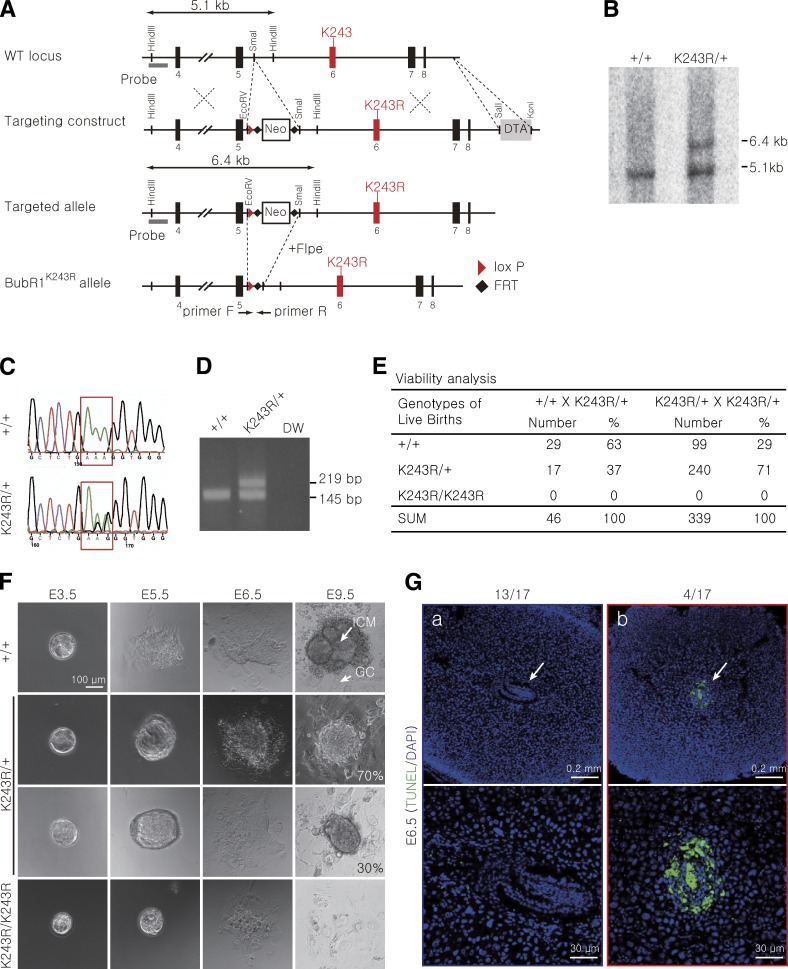
Failure of chromosome–spindle attachment and a weakened spindle assembly checkpoint lead to genetic instability and cancer in mice expressing acetylation-deficient BubR1.
Abstract
BubR1 acetylation is essential in mitosis. Mice heterozygous for the acetylation-deficient BubR1 allele (K243R/+) spontaneously developed tumors with massive chromosome missegregations. K243R/+ mouse embryonic fibroblasts (MEFs) exhibited a weakened spindle assembly checkpoint (SAC) with shortened mitotic timing. The generation of the SAC signal was intact, as Mad2 localization to the unattached kinetochore (KT) was unaltered; however, because of the premature degradation of K243R-BubR1, the mitotic checkpoint complex disassociated prematurely in the nocodazole-treated condition, suggesting that maintenance of the SAC is compromised. BubR1 acetylation was also required to counteract excessive Aurora B activity at the KT for stable chromosome–spindle attachments. The association of acetylation-deficient BubR1 with PP2A-B56α phosphatase was reduced, and the phosphorylated Ndc80 at the KT was elevated in K243R/+ MEFs. In relation, there was a marked increase of micronuclei and p53 mutation was frequently detected in primary tumors of K243R/+ mice. Collectively, the combined effects of failure in chromosome–spindle attachment and weakened SAC cause genetic instability and cancer in K243R/+ mice.
Introduction
The climax of mitosis is the poleward separation of sister chromatids into two daughter cells. For genetic integrity, the chromosome separation of the duplicated genome must be precisely controlled. Accurate chromosomal segregation in mitosis is ensured by the spindle assembly checkpoint (SAC), a process that ultimately inhibits anaphase-promoting complex/cyclosome (APC/C), which is the multisubunit E3 ligase responsible for the destruction of cyclin B and securin during mitosis, until all of the chromosomes are attached to microtubule (MT) spindles in a bipolar manner (Lara-Gonzalez et al., 2012).
BubR1 is a core component of the SAC, which constitutes the mitotic checkpoint complex (MCC) with Mad2, Bub3, and Cdc20 (Sudakin et al., 2001). The MCC inhibits APC/C in mitosis (Kim and Yu, 2011). BubR1 is also involved in the regulation of chromosome–spindle attachments (Lampson and Kapoor, 2005). BubR1 binds to KNL1/Blinkin, which constitutes the KMN (KNL1/Mis12/Ndc80) network at kinetochores (KTs), where MTs attach (Kiyomitsu et al., 2007; Bolanos-Garcia et al., 2011). Furthermore, BubR1, when phosphorylated by Plk1, recruits the B56α subunit of PP2A phosphatase to KTs and counteracts the excessive Aurora B activity on the KMN network. This dephosphorylation of the KMN network stabilizes the KT–MT attachment (Suijkerbuijk et al., 2012; Kruse et al., 2013). And, BubR1 binds to centromere-associated protein E (CENP-E; Mao et al., 2003; Guo et al., 2012), a plus end–directed motor that enables the gliding of monoorientated chromosomes along the existing KT fiber (K-fiber) to the cell equator (Kapoor et al., 2006). Therefore, in addition to SAC activity, BubR1 function is crucial in chromosome congression, the bipolar spindle attachment that forms the metaphase plate.
We reported previously that BubR1 is acetylated at a single lysine residue, K250, in prometaphase and that acetylated BubR1 inhibits APC/C-Cdc20, whereas deacetylated BubR1 becomes an APC/C substrate (Choi et al., 2009). Furthermore, BubR1 acetylation requires the breast cancer susceptibility gene BRCA2, suggesting that BubR1 acetylation is a tumor-suppressive mechanism (Choi et al., 2012).
To elucidate the physiological consequences of BubR1 acetylation, we generated an acetylation-deficient BubR1 allele (K243R) in mice. Here, we report that mice heterozygous for the K243R allele are predisposed to various types of cancers and that BubR1 acetylation is required for both SAC maintenance and chromosome congression.
Results
Generation of an acetylation-defective BubR1 allele in mice
We generated an acetylation-defective BubR1 allele in mice via substitution of the lysine 243 residue, which corresponds to lysine 250 in humans, with an arginine (K243R) using knock-in technology (Fig. 1, A–D). We monitored 339 live animals from BubR1K243R/+ intercrosses and failed to identify any homozygous mutant (BubR1K243R/K243R) newborn pups. In comparison, the heterozygous mutant mice (BubR1K243R/+; referred to here as K243R/+) survived to birth at the correct Mendelian frequency without apparent developmental defects (Fig. 1 E). Moreover, we did not detect early aging phenotypes, which were observed previously in hypomorphic BubR1 mice (Baker et al., 2004). BubR1 expression and the overall protein levels were unaltered in the thymus and testis of the K243R/+ mice (Fig. S1 A); therefore, these mice differ from BubR1-haploinsufficient mice (Wang et al., 2004) and hypomorphic BubR1 mice (Baker et al., 2004). The body weights and growth rates of the heterozygous mice were comparable to those of wild-type (WT) mice (Fig. S1 B).
Figure 1.
BubR1 acetylation is essential for embryonic development. (A) Schematic representation of the BubR1 gene-targeting strategy. Shown are the structures of the WT BubR1 locus, the targeting construct, the targeted locus, and the disrupted locus after the Flpe-mediated deletion. The neomycin-resistance gene in the targeting construct is removed when crossed with the Flpe transgenic mouse to generate the BubR1K243R (K243R) allele. The probe for Southern blot analysis and the primers for PCR genotyping are marked with gray bars and arrows, respectively. (B) Southern blot analysis of the WT and targeted ES cell lines using the probe indicated in A. HindIII-digested genomic DNA generated a 5.1-kb fragment in the WT and a 6.4-kb band in the targeted allele before the removal of the neo-resistance cassette. (C) Sequence analysis of genomic DNA from WT and K243R/+ ES cells. (D) Genotyping of WT and K243R/+ mice using PCR. (E) Summary of the crosses and progeny. The K243R/+ heterozygous mice were intercrossed, and the newborn pups were scored. (F) In vitro culture of the WT and K243R/+ embryos. The E3.5 embryos from the K243R/+ intercrosses were cultured in vitro. The inner cell mass (ICM) and the trophoblast giant cells (GC) are marked. (G) Sagittal sections of the E6.5 embryos in the uterus were examined for apoptosis using TUNEL staining. Low (top) and high (bottom) power magnification images are shown. The arrows indicate embryos or embryonic remnants.
To determine when BubR1K243R/K243R embryos die, 20 blastocysts at E3.5 from four K243R/+ intercrosses were subjected to in vitro culture and monitoring (Fig. 1 F). After 3 d, the WT embryos in culture (n = 4) were able to hatch, the trophoblast giant cells spread out in the culture dish, and the inner cell mass was well formed. Out of twelve heterozygous K243R/+ embryos, eight of them proliferated and exhibited a morphology and size comparable to WT, and four of them were able to grow in culture with growth retardation (Fig. 1 F). In comparison, four blastocysts stopped proliferating from E6.5: these embryos were obtained at a frequency similar to that of homozygous mutant BubR1K243R/K243R.
We were unable to obtain DNA that was adequate for genotyping the presumed homozygous mutant embryos. Therefore, as an alternative approach, 17 E6.5 embryos, which were derived from three intercrosses of K243R/+ parents, were fixed and embedded in paraffin and apoptosis was examined. Four out of seventeen embryos exhibited an abnormal morphology with massive TUNEL staining (Fig. 1 G). These phenotypes and apoptosis were observed only in K243R/+ intercrosses and were not observed when the K243R/+ mice were mated with WT mice. Therefore, these data suggest that BubR1K243R/K243R embryos die as a result of apoptosis at approximately E6.5.
Robust cancer development in K243R/+ mice
Progeny of the K243R/+ intercrosses, WT mice (n = 41) and K243R/+ mice (n = 121) on a mixed 129 × C57BL/6 background, were monitored until 38 mo of age. Although no overt developmental defects were noted, a significant incidence of spontaneous tumor development was observed between 60 and 70 wk after birth in the K243R/+ mice (Fig. 2 A). A marked increase in malignant tumor development (23.1%, n = 28) was observed (Table 1 and Fig. 2 A, right) for both solid (10.7%, n = 13) and hematologic (12.4%, n = 15) tumors. The tumors were found in a variety of tissues (Fig. 2 B), with an overall tumor incidence of 38% (Table 1 and Fig. 2 A, left).
Figure 2.
Spontaneous tumorigenesis in K243R/+ mice. (A) Tumor incidence was assessed using Kaplan-Meier graphs. The tumor-free survival analysis (left) includes both benign and malignant tumors; the cancer-free survival analysis (right) includes only the malignant cancers. The statistical analyses were performed using SPSS software. Total number of mice analyzed cumulatively: WT, n = 41; K243R/+, n = 121. (B) Representative H&E-stained sections of the major pathologies found in the K243R/+ mice. The insets show high-magnification images. (C) H&E staining of spleens. (D–F) Analysis of lymph node cells. (D) Freshly isolated lymph node cells were immunostained using the indicated antibodies and subjected to flow cytometric analysis. (E) Lymph node sections from the K243R/+ mice were stained using an anti-B220 antibody and were counterstained with hematoxylin. (F) Wright-Giemsa staining of cytocentrifuged lymph node cells. (G) Megakaryocytic leukemia in the K243R/+ mice. The spleens were stained with H&E (left) or anti–von Willebrand factor (vWF) antibody and hematoxylin (right).
Table 1.
Tumor spectrum and analysis
| Tumor types | BubR1K243R/+ (n = 121) | BubR1+/+ (n = 41) |
| Malignant tumors | 28 (23.1%) | 2 (4.8%) |
| Malignant solid tumors | 13 (10.7%) | 1 (2.4%) |
| Hepatocellular carcinoma | 6 (5.0%) | |
| Sarcomaa | 4 (3.3%) | 1 (2.4%) |
| Adenocarcinomab | 2 (1.7%) | |
| Other cancerc | 1 (0.8%) | |
| Hematologic malignancies | 15 (12.4%) | 1(2.4%) |
| Megakaryocyte leukemia | 1 (0.8%) | |
| B cell lymphoma | 11 (9.1%) | 1 (2.4%) |
| Metastatic lymphoma/leukemia | 3 (2.5%) | |
| Benign tumorsd | 18 (14.9%) | |
| Total tumor incidence | 46 (38.0%) | 2 (4.8%) |
Sarcomas included smooth muscle sarcoma (n = 2), endometrial stromal sarcoma (n = 1), and angiosarcoma (n = 1) in K243R/+ mice. A single fibrosarcoma was found in the WT.
Adenocarcinomas included lung adenocarcinoma (n = 1) and rectal adenocarcinoma (n = 1) in K243R/+.
Other cancer included skin cancer.
Benign tumors included simple cysts in the genital tract (n = 10), hemangioma (n = 5), epidermal cyst (n = 1), acrochordon (n = 1), and mesenchymal fibroma (n = 1).
Splenomegaly was frequently observed when the K243R/+ mice were killed at ∼60 wk after birth (Fig. 2 C, right). These splenomegalies included B cell lymphomas, which exhibited white pulp expansion with B220 positivity in either a follicular or diffuse pattern. The level of B220 varied, and a distinction between the B and the presumed T cell area was not observed among the B cell lymphomas examined (9.1%, n = 11; Fig. 2, B and C). In the lymph nodes, B cell hyperproliferation was observed (Fig. 2 D) and consisted of large, often fragmented, nuclei with irregular shapes (Fig. 2, E and F). Notably, in two cases, we observed the invasion of B cells into the liver (Fig. 2 B, bottom) and into the lung (not depicted). In one of these samples, nodules measuring 0.7 cm in diameter were detected in the mesenteric lymph nodes, which were populated with leukemic B and activated T cells. In addition, several of the lymphomas developed into B cell leukemias (2.5%, n = 3). In the other sample, large granular cells populated the spleen, which displayed von Willebrand factor positivity (Fig. 2 G), suggestive of megakaryocytic leukemia (0.8%, n = 1; Hao et al., 2006).
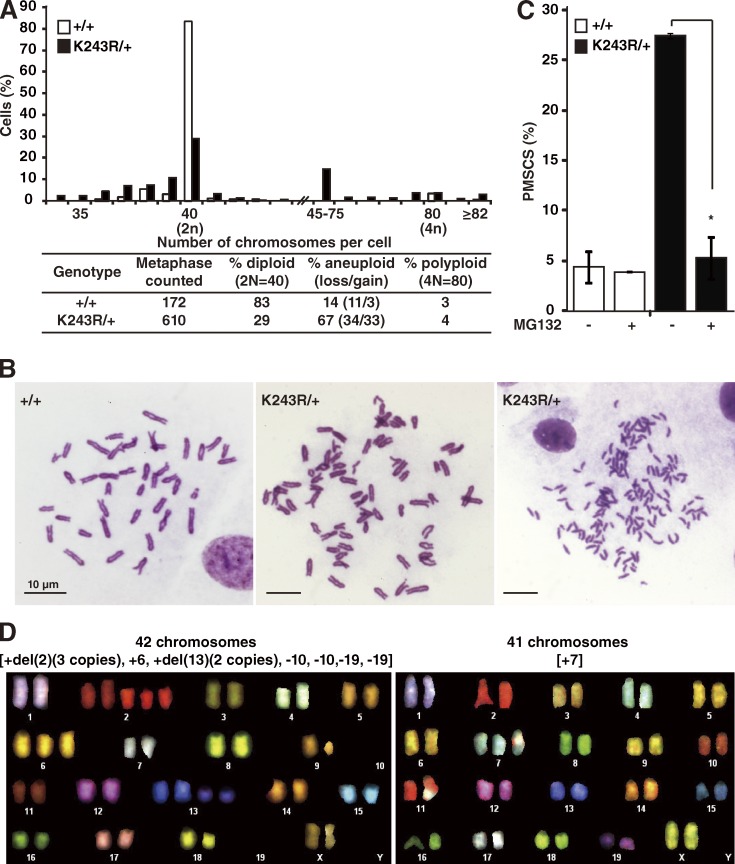
Aneuploidy and premature sister chromatid separations (PMSCS) in K243R/+ mouse embryonic fibroblasts (MEFs)
Analysis of metaphase chromosome spreads revealed near-diploid-type aneuploidy in the K243R/+ MEFs. In fact, 67% of the cells exhibited aneuploidy with chromosome loss or gain. In comparison, the rate of polyploidy was comparable to that of WT MEFs (Fig. 3 A). PMSCS (Fig. 3 B, right) was increased 5.5-fold in K243R/+ MEFs compared with controls (Fig. 3 C). PMSCS result from a compromised SAC (Michel et al., 2001) or from the failure to maintain the centromeric cohesion that is mediated by Shugosin and phosphatase PP2A (Tang et al., 2006; Rivera and Losada, 2009). Addition of MG132 to cells before preparing the chromosome spreads restored the incidence of PMSCS in the K243R/+ MEFs to a level comparable to that of the WT control (Fig. 3 C). Because MG132 treatment preferentially interferes with the destruction of securin and cyclin B, these data suggest that the PMSCS caused by the partial loss of BubR1 acetylation is a response to premature activation of APC/C.
Figure 3.
K243R/+ MEFs exhibit aneuploidy with a high incidence of PMSCS. (A) The percentage of cells displaying the indicated number of chromosomes. The results are from three and five different WT and K243R/+ MEFs, respectively. (bottom) The table summarizes the analysis of the chromosome numbers in the histogram. (B) Representative images of the metaphase chromosome spreads. (C) Incidence of PMSCS. MEFs were treated with MG132 for 2 h or left untreated, and chromosome spreads were analyzed (WT, n = 96; WT treated with MG132, n = 51; K243R/+, n = 91; K243R/+ treated with MG132, n = 70; *, P = 0.04). Cells containing more than one chromosome featuring PMSCS were scored. (D) Two examples of SKY analysis of the K243R/+ MEFs cultured for 27 passages are shown.
Next, chromosomes from K243R/+ MEFs at a late passage were subjected to spectral karyotyping (SKY). Aneuploidy was significant, and chromosome translocations were also detected in one case out of 14 (Fig. 3 D and Table 2). Signs of chromosome pulverization, like those from errors in mitosis (Crasta et al., 2012), were also detected (Fig. 3 D, left, chromosomes 2 and 13). As shown in the representative data, the cells exhibited heterogeneity in chromosome instability until MEFs reached passage 27 (Table 2). Aneuploidy was detected consistently in lymphoma/leukemia cells. Indeed, >50% of the splenic cells and lymphomas exhibited aneuploidy, and PMSCS were apparent in tumors as well (Table 3).
Table 2.
SKY analysis in K243R/+ MEFs at passage 27
| Metaphases | Karyotypes |
| 1 | 42 [XX, +del(2) (three copies), +6, +del(13) (two copies), −10, −10, −19, −19] |
| 2 | 41 [XX, +7] |
| 3 | 40 [X, −X, t (4:X)] |
| 4 | 42 [XX, +17, +17] |
| 5 | 42 [XX, +9, +18] |
| 6 | 41 [XX, +2] |
| 7 | 40 [XXX, +5, −17, −17] |
| 8 | 39 [−X, −X, +del(10), +8] |
| 9 | 41 [XX, +3] |
| 10 | 40 [XX] |
| 11 | 40 [XX] |
| 12 | 40 [XX] |
| 13 | 40 [XX] |
| 14 | 40 [XX] |
The karyotypes of 14 metaphases are presented. del, deletion; t, translocation.
Table 3.
Chromosome analysis of lymphoma/leukemia derived from K243R/+ mice
| Genotype | Tissue/tumor | Age (n) | Metaphase counted | Diploid (2N = 40) | Aneuploid (loss/gain) | PMSCS |
| wk | % | % | % | |||
| BubR1+/+a | Spleen | 60 (3) | 65 | 95.5 | 4.5 (2.25/2.25) | 0 |
| BubR1K243R/+a | Spleen | 60 (6) | 319 | 48 | 52 (42/10) | 5 |
| BubR1K243R/+ | Lymphoma | 57 | 98 | 46 | 54 (33/19) | 31 |
| BubR1K243R/+ | Lymphoma/leukemia 1 | 65 | 76 | 62 | 38 (22/16) | 14 |
| BubR1K243R/+ | Lymphoma/leukemia 2 | 104 | 137 | 51 | 49 (30/19) | 19 |
Normal splenocytes from adult mice of the indicated genotypes and ages. Splenocytes were activated using 10 µg/ml lipopolysaccharides, and a chromosome spread was performed.
Weakened SAC and chromosome missegregation in K243R/+ MEFs
Next, we assessed the integrity of the SAC. First, the mitotic index was measured after challenge with a MT polymerization inhibitor nocodazole (Noc) or the depolymerization inhibitor paclitaxel (Taxol). The MEFs in culture were treated with Noc or Taxol, and cell aliquots collected at the indicated times were subjected to immunostaining using MPM-2 (mitotic monoclonal 2) and propidium iodide staining. The mitotic cells that were positive for MPM-2 staining with 4C DNA content were analyzed by flow cytometry (Choi et al., 2009). This analysis demonstrated that the K243R/+ MEFs exhibited a weakened SAC (Fig. 4 A).
Figure 4.
Weakened SAC and congression failure in K243R/+ MEFs. (A) WT and K243R/+ MEFs were treated with 200 ng/ml nocodazole (Noc; left) or 2 µM paclitaxel (Taxol; right). The cells were collected at the indicated time points, stained with MPM-2 and 7-amino-actinomycid D, and analyzed by flow cytometry. The results and SDs are from three independent experiments. (B) WT, K243R/+, and BubR1+/− MEFs were treated with 3.3 µM Noc or 2 µM Taxol or left untreated. Cells were subjected to coimmunostaining with anti-Mad2 or -Mad1 antibody and FITC-conjugated anti–α-tubulin antibodies. Mad2 or Mad1 at the KTs with similar intensities were scored in each setting (Asyn; prometaphase cell in untreated sample). 10 unaligned/unattached KTs were analyzed per cell. The result is from three independent experiments of 35 different cells each. Relative value compared with the untreated prometaphase cell (Asyn) in WT is depicted by bar graphs (mean ± SEM; n > 350 KTs each). (C) The asynchronous and mitotic MEFs of WT and K243R/+, respectively, were subjected to IP and WB as indicated. For the mitotic extracts, the cells were serum starved for 26 h, released for 20 h, and treated with nocodazole for 7 h. In Noc + MG132-treated MEFs, Noc treatment was followed by MG132 treatment for 2 h before lysis. IP with anti-HA antibody was included as a negative control (Neg.). A sample representing 3% of the total cell lysate (TCL) was loaded as a control. (D) Assessment of BubR1 acetylation levels in WT and K243R/+ MEFs. The MEFs were treated with Noc and MG132 as in C and subjected to IP and WB as indicated. A sample representing 1% of the TCL was loaded as a control. The ratio of AcK243/total BubR1, when the level in +/+ cells is normalized to 1, was the same for both the anti-BubR1 and anti-AcK IPs and is marked below each lane. IP with a mixture of rabbit serum and 12CA5 anti-HA antibody is shown as a negative control for each panel (Neg.). Only a non-specific band migrating just below the band recognized by anti-AcK antibody (top left two lanes) was detected. Black lines in the top four panels indicate the removal of an intervening lane for presentation purposes. (E) Quantification of BubR1 at KTs in chromosome spreads after treatment with 200 ng/ml Noc. The level of BubR1, determined by anti-BubR1 immunofluorescence in Noc-treated prometaphase MEFs, was scored in WT, BubR1+/−, and K243R/+ MEFs. Each dot represents the mean BubR1 intensity calculated from 20 randomly picked KTs per cell. 40 chromosome spreads from each genotype were scored in two independent experiments. Mean value is indicated with a line (mean ± SEM; n > 800 KTs each). (F) Statistical mitotic timing from the NEBD to anaphase onset. The MEFs were subjected to time-lapse microscopy with or without treatment with 200 ng/ml Noc or 2 µM Taxol. Images were captured every 5 min, and the live images were processed for 36 h. Without a spindle poison, mitosis required an average of 25 min in the WT cells (n = 170) and ∼20 min in the K243R/+ MEFs (n = 153). After spindle poison exposure, the WT cells remained in mitosis for 331 min with Noc (n = 66) and 106 min with Taxol (n = 218). The K243R/+ MEFs exited mitosis (chromosome decondensation) within 117 min with Noc treatment (n = 144) and within 87 min with Taxol treatment (n = 191). The bars in the box represent the median values. The outliers (open circles) and suspected outliers (asterisks), as determined by statistical analysis, are marked. (G) Representative images captured at the indicated time points from NEBD without Noc treatment. Of the 20 K243R/+ MEFs, 17 displayed congression failure (second row; Video 2) and 3 exited mitosis without segregation (third row; Video 3). See Video 1 for the WT MEF control (first row). The timing of the onset of anaphase is marked. The white arrow indicates the lagging chromosome. NEBD: 00:00. Bar, 5 µm. (H) WT, BubR1+/−, and K243R/+ MEFs were treated with MG132 for 2 h and subjected to cold MT assay, followed by staining with anti–α-tubulin and CREST. Enlarged images of the insets show the properly attached MT in WT cells; syntelic attachment in BubR1+/− cells; and unattached (c), syntelic (a and b), and monotelic attachment (d) in K243R/+ cells. Green, α-tubulin; red, CREST immunostaining. Bars: (yellow) 5 µm; (white) 1 µm. (I) The congression defects were scored in cells from each mouse strain in the absence of MT poison, and data are shown as bar graphs (mean ± SEM). Number of cells scored: WT, n = 75; K243R/+, n = 102; BubR1+/−, n = 60.
We assessed the localization of Mad2 at the unattached KTs in untreated prometaphase and Noc- or Taxol-treated WT, K243R/+, and haploinsufficient BubR1 MEFs (BubR1+/−; Wang et al., 2004). The number of Mad2 foci at the KTs was highest when MTs were completely depolymerized after treatment with 3.3 µM Noc. Slightly lower but similar levels of Mad2 were detected at KTs in untreated prometaphase cells. Upon Taxol treatment, the number of Mad2 at KTs decreased to ∼1/3 of that in Noc-treated cells (Fig. 4 B). Nevertheless, the pattern and extent of Mad2 localization at KTs were similar between WT, K243R/+, and haploinsufficient BubR1 cells. Similar results were obtained with Mad1 (Fig. 4 B). According to the Mad2 template model, the presence of the Mad1/Mad 2 complex at unattached KTs predicts the cytosolic propagation of the SAC signal (De Antoni et al., 2005). Therefore, these results suggest that the establishment of the initial SAC signal is intact in K243R/+ cells.
We then analyzed the formation of MCC. In Noc-treated MEFs, BubR1 levels were lower in K243R/+ cell lysates, confirming that nonacetylated BubR1 is unstable during mitosis because this protein is a substrate of APC/C. When proteolysis was inhibited (Fig. 4 C, +Noc+MG132), BubR1 levels were restored in the lysates. Immunoprecipitation (IP) of BubR1 immune complex demonstrated that Cdc20, Bub3, and Mad2 levels were decreased in K243R/+ cells compared with WT cells, even though similar amounts of BubR1 were immunoprecipitated in Noc-treated condition (Fig. 4 C, +Noc). Similarly, Cdc20, Bub3 and Mad2 in immunoprecipitated BubR1 complexes from K243R/+ cells were restored to levels comparable to those from complexes from WT cells when proteolysis was inhibited (Fig. 4 C, +Noc+MG132). As the ratio of WT BubR1 to K243R was 1:1 in K243R/+ MEFs (Fig. 4 D), these results suggest that acetylation-deficient BubR1 (K243R) is capable of binding to the other MCC components and to APC. However, the degradation of K243R (Choi et al., 2009) in prometaphase (Noc-treated condition) results in a failure to maintain MCC.
The amount of BubR1 at the KTs was assessed in chromosome spreads after Noc treatment. The result showed that the level of BubR1 at KT was lowest in BubR1+/− cells with higher levels in K243R/+ cells and the highest in WT cells (Fig. 4 E). As the antibody used in this assay recognizes both WT BubR1 and K243R, this result indicates that K243R localizes to KT and is degraded during mitosis (Fig. 4 E).
Next, we measured the mitotic timing in K243R/+ cells. K243R/+ mice were crossed with GFP-H2B transgenic mice. MEFs from WT mice expressing GFP-H2B (+/+; GFP-H2B) and from K243R/+ mice expressing GFP-H2B (K243R/+; GFP-H2B) were subjected to time-lapse microscopy to measure the mitotic timing from nuclear envelope breakdown (NEBD) to the onset of anaphase. Mitotic timing was ∼5 min shorter in the K243R/+ MEFs (Fig. 4 F, left) than in the WT MEFs. In the presence of either Noc or Taxol, the duration of mitotic arrest was greatly reduced in the K243R/+ MEFs (Fig. 4 F, right); however, the effect on mitotic timing in response to Noc was more profound than to Taxol.
In the captured live-cell imaging, chromosome missegregation was apparent in the K243R/+ MEFs, compared to WT (Fig. 4 G, first row; and Video 1), even in the absence of spindle poisons. Indeed, 85% of the K243R/+ MEFs exhibited chromosome segregation without forming a distinct metaphase plate, yielding lagging chromosomes and chromosome bridges (Fig. 4 G, second row; and Video 2), and 15% of K243R/+ MEFs exited mitosis without proper chromosome alignment (Fig. 4 G, third row; and Video 3). This result indicates that K243R/+ MEFs have problems in chromosome alignment.
The absence of BubR1 results in failure to maintain stable KT–MT attachments (Lampson and Kapoor, 2005), and one study suggested that the BubR1 N terminus is required for the KT–MT interaction in metaphase (Malureanu et al., 2009). To assess whether BubR1 acetylation is involved in the stable attachments of K-fibers to the chromosomes at metaphase, a cold-stable MT assay (Rieder, 1981) was performed. As MG132 treatment was included in the assay, it blocks the degradation of K243R. Therefore, both K243R and WT BubR1 are present in K243R/+ MEFs. The K243R/+ MEFs exhibited severe defects in KT–MT attachment (Fig. 4 H, K243R/+). Interestingly, defects in congression were more profound in K243R/+ than in BubR1+/− (Fig. 4 I). Thus, these results confirm that acetylation of BubR1 is required for chromosome congression.
Acetylation of BubR1 is required for recruiting PP2A-B56α phosphatase to KT
We assessed whether binding of BubR1 to CENP-E or Blinkin (KNL1) was affected by the acetylation status. BubR1 associates with CENP-E (Chan et al., 1999; Abrieu et al., 2000; Mao et al., 2003; Weaver et al., 2003), which is the kinesin-7 motor protein at KT that enables the monotelic polar chromosomes to glide along the existing K-fiber toward the cell center (Kapoor et al., 2006) to aid chromosome congression (Kim et al., 2010). BubR1 also binds to KNL1/Blinkin (Kiyomitsu et al., 2007; Bolanos-Garcia et al., 2011), which is a constituent of the KMN network that provides the MT-binding interface in the KT required for the KT–MT interaction.
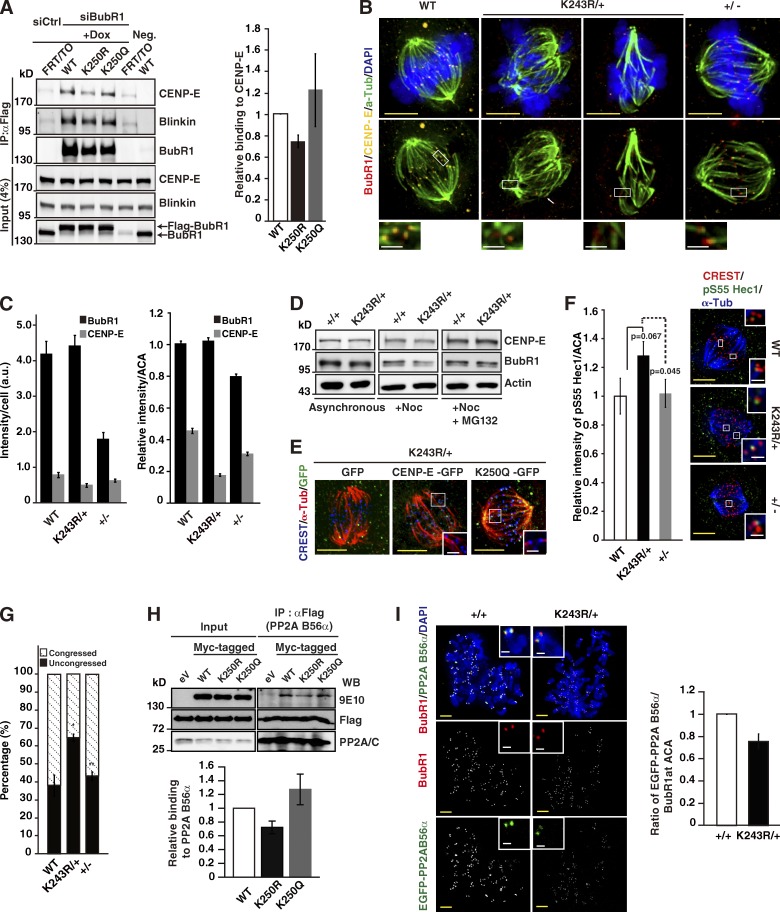
For this study, we took advantage of the stable inducible HeLa cell lines in which a single copy of FLAG-tagged WT-BubR1, K250R (acetylation deficient), or K250Q (acetylation mimetic) is integrated into the genome at a flippase recognition target (FRT) site. The endogenous BubR1 was depleted in these cells using siRNA transfection, and the expression of the ectopic BubR1, K250R, or K250Q was induced. After IP and Western blot (WB) of cell lysates, we found that the acetylation status of BubR1 affected CENP-E binding. The K250Q protein binding to CENP-E was increased ∼1.2-fold, whereas binding of the K250R to CENP-E was decreased ∼0.7-fold (Fig. 5 A, right). Meanwhile, KNL1/Blinkin binding was unaltered (Fig. 5 A).
Figure 5.
How BubR1 acetylation is involved in KT–MT interaction. (A) The HeLa-FRT-BubR1, -K250R, or -K250Q cells were depleted of endogenous BubR1, and the expression of the indicated FLAG-tagged BubR1 constructs was induced by doxycycline treatment. The cells were synchronized in mitosis using the thymidine block and treated with nocodazole/MG132. BubR1 complexes were immunopurified using an anti-FLAG (M2) antibody, followed by WB using the indicated antibodies. The total lysates were included in the WB as an input control. Immunoprecipitated CENP-E bands were normalized to BubR1, and the binding affinities of each BubR1 to CENP-E were determined. The relative binding to CENP-E and the SDs in the graph are from four independent experiments (right). (B) Immunostaining of CENP-E and CREST in WT, K243R/+, and BubR1+/− MEFs after the cold MT assay. The cells were treated with MG132 before fixation. Enlarged images are shown at the bottom. Red, BubR1; yellow, CENP-E; green, α-tubulin. Bars: (yellow) 5 µm; (white) 1 µm. (C) Relative levels of BubR1 and CENP-E from B, in the presence of MG132, as measured by fluorescence intensities at the cellular level (left) and at KTs (right) are represented by bar graphs. Fifty cells in each genotype were scored in two independent experiments (mean ± SEM; n > 500 KTs). (D) WB analysis of CENP-E and BubR1 from lysates prepared from untreated, Noc-treated, or Noc + MG132-treated MEFs. The blot was reprobed with anti-actin antibody as a loading control. (E) GFP-tagged CENP-E expression construct (Kim et al., 2010) or K250Q expression construct was transfected into K243R/+ MEFs. 1 d after transfection, MEFs were synchronized via serum starvation and released into mitosis with change with fresh media supplemented with 16% FBS. 24 h later, cells were treated with MG132 for 2 h and subjected to cold MT assay. Representative images are shown. Ectopic expression of CENP-E-GFP or K250Q-EGFP was detected by green fluorescence. EGFP-expressing construct was transfected for control. Enlarged images of the insets (excluding GFP) are shown at the right. Red, α-tubulin; blue, CREST staining; green, GFP. Bars: (white) 1 µm; (yellow) 5 µm. (F) Comparison of phosphorylated Hec1 in metaphase cells. MEFs from WT, K243R/+, and BubR1+/− mice were treated with 10 µM MG132 for 2 h and subjected to cold MT assay. The levels of pHec1 scored in two independent experiments are shown in bar graphs (mean ± SEM; n > 450 KTs; number of cells scored: WT, n = 23; K243R/+, n = 34; BubR1+/−, n = 25). Representative images are shown at the right with enlarged images of individual KTs in the insets. Bars: (white) 1 µm; (yellow) 5 µm. (G) MEFs were treated with Monastrol for 3 h, washed extensively, and released 1 h in the presence of MG132 and subjected to cold MT assay. Congressed and uncongressed chromosomes were scored in two independent experiments and presented in the bar graphs (mean ± SEM; number of cells scored: WT, n = 71; K243R/+, n = 78; BubR1+/−, n = 73). Asterisks mark significant p-values when compared with WT (*, P = 0.1; **, P = 0.5). (H) Myc-tagged WT-BubR1, K250R, or K250Q expression constructs were transfected into PP2A-B56α–expressing HeLa-FRT cells (Kruse et al., 2013). Expression of PP2A-B56α was induced with doxycycline followed by cell synchronization and mitotic arrest with the treatment of Noc and MG132 to inhibit proteolysis. The cells were subjected to IP and WB. WB of the catalytic subunit of PP2A (PP2A/C) was included for normalization of IP (top). Relative binding affinity to PP2A-B56α was calculated by measuring 9E10 band intensities (WT, K250R, and K250Q, respectively) in FLAG immune complex (PP2A-B56α). SDs in bar graphs are from four independent experiments (bottom). eV, empty vector. (I) EGFP-PP2A-B56α–expressing construct was transfected into K243R/+ MEFs. 24 h later, MEFs were treated with 200 ng/ml Noc for 4 h, and then subjected to chromosome spread and immunofluorescence assay. BubR1 was detected by immunofluorescence and PP2A-B56α by the fluorescence of GFP (left). This experiment was done in the absence of MG132. Fluorescence intensity of PP2A-B56α relative to BubR1 at KT was scored in WT and K243R/+ cells and depicted as a histogram (right). The results are from two independent experiments of >600 KTs (mean ± SEM; number of cells: WT, n = 33; K243R/+, n = 32; P = 0.061). Red, BubR1; green, PP2A-B56α; blue, DAPI. Bars: (white) 1 µm; (yellow) 5 µm.
CENP-E assembly at the KT requires BubR1 (Johnson et al., 2004). Therefore, we determined whether BubR1 acetylation deficiency affected CENP-E localization. WT, K243R/+, and BubR1+/− MEFs were subjected to the cold MT assay combined with immunofluorescence with an anti–CENP-E antibody (Fig. 5 B). The KT–MT attachment was severely impaired in the K243R/+ cells (Fig. 5 B). The CENP-E to BubR1 ratio at the KT was similar in the WT and the BubR1+/− cells, although BubR1 and CENP-E at the KT was lower in the BubR1+/− MEFs (Fig. 5, B and C, right). In comparison, the level of CENP-E at KT was reduced to ∼50% of the WT level in K243R/+ cells (Fig. 5, B and C, right), although the total BubR1 level was comparable to that in WT cells as a result of the treatment with MG132 (Fig. 5 C, left). As the total level of CENP-E in K243R/+ MEFs was unaltered (Fig. 5 D), the observed decrease of CENP-E at KTs in K243R/+ MEFs suggests that binding to BubR1 is required for CENP-E localization at KTs.
Upon depletion of CENP-E in HeLa cells, most of the chromosomes align at the metaphase plate with some near the poles (Lampson and Kapoor, 2005). In comparison, the congression failure induced by the BubR1 acetylation deficiency was much more severe. Overexpression of CENP-E-GFP in K243R/+ MEFs did not rescue the congression defects (Fig. 5 E, CENP-E), whereas ectopic expression of the human form of acetylation-mimetic BubR1 (K250Q) rescued the KT–MT attachment in K243R/+ MEFs (Fig. 5 E, K250Q) and in BubR1-depleted HeLa cells (Fig. S2 A, K250Q). These results suggest that the lowered binding of acetylation-deficient BubR1 to CENP-E does not fully explain the congression failure in K243R/+.
MTs are bound to the KT through the conserved KMN network (Cheeseman et al., 2006). In this network, the Ndc80 (Hec1 in human) complex is the primary contact point for KT–MT attachment (DeLuca et al., 2006; Wei et al., 2007; Sundin and Deluca, 2010). During the error correction process of mitosis, Aurora B phosphorylates Ndc80 and other KMN proteins in sequence (Welburn et al., 2010), resulting in the disassembly of incorrectly attached MTs (Lampson and Cheeseman, 2011). As the chromosomes biorient, excess Aurora B activity is counteracted and Ndc80 is dephosphorylated, establishing the stable KT–MT attachment before chromosome segregation.
As the number of K-fiber was reduced in K243R/+ cells (and K250R-expressing cells), we determined the phosphorylation of Hec1 (human Ndc80) at the KT using phospho-S55–specific Hec1 antibody (anti-pHec1; DeLuca et al., 2011). After 2 h in MG132, MEFs were subjected to cold MT assays and immunofluorescence with anti-pHec1 antibody (Fig. 5 F, right). K243R/+ MEFs displayed a 1.3-fold higher level of pHec1 than either WT or BubR1+/− cells (Fig. 5 F, left). When assessed for the error correction capability from the treatment of Monastrol (Lampson et al., 2004), K243R/+ MEFs displayed a markedly higher percentage of uncongressed chromosomes compared with WT or BubR1+/− MEFs (Fig. 5 G).
Recently, BubR1 was found to bind to B56α subunit of PP2A phosphatase after phosphorylation. BubR1 binding to PP2A-B56α recruits PP2A phosphatase activity to KTs and counteracts Aurora B activity on KMN (Suijkerbuijk et al., 2012; Kruse et al., 2013). As the hyperphosphorylation of Hec1 in K243R/+ cells indicated excessive Aurora B activity, we assessed the binding properties of acetylation-deficient BubR1 to PP2A-B56α. Myc-tagged WT-BubR1, K250R, or K250Q were ectopically expressed in HeLa-FRT cells that stably express PP2A-B56α (Kruse et al., 2013). In IP and WB, we found that binding to PP2A-B56α was reduced to ∼75% by the BubR1 acetylation deficiency (Fig. 5 H). Immunofluorescence analysis in Noc-treated chromosome spreads confirmed that PP2A-B56α binding to BubR1 and recruitment to KTs were reduced to ∼75% in K243R/+ MEFs (Fig. 5 I and Fig. S2 B). Immunostaining of Noc and MG132–treated cells with an antibody that recognizes the phosphorylation of BubR1 at T680 (p680), the site phosphorylated by Plk1 that is involved in the recruitment of PP2A-B56α (Suijkerbuijk et al., 2012), revealed that K243R/+ prometaphase cells exhibited lower p680 at KTs (Fig. S2 C); however, the level of p680 correlated with the level of BubR1 at KTs. Collectively, BubR1 acetylation is involved in the recruitment of PP2A-B56α to KTs for chromosome congression.
Micronuclei in MEFs, regenerating hepatocytes, and hepatocellular carcinoma
Failure in bipolar spindle attachment activates SAC, arresting the cells in mitosis. Delay in mitosis was observed in cells expressing BubR1 mutants that are defective in binding to PP2A-B56α (Kruse et al., 2013). In comparison, K243R/+ cells display shortened mitotic timing. Because of the weakened SAC despite the failure in chromosome–spindle attachment, K243R/+ cells exhibited marked increase of micronuclei, which result from lagging chromosomes (Fig. 6, A and B). The incidence of micronuclei was found to be ∼18% in K243R/+ MEFs (Fig. 6 B). An interesting paper showed that micronuclei do not degenerate but persist through several cell divisions (Crasta et al., 2012). It showed that because micronuclei are uncoupled with DNA replication and repair, micronuclei contain damaged DNA and pulverized chromosomes (Crasta et al., 2012).
Figure 6.
Marked increase of micronuclei in K243R/+ mice. (A) The frequency of MEFs that display lagging chromosomes (WT, n = 52; K243R/+, n = 153). (B) The frequency of micronuclei shown in bar graphs (WT, n = 52; K243R/+, n = 1,976). (C) γ-H2AX–positive micronuclei (arrows) in K243R/+ MEFs. The cells were stained with a γ-H2AX antibody, and DNA was counterstained with DAPI. (left) The incidence of γ-H2AX–positive micronuclei (WT, n = 1,463; K243R/+, n = 1,132). (right) Cellular DNA damage in WT and K243R/+ MEFs (WT, n = 1,463; K243R/+, n = 1,132). The results in A–C are from two independent experiments (mean ± SEM). (D) Paraffin-embedded sections of livers from WT or K243R/+ mice after partial hepatectomy were stained with H&E. Three animals each were subjected to partial hepatectomy. Micronuclei in regenerating hepatocytes (arrows) are compared as fold increase on the right: WT, 0.4% (3 out of 693 cells); K243R/+, 2.0% (21 out of 1,069). (E) Micronuclei or nuclear blebs (top, arrows) and lagging chromosomes (bottom) observed in the paraffin-embedded sections of hepatocellular carcinoma from the K243R/+ mice. Bars, 10 µm.
Assessment of the DNA damage in the micronuclei revealed γ-H2AX positivity (Fig. 6 C, left). Notably, general DNA damage was not altered in K243R/+ cells (Fig. 6 C, right). The micronuclei were consistently found in the regenerating hepatocytes after hepatectomy (Mitchell and Willenbring, 2008) with a marked increase in K243R/+ mouse livers (Fig. 6 D). Paraffin sections of hepatocellular carcinoma from K243R/+ mice also contained micronuclei and lagging chromosomes (Fig. 6 E). Combined with the observation that SKY revealed signs of chromosome pulverization (Fig. 3 D), the persistence of micronuclei from primary cells to cancer suggests an interesting possibility that the micronuclei facilitated DNA breakage and chromosome pulverization that contributed to tumorigenesis in K243R/+ mice.
The inefficient DNA repair in the micronuclei indicates that they could be the source of genetic alterations. In this line of thinking, it is interesting that 8 out of 11 tumors possessed mutant p53: the mutations were predominantly missense mutations, small deletions, and insertions (Table 4). Using the International Agency for Research on Cancer TP53 database, we determined that several of the mutations affected p53 function (e.g., impaired transcriptional or DNA-binding activities) and were correlated to mutations found in human cancers (Table 4). Collectively, these findings suggest that K243R/+ cells are susceptible to acquisition of additional mutations.
Table 4.
p53 status in K243R/+ tumors
| Tumor | Sex | Age | Mutationa | Mutanta | Effect | p_domainb |
| wk | ||||||
| Hepatocellular carcinoma 1 | Male | 61 | c.163ins_T | STOP 55 | Insertion | TAD |
| Hepatocellular carcinoma 2 | Male | 117 | c.242C>Tc.301C>T | p.A81Vp.Q101X | MissenseNonsense | PRDDBD |
| Sarcoma 1 | Male | 69 | c.163ins_Tc.296ins_A | STOP 55STOP 99 | InsertionInsertion | TADDBD |
| Sarcoma 2 | Female | 122 | c.296ins_Ac.467C>T | STOP 99p.A156V | InsertionMissense | DBDDBD |
| B cell lymphoma 1 | Female | 62 | c.359-365del7 | STOP 128 | Deletion | DBD |
| B cell lymphoma 2 | Female | 119 | c.109T>C;980T>Cc.429G>A | p.S37P;L327Pp.W143X | MissenseNonsense | TAD, 4DDBD |
| B cell lymphoma 3 | Female | 139 | c.443C>T | p.P148L | Missense | DBD |
| Leukemia 1 | Female | 57 | c.1064A>Gc.841A>G | p.E355Gp.T281A | MissenseMissense | DBDDBD |
Mutation nomenclatures follow the Human Genome Variation Society recommendations. c, coding sequence; ins, insertion; del, deletion; p, protein sequence. p53 status was determined by the nucleotide sequencing of four to five clones derived from each tumor.
Protein domain with mutation indicated. TAD, transactivation domain; PRD, proline-rich domain; DBD, DNA-binding domain; 4D, tetramerization domain.
Discussion
BubR1 haploinsufficiency in mice (BubR1+/−) resulted in abnormal megakaryopoiesis, but spontaneous cancer development was not observed (Wang et al., 2004). Unexpectedly, BubR1 insufficiency in hypomorphic mice (BubR1H allele) exhibited a premature aging phenotype (Baker et al., 2004). In comparison, in K243R/+ mice, premature aging was not observed and, strikingly, a high incidence of spontaneous tumorigenesis, both solid and hematologic malignancies, was observed (Table 1). These differences suggest that the effects of a BubR1 acetylation deficiency are not simply because of a decrease in protein level. Indeed, in assays for chromosome congression, K243R/+ MEFs displayed significant defects in the KT–MT interaction, and these defects were even worse than those observed in the haploinsufficient BubR1 MEFs. As BubR1+/− and hypomorphic BubR1 MEFs also exhibit aneuploidy, whole chromosome aneuploidy alone cannot be the cause of spontaneous tumorigenesis.
In dividing cells, chromosome movement is coupled with MT dynamics and attachment. Aurora B kinase is essential in correcting the improperly attached chromosomes (Lampson et al., 2004) via phosphorylation of the contact point of the MTs at the KT in the KMN network and disassembling the improperly attached KT–MT fibers (Lampson et al., 2004; Cheeseman and Desai, 2008). Dynein drives the poleward movement of the chromosomes, and CENP-E enables the monooriented chromosome at the pole to glide along the existing K-fiber toward the spindle equator. The unattached KT of the chromosome attaches to the MT that is growing from the opposite pole (Cheeseman and Desai, 2008). When bioriented at the equator, the Ndc80 complex is dephosphorylated to stably attach to the K-fibers. Two phosphatases have been suggested to be involved in chromosome congression: PP1 (Kim et al., 2010; DeLuca et al., 2011) and the B56α subunit of PP2A (Suijkerbuijk et al., 2012; Kruse et al., 2013). We found here that BubR1 acetylation is required to bind PP2A-B56α, suggesting that BubR1 acetylation is a signal cross talking with phosphorylation at the KARD domain at the C terminus (Suijkerbuijk et al., 2012; Kruse et al., 2013) that integrates the phosphatase to the KMN network for chromosome alignment. As the phosphorylation of T680 was not altered but coincided with total BubR1 level in K243R/+ prometaphase cells, it is possible that sites other than T680 in the KARD domain may be affected by acetylation of BubR1. Or, acetylation may act in concert with all three phosphorylation sites in the KARD domain (Suijkerbuijk et al., 2012; Kruse et al., 2013).
Together, these findings indicate that chromosome congression failure in K243R/+ cells is a result of the unstable interaction between KT and MT. Malorientation of chromosomes cannot be corrected, and the reduced association of CENP-E at KTs may exacerbate the problem (Fig. 7, left). Future studies should examine whether an acetylation–phosphorylation signaling code in BubR1 is involved in sensing tension and linking it to SAC.
Figure 7.
Model for how tumor develops in K243R/+ mice. BubR1 acetylation plays dual roles: it is required for stable maintenance of the KT–MT interaction and for SAC maintenance. K243R/+ cells exhibit congression failure because 50% of BubR1 is acetylation deficient. Acetylation-deficient BubR1 (K243R) is incapable of recruiting PP2A-B56α to counteract excessive Aurora B kinase activity at the KMN network, and this process is crucial for stable KT–MT interaction. K243R also has reduced CENP-E binding, which may contribute to the problem in chromosome congression (left). Half of the SAC complex contains K243R-BubR1 and fails to maintain SAC activity. K243R associates with other MCC components; however, the protein is readily ubiquitinated by the APC/C, resulting in disassociation of MCC. As initial SAC signal generation is intact, premature disassembly of MCC leads to failure in the maintenance of SAC, effectively weakening SAC activity. The combined effects of chromosome–spindle attachment failure and weakened SAC lead to massive chromosome missegregation and initiate tumorigenesis in K243R/+ mice.
SAC signal establishment was intact in K243R/+ cells. This signal establishment was also unaffected in haploinsufficient BubR1 MEFs, indicating that neither reduced BubR1 levels or BubR1 acetylation deficiency affects Mad1/Mad2 localization to the unattached KTs. Blockade of proteolysis led to association of acetylation-deficient BubR1 with other MCC components, similar to WT BubR1. In comparison, the acetylation-deficient BubR1-containing MCC was not intact in Noc-treated MEFs, because of the premature K243R ubiquitination and degradation, which led to premature disassembly of the MCC and consequent failure to sustain the SAC (Fig. 7, right). Thus, the acetylation-deficient BubR1 allele (K243R) functions as a dominant allele both in chromosome–spindle attachment and in SAC maintenance. The fact that both the K243R and WT alleles are present in primary tumors of K243R/+ mice supports this notion (Fig. S3).
The spontaneous tumorigenesis in K243R/+ mice is because of the dual roles of BubR1 acetylation in mitosis: chromosome–spindle attachment and SAC manintenance. In K243R/+ cells, malorientated chromosomes are not corrected and before SAC satisfaction, cells proceed through mitosis and produce massive chromosome missegregations.
Chromosome missegregation can give rise to aneuploidy. Although aneuploidy can be oncogenic, this condition can also inhibit growth (Weaver et al., 2007; Sheltzer et al., 2011; Oromendia et al., 2012). Considering that cancers are the result of multiple mutations, the high incidence of spontaneous cancer development in K243R/+ mice suggests that BubR1 acetylation deficiency not only promotes aneuploidy but also induces multiple genetic changes. General DNA damage, however, was not increased in the K243R/+ mice, even when compared with Mad2-overexpressing mice (Sotillo et al., 2007). Therefore, it is conceivable to think that chromosome missegregation that results from the failure of chromosome–spindle attachment combined with weakened SAC in K243R/+ mice yields genetic alterations, in addition to chromosome number instability.
Errors in mitosis and the resulting micronuclei have been suggested by Crasta et al. (2012) as a source of chromothripsis, the shattering of chromosomes and one-off genetic rearrangement in cis (Stephens et al., 2011; Kloosterman et al., 2012). We have not yet analyzed the genome-wide sequencing of tumors to support the idea that chromothripsis accompanies the tumorigenesis in K243R/+ mice; yet, the signs of chromosome pulverizations with a marked increase of micronuclei from primary cells to tumors suggest the possibility of a chromothripsis-like event taking place in the tumorigenesis of K243R/+. A previous paper suggested that p53 mutation is linked with chromothripsis in sonic hedgehog–associated pediatric medulloblastoma (Rausch et al., 2012). That the p53 mutation is highly associated with the tumorigenesis of K243R/+ mice is interesting in this line of thinking.
It is also worth looking into the mutation types we observed in the case of p53. As found in p53, we propose that small insertions, deletions, and missense mutations can also result from DNA replication–uncoupled micronuclei. As the homozygous mutant mice (BubR1K243R/K243R) exhibited early embryonic lethality, we propose that the degree of chromosome missegregation in K243R/+ cells was just enough to be tolerated through embryogenesis but generated a mutation-prone cellular environment that led to tumorigenesis. The accompanying aneuploidy in these cells may have contributed to the process of cancer development.
Lysine 250, which is the acetylation site in human BubR1 (Choi et al., 2009), was also suggested for sumoylation after we reported the acetylation of this site (Yang et al., 2012a). Yang et al. (2012b) proposed that sumoylation occurs after BubR1 deacetylation, suggesting that deacetylation and sumoylation may be involved in mitotic exit. One might argue that the phenotype exhibited in K250R/+ cells is the result of the combination of acetylation and sumoylation deficiency. We cannot completely rule out this possibility. However, because the consequences of the K250Q mutation, which mimics acetylated BubR1, is completely opposite to those observed in K250R with respect to the SAC activity and congression, it is more likely that the tumorigenesis stems from the problems in mitosis that resulted from the acetylation deficiency for the following reasons. First, K250Q expression rescues the congression defects in BubR1-depleted cells (Fig. 5 E and Fig. S2 A). Second, K250Q-expressing cells do not segregate for long (Choi et al., 2009). Finally, K250Q expression rescues BRCA2 deficiency, whereas K250R expression does not (Choi et al., 2012).
Although a mutation at K250 in BubR1 has not been identified in human cancers, it is possible that BubR1 mutations at sites other than K250 interfere with BubR1 acetylation. Furthermore, the BRCA2 tumor suppressor is required for BubR1 acetylation during mitosis (Choi et al., 2012). We proposed that BRCA2, which is a regulator of mitosis but not a critical component of the SAC, may be the preferred target in tumorigenesis (Choi et al., 2012). The analysis of the K243R/+ mice suggests that BubR1 acetylation is indeed a tumor-suppressive mechanism.
Materials and methods
Generation of the BubR1 acetylation-deficient (K243R) allele and BubR1K243R/+; GFP-H2B mice
Mouse BubR1 genomic DNA was isolated by screening the mouse 129/Sv library (Agilent Technologies) using a probe from mouse BubR1 cDNA, and two BAC clones were mapped and sequenced. To generate the targeting vector, an 8-kb fragment containing exons 5, 6, 7, and 8 was cloned into the pBluescript KS (+) vector. The neo cassette and diphtheria toxin A fragment were also inserted into the vector (Fig. 1). The lysine at position 243 in BubR1 was substituted with an arginine using site-directed mutagenesis in the targeting construct. The targeting construct was linearized and electroporated into 129/Sv embryonic stem (ES) cells. The accurately targeted ES cell clones were selected using Southern blotting and DNA sequencing analysis. The heterozygous ES cells were injected into the blastocysts of C57BL/6 mice to generate the K243R/+ mice. The neo cassette was removed by crossing the K243R/+ mice with Flpe transgenic mice. The primers used for PCR genotyping were as follows: 5′-GAGGTAAAGGCAGGGGAATC-3′ (forward) and 5′-GAGAAAGCGGGGGTCATTAT-3′ (reverse).
The K243R/+ mice were crossed with EGFP-H2B transgenic mice (B6.Cg-Tg[HIST1H2BB/EGFP]1 pa/J; The Jackson Laboratory) for monitoring cell division. The BubR1+/− mice were a gift from W. Dai (New York University School of Medicine, Tuxedo, NY). The mice were housed in a specific pathogen-free facility or semi-conventional (virus antibody–free) facility. The Institutional Animal Care and Use Committee of Seoul National University (SNU-090630-3) approved the animal experimental protocols. The guidelines, policies, and regulations for the Care and Use of Laboratory Animals in Seoul National University were followed.
Genotyping
PCR was used to genotype the adult BubR1 acetylation-deficient mice using genomic DNA extracted from the tail. The genomic DNA from the in vitro cultured ES cells and embryos was extracted using the DirectPCR reagent (Viagen Biotech, Inc.). A 3–5-µl sample of the genomic DNA was used for the PCR. The primers for BubR1 were as follows: 5′-CCCTCACAAACGCCTACC-3′ (forward) and 5′-CATCTCACCAGCCCAGAAGA-3′ (reverse). The resulting PCR products were separated on a 1.5% agarose gel. Using the primers indicated, the WT DNA produced a 145-bp PCR product, and the homozygote mutant allele produced a slower migrating 219-bp band. We verified the PCR genotyping by sequencing of the targeted region. The sequencing primers used were as follows: 5′-GGTTGTCCTTTGTCTTGCT-3′ and 5′-TGCTTACCATCTCTTGGGT-3′ for reverse sequence confirmation.
Statistical analysis
The relationships between categorical variables were assessed using the χ2 test. Tumor- and cancer-free survival were estimated using the Kaplan-Meier method. For the statistical analyses, t test was used unless otherwise stated. The means ± SEM or SD are shown. The statistical data were analyzed using SPSS software (version 15.0; SPSS Inc.).
Embryo culture
E3.5 blastocysts from intercrosses of the K243R/+ mice were harvested into gelatinized 4-well plates. The embryos were cultured in DMEM supplemented with 1,000 U/ml leukemia inhibitor factor (ESGRO), 15% vol/vol FBS, 100 mM sodium pyruvate, 10,000 U/ml of penicillin, 10,000 µg/ml of streptomycin, 200 mM l-glutamine, 100 µM MEM nonessential amino acids, and 55 µM β-mercaptoethanol for 6 d. The embryos were examined using a phase-contrast microscope (Axio Observer.Z1; Carl Zeiss) before genotyping.
Constructs and antibodies
PP2A-B56α-expressing constructs were gifts from G. Kops (University Medical Center, Utrecht, Netherlands) and J. Nilsson (University of Copenhagen, Copenhagen, Denmark). EGFP-PP2A-B56α–expressing construct was generated by subcloning PP2A-B56α into EGFP-N1 vector (Invitrogen). The following antibodies were used: anti-cyclin A (H-432), anti–cyclin B (H-433), anti-Mad2 (C-19), anti-PCAF (H369 and E-8), anti-CDC20 (H-175), and anti-APC3 (Santa Cruz Biotechnology, Inc.); anti–α-tubulin (DM1A), anti–γ-tubulin, and anti-actin (AC-15; Sigma-Aldrich); anti-BubR1 and anti–PP2A-B56α (BD); anti–phospho-histone H3, anti–acetyl-lysine monoclonal, and anti–γ-H2AX (EMD Millipore); CREST (Cortex Biochem); anti-CD3 (2C11) and anti-B220 (RA3-6B2; BioLegend); anti–von Willebrand factor and anti–Ki-67 (Dako); anti–acetyl-lysine polyclonal (Cell Signaling Technology); Alexa Fluor 488–phalloidin (Invitrogen); and Alexa Fluor 488 goat anti–mouse IgG (Molecular Probes). The anti-Bub3 antibody was a gift from S. Taylor (University of Manchester, Manchester, UK). The anti-Mad2 and anti-Mad1 antibodies were obtained from H. Yu (University of Texas Southwestern Medical Center, Dallas, TX) and the anti-Blinkin antibody was obtained from I. Cheeseman (Whitehead Institute for Biomedical Research, Cambridge, MA). The anti–phospho-Hec1 antibody was obtained from J. Deluca (Colorado State University, Fort Collins, CO), the anti-pT680 antibody from G. Kops, and the anti–CENP-E (Hpx-1) antibody from D. Cleveland (University of California Ludwig Institute, San Diego, CA).
Stable inducible HeLa-FRT cell lines, transfection, and depletion of endogenous BubR1
The WT hBubR1 was cloned into pcDNA5/FRT/TO (a gift from J. Pines, Gurdon Institute, Cambridge, UK), and mutagenesis was performed to generate BubR1-K250R and -K250Q mutants. The HeLa-FRT/TO cells (a gift from S. Taylor) were cotransfected with the pOG44 and pcDNA5/FRT/TO-BubR1 constructs, and the cells were selected according to the FLIP-in protocol (Invitrogen). The PP2A-B56α–expressing HeLa-FRT/TO cell line was a gift from J. Nilsson. HeLa-FRT stable cells were treated with 1 µg/ml of doxycycline to induce expression of BubR1 mutants or PP2A-B56α, followed by treatment with 200 ng/ml Noc and 10 µM MG132 to enrich cells in mitosis. Cell lysates were subjected to IP and WB.
IP and WB analysis
Cells were lysed in NETN buffer (150 mM NaCl, 1 mM EDTA, 20 mM Tris, pH 8.0, and 0.5% NP-40) supplemented with protease inhibitors for IP and WB analysis. The mouse tissues were resuspended in lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris, pH 8.0, and 1 mM EGTA) plus the complete EDTA-free proteinase inhibitor cocktail (Roche), which was dissolved in PBS, pH 7.4, and homogenized. For detection of the acetylated BubR1, 20 µM of trichostatin A was added to the NETN buffer.
Immunofluorescence assay
The immunofluorescence assays were performed as previously described with slight modifications for staining the paraffin-embedded tissues (Choi and Lee, 2008; Choi et al., 2009, 2012). In brief, cells were grown on coverslips, fixed in 4% paraformaldehyde, and permeabilized by two incubations in PBS-0.5% Triton X-100 (0.5% PBST) for 15 min at RT before being subjected to indirect immunofluorescence microscopy. Goat serum (20%) in 0.1% PBST was used as the blocking agent and the antibody dilution solution. After permeabilization, the fixed cells were incubated in blocking buffer for 1 h at RT. The cells were incubated with the indicated primary antibodies for 1 h at RT or overnight at 4°C followed by incubation with fluorescence-conjugated secondary antibodies.
Histopathology
Tissue specimens were collected from all organs that exhibited an abnormal appearance. Hematoxylin and eosin (H&E) staining was conducted following the standard procedures. First, paraffinized tissue block was deparaffinization in xylene, followed by rehydration in ethanol in a serial manner (100%, 95%, and 70%), and subjected to hematoxylin staining. Then the specimen was decolorized in acid alcohol and counterstained with eosin. H&E or immunohistochemical stains and were reviewed by the pathologist (G. Gong). The immunohistochemical analyses were performed as previously described with slight modifications (Lee et al., 2009). The sections were incubated with an anti-B220 antibody at a 1:100 dilution in a humidifying chamber at 4°C overnight.
Cytogenetics
For metaphase spreads, MEFs and lymphocytes were treated with 0.1 and 10 µg/ml colcemid for 7 and 2 h, respectively, and were collected by centrifugation. The cells were incubated in a hypotonic solution (0.075 M KCl) for 20 min at 37°C and were centrifuged slowly at 800 rpm for 5 min. The supernatant was removed, and the cells were rinsed once with fixation solution (methanol/acetic acid = 3:1). The cells were incubated in fixation solution overnight at 4°C and placed onto humidified, clean glass slides in a 65°C water bath. The slides were dried and stained in 5% Giemsa solution (Gibco) in phosphate buffer, pH 6.8. The stained slides were mounted with glycerol and were examined under a light microscope (Axio Imager.A1; Carl Zeiss). For the SKY analysis, the K243R/+ MEFs (at passage 27) were treated with 5 µg/ml colcemid for 7 h before fixation. The SKY analysis was performed at the Molecular Cytogenetics Core at the University of Texas M.D. Anderson Cancer Center.
For chromosome spreads coupled with immunofluorescence assays, the basic protocol by Choi et al. (2012) was followed, except that cells were incubated with 200 ng/ml Noc for 4 h instead of with colcemid. Cells were swelled in tap water and cytospinned at 850 rpm for 5 min. The slides were subjected to immunofluorescence assay as described.
Mitotic index measurements in MEFs
The cells were fixed in cold 70% ethanol overnight. The fixed cells were incubated for 1 h at RT with MPM2 antibodies (EMD Millipore) diluted in PBS, 0.02% Triton X-100, and 2% BCS. The cells were washed twice in PBS and incubated with Alexa Fluor 488–conjugated goat anti–mouse antibody (Molecular Probes) for 1 h at RT. The cells were treated with RNase, and the DNA was stained with 7-amino-actinomycid D or propidium iodide and analyzed in a FACS Canto machine (BD).
Partial hepatectomy
10–15-wk-old mice were anesthetized using a mixture of isoflurane/oxygen, and a 70% partial hepatectomy and excision of the medial and left lateral lobes was performed (Mitchell and Willenbring, 2008). At least three animals were used for each time point. After the procedure, the mice were maintained in ad libitum conditions and were killed by cervical dislocation at the indicated time points. The livers were harvested and processed for subsequent analyses.
Microscope image acquisition and processing
Fixed cell images were acquired with a microscope (DeltaVision; Applied Precision) equipped with a 100× objective lens (Olympus). The images were obtained with 0.2-µm-distanced optical sections in z-axis. Each section was deconvoluted and projected into one image using the softWoRx software (Applied Precision) for image display.
For live-cell imaging, cells were monitored using the UPlanFL N 40×/NA 1.30 oil lens on a microscope (DeltaVision Cpre; GE Healthcare) equipped with a charge-coupled device camera (Photometrics) in a CO2 chamber at 37°C (Applied Precision) as previously described (Choi and Lee, 2008; Choi et al., 2009). The cells were seeded in a glass-bottom dish containing DMEM supplemented with 20% FBS and 2 mM l-glutamine and analyzed after 6 h. The images were acquired every 5 min using a 20× objective lens (Olympus).
Cold stable MT assay and scoring of the immunofluorescence intensity
The cells were treated with 10 µM MG132 for 2 h and incubated with serum-free DMEM containing 20 mM Hepes, pH 7.3, for 10 min on ice. When cold MT assay was performed to assess recovery from Monastrol treatment, cells were treated with 100 µM Monastrol for 3 h. In the final hour, cells were incubated with 10 µM MG132 as well. Monastrol-arrested cells were washed at least three times with fresh media containing 10 µM MG132. The cells were released into media with 10 µM MG132 for 1 h. The cells were fixed in 4% paraformaldehyde for 10 min and the immunofluorescence assays were performed as needed.
Images were acquired and processed using DetaVision Softworx software (GE Healthcare), and the images for display were generated by projecting the sum of the optical sections. Quantitative analysis of the immunofluorescence was performed in the projected images using the Image J software (National Institutes of Health). For quantification of intensities at KTs, a mask covering each KT foci was created within a circular region, and the mean pixel intensities were obtained. More than 300 KTs were scored for the intensities in each experiment for statistical analysis.
Mutation analysis of p53 in tumors
The total RNA was extracted from the tumors using the TRIzol reagent (Invitrogen), and the cDNA was synthesized using Superscript II reverse transcription (Invitrogen). The PCR primers were designed to cover the entire mouse p53 sequence as follows: 5′-ATGGAGGAGTCACAGTCGGATATC-3′ (forward) and 5′-TCAGTCTGAGTCAGGCCCC-3′ (reverse). The p53 status was analyzed by the nucleotide sequencing of at least four clones derived from each tumor.
Online supplemental material
Fig. S1 shows that there is no difference in BubR1 expression in the thymus and testis, as well as body weight change between K243R/+ and WT mice. Fig. S2 shows that BubR1 acetylation is involved in proper chromosome congression. Fig. S3 shows that both WT BubR1 and K243R-BubR1 alleles are detected in K243R/+ primary tumors by PCR. Videos related to Fig. 4 G are also available: Video 1 is the live imaging of mitosis in WT MEFs and Videos 2 and 3 are the live imaging of K243R/+ MEFs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201210099/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201210099.dv.
Supplementary Material
Acknowledgments
Animal care and breeding were carried out in the Specific Pathogen Free Mouse Facility of Seoul National University. Imaging and flow cytometry analysis were carried out at the Institute of Molecular Biology and Genetics Imaging Facility. We are very grateful to W. Dai for generously providing the BubR1+/− mice. We thank H.S. Min for the help with several of the pathological analyses. We thank D. Cleveland, J. Deluca, J. Nilsson, G. Kops, J. Pines, H. Yu, S. Taylor, and I. Cheeseman for valuable reagents.
The Basic Science Research Program (2008-0059996 and 2013R1A2A1A01009184), the Novel Drug Target Validation by the Korean National Research Foundation (2012M3A9A8052), and the Cancer Control Program of the Korean Ministry of Health (1220210) funded this research.
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- CENP-E
- centromere-associated protein E
- ES
- embryonic stem
- FRT
- flippase recognition target
- H&E
- hematoxylin and eosin
- IP
- immunoprecipitation
- KT
- kinetochore
- MCC
- mitotic checkpoint complex
- MEF
- mouse embryonic fibroblast
- MT
- microtubule
- NEBD
- nuclear envelope breakdown
- PMSCS
- premature sister chromatid separations
- SAC
- spindle assembly checkpoint
- SKY
- spectral karyotyping
- WB
- Western blot
- WT
- wild type
References
- Abrieu A., Kahana J.A., Wood K.W., Cleveland D.W. 2000. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell. 102:817–826. 10.1016/S0092-8674(00)00070-2 [DOI] [PubMed] [Google Scholar]
- Baker D.J., Jeganathan K.B., Cameron J.D., Thompson M., Juneja S., Kopecka A., Kumar R., Jenkins R.B., de Groen P.C., Roche P., van Deursen J.M. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36:744–749. 10.1038/ng1382 [DOI] [PubMed] [Google Scholar]
- Bolanos-Garcia V.M., Lischetti T., Matak-Vinković D., Cota E., Simpson P.J., Chirgadze D.Y., Spring D.R., Robinson C.V., Nilsson J., Blundell T.L. 2011. Structure of a Blinkin-BUBR1 complex reveals an interaction crucial for kinetochore-mitotic checkpoint regulation via an unanticipated binding Site. Structure. 19:1691–1700. 10.1016/j.str.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G.K., Jablonski S.A., Sudakin V., Hittle J.C., Yen T.J. 1999. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 146:941–954. 10.1083/jcb.146.5.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46. 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Choi E., Lee H. 2008. Chromosome damage in mitosis induces BubR1 activation and prometaphase arrest. FEBS Lett. 582:1700–1706. 10.1016/j.febslet.2008.04.028 [DOI] [PubMed] [Google Scholar]
- Choi E., Choe H., Min J., Choi J.Y., Kim J., Lee H. 2009. BubR1 acetylation at prometaphase is required for modulating APC/C activity and timing of mitosis. EMBO J. 28:2077–2089. 10.1038/emboj.2009.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E., Park P.G., Lee H.O., Lee Y.K., Kang G.H., Lee J.W., Han W., Lee H.C., Noh D.Y., Lekomtsev S., Lee H. 2012. BRCA2 fine-tunes the spindle assembly checkpoint through reinforcement of BubR1 acetylation. Dev. Cell. 22:295–308. 10.1016/j.devcel.2012.01.009 [DOI] [PubMed] [Google Scholar]
- Crasta K., Ganem N.J., Dagher R., Lantermann A.B., Ivanova E.V., Pan Y., Nezi L., Protopopov A., Chowdhury D., Pellman D. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 482:53–58. 10.1038/nature10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A., Pearson C.G., Cimini D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., Musacchio A. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15:214–225. 10.1016/j.cub.2005.01.038 [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- DeLuca K.F., Lens S.M., DeLuca J.G. 2011. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 124:622–634. 10.1242/jcs.072629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Kim C., Ahmad S., Zhang J., Mao Y. 2012. CENP-E–dependent BubR1 autophosphorylation enhances chromosome alignment and the mitotic checkpoint. J. Cell Biol. 198:205–217. 10.1083/jcb.201202152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X., Shin M.S., Zhou J.X., Lee C.H., Qi C.F., Naghashfar Z., Hartley J.W., Fredrickson T.N., Ward J.M., Morse H.C., III 2006. Histologic and molecular characterizations of megakaryocytic leukemia in mice. Leuk. Res. 30:397–406. 10.1016/j.leukres.2005.08.021 [DOI] [PubMed] [Google Scholar]
- Johnson V.L., Scott M.I., Holt S.V., Hussein D., Taylor S.S. 2004. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 117:1577–1589. 10.1242/jcs.01006 [DOI] [PubMed] [Google Scholar]
- Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., Khodjakov A. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science. 311:388–391. 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Yu H. 2011. Mutual regulation between the spindle checkpoint and APC/C. Semin. Cell Dev. Biol. 22:551–558. 10.1016/j.semcdb.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Holland A.J., Lan W., Cleveland D.W. 2010. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 142:444–455. 10.1016/j.cell.2010.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 13:663–676. 10.1016/j.devcel.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Tavakoli-Yaraki M., van Roosmalen M.J., van Binsbergen E., Renkens I., Duran K., Ballarati L., Vergult S., Giardino D., Hansson K., et al. 2012. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep. 1:648–655. 10.1016/j.celrep.2012.05.009 [DOI] [PubMed] [Google Scholar]
- Kruse T., Zhang G., Larsen M.S., Lischetti T., Streicher W., Kragh Nielsen T., Bjørn S.P., Nilsson J. 2013. Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J. Cell Sci. 126:1086–1092. 10.1242/jcs.122481 [DOI] [PubMed] [Google Scholar]
- Lampson M.A., Cheeseman I.M. 2011. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21:133–140. 10.1016/j.tcb.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Kapoor T.M. 2005. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 7:93–98. 10.1038/ncb1208 [DOI] [PubMed] [Google Scholar]
- Lampson M.A., Renduchitala K., Khodjakov A., Kapoor T.M. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237. 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- Lara-Gonzalez P., Westhorpe F.G., Taylor S.S. 2012. The spindle assembly checkpoint. Curr. Biol. 22:R966–R980. 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Lee Y.K., Choi E., Kim M.A., Park P.G., Park N.H., Lee H. 2009. BubR1 as a prognostic marker for recurrence-free survival rates in epithelial ovarian cancers. Br. J. Cancer. 101:504–510. 10.1038/sj.bjc.6605161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malureanu L.A., Jeganathan K.B., Hamada M., Wasilewski L., Davenport J., van Deursen J.M. 2009. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev. Cell. 16:118–131. 10.1016/j.devcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Abrieu A., Cleveland D.W. 2003. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 114:87–98. 10.1016/S0092-8674(03)00475-6 [DOI] [PubMed] [Google Scholar]
- Michel L.S., Liberal V., Chatterjee A., Kirchwegger R., Pasche B., Gerald W., Dobles M., Sorger P.K., Murty V.V., Benezra R. 2001. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 409:355–359. 10.1038/35053094 [DOI] [PubMed] [Google Scholar]
- Mitchell C., Willenbring H. 2008. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat. Protoc. 3:1167–1170. 10.1038/nprot.2008.80 [DOI] [PubMed] [Google Scholar]
- Oromendia A.B., Dodgson S.E., Amon A. 2012. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 26:2696–2708. 10.1101/gad.207407.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T., Jones D.T., Zapatka M., Stütz A.M., Zichner T., Weischenfeldt J., Jäger N., Remke M., Shih D., Northcott P.A., et al. 2012. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 148:59–71. 10.1016/j.cell.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L. 1981. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 84:145–158. 10.1007/BF00293368 [DOI] [PubMed] [Google Scholar]
- Rivera T., Losada A. 2009. Shugoshin regulates cohesion by driving relocalization of PP2A in Xenopus extracts. Chromosoma. 118:223–233. 10.1007/s00412-008-0190-4 [DOI] [PubMed] [Google Scholar]
- Sheltzer J.M., Blank H.M., Pfau S.J., Tange Y., George B.M., Humpton T.J., Brito I.L., Hiraoka Y., Niwa O., Amon A. 2011. Aneuploidy drives genomic instability in yeast. Science. 333:1026–1030. 10.1126/science.1206412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R., Hernando E., Díaz-Rodríguez E., Teruya-Feldstein J., Cordón-Cardo C., Lowe S.W., Benezra R. 2007. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 11:9–23. 10.1016/j.ccr.2006.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A., et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 144:27–40. 10.1016/j.cell.2010.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan G.K., Yen T.J. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936. 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk S.J., Vleugel M., Teixeira A., Kops G.J. 2012. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev. Cell. 23:745–755. 10.1016/j.devcel.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Sundin L.J., Deluca J.G. 2010. Kinetochores: NDC80 toes the line. Curr. Biol. 20:R1083–R1085. 10.1016/j.cub.2010.11.033 [DOI] [PubMed] [Google Scholar]
- Tang Z., Shu H., Qi W., Mahmood N.A., Mumby M.C., Yu H. 2006. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev. Cell. 10:575–585. 10.1016/j.devcel.2006.03.010 [DOI] [PubMed] [Google Scholar]
- Wang Q., Liu T., Fang Y., Xie S., Huang X., Mahmood R., Ramaswamy G., Sakamoto K.M., Darzynkiewicz Z., Xu M., Dai W. 2004. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 103:1278–1285. 10.1182/blood-2003-06-2158 [DOI] [PubMed] [Google Scholar]
- Weaver B.A., Bonday Z.Q., Putkey F.R., Kops G.J., Silk A.D., Cleveland D.W. 2003. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162:551–563. 10.1083/jcb.200303167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B.A., Silk A.D., Montagna C., Verdier-Pinard P., Cleveland D.W. 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 11:25–36. 10.1016/j.ccr.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Wei R.R., Al-Bassam J., Harrison S.C. 2007. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 14:54–59. 10.1038/nsmb1186 [DOI] [PubMed] [Google Scholar]
- Welburn J.P., Vleugel M., Liu D., Yates J.R., III, Lampson M.A., Fukagawa T., Cheeseman I.M. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392. 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Hu L., Chen C., Yu J., O’Connell C.B., Khodjakov A., Pagano M., Dai W. 2012a. BubR1 is modified by sumoylation during mitotic progression. J. Biol. Chem. 287:4875–4882. 10.1074/jbc.M111.318261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Huang Y., Dai W. 2012b. Sumoylated BubR1 plays an important role in chromosome segregation and mitotic timing. Cell Cycle. 11:797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.