TRP1 (Sung and Huh, 2007) as a template and incorporating an I to L mutation at aa 153; the PCR used an F2 primer (Longtine et al., 1998) with a BamHI site upstream of the coding sequence and a reverse primer (5′-ATTTAGAAGTGGCGCGCCCTAGGCGGTGAGATAGACGTTGTGGCT-3′) that included an AscI site (underlined). The PCR product was digested with BamHI and AscI and then ligated into the corresponding sites of pFA6a-VC155-TRP1. Plasmid pRS315-VC-GGS2-RHO1 was constructed by PCR amplifying a sequence encoding the C-terminal portion of Venus using pFA6a-VC155-TRP1 as a template and primers (5′-GAAAGATGTGCGGCCGCCGTCCGGCGTGCAAAATCCCG-3′ and 5′-GGGGATCCCTTGTACAGCTCGTCCATGCCGAGA-3′) that included NotI and BamHI sites (underlined). The PCR product was digested with NotI and BamHI and then ligated into the corresponding sites of pRS315-GFP-GGS2-RHO1 (this study).
To construct plasmid pRS315-CYK3TGcΔ, two DNA fragments containing genomic regions around the CYK3 ORF were PCR amplified using plasmid pRS315-CYK3 (this study) as a template and primers 5′-ATTAACCCTCACTAAAGGGA-3′ and 5′-CATTTCCTAAGTATAGTATTTTCTCTAAATTTTGG-3′ (fragment 1) and 5′-AATACTATACTTAGGAAATGTAACCAACCCAATTC-3′ and 5′-TAATACGACTCACTATAGGG-3′ (fragment 2). Fragment 1 contained the promoter and base pairs 1–1,555 of the CYK3 ORF, and fragment 2 contained base pairs from 1,740 of the ORF through the stop codon and terminator. These two fragments have 20 bp of homologous sequence at their 1,555 and 1,740 ends and were integrated into ApaI–SacI-digested pRS315 (Sikorski and Hieter, 1989) by gap repair.
Identification of multicopy suppressors for cyk3Δ hof1Δ
A cyk3Δ hof1Δ strain carrying a low-copy, URA3-marked HOF1 plasmid (MOY66) was transformed with a LEU2-marked high-copy (2µ) library in plasmid YEp13 (DeMarini et al., 1997), grown on a SC-Ura-Leu plate to form colonies, and replica plated onto SC-Leu + 5-FOA to select for ability to lose the HOF1 plasmid. From 35 transformants identified initially, plasmid recovery and retransformation yielded 22 plasmids that reproducibly rescued strain MOY66. Subcloning and/or deletion analyses identified the suppressor genes in 16 of these plasmids as (numbers of isolates in parentheses) CYK3 (3), HOF1 (2), RHO1 (2), KDX1 (2), RGD2 (1), cdc24* (1), PAP1 (2), NAB6 (2), and the 5-FOA resistance gene YJL055W (1; Ko et al., 2008). The remaining six plasmids contained distinct genome fragments in which the suppressor genes have not been determined. A parallel screen used a yeast genomic tiling library (Thermo Fisher Scientific) that was pooled from 384-well plates to five sublibraries and transformed separately; it yielded 26 plasmids that contained HOF1 (five plasmids), RHO1 (five plasmids), RHO2 (two plasmids), MSS4 (three plasmids), and RGD2 (11 plasmids), plus other plasmids that were isolated once each and not analyzed further.
Light microscopy and EM
Differential interference contrast and fluorescence microscopy were performed using a microscope (Eclipse 600-FN; Nikon), an Apochromat 100×/1.40 NA oil immersion objective, a cooled charge-coupled device camera (ORCA-2; Hamamatsu Photonics), and MetaMorph version 7.0 software (Molecular Devices). Most time-lapse microscopy was performed using the same equipment, by sandwiching cells suspended in SC medium between a coverslip and a slide and sealing with VALAP, as described previously (Nishihama et al., 2009). In one case (Fig. 5), the cells were spotted on a thin layer of YPD medium + 1% low-melting agarose (SeaPlaque; FMC Corporation), which was then placed on a coverslip. EM was performed essentially as described by Pollard et al. (2012): cells were collected by filtration, fixed with glutaraldehyde and potassium permanganate, embedded in LR white resin (Fluka; Sigma-Aldrich), and stained with uranyl acetate and lead citrate. Images were obtained using a microscope (JEM-1230; JEOL) equipped with a cooled charge-coupled device camera (967 or Orius SC1000A; Gatan). Images were postprocessed using the MetaMorph, Photoshop (Adobe), and/or ImageJ (National Institutes of Health) software.
Biochemistry
Expression and purification of GST-tagged proteins from Escherichia coli BL21 (DE3 and pLysS) were performed as described by Frangioni and Neel (1993). In brief, E. coli strains transformed with the expression vectors were cultured in LB medium at 37°C to OD600 ≈ 0.3, and IPTG was added at 0.1 mM. The cells were collected by centrifugation after 3 h of incubation at 37°C, resuspended in STE (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1 mM EDTA) + 0.1 mg/ml lysozyme, and incubated for 15 min on ice. The cells were lysed by addition of 1.5% (final concentration) sarkosyl and brief sonication on ice, and the lysate was cleared by centrifugation at 12,000 g for 5 min at 4°C. Supernatants were supplemented with 2% (final concentration) Triton X-100, incubated with glutathione–Sepharose beads for 1 h at 4°C and then washed three times with Rho pull-down (RPD) or RIPA buffer (see below) depending on the downstream application. The beads were then resuspended in the same buffer supplemented with 10% glycerol and stored at −80°C. The pull-down assays using the RBD from Pkc1 was performed as described by Kono et al. (2008) with modifications. In brief, protein extracts were prepared from frozen yeast cells using glass beads in RPD buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 12 mM MgCl2, 1 mM DTT, and 1% NP-40) supplemented with a complete protease inhibitor cocktail (Roche), and centrifuged at 12,000 g for 10 min. 3 mg of each cleared extract was incubated with 10 µl GST-RBD beads for 1 h at 4°C, washed three times with RPD buffer, and eluted by boiling in SDS sample buffer for 3 min. Samples were analyzed by SDS-PAGE (14%) and Western blotting using a peroxidase-conjugated mouse anti-HA antibody (3F10; Roche). 15 µg of the cleared extract was loaded in the input lanes. The pull-down assay using GST-Cyk3 and GST-Cyk3(480–885) was performed similarly but using RIPA buffer (6 mM Na2HPO4, 4 mM NaH2PO4, 150 mM NaCl, 2 mM EDTA, 1 mM DTT, 50 mM NaF, 0.1 mM NA3VO4, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) for protein extraction and incubation with the beads. Samples were analyzed by SDS-PAGE (14% for 3HA-Rho1 and 10% for Hof1–tandem affinity purification (TAP) and Western blotting using the anti-HA antibody or (for Hof1-TAP) peroxidase antiperoxidase soluble complex (Sigma-Aldrich).
Online supplemental material
Fig. S1 shows an expanded version of the data shown in Fig. 2 A, and also includes the iqg1Δ suppression results and the suppression by some GEFs and GAPs. Fig. S2 shows EM images of cyk3Δ hof1Δ and iqg1Δ cells expressing some of the suppressors. Fig. S3 shows a detailed characterization of cdc24*, which lead to the conclusion that it inhibits the Cdc42 pathway. Fig. S4 shows data implicating Pxl1 in regulation of SS formation. Fig. S5 shows supporting evidence of cytokinesis defects caused by ectopically activated Rho1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201302001/DC1.
Supplementary Material
Acknowledgments
We thank M. Wang of our laboratory for strains and permission to cite her unpublished data; K. Nakashima, R. Schekman, M. Hall, D. Lew, W.-K. Huh, E. Bi, Y. Ohya, K. Kono, S. Yoshida, and D. Pellman for strains and plasmids; J. Mulholland and J. Perrino (Stanford University Cell Sciences Imaging Facility) for assistance with EM; and B. Atkins, S. Yoshida, and D. Pellman for sharing unpublished information.
M. Onishi was supported in part by a postdoctoral fellowship from the Uehara Memorial Foundation. This work was supported by National Institutes of Health grant GM31006 (to J.R. Pringle).
Footnotes
Abbreviations used in this paper:
- 5-FOA
- 5-fluoroorotic acid
- AMR
- actomyosin ring
- BiFC
- bimolecular fluorescence complementation
- GAP
- GTPase-activating protein
- GEF
- guanine nucleotide exchange factor
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PS
- primary septum
- RBD
- Rho-binding domain
- RPD
- Rho pull-down
- SC
- synthetic complete
- SS
- secondary septum
- TAP
- tandem affinity purification
- TGc
- transglutaminase core
References
- Albertson R., Riggs B., Sullivan W. 2005. Membrane traffic: a driving force in cytokinesis. Trends Cell Biol. 15:92–101 10.1016/j.tcb.2004.12.008 [DOI] [PubMed] [Google Scholar]
- Atkins B.D., Yoshida S., Saito K., Wu C.-F., Lew D.J., Pellman D. 2013. Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J. Cell Biol. 202:231–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A., Emr S.D. 2002. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell. 2:593–605 10.1016/S1534-5807(02)00168-5 [DOI] [PubMed] [Google Scholar]
- Babour A., Bicknell A.A., Tourtellotte J., Niwa M. 2010. A surveillance pathway monitors the fitness of the endoplasmic reticulum to control its inheritance. Cell. 142:256–269 10.1016/j.cell.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M.K., Bi E., Glotzer M. 2004. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 14:R806–R818 10.1016/j.cub.2004.09.022 [DOI] [PubMed] [Google Scholar]
- Barr F.A., Gruneberg U. 2007. Cytokinesis: placing and making the final cut. Cell. 131:847–860 10.1016/j.cell.2007.11.011 [DOI] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329–3330 10.1093/nar/21.14.3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement W.M., Miller A.L., von Dassow G. 2006. Rho GTPase activity zones and transient contractile arrays. Bioessays. 28:983–993 10.1002/bies.20477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender L., Lo H.S., Lee H., Kokojan V., Peterson V., Bender A. 1996. Associations among PH and SH3 domain-containing proteins and Rho-type GTPases in Yeast. J. Cell Biol. 133:879–894 10.1083/jcb.133.4.879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Pringle J.R. 1996. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:5264–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Maddox P., Lew D.J., Salmon E.D., McMillan J.N., Yeh E., Pringle J.R. 1998. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142:1301–1312 10.1083/jcb.142.5.1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck D.C., Bloom K.S. 2005. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc. Natl. Acad. Sci. USA. 102:5408–5413 10.1073/pnas.0405925102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Abramson D.H. 1968. Cytokinesis in HeLa: post-telophase delay and microtubule-associated motility. Protoplasma. 66:413–435 10.1007/BF01255868 [DOI] [PubMed] [Google Scholar]
- Cabib E. 2004. The septation apparatus, a chitin-requiring machine in budding yeast. Arch. Biochem. Biophys. 426:201–207 10.1016/j.abb.2004.02.030 [DOI] [PubMed] [Google Scholar]
- Caviston J.P., Longtine M., Pringle J.R., Bi E. 2003. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell. 14:4051–4066 10.1091/mbc.E03-04-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalamalasetty R.B., Hümmer S., Nigg E.A., Silljé H.H. 2006. Influence of human Ect2 depletion and overexpression on cleavage furrow formation and abscission. J. Cell Sci. 119:3008–3019 10.1242/jcs.03032 [DOI] [PubMed] [Google Scholar]
- Christianson T.W., Sikorski R.S., Dante M., Shero J.H., Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene. 110:119–122 10.1016/0378-1119(92)90454-W [DOI] [PubMed] [Google Scholar]
- Chuang J.S., Schekman R.W. 1996. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 135:597–610 10.1083/jcb.135.3.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner A., Chin T.E., Brent R. 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 107:739–750 10.1016/S0092-8674(01)00596-7 [DOI] [PubMed] [Google Scholar]
- DeMarini D.J., Adams A.E., Fares H., De Virgilio C., Valle G., Chuang J.S., Pringle J.R. 1997. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139:75–93 10.1083/jcb.139.1.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J., Barral Y. 2004. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 305:393–396 10.1126/science.1099892 [DOI] [PubMed] [Google Scholar]
- Echard A., Hickson G.R., Foley E., O’Farrell P.H. 2004. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 14:1685–1693 10.1016/j.cub.2004.08.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp J.A., Chant J. 1997. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr. Biol. 7:921–929 10.1016/S0960-9822(06)00411-8 [DOI] [PubMed] [Google Scholar]
- Estey M.P., Di Ciano-Oliveira C., Froese C.D., Bejide M.T., Trimble W.S. 2010. Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. J. Cell Biol. 191:741–749 10.1083/jcb.201006031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Luo J., Nishihama R., Wloka C., Dravis C., Travaglia M., Iwase M., Vallen E.A., Bi E. 2010. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J. Cell Biol. 191:1333–1350 10.1083/jcb.201005134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fededa J.P., Gerlich D.W. 2012. Molecular control of animal cell cytokinesis. Nat. Cell Biol. 14:440–447 10.1038/ncb2482 [DOI] [PubMed] [Google Scholar]
- Foreman P.K., Davis R.W. 1994. Cloning vectors for the synthesis of epitope-tagged, truncated and chimeric proteins in Saccharomyces cerevisiae. Gene. 144:63–68 10.1016/0378-1119(94)90204-6 [DOI] [PubMed] [Google Scholar]
- Frangioni J.V., Neel B.G. 1993. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210:179–187 10.1006/abio.1993.1170 [DOI] [PubMed] [Google Scholar]
- Futcher B. 1999. Cell cycle synchronization. Methods Cell Sci. 21:79–86 10.1023/A:1009872403440 [DOI] [PubMed] [Google Scholar]
- Gai M., Camera P., Dema A., Bianchi F., Berto G., Scarpa E., Germena G., Di Cunto F. 2011. Citron kinase controls abscission through RhoA and anillin. Mol. Biol. Cell. 22:3768–3778 10.1091/mbc.E10-12-0952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X.D., Caviston J.P., Tcheperegine S.E., Bi E. 2004. Pxl1p, a paxillin-like protein in Saccharomyces cerevisiae, may coordinate Cdc42p and Rho1p functions during polarized growth. Mol. Biol. Cell. 15:3977–3985 10.1091/mbc.E04-01-0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Sugino A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 74:527–534 10.1016/0378-1119(88)90185-0 [DOI] [PubMed] [Google Scholar]
- Gladfelter A.S., Bose I., Zyla T.R., Bardes E.S., Lew D.J. 2002. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J. Cell Biol. 156:315–326 10.1083/jcb.200109062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Tamanoi F., Novick P. 2001. Spatial regulation of the exocyst complex by Rho1 GTPase. Nat. Cell Biol. 3:353–360 10.1038/35070029 [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G., 1991. Guide to yeast genetics and molecular biology. Methods in Enzymology, Vol. 194. San Diego: Academic Press; [PubMed] [Google Scholar]
- Hakoshima T., Shimizu T., Maesaki R. 2003. Structural basis of the Rho GTPase signaling. J. Biochem. 134:327–331 10.1093/jb/mvg149 [DOI] [PubMed] [Google Scholar]
- Horiguchi Y., Inoue N., Masuda M., Kashimoto T., Katahira J., Sugimoto N., Matsuda M. 1997. Bordetella bronchiseptica dermonecrotizing toxin induces reorganization of actin stress fibers through deamidation of Gln-63 of the GTP-binding protein Rho. Proc. Natl. Acad. Sci. USA. 94:11623–11626 10.1073/pnas.94.21.11623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H.Y., Olson S.K., Esko J.D., Horvitz H.R. 2003. Caenorhabditis elegans early embryogenesis and vulval morphogenesis require chondroitin biosynthesis. Nature. 423:439–443 10.1038/nature01634 [DOI] [PubMed] [Google Scholar]
- Jank T., Pack U., Giesemann T., Schmidt G., Aktories K. 2006. Exchange of a single amino acid switches the substrate properties of RhoA and RhoD toward glucosylating and transglutaminating toxins. J. Biol. Chem. 281:19527–19535 10.1074/jbc.M600863200 [DOI] [PubMed] [Google Scholar]
- Jendretzki A., Ciklic I., Rodicio R., Schmitz H.P., Heinisch J.J. 2009. Cyk3 acts in actomyosin ring independent cytokinesis by recruiting Inn1 to the yeast bud neck. Mol. Genet. Genomics. 282:437–451 10.1007/s00438-009-0476-0 [DOI] [PubMed] [Google Scholar]
- Ji Y., Rath U., Girton J., Johansen K.M., Johansen J. 2005. D-Hillarin, a novel W180-domain protein, affects cytokinesis through interaction with the septin family member Pnut. J. Neurobiol. 64:157–169 10.1002/neu.20131 [DOI] [PubMed] [Google Scholar]
- Kim K.Y., Truman A.W., Levin D.E. 2008. Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol. Cell. Biol. 28:2579–2589 10.1128/MCB.01795-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko N., Nishihama R., Pringle J.R. 2008. Control of 5-FOA and 5-FU resistance by Saccharomyces cerevisiae YJL055W. Yeast. 25:155–160 10.1002/yea.1554 [DOI] [PubMed] [Google Scholar]
- Kodama Y., Hu C.D. 2010. An improved bimolecular fluorescence complementation assay with a high signal-to-noise ratio. Biotechniques. 49:793–805 10.2144/000113519 [DOI] [PubMed] [Google Scholar]
- Kono K., Nogami S., Abe M., Nishizawa M., Morishita S., Pellman D., Ohya Y. 2008. G1/S cyclin-dependent kinase regulates small GTPase Rho1p through phosphorylation of RhoGEF Tus1p in Saccharomyces cerevisiae. Mol. Biol. Cell. 19:1763–1771 10.1091/mbc.E07-09-0950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono K., Saeki Y., Yoshida S., Tanaka K., Pellman D. 2012. Proteasomal degradation resolves competition between cell polarization and cellular wound healing. Cell. 150:151–164 10.1016/j.cell.2012.05.030 [DOI] [PubMed] [Google Scholar]
- Korinek W.S., Bi E., Epp J.A., Wang L., Ho J., Chant J. 2000. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr. Biol. 10:947–950 10.1016/S0960-9822(00)00626-6 [DOI] [PubMed] [Google Scholar]
- Kuranda M.J., Robbins P.W. 1991. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J. Biol. Chem. 266:19758–19767 [PubMed] [Google Scholar]
- Labedzka K., Tian C., Nussbaumer U., Timmermann S., Walther P., Müller J., Johnsson N. 2012. Sho1p connects the plasma membrane with proteins of the cytokinesis network through multiple isomeric interaction states. J. Cell Sci. 125:4103–4113 10.1242/jcs.105320 [DOI] [PubMed] [Google Scholar]
- Lerm M., Schmidt G., Goehring U.M., Schirmer J., Aktories K. 1999. Identification of the region of rho involved in substrate recognition by Escherichia coli cytotoxic necrotizing factor 1 (CNF1). J. Biol. Chem. 274:28999–29004 10.1074/jbc.274.41.28999 [DOI] [PubMed] [Google Scholar]
- Levin D.E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262–291 10.1128/MMBR.69.2.262-291.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie S.H., Pringle J.R. 1980. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol. 143:1384–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Li R. 1998. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140:355–366 10.1083/jcb.140.2.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Lorberg A., Schmitz H.P., Jacoby J.J., Heinisch J.J. 2001. Lrg1p functions as a putative GTPase-activating protein in the Pkc1p-mediated cell integrity pathway in Saccharomyces cerevisiae. Mol. Genet. Genomics. 266:514–526 10.1007/s004380100580 [DOI] [PubMed] [Google Scholar]
- Lord M., Laves E., Pollard T.D. 2005. Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol. Biol. Cell. 16:5346–5355 10.1091/mbc.E05-07-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackin N.A., Sousou T.J., Erdman S.E. 2004. The PXL1 gene of Saccharomyces cerevisiae encodes a paxillin-like protein functioning in polarized cell growth. Mol. Biol. Cell. 15:1904–1917 10.1091/mbc.E04-01-0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Aravind L., Koonin E.V. 1999. A superfamily of archaeal, bacterial, and eukaryotic proteins homologous to animal transglutaminases. Protein Sci. 8:1714–1719 10.1110/ps.8.8.1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y., Matsui R., Akada R., Toh-e A. 1996. Yeast src homology region 3 domain-binding proteins involved in bud formation. J. Cell Biol. 133:865–878 10.1083/jcb.133.4.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura F. 2005. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 15:371–377 10.1016/j.tcb.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Mazur P., Baginsky W. 1996. In vitro activity of 1,3-beta-D-glucan synthase requires the GTP-binding protein Rho1. J. Biol. Chem. 271:14604–14609 10.1074/jbc.271.24.14604 [DOI] [PubMed] [Google Scholar]
- McKay H.F., Burgess D.R. 2011. ‘Life is a highway’: membrane trafficking during cytokinesis. Traffic. 12:247–251 10.1111/j.1600-0854.2010.01139.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F., Petrova B., Lombardi I.M., Bertazzi D.T., Hub B., Zentgraf H., Pereira G. 2010. Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J. Cell Sci. 123:1851–1861 10.1242/jcs.063891 [DOI] [PubMed] [Google Scholar]
- Meitinger F., Boehm M.E., Hofmann A., Hub B., Zentgraf H., Lehmann W.D., Pereira G. 2011. Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev. 25:875–888 10.1101/gad.622411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M., Norden C., Durrer K., Rauter H., Uhlmann F., Barral Y. 2009. A mechanism for chromosome segregation sensing by the NoCut checkpoint. Nat. Cell Biol. 11:477–483 10.1038/ncb1855 [DOI] [PubMed] [Google Scholar]
- Mizuguchi S., Uyama T., Kitagawa H., Nomura K.H., Dejima K., Gengyo-Ando K., Mitani S., Sugahara K., Nomura K. 2003. Chondroitin proteoglycans are involved in cell division of Caenorhabditis elegans. Nature. 423:443–448 10.1038/nature01635 [DOI] [PubMed] [Google Scholar]
- Neto H., Gould G.W. 2011. The regulation of abscission by multi-protein complexes. J. Cell Sci. 124:3199–3207 10.1242/jcs.083949 [DOI] [PubMed] [Google Scholar]
- Ng M.M., Chang F., Burgess D.R. 2005. Movement of membrane domains and requirement of membrane signaling molecules for cytokinesis. Dev. Cell. 9:781–790 10.1016/j.devcel.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Nishihama R., Schreiter J.H., Onishi M., Vallen E.A., Hanna J., Moravcevic K., Lippincott M.F., Han H., Lemmon M.A., Pringle J.R., Bi E. 2009. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J. Cell Biol. 185:995–1012 10.1083/jcb.200903125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden C., Mendoza M., Dobbelaere J., Kotwaliwale C.V., Biggins S., Barral Y. 2006. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 125:85–98 10.1016/j.cell.2006.01.045 [DOI] [PubMed] [Google Scholar]
- O’Connell C.B., Wheatley S.P., Ahmed S., Wang Y.L. 1999. The small GTP-binding protein rho regulates cortical activities in cultured cells during division. J. Cell Biol. 144:305–313 10.1083/jcb.144.2.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y., Chang K.J., Orlean P., Wloka C., Deshaies R., Bi E. 2012. Mitotic exit kinase Dbf2 directly phosphorylates chitin synthase Chs2 to regulate cytokinesis in budding yeast. Mol. Biol. Cell. 23:2445–2456 10.1091/mbc.E12-01-0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palani S., Meitinger F., Boehm M.E., Lehmann W.D., Pereira G. 2012. Cdc14-dependent dephosphorylation of Inn1 contributes to Inn1-Cyk3 complex formation. J. Cell Sci. 125:3091–3096 10.1242/jcs.106021 [DOI] [PubMed] [Google Scholar]
- Piekny A., Werner M., Glotzer M. 2005. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 15:651–658 10.1016/j.tcb.2005.10.006 [DOI] [PubMed] [Google Scholar]
- Pollard L.W., Onishi M., Pringle J.R., Lord M. 2012. Fission yeast Cyk3p is a transglutaminase-like protein that participates in cytokinesis and cell morphogenesis. Mol. Biol. Cell. 23:2433–2444 10.1091/mbc.E11-07-0656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouzet-Mauléon V., Lefebvre F., Thoraval D., Crouzet M., Doignon F. 2008. Phosphoinositides affect both the cellular distribution and activity of the F-BAR-containing RhoGAP Rgd1p in yeast. J. Biol. Chem. 283:33249–33257 10.1074/jbc.M805161200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H., Python C.P., Inoue S.B., Arisawa M., Anraku Y., Zheng Y., Watanabe T., Levin D.E., Ohya Y. 1996. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science. 272:279–281 10.1126/science.272.5259.279 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Quiñones J.F., Irizarry R.A., Díaz-Blanco N.L., Rivera-Molina F.E., Gómez-Garzón D., Rodríguez-Medina J.R. 2008. Global mRNA expression analysis in myosin II deficient strains of Saccharomyces cerevisiae reveals an impairment of cell integrity functions. BMC Genomics. 9:34 10.1186/1471-2164-9-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A.B., Broach J.R. 1990. Propagation and expression of cloned genes in yeast: 2-microns circle-based vectors. Methods Enzymol. 185:234–279 10.1016/0076-6879(90)85024-I [DOI] [PubMed] [Google Scholar]
- Sanchez-Diaz A., Marchesi V., Murray S., Jones R., Pereira G., Edmondson R., Allen T., Labib K. 2008. Inn1 couples contraction of the actomyosin ring to membrane ingression during cytokinesis in budding yeast. Nat. Cell Biol. 10:395–406 10.1038/ncb1701 [DOI] [PubMed] [Google Scholar]
- Schenkman L.R., Caruso C., Pagé N., Pringle J.R. 2002. The role of cell cycle–regulated expression in the localization of spatial landmark proteins in yeast. J. Cell Biol. 156:829–841 10.1083/jcb.200107041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel J.A., Prekeris R. 2010. Making the final cut - mechanisms mediating the abscission step of cytokinesis. ScientificWorldJournal. 10:1424–1434 10.1100/tsw.2010.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt G., Sehr P., Wilm M., Selzer J., Mann M., Aktories K. 1997. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 387:725–729 10.1038/42735 [DOI] [PubMed] [Google Scholar]
- Shaw J.A., Mol P.C., Bowers B., Silverman S.J., Valdivieso M.H., Durán A., Cabib E. 1991. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 114:111–123 10.1083/jcb.114.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U.S., Kunar M.T., Kao Y.L., Baker K.M. 2001. Role of transglutaminase II in retinoic acid-induced activation of RhoA-associated kinase-2. EMBO J. 20:2413–2423 10.1093/emboj/20.10.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M.K., Huh W.K. 2007. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 24:767–775 10.1002/yea.1504 [DOI] [PubMed] [Google Scholar]
- Svarovsky M.J., Palecek S.P. 2005. Disruption of LRG1 inhibits mother-daughter separation in Saccharomyces cerevisiae. Yeast. 22:1117–1132 10.1002/yea.1301 [DOI] [PubMed] [Google Scholar]
- Tolliday N., VerPlank L., Li R. 2002. Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr. Biol. 12:1864–1870 10.1016/S0960-9822(02)01238-1 [DOI] [PubMed] [Google Scholar]
- Tully G.H., Nishihama R., Pringle J.R., Morgan D.O. 2009. The anaphase-promoting complex promotes actomyosin-ring disassembly during cytokinesis in yeast. Mol. Biol. Cell. 20:1201–1212 10.1091/mbc.E08-08-0822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda T.Q., Nagasaki A. 2004. Variations on a theme: the many modes of cytokinesis. Curr. Opin. Cell Biol. 16:55–60 10.1016/j.ceb.2003.11.004 [DOI] [PubMed] [Google Scholar]
- Valdivia R.H., Schekman R. 2003. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA. 100:10287–10292 10.1073/pnas.1834246100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas J.D., Culetto E., Ponting C.P., Miguel-Aliaga I., Davies K.E., Sattelle D.B. 2002. Cloning and developmental expression analysis of ltd-1, the Caenorhabditis elegans homologue of the mouse kyphoscoliosis (ky) gene. Mech. Dev. 117:289–292 10.1016/S0925-4773(02)00182-X [DOI] [PubMed] [Google Scholar]
- VerPlank L., Li R. 2005. Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis. Mol. Biol. Cell. 16:2529–2543 10.1091/mbc.E04-12-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe D., Abe M., Ohya Y. 2001. Yeast Lrg1p acts as a specialized RhoGAP regulating 1,3-beta-glucan synthesis. Yeast. 18:943–951 10.1002/yea.742 [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Takaesu G., Hagiwara M., Irie K., Matsumoto K. 1997. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:2615–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E.L. 2012. Mitotic exit and separation of mother and daughter cells. Genetics. 192:1165–1202 10.1534/genetics.112.145516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C., Bi E. 2012. Mechanisms of cytokinesis in budding yeast. Cytoskeleton (Hoboken). 69:710–726 10.1002/cm.21046 [DOI] [PubMed] [Google Scholar]
- Wloka C., Nishihama R., Onishi M., Oh Y., Hanna J., Pringle J.R., Krauss M., Bi E. 2011. Evidence that a septin diffusion barrier is dispensable for cytokinesis in budding yeast. Biol. Chem. 392:813–829 10.1515/BC.2011.083 [DOI] [PubMed] [Google Scholar]
- Wu H., Brennwald P. 2010. The function of two Rho family GTPases is determined by distinct patterns of cell surface localization. Mol. Cell. Biol. 30:5207–5217 10.1128/MCB.00366-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Vogel B.E. 2011. A secreted protein promotes cleavage furrow maturation during cytokinesis. Curr. Biol. 21:114–119 10.1016/j.cub.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Kono K., Lowery D.M., Bartolini S., Yaffe M.B., Ohya Y., Pellman D. 2006. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science. 313:108–111 10.1126/science.1126747 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Bartolini S., Pellman D. 2009. Mechanisms for concentrating Rho1 during cytokinesis. Genes Dev. 23:810–823 10.1101/gad.1785209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki H., Ohba Y., Parrini M.C., Dulyaninova N.G., Bresnick A.R., Mochizuki N., Matsuda M. 2004. Cell type-specific regulation of RhoA activity during cytokinesis. J. Biol. Chem. 279:44756–44762 10.1074/jbc.M402292200 [DOI] [PubMed] [Google Scholar]
- Young S.-H., Jacobs R.R. 1998. Sodium hydroxide-induced conformational change in schizophyllan detected by the fluorescence dye, aniline blue. Carbohydr. Res. 310:91–99 10.1016/S0008-6215(98)00167-0 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.