The direct interaction between the ERM protein Moesin and microtubules is required for spindle organization in metaphase and cell shape transformation after anaphase onset.
Abstract
Ezrin, Radixin, and Moesin (ERM) proteins play important roles in many cellular processes including cell division. Recent studies have highlighted the implications of their metastatic potential in cancers. ERM’s role in these processes is largely attributed to their ability to link actin filaments to the plasma membrane. In this paper, we show that the ERM protein Moesin directly binds to microtubules in vitro and stabilizes microtubules at the cell cortex in vivo. We identified two evolutionarily conserved residues in the FERM (4.1 protein and ERM) domains of ERMs that mediated the association with microtubules. This ERM–microtubule interaction was required for regulating spindle organization in metaphase and cell shape transformation after anaphase onset but was dispensable for bridging actin filaments to the metaphase cortex. These findings provide a molecular framework for understanding the complex functional interplay between the microtubule and actin cytoskeletons mediated by ERM proteins in mitosis and have broad implications in both physiological and pathological processes that require ERMs.
Introduction
Achieving a particular cell shape optimized for a specific function requires hundreds of proteins that are properly activated, localized, and assembled into complexes. The actin and microtubule cytoskeletons play essential roles in orchestrating this complexity (Hall, 2009). During cell division, cell shape changes are controlled by the reorganization of F-actin linked to the plasma membrane (Green et al., 2012). This needs to be coordinated with the assembly of the microtubule-based mitotic spindle to ensure faithful transmission of genetic material. Here, we report that the well-characterized actin-binding proteins of the Ezrin, Radixin, and Moesin (ERM) family directly interact with microtubules, and this interaction is required for specific ERM-dependent functions in mitosis.
ERMs transition between an active form that bridges F-actin to the plasma membrane and an inactive form that localizes in the cytosol (Fehon et al., 2010). ERM proteins are kept dormant by an intramolecular interaction between the N-terminal domain (4.1 protein and ERM [FERM]) and the C-terminal tail (COOH-ERM association domain [CERMAD]). Their activation requires the disruption of this interaction to unmask the CERMAD actin binding region and the FERM plasma membrane binding domain. This begins with the opening of ERMs through binding of the FERM domain to phosphatidylinositol 4,5-bisphosphate (Pi(4,5)P2) at the plasma membrane (Fievet et al., 2004; Roch et al., 2010; Roubinet et al., 2011). Then, phosphorylation of a CERMAD conserved threonine residue stabilizes the active open conformation (Pearson et al., 2000). Drosophila melanogaster expresses a unique ERM protein (Moesin) and thus offers a powerful system to study ERM functions (Hughes and Fehon, 2007). We and others reported that Moesin regulates cell shape changes during cell division (Carreno et al., 2008; Kunda et al., 2008). Moesin is phosphorylated at mitosis entry and dictates mitotic cell shape changes. In prometaphase, activated Moesin spreads around the cortex and helps to increase cortical rigidity, thus contributing to metaphase cell rounding. At anaphase onset, redistribution of active Moesin at the cell equator controls cell elongation and subsequent cytokinesis (Roubinet et al., 2011; Kunda et al., 2012). Throughout cell division, lack of Moesin or deregulation of its activation, through depletion of its activating kinase Slik, causes severe cell shape deformations. Moesin inactivation also disrupts spindle organization. These phenotypes are generally thought to result from defects in the organization of actin at the cell cortex (Théry and Bornens, 2008). Here, we present data to illustrate the direct involvement of microtubules in these mitotic processes mediated through Moesin.
Results and discussion
Moesin modulates microtubule dynamics in cells and can bind directly to microtubules in vitro
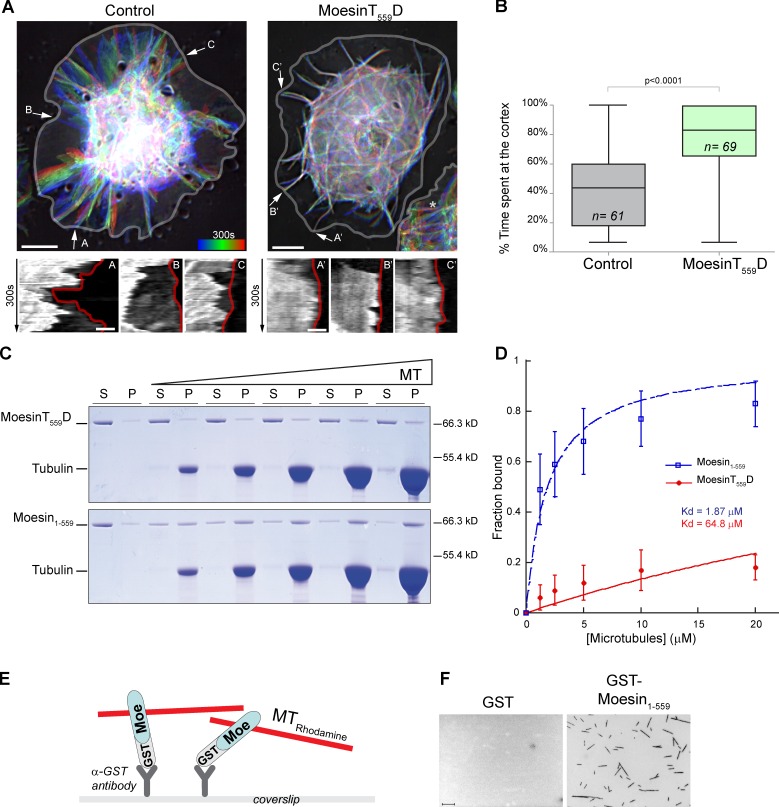
To investigate whether Moesin influences microtubule dynamics, we performed time-lapse imaging of Drosophila S2 cells coexpressing Tubulin-GFP and MoesinT559D, a phosphomimetic, constitutively active form of Moesin. As previously reported, MoesinT559D is almost exclusively at the plasma membrane and causes rounding in ∼90% of cells plated on concanavalin A (Kunda et al., 2008). We thus examined microtubule dynamics in the remaining spread cells that express low levels of MoesinT559D. We found that MoesinT559D increases the time spent by microtubule plus ends at the cortex when compared with control (by approximately twofold; Fig. 1, A and B; and Videos 1 and 2). This raises the possibility of a direct Moesin–microtubule interaction. To test this, we performed a microtubule cosedimentation assay using recombinant Moesin constructs: MoesinT559D, which without the activation by Pi(4,5)P2 is partially open in vitro, and Moesin1–559 (with the C-terminal 19 amino acids deleted), which is fully open (Chambers and Bretscher, 2005; Jayasundar et al., 2012). We found that Moesin1–559 associates with microtubules, whereas MoesinT559D barely interact with these filaments (Fig. 1, C and D). We determined the dissociation constant (Kd) of Moesin1–559 to microtubules to be ∼1.9 µM. The binding affinity is relatively weak compared with other typical microtubule associated proteins, such as kinesins (submicromolar), but is not uncharacteristic of F-actin–microtubule cross-linkers. For example, the protein coronin has a Kd of 6 nM to actin but only weakly binds to microtubules (Kd of 15–20 µM; Goode et al., 1999). To directly visualize Moesin–microtubule interactions, we performed a microscopy-based flow chamber assay with surface-immobilized GST-Moesin1–559. We observed the capture of microtubules by GST-Moesin1–559, but not GST, demonstrating a direct Moesin–microtubule interaction (Fig. 1, E and F). Similar results were obtained for Homo sapiens Ezrin (hsEzrin), indicating functional conservation across species (Fig. S1). Therefore, in addition to bridging F-actin with the plasma membrane, ERMs can bind directly to microtubules in vitro and modulate cortical microtubule dynamics in cells.
Figure 1.
Moesin binds microtubules in vitro and regulates their cortical dynamics in cultured cells. (A) Time-lapse projection of microtubules in Tubulin-GFP cells transfected with mCherry (left) or MoesinT559D-mCherry (right). (top) Time frames of the Tubulin-GFP channel is overlaid using heat map (blue to red, 60 frames every 5 s). The asterisk indicates a nontransfected cell, and arrows show representative microtubules used for kymography (bottom). (bottom) Kymographs of microtubules. The cell edge is outlined in red. (B) A plot showing the percentage of time spent by individual microtubules at the cortex (<500 nm from the cell edge). Boxes show top and bottom quartiles, horizontal lines show median values, and vertical lines show minimal and maximal values. (C) Constructs were titrated against increasing concentrations of microtubules (MT) in a cosedimentation assay. Coomassie blue–stained gels of supernatant (S) and pellet (P) fractions are shown. (D) Fractions of microtubule-bound Moesin from four independent cosedimentation experiments were plotted against microtubule concentrations and fitted to a hyperbola to determine the Kd. Error bars represent SDs. (E) An illustration depicting the microscopy-based microtubule-binding assay. Moe, Moesin. (F) Microscopy images of X-rhodamine–microtubules captured by surface-anchored GST (left) or GST-Moesin1–559 (right). Bars: (A [top] and F) 5 µm; (A, bottom) 1 µm.
Two conserved lysine residues mediate Moesin–microtubule interaction
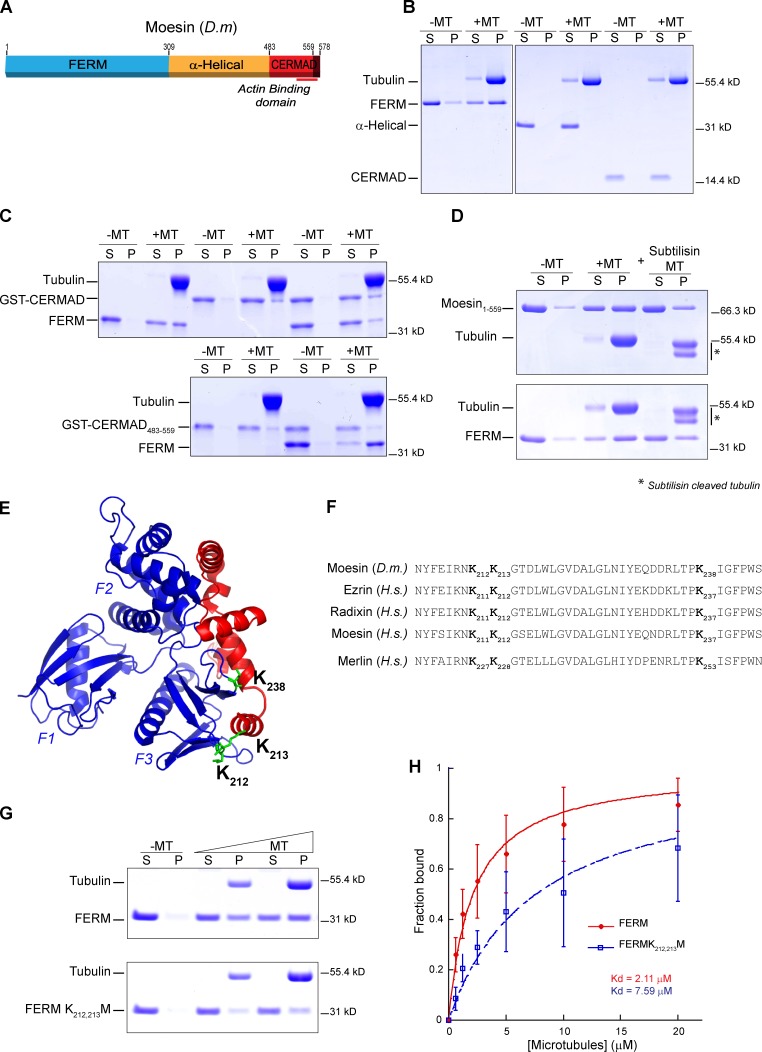
To map the microtubule-binding region of ERMs, we purified and tested each of the three conserved domains of Drosophila Moesin and hsEzrin. Among them, only the FERM domains interact with microtubules (Fig. 2, A and B; and Fig. S2), indicating that ERMs bind to microtubules and F-actin via two distinct domains. Preincubation with the CERMAD domain severely perturbed the interaction between FERM and microtubules, whereas the truncated CERMAD (residues 483–559) that cannot bind to FERM did not (Fig. 2 C). This indicates that the ERM–microtubule binding domain is masked when ERM proteins are in their closed conformation. We also found that subtilisin digestion, which removes the C-terminal negatively charged tails of tubulins, impairs Moesin–microtubule interaction (Fig. 2 D). This implicates the involvement of positively charged residues on Moesin. Examination of the FERM–CERMAD crystal structure (Pearson et al., 2000) revealed three candidate positively charged surface residues in the FERM domain (K212,213 and K238 in Moesin). They are situated along the FERM–CERMAD binding interface and could be sterically masked in the closed conformation (Fig. 2, E and F). Via characterization of point mutants, we found that only mutations at K212 and K213 in Drosophila Moesin or at the corresponding residues in hsEzrin (K211 and K212) perturb microtubule binding (∼3.6- and ∼9-fold reduction, respectively in the apparent Kd; Fig. 2, G and H; and Fig. S2). This observed change in microtubule binding affinity is not a result of protein misfolding, as evident by the overlapping circular dichroism spectra with almost identical secondary characteristics between FERM-K212,213M and FERM–wild type (Fig. S2). Sequence alignment analysis shows that the two lysines mediating microtubule binding are conserved among ERM proteins but not found in other FERM domain–containing proteins except for the ERM-related protein Merlin (Fig. 2 F and Fig. S2), which interestingly has been reported to bind microtubules (Muranen et al., 2007). In contrast, myosin X, which was reported to interact with microtubules, does not have these two lysines in its FERM domain, which alone is insufficient to bind to microtubules (Weber et al., 2004).
Figure 2.
Moesin binds to microtubules via its N-terminal FERM domain. (A) A schematic depicting the three domains of Moesin. D.m, Drosophila melanogaster. (B–D) Microtubule cosedimentation assay. Samples from protein alone (−MT) and protein with microtubules (+MT) are shown in Coomassie blue–stained gels. S, supernatant; P, pellet. (B) Sedimentation assay with purified domains of Moesin. (C) FERM domain was preincubated with GST-CERMAD of wild-type Moesin (residues 483–578; top) or with GST-CERMAD of Moesin483–559 (residues 483–559; bottom) before performing microtubule cosedimentation. (D) Untreated or subtilisin-treated microtubules were tested in sedimentation assay with Moesin1–559 or FERM of Moesin. (E) A crystal structure of FERM (blue) and CERMAD (red) complex of Moesin. Candidate lysine residues (K238, K212, and K213) are highlighted in green. (F) Sequence alignment of the microtubule binding region of ERM proteins and Merlin. H.s, H. sapiens. (G) Comparison of binding affinities to microtubules between FERM wild type and K212,213M. (H) Fractions of microtubule-bound Moesin FERM from three independent microtubule sedimentation experiments were plotted against microtubule concentrations and fitted to a hyperbola to determine the Kd. Error bars represent SDs.
Moesin’s microtubule binding is separable from other Moesin functions
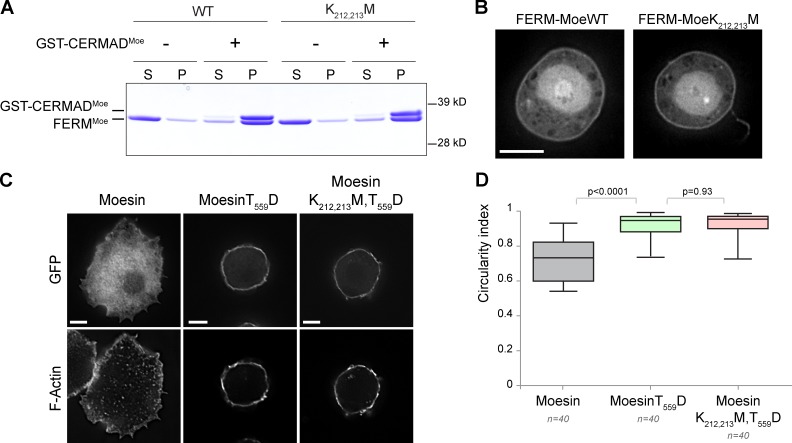
To assess whether the K212,213M mutations perturb other aspects of Moesin functions, we examined the intramolecular interaction between FERM and CERMAD as well as its association with the plasma membrane and the cortical actin network. First, we found that the K212,213M mutations do not interfere with the intramolecular association of the FERM domain with the CERMAD using an in vitro binding assay (Fig. 3 A). FERM-K212,213M, like the wild-type protein, still binds to CERMAD at a 1:1 stoichiometric ratio. Second, FERM-K212,213M-GFP associates with the plasma membrane, indicating that K212,213M mutations do not affect binding to Pi(4,5)P2 (Fig. 3 B). This is consistent with the fact that K212 and K213 are located in a subdomain (F3) away from the Pi(4,5)P2 binding site (linker between F2 and F3 lobes; Fehon et al., 2010). Third, we tested whether K212,213M mutations affect Moesin ability to cross-link F-actin to the plasma membrane in a cell rounding assay. As reported, interphase cells expressing MoesinT559D-GFP exhibit a distinct rounding phenotype, triggered by cross-linking of F-actin to the plasma membrane (Kunda et al., 2008). K212,213M mutations on MoesinT559D-GFP do not prevent this cell rounding (Fig. 3, C and D). Thus, Moesin microtubule binding ability is functionally distinct from its association with the plasma membrane or F-actin.
Figure 3.
Microtubule-binding mutations on Moesin do not perturb FERM–CERMAD interaction or Moesin’s association with the plasma membrane and cortical actin network. (A) The GST fusion protein bound to magnetic glutathione beads was allowed to interact with the FERM domain in solution. Bead-bound fraction (P, pellet) was separated from the unbound fraction (S, supernatant) using a magnet. Both FERM wild type and FERM K212,213M associate with CERMAD in a 1:1 stoichiometric ratio. (B) FERM wild type and FERM K212,213M domain of Moesin fused to GFP were expressed in S2 cells. Bar, 10 µm. (C) Cells expressing the indicated Moesin constructs were plated on concanavalin A–treated glass coverslips to allow firm spreading. GFP expression and F-actin staining with Texas red–phalloidin are shown. Bars, 5 µm. (D) Quantification of rounding in cells transfected with the indicated constructs. Boxes show top and bottom quartiles, horizontal lines show median values, and vertical lines show minimal and maximal values. Moe, Moesin; WT, wild type.
Moesin–microtubule interaction is required for regulating cortical microtubule dynamics
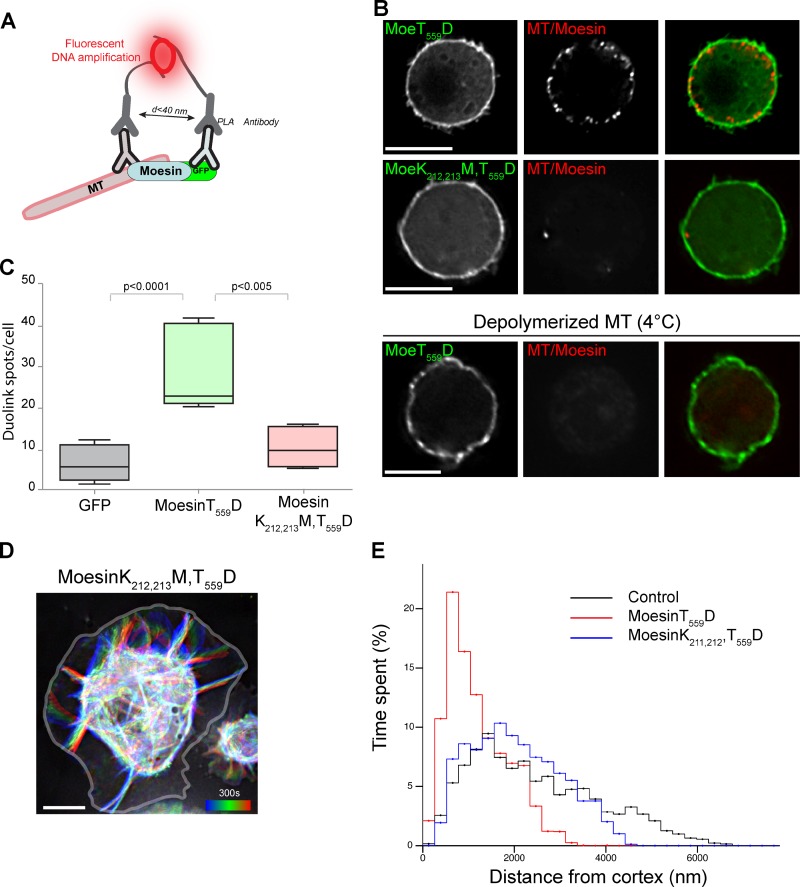
To test the possibility that Moesin stabilizes microtubules via a direct interaction at the cell cortex, we used Duolink, a proximity ligation method that generates fluorescent signals at sites of protein–protein interactions (Fig. 4 A; Fredriksson et al., 2002; Söderberg et al., 2006). In cells expressing MoesinT559D, we observed fluorescent Duolink signals at the cortex indicative of direct interactions between Moesin and microtubules. Importantly, Duolink signals disappeared when microtubules were depolymerized by cold treatment. We also found that K212,213M mutations drastically perturbed Moesin–microtubule interactions in cells (Fig. 4, B and C), consistent with our biochemical data (Fig. 2). To determine whether microtubule interaction is needed for controlling microtubule dynamics, we analyzed the effect of K212,213M mutations by tracking microtubule ends over time using an automated algorithm (Currie et al., 2011; Zhang et al., 2011). We found that MoesinT559D expression increased the time microtubule ends spent at the proximity of the cell periphery (Fig. 4 E), consistent with our manual measurements (Fig. 1 B). In comparison, MoesinK212,213M,T559D failed to exert this effect on microtubules (Fig. 4, D and E; and Video 3). In addition, MoesinT559D expression significantly reduced the frequencies of catastrophes and rescues, likely as a result of suppressing exit from the pause state (Table 1). Microtubule binding mutations (K212,213M) abolished this effect, indicating that direct Moesin–microtubule interaction is crucial for regulating cortical microtubule dynamics (Table 1).
Figure 4.
Moesin directly anchors microtubules to the cortex and regulate their dynamics. (A) An illustration depicts a proximity ligation in situ assay (PLA; Duolink), which allows the detection of protein–protein interactions as bright fluorescent spots in situ. (B) MoesinT559D-GFP (MoeT559D) interacts with microtubules at the cortex of S2 cells, as evident by the presence of fluorescent Duolink spots at the periphery of the cell (red, top). Interphase cells plated on glass are shown. MT, microtubule. (C) Quantification of Tubulin/Moesin-GFP Duolink spots in cells expressing the indicated constructs. Boxes show top and bottom quartiles, horizontal lines show median values, and vertical lines show minimal and maximal values (control, n = 12; MoesinT559D, n = 12; MoesinK212,213M,T559D, n = 10). (D) Time-lapse projection of Tubulin-GFP in S2 cells transfected with MoesinK212,213M,T559D-mCherry construct. Heat map representation is shown as in Fig. 1. (E) Ends of GFP-labeled microtubules were tracked using an automated tracking algorithm. The microtubule ends in MoesinT559D-expressing cells (two cells; 203 microtubules) spent significantly more time in the vicinity of the cortex compared with control cells (three cells; 310 microtubules) or cells expressing MoesinK212,213M,T559D-GFP (two cells; 164 microtubules). Bars: (B) 10 µm; (C and D) 5 µm.
Table 1.
Comparison of state-to-state frequency and growth/shrinkage rate between control and MoesinT559D- and MoesinK212,213M,T559D-expressing cells
| State to state and rates | Control | MoesinT559D | MoesinK211,212M,T559D |
| G → S | |||
| (ns−1) | 0.0114 (44) | 0.0055 (13) | 0.0125 (20) |
| (ns−1) | 0.0085–0.0148 | 0.0031–0.0091 | 0.0079–0.0184 |
| P = 0.0139a | P = 0.714 | ||
| G → P | |||
| (ns−1) | 0.0569 (220) | 0.062 (146) | 0.0579 (93) |
| (ns−1) | 0.0527–0.0609 | 0.0569–0.0665 | 0.0525–0.0635 |
| P = 0.134 | P = 0.783 | ||
| S → G | |||
| (ns−1) | 0.0110 (37) | 0.0142 (22) | 0.0062 (12) |
| (ns−1) | 0.0079–0.0147 | 0.0092–0.0203 | 0.0034–0.0104 |
| P = 0.298 | P = 0.0788 | ||
| S → P | |||
| (ns−1) | 0.0564 (190) | 0.0544 (84) | 0.0543 (105) |
| (ns−1) | 0.0521–0.0605 | 0.0481–0.0602 | 0.0483–0.0595 |
| P = 0.589 | P = 0.566 | ||
| P → G | |||
| (ns−1) | 0.0271 (148) | 0.0177 (85) | 0.0192 (54) |
| (ns−1) | 0.0229–0.0318 | 0.0139–0.0218 | 0.0143–0.0248 |
| P = 0.0047b | P = 0.037a | ||
| P → S | |||
| (ns−1) | 0.0216 (118) | 0.0121 (58) | 0.0214 (60) |
| (ns−1) | 0.018–0.0256 | 0.009–0.0158 | 0.0161–0.0275 |
| P = 0.0014c | P = 0.9401 | ||
| Pause state | |||
| (%) | 67.3 (3,421) | 78.2 (3,235) | 69.6 (1,863) |
| (%) | 66.0–68.6 | 76.9–79.4 | 67.8–71.3 |
| Shrinkage state | |||
| (%) | 14.8 (752) | 8.3 (343) | 15.8 (422) |
| (%) | 13.8–15.8 | 7.5–9.2 | 14.4–17.2 |
| Growth state | |||
| (%) | 17.9 (912) | 13.5 (558) | 14.6 (391) |
| (%) | 16.9–19.0 | 12.5–14.6 | 13.3–16.0 |
| Growth rate | |||
| (µm/min) | 2.68 | 2.39 | 2.55 |
| (µm/min) | ±0.052 | ±0.063 | ±0.078 |
| P = 4.3.10−4c | P=0.141 | ||
| Shrinkage rate | |||
| (µm/min) | 2.61 | 2.54 | 2.46 |
| (µm/min) | ±0.058 | ±0.09 | ±0.078 |
| P = 0.552 | P = 0.159 |
Bold numbers denote statistically significant different values from controls. Numbers in parentheses indicate the event count. Intervals indicate 95% confidence. Statistical significance for sate-to-state transition frequencies was determined using a two-tailed permutation test with 10,000 resamples. For growth and shrinkage rates, a two-sample t test was used to determine statistical significance (±SEM).
Statistically different from control if P < 0.05.
P < 0.01.
P < 0.001.
Moesin–microtubule interaction regulates mitotic spindle organization and specific cell morphogenesis
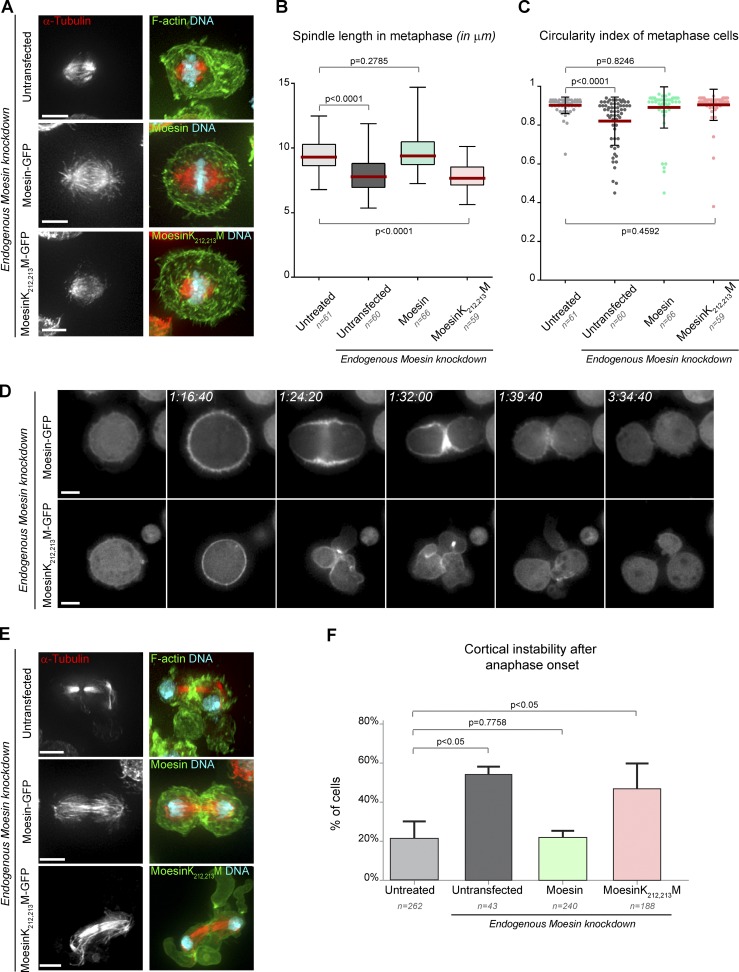
To explore the physiological importance of ERM–microtubule interaction, we tested the ability of the microtubule binding mutant of Moesin to substitute endogenous protein function in mitosis. We established a stable cell line expressing MoesinK212,213M-GFP that is not targeted by a Moesin 3′UTR double-stranded RNA (dsRNA; Fig. S3). Moesin-GFP, which rescues dsRNA depletion of endogenous Moesin (Roubinet et al., 2011), served as a control. We focused on three characterized mitotic defects resulting from Moesin depletion: spindle length reduction, cell rounding defects at metaphase entry, and cortical instability after anaphase onset (Carreno et al., 2008; Kunda et al., 2008). First, in light of our finding, we hypothesize that Moesin regulates spindle length by providing a cortical anchor for astral microtubules. Consistent with this, we found that although MoesinK212,213M-GFP was properly recruited to the cortex of metaphase cells, it did not rescue the reduction in spindle length caused by Moesin depletion (Fig. 5, A and B). Second, metaphase cell rounding is controlled by an isotropic activation of Moesin at the cortex of cells entering mitosis (Carreno et al., 2008; Kunda et al., 2008). MoesinK212,213M was able to rescue metaphase cell rounding (Fig. 5, A, C, and D), demonstrating that K212,213M mutations do not impair Moesin’s ability to organize F-actin at the cortex in metaphase. This result is in line with a study showing that microtubules are dispensable in metaphase cell rounding (Matthews et al., 2012). Third, at anaphase onset, ingression of the cleavage furrow and cell elongation are precisely coordinated to ensure robust cytokinesis. Spindle microtubules mediate these transformations by controlling reorganization of actin filaments at specific regions of the plasma membrane. For instance, microtubules activate RhoA at the cell equator, which in turn promotes anaphase elongation and the formation of the actomyosin contractile ring (Rappaport, 1971; Green et al., 2012). As we previously reported (Carreno et al., 2008), depletion of Moesin (or depletion of Slik, its activating kinase) disrupts this coordination and induces cortical instability, signified by uncontrolled membrane blebbing (Video 4). We found that MoesinK212,213M was not able to prevent cortical instability in anaphase and telophase caused by endogenous Moesin depletion (Fig. 5, D–F; and Videos 5 and 6). This indicates that Moesin’s binding to microtubules is needed for mediating the coordination between the mitotic spindle and the actin cortex, which in turn regulates cell morphogenesis after anaphase onset.
Figure 5.
Moesin–microtubule interaction is required for spindle organization in metaphase and cell morphogenesis after the anaphase onset. (A) Microtubule organization and cortical organization in metaphase cells upon depletion of endogenous Moesin (top: wild-type cell; middle: Moesin-GFP cell; bottom: MoesinK212,213M-GFP cell). Images show maximum projections. Microtubules (red), F-actin or Moesin constructs, and DNA are shown. (B) Spindle length in metaphase (in micrometers). Boxes show top and bottom quartiles, red horizontal lines show median values, and vertical lines show minimal and maximal values. (C) Circularity index in metaphase (in micrometers). Each dot represents the calculated value for one cell. Red horizontal bars show mean values. Error bars represent SDs. (D) Dynamics of Moesin-GFP and MoesinK212,213M-GFP observed upon depletion of endogenous Moesin. Numbers at the top show hours, minutes, and seconds. (E) Microtubule organization and cortical organization in telophase cells upon depletion of endogenous Moesin (top: wild-type cell; middle: Moesin-GFP cell; bottom: MoesinK212,213M-GFP cell). Images show maximum projections. Microtubules (red), F-actin or Moesin constructs, and DNA are shown. (F) Percentage of cells with cortical instability after anaphase onset in the indicated treated cells (from live-cell recordings; three independent experiments). Error bars represent SDs. Bars, 5 µm.
In this study, we show that ERMs bind to microtubules via a conserved motif in their FERM domains. We demonstrate that Moesin–microtubule interaction is essential in mitosis for regulating spindle length in metaphase and cell shape changes after anaphase onset. We showed that Moesin controls polymerization dynamics of cortical microtubules by modulating their exits from pauses. This could be a direct consequence of Moesin binding to microtubules. Alternatively, Moesin could prolong the dwell time of microtubules at the cortex to allow stabilization (or prevent destabilization) by other factors such as NuMa (nuclear mitotic apparatus)/MUD (Mushroom Body Defect), dynein, EB1, and kinesin-13 proteins. Our data indicate that perturbing Moesin-mediated anchoring of astral microtubules to the cortex leads to reduction in spindle size, possibly by disrupting the pulling forces exerted by the Gα/PINS (partner of Insc)/MUD/dynein system (McNally, 2013). They also suggest that by stabilizing microtubule ends at the cortex, Moesin plays important roles in mediating the signaling between microtubules and the actin cortex to control mitotic morphogenesis after anaphase onset. In sum, our finding provides a novel framework for studying how ERM proteins mediate the coordination between cortical actin and spindle microtubule dynamic reorganizations during cell division. The notion that ERM proteins can bind microtubules will lead to a better understanding of the physiological and pathological processes requiring ERM functions.
Materials and methods
Protein expression and purification
Recombinant Moesin constructs were cloned into pGEX-6P1 vector expressed in BL21-pLys Escherichia coli cells. For most constructs, expressions were induced by 0.5 mM IPTG overnight at 18°C. Cells were lysed in TBS/0.5% Triton X-100 buffer. GST fusion proteins were purified by affinity chromatography using glutathione–Sepharose resins according to the manufacturer’s instructions (GE Healthcare). To obtain GST fusions, bound proteins were eluted with 5 mM reduced glutathione and then dialyzed into appropriate buffers for different assays (see following paragraphs) in the presence of 10% sucrose as a cryoprotectant. To obtain the untagged constructs, proteins bound to the resin were cleaved with PreScission Protease (GE Healthcare). Sucrose was added to 10% before flash freezing in liquid N2.
Microtubule cosedimentation assay and microscopy-based binding assay
Microtubules were polymerized in the presence of 0.5 mM GMPCPP (guanosine-5′-[(α,β)-methyleno]triphosphate; Jena Bioscience) in BRB80 (80 mM Pipes, pH 6.8, 1 mM EGTA, and 1 mM MgCl2) with 1 mM DTT at 37°C. For the cosedimentation assay, microtubules were diluted to the desired concentrations in BRB80 + 50 mM KCl + 10 µM taxol (Sigma-Aldrich) and incubated with the indicated ERM constructs (∼1 µM) at room temperature for 20 min before subjecting the mixtures to ultracentrifugation (60,000–80,000 RPM for 5 min; Sorvall S120-AT3; Thermo Fisher Scientific). Supernatant and pellet fractions were recovered, resuspended in Laemmli buffer, and resolved by SDS-PAGE. Gels were stained in Coomassie blue R250 dye, destained, and scanned with a digital scanner. Protein bands are quantified using ImageJ (National Institutes of Health). Kd measurements were made by plotting fractions bound to microtubules after cosedimentation (pellet) against concentrations of Tubulin and fitting the plots to a hyperbola using KaleidaGraph (Synergy). For the microscopy-based binding assay, X-rhodamine–labeled Tubulin was added during microtubule polymerization. A custom-made flow chamber, by adhering a coverslip onto a glass slide with double-sided adhesive tape, was used. First, the chamber was filled with ∼2 µg/ml α-GST antibody to allow immobilization onto the coverslip followed by the addition of 0.5 mg/ml casein to block the surface. Then, the GST fusion ERM construct was flowed into the chamber to bind to the surface-anchored GST antibody. After a 5-min incubation, X-rhodamine–labeled microtubules were flowed into the chamber and incubated for another 5 min. Unbound microtubules were then flushed out with assay buffer (BRB80 + oxidation mix: 22.5 mM glucose, 0.22 mg/ml glucose oxidase [Sigma-Aldrich], and 0.036 mg/ml catalase [Sigma-Aldrich]). Images of captured microtubules were recorded on a microscope (Axio Imager Z1; Carl Zeiss) equipped with a 100×, 1.4 NA Plan Apochromat objective.
Cell culture and dsRNA treatment
Moesin 3′UTR dsRNA were produced using the large-scale RNA production system (T7 RiboMAX; Promega) with the following primers: forward, 5′-TAATACGACTCACTATAGGGAGACGGTGAGACTCCAGAAAGAAAA-3′; and reverse, 5′-TAATACGACTCACTATAGGGAGATACAAAAGCTGCGAGACAAAAC-3′. Drosophila S2 cells were grown in FCS-supplemented Schneider’s medium (Invitrogen), and dsRNA were directly added in the culture medium for 6 d. cDNA Moesin point mutants were produced using inverse PCR (Phusion polymerase; New England Biolabs, Inc.) and fused with GFP or mCherry into pAc5.1 (Invitrogen). Stable cell lines were obtained using hygromycin selection and sorted by FACS (FACSAria; BD) to select cell lines expressing equivalent levels of endogenous and exogenous Moesin. Dynamics of microtubules were analyzed after transient transfection (FuGENE HD; Roche) of the corresponding constructs.
Imaging of fixed samples and time-lapse recording
Images of fixed cells were acquired using a microscope (DeltaVision; Olympus) as previously described (Ben El Kadhi et al., 2011). In brief, for immunofluorescence analysis, cells were plated on glass coverslips fixed in 4% formaldehyde before immunostaining or Duolink detection following the manufacturer’s instructions (Olink Bioscience). Antibodies were used at the following dilutions: anti–P-Moesin (Roubinet et al., 2011) at 1:500, anti-GFP at 1:500 (Sigma-Aldrich), anti–α-Tubulin (Sigma-Aldrich) at 1:200, Alexa Fluor 488– and Alexa Fluor 546–conjugated secondary antibodies (Molecular Probes) at 1:500, and Texas red–X phalloidin (Molecular Probes) at 1:200. Coverslips were mounted using Vectashield mounting medium with DAPI (Vector Laboratories). Multichannel images were acquired with an elite inverted microscope (DeltaVision; Applied Precision) controlled by the softWoRx software (Applied Precision), using a 60×, 1.42 NA oil objective, with voxels collected at 107-nm lateral and 200-nm axial intervals. For live imaging, cell lines were cultured in glass-bottom plates coated with concanavalin A (for imaging microtubule dynamics) or uncoated (for observing cell divisions) in a temperature-controlled chamber (27°C) with a camera (CoolSNAP HQ2; Photometrics) every 5 s for 5 min (for recording microtubule dynamics) and every 7 min and 40 s for 16 h (for imaging cell divisions). Images were analyzed using softWoRx Explorer and ImageJ packages. Images were deconvolved using softWoRx and processed with Photoshop (Adobe). To assess microtubule dynamics, time spent by an individual microtubule near the cortex (<500 nm from cell edge) was measured by kymography. A minimum of 46 individual microtubules and four individual cells was measured for each condition (Fig. 1 B). Automated tracking (Fig. 4 E and Table 1) was performed as described previously (Currie et al., 2011; Díaz-Valencia et al., 2011; Zhang et al., 2011). In brief, a region of interest is manually specified, consisting of an outer boundary of the cell and an inner boundary that is several micrometers away to avoid tracking the extremely crowded interior region. Microtubules in the region of interest are identified automatically by the algorithm using a Gaussian filter. Then, the algorithm links microtubules in consecutive time frames that are the closest to one another based on a spatial distance metric for extended objects to form trajectories. From the tracking trajectories, the growth, shrinking, or paused behaviors are determined using the displacement of a microtubule tip between two consecutive time points and the angle between this displacement vector and the microtubule body.
Online supplemental material
Fig. S1 shows that human Ezrin binds microtubules in vitro. Fig. S2 shows that K211,212 of hsEzrin FERM domain are essential for microtubule binding. Fig. S3 shows a Western blot analysis for Moesin depletion and rescue experiments. Video 1 shows α-Tubulin–GFP–expressing mCherry in interphase S2 cells. Video 2 shows α-Tubulin–GFP–expressing MoesinT559D-mCherry in interphase S2 cells. Video 3 shows α-Tubulin–GFP–expressing MoesinK211,212T559D-mCherry in interphase S2 cells. Video 4 shows Moesin-GFP in dividing Slik-depleted S2 cells. Video 5 shows Moesin-GFP in dividing endogenous Moesin-depleted S2 cells. Video 6 shows MoesinK212,213M-GFP in dividing endogenous Moesin-depleted S2 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201304052/DC1.
Supplementary Material
Acknowledgments
We would like to dedicate this study to all people suffering from and fighting against cancer.
We thank the Institute for Research in Immunology and Cancer (IRIC) Bio-imaging, Biophysics, and Flow Cytometry core facilities for technical assistance and P. Roux and A. Maddox for critical review of the manuscript.
K. Ben El Kadhi is a recipient of a Fonds de Recherche du Québec en Santé (FRQS) fellowship. S. Solinet and A. Ma are supported by a National Institutes of Health grant (R01GM086536 to A. Ma); B.H. Kwok is supported by a FRQS Junior 1 award and a Canadian Institutes of Health Research (CIHR) New Investigator Award. S. Carreno holds a CIHR New Investigator Award. This work was supported by grants from the CIHR (MOP-89877 to S. Carreno and MOP-97928 to B.H. Kwok) and from the Natural Sciences and Engineering Research Council of Canada (S. Carreno). IRIC is supported in part by the Canadian Center of Excellence in Commercialization and Research, the Canada Foundation for Innovation, and by the FRQS.
Author contributions: S. Solinet, B.H. Kwok, and S. Carreno designed the research project. S. Solinet, K. Mahmud, K. Ben El Kadhi, B. Decelle, and L. Talje performed the experiments. S.F. Stewman designed the automated tracking algorithm under the direction of A. Ma and used it to track microtubules in live-imaging videos provided by S. Solinet. B.H. Kwok, S. Carreno and S. Solinet analyzed the data. B.H. Kwok and S. Carreno wrote the manuscript.
Footnotes
Abbreviations used in this paper:
- CERMAD
- COOH-ERM association domain
- dsRNA
- double-stranded RNA
- ERM
- Ezrin, Radixin, and Moesin
- FERM
- 4.1 protein and ERM
- hsEzrin
- Homo sapiens Ezrin
- Pi(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
References
- Ben El Kadhi K., Roubinet C., Solinet S., Emery G., Carréno S. 2011. The inositol 5-phosphatase dOCRL controls PI(4,5)P2 homeostasis and is necessary for cytokinesis. Curr. Biol. 21:1074–1079 10.1016/j.cub.2011.05.030 [DOI] [PubMed] [Google Scholar]
- Carreno S., Kouranti I., Glusman E.S., Fuller M.T., Echard A., Payre F. 2008. Moesin and its activating kinase Slik are required for cortical stability and microtubule organization in mitotic cells. J. Cell Biol. 180:739–746 10.1083/jcb.200709161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers D.N., Bretscher A. 2005. Ezrin mutants affecting dimerization and activation. Biochemistry. 44:3926–3932 10.1021/bi0480382 [DOI] [PubMed] [Google Scholar]
- Currie J.D., Stewman S., Schimizzi G., Slep K.C., Ma A., Rogers S.L. 2011. The microtubule lattice and plus-end association of Drosophila Mini spindles is spatially regulated to fine-tune microtubule dynamics. Mol. Biol. Cell. 22:4343–4361 10.1091/mbc.E11-06-0520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Valencia J.D., Morelli M.M., Bailey M., Zhang D., Sharp D.J., Ross J.L. 2011. Drosophila katanin-60 depolymerizes and severs at microtubule defects. Biophys. J. 100:2440–2449 10.1016/j.bpj.2011.03.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon R.G., McClatchey A.I., Bretscher A. 2010. Organizing the cell cortex: the role of ERM proteins. Nat. Rev. Mol. Cell Biol. 11:276–287 10.1038/nrm2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fievet B.T., Gautreau A., Roy C., Del Maestro L., Mangeat P., Louvard D., Arpin M. 2004. Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J. Cell Biol. 164:653–659 10.1083/jcb.200307032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S.M., Ostman A., Landegren U. 2002. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20:473–477 10.1038/nbt0502-473 [DOI] [PubMed] [Google Scholar]
- Goode B.L., Wong J.J., Butty A.C., Peter M., McCormack A.L., Yates J.R., Drubin D.G., Barnes G. 1999. Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J. Cell Biol. 144:83–98 10.1083/jcb.144.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.A., Paluch E., Oegema K. 2012. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28:29–58 10.1146/annurev-cellbio-101011-155718 [DOI] [PubMed] [Google Scholar]
- Hall A. 2009. The cytoskeleton and cancer. Cancer Metastasis Rev. 28:5–14 10.1007/s10555-008-9166-3 [DOI] [PubMed] [Google Scholar]
- Hughes S.C., Fehon R.G. 2007. Understanding ERM proteins—the awesome power of genetics finally brought to bear. Curr. Opin. Cell Biol. 19:51–56 10.1016/j.ceb.2006.12.004 [DOI] [PubMed] [Google Scholar]
- Jayasundar J.J., Ju J.H., He L., Liu D., Meilleur F., Zhao J., Callaway D.J., Bu Z. 2012. Open conformation of ezrin bound to phosphatidylinositol 4,5-bisphosphate and to F-actin revealed by neutron scattering. J. Biol. Chem. 287:37119–37133 10.1074/jbc.M112.380972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P., Pelling A.E., Liu T., Baum B. 2008. Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr. Biol. 18:91–101 10.1016/j.cub.2007.12.051 [DOI] [PubMed] [Google Scholar]
- Kunda P., Rodrigues N.T., Moeendarbary E., Liu T., Ivetic A., Charras G., Baum B. 2012. PP1-mediated moesin dephosphorylation couples polar relaxation to mitotic exit. Curr. Biol. 22:231–236 10.1016/j.cub.2011.12.016 [DOI] [PubMed] [Google Scholar]
- Matthews H.K., Delabre U., Rohn J.L., Guck J., Kunda P., Baum B. 2012. Changes in Ect2 localization couple actomyosin-dependent cell shape changes to mitotic progression. Dev. Cell. 23:371–383 10.1016/j.devcel.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F.J. 2013. Mechanisms of spindle positioning. J. Cell Biol. 200:131–140 10.1083/jcb.201210007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranen T., Grönholm M., Lampin A., Lallemand D., Zhao F., Giovannini M., Carpén O. 2007. The tumor suppressor merlin interacts with microtubules and modulates Schwann cell microtubule cytoskeleton. Hum. Mol. Genet. 16:1742–1751 10.1093/hmg/ddm122 [DOI] [PubMed] [Google Scholar]
- Pearson M.A., Reczek D., Bretscher A., Karplus P.A. 2000. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 101:259–270 10.1016/S0092-8674(00)80836-3 [DOI] [PubMed] [Google Scholar]
- Rappaport R. 1971. Cytokinesis in animal cells. Int. Rev. Cytol. 31:169–213 10.1016/S0074-7696(08)60059-5 [DOI] [PubMed] [Google Scholar]
- Roch F., Polesello C., Roubinet C., Martin M., Roy C., Valenti P., Carreno S., Mangeat P., Payre F. 2010. Differential roles of PtdIns(4,5)P2 and phosphorylation in moesin activation during Drosophila development. J. Cell Sci. 123:2058–2067 10.1242/jcs.064550 [DOI] [PubMed] [Google Scholar]
- Roubinet C., Decelle B., Chicanne G., Dorn J.F., Payrastre B., Payre F., Carreno S. 2011. Molecular networks linked by Moesin drive remodeling of the cell cortex during mitosis. J. Cell Biol. 195:99–112 10.1083/jcb.201106048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Théry M., Bornens M. 2008. Get round and stiff for mitosis. HFSP J. 2:65–71 10.2976/1.2895661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K.L., Sokac A.M., Berg J.S., Cheney R.E., Bement W.M. 2004. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature. 431:325–329 10.1038/nature02834 [DOI] [PubMed] [Google Scholar]
- Zhang D., Grode K.D., Stewman S.F., Diaz-Valencia J.D., Liebling E., Rath U., Riera T., Currie J.D., Buster D.W., Asenjo A.B., et al. 2011. Drosophila katanin is a microtubule depolymerase that regulates cortical-microtubule plus-end interactions and cell migration. Nat. Cell Biol. 13:361–370 10.1038/ncb2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.