Abstract
Approaches to evaluate T-cell responses to Epstein–Barr virus (EBV) include enzyme-linked immunospot (ELISPOT), which quantifies cells capable of immediate interferon-γ secretion upon antigen stimulation. However, evaluation of expandable EBV-specific memory T cells in an ELISPOT format has not been described previously. We quantified EBV-specific T-cell precursors with high proliferative capacity by using a peptide-based cultured interferon-γ ELISPOT assay. Standard and cultured ELISPOT responses to overlapping peptide pools (15-mers overlapping by 11 amino acids) covering the lytic (BZLF1 and BMRF1) and latent (EBNA1, EBNA3a, EBNA3b, EBNA3c, LMP1 and LMP2) EBV proteins were evaluated in 20 healthy subjects with remote EBV infection and, for comparison, in four solid organ transplant recipients. Cultured ELISPOT responses to both lytic and latent EBV antigens were significantly higher than standard ELISPOT responses. The distribution of EBV-specific T-cell responses detected in healthy virus carriers showed more consistent cultured ELISPOT responses compared with standard ELISPOT responses. T-cell responses quantified by cultured ELISPOT were mainly mediated by CD4+ T cells and a marked pattern of immunodominance to latent-phase antigens (EBNA1 > EBNA3 family antigens > LMP2 > LMP1) was shown. Both the magnitude and distribution of EBV-specific T-cell responses were altered in solid organ transplant recipients; in particular, cultured ELISPOT responses were almost undetectable in a lung-transplanted patient with EBV-associated diseases. Analysis of T-cell responses to EBV by ELISPOT assays might provide new insights into the pathogenesis of EBV-related diseases and serve as new tools in the monitoring of EBV infection in immunocompromised patients.
Keywords: ELISPOT, Epstein–Barr virus, immunocompetent subjects, memory T cells, solid organ transplant recipients
Introduction
Epstein–Barr virus (EBV) is a gamma-herpes virus that infects > 90% of the population worldwide. Primary EBV infection usually occurs during childhood and is generally asymptomatic; however, acute infectious mononucleosis may manifest in some individuals.1 After primary infection, the virus persists life-long within the B-cell compartment. Replication and persistence of EBV are maintained through a balance between productive (lytic) and non-productive (latent) infections. In healthy individuals, EBV-specific T-cell responses play a key role in controlling viral replication and latency establishment during primary infection, and through the life-long carrier state, preventing EBV-associated diseases.2,3 However, EBV has been implicated in the development of malignancies such as Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's lymphoma and post-transplant lymphoproliferative disorders (PTLD).4
Epstein–Barr virus-specific T-cell responses have been mostly determined by MHC-peptide tetramer staining,5 intracellular cytokine staining6,7 and the enzyme-linked immunospot (ELISPOT) assay.8,9 These assays have relied mainly on the use of autologous EBV-transformed B-lymphoblastoid cell lines (B-LCL) as a source of antigen for stimulation of lymphocytes. The B-LCL express not only the latent proteins of EBV but also the lytic proteins, owing to the fact that a small proportion of cells (< 5%) in the B-LCL enter into the lytic phase of infection.10 Additionally, B-LCL stimulation can also activate both CD8+ and CD4+ lytic antigen-specific T cells.11,12 However, attempts to generate autologous B-LCL are not always successful. Although the tetramer approach allows direct quantification of antigen-specific T cells, both the viral peptide and its restricting MHC molecule must be known in advance, limiting the use of this approach to the clinical setting. The ELISPOT and intracellular cytokine staining assays are attractive alternatives because they use an entire protein-spanning mixture of overlapping peptides, presented by a variety of HLA alleles, which is efficient for in vitro stimulation of T lymphocytes. The interferon-γ (IFN-γ) ELISPOT assay is widely used to quantify human antigen-specific immune responses. This assay quantifies T cells capable of immediate secretion of IFN-γ upon antigen stimulation. These cells are thought to represent mainly effector memory T cells.13 On the other hand, the cultured ELISPOT assay quantifies expandable memory T cells,14 probably representing central memory T cells.15 The assay is performed by culturing lymphocytes with specific antigens for 10 days allowing T cells to expand in response to the antigen. Then, the standard ELISPOT procedure is applied in response to the corresponding antigens used for the 10-day stimulation period. So far, the dynamics of EBV-specific T-cell responses simultaneously analysed by standard and cultured ELISPOT assays have not been described.
In the present study, we evaluated virus-specific T-cell responses to overlapping peptide pools of both lytic (BZLF1 and BMRF1) and latent [EBV nuclear antigen 1 (EBNA1), EBNA3a, EBNA3b, EBNA3c, latent membrane protein 1 (LMP1) and LMP2] EBV proteins by standard ELISPOT in healthy virus carriers. At the same time, EBV-specific T-cell responses were evaluated by the cultured ELISPOT assay. Subsequently, we examined memory T-cell responses against EBV in immunocompromised transplant patients.
Materials and methods
Immunocompetent healthy subjects and immunocompromised patients
Following approval by the local ethics committee and after obtaining written informed consent, 23 healthy laboratory personnel (17 women and six men) from the Fondazione IRCCS Policlinico San Matteo, Pavia, Italy, volunteered to give blood samples. The mean subject age at sample collection was 37·1 (standard deviation, SD: 8·1) years. Twenty subjects were EBV-seropositive and three were EBV-seronegative. The EBV-specific serological profile indicated that the 20 EBV-positive subjects were individuals with remote EBV infection (VCA IgG and EBNA IgG positive and VCA IgM negative). Four additional EBV-seropositive individuals, with remote EBV infection, were recruited from among the blood bank donors at the Fondazione IRCCS Policlinico San Matteo, Pavia, Italy, providing a sufficient number of cells for depletion studies as well as reproducibility studies. Blood samples from four solid organ transplant recipients were analysed for comparison. Three patients (P01 and P02, males aged 68 and 42 years, respectively; P03, female aged 54 years) underwent heart transplantation, and one patient (P04, male aged 69 years) underwent lung transplantation. The four patients were seropositive for remote EBV infection before transplantation; three (P01–03) had not experienced EBV-related diseases in the post-transplant period, whereas the fourth (P04) experienced a PTLD and an EBV-related lymphoma at 4 and 6 months after transplantation, respectively. At the time of blood collection, patient P01 was receiving cyclosporine (175 mg/day), everolimus (1·25 mg/day) and steroid (15 mg/day), patient P02 was receiving cyclosporine (200 mg/day) and steroid (5 mg/day), patient P03 was receiving mycophenolate mofetil (1500 mg/day), tacrolimus (7 mg/day) and steroid (10 mg/day), and patient P04 was receiving prednisone (25 mg/day) without other immunosuppressive drugs because of lymphopenia.
Isolation of peripheral blood mononuclear cells
Peripheral blood was collected into tubes containing heparin (BD Vacutainer, Plymouth, UK) or into a blood collection bag. Whole blood was used for determination of T-cell subsets by flow cytometry. Peripheral blood mononuclear cells (PBMC) were isolated by standard density gradient centrifugation using Lymphoprep (Axis-Shield, Oslo, Norway). Isolated PBMC were cryopreserved in freezing medium [10% DMSO (Sigma-Aldrich, St Louis, MO), 25% human albumin (Grifolds Biologicals, Los Angeles, CA) and 65% RPMI-1640 supplemented with 2 mm l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin (all from Euroclone, Milan, Italy)] and kept in liquid nitrogen until ELISPOT analyses.
Determination of T-cell subsets
Fresh whole blood was stained with anti-CD3-PC5 (phycoerythrin-Cy5), anti-CD45-FITC, anti-CD4-RD1 (phycoerythrin) and anti-CD8-ECD (phycoerythrin-Texas Red-X) monoclonal antibodies (all from Beckman Coulter, Milan, Italy). After lysis of red cells, CD4 and CD8 T-cell subsets were analysed by flow cytometry (Navios, Beckman Coulter) using Flow-Count Fluorospheres (Beckman Coulter).
Synthetic peptides
Peptide pools spanning full-length EBV lytic proteins, BZLF1 (59 peptides) and BMRF1 (99 peptides), as well as full-length EBV latent proteins EBNA1 (158 peptides), EBNA3a (234 peptides), EBNA3b (279 peptides), EBNA3c (265 peptides), LMP1 (94 peptides) and LMP2 (122 peptides) were purchased from JPT Peptide Technologies, Berlin, Germany. Peptides were 15 amino acids in length with an 11-amino-acid overlap. Each lyophilized peptide pool was dissolved in DMSO (Sigma-Aldrich) and diluted with RPMI-1640 medium supplemented with 2 mm l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin to generate stock solutions. Aliquots were stored at −20° until use. A final concentration of 0·25 µg/ml for each individual peptide in the corresponding pool was used in all experiments. Additionally, an EBV peptide pool containing 15 peptides, 8 to 10 amino acids in length (Anaspec, Fremont, CA), each corresponding to a defined HLA class I-restricted T-cell epitope from EBV that are part of the CEF control peptide pool,16 was used at a final concentration of 2 µg/ml for each individual peptide.
ELISPOT assays
The PBMC were thawed, washed and resuspended in culture medium [RPMI-1640 medium supplemented with 2 mm l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin and 10% heat-inactivated fetal calf serum (Euroclone)]. Cells were kept overnight at 37° in a humidified 5% CO2 atmosphere. Cells were then used in the standard ELISPOT or were transferred to a 48-well flat-bottom plate (5 × 105 cells/ml per well), stimulated with the corresponding EBV peptide pool (one pool per well) or culture medium only or phytohaemagglutinin (PHA; 5 μg/ml; Sigma-Aldrich), and cultured at 37° in a humidified 5% CO2 atmosphere for 10 days. On days 3 and 7, half of the supernatant from each well was removed and replaced with fresh culture medium supplemented with 20 IU/ml recombinant human interleukin-2 (Peprotech, London, UK). After 10 days, cells from each well were harvested, washed three times with culture medium and resuspended at a concentration of 4 × 105 cells/ml before their use in ELISPOT assays. Human IFN-γ ELISPOT kits were purchased from Diaclone (Cedex, France). MultiScreen-IP 96-well plates (Merck Millipore, Darmstadt, Germany) were coated with IFN-γ capture antibody and incubated overnight at 4°. Plates were then washed five times with PBS (Euroclone) and blocked with culture medium for 2 hr at room temperature. For the standard ELISPOT, PBMC (1 × 105 cells/well) were stimulated (in duplicate) with the corresponding EBV peptide pool, or PHA (5 µg/ml), or culture medium only. For the cultured ELISPOT, cells stimuated with antigen for 10 days were added in duplicate (4 × 104 cells/well) and re-stimulated with the corresponding antigen used for stimulation during the 10-day period or staphylococcal enterotoxin B (SEB; 2 µg/ml; Sigma-Aldrich, for cells stimulated with PHA during the 10-day period). After an incubation at 37° in a humidified 5% CO2 atmosphere for 24 hr, plates were washed five times with PBS supplemented with 0·05% Tween-20 (PBST; Sigma-Aldrich). Biotinylated detection antibody for IFN-γ was added and incubated overnight at 4°. After five washes with PBST, strepatavidin–alkaline phosphatase conjugate was added and plates were incubated at 37° in 5% CO2 atmosphere for 1 hr. Plates were then washed, 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium was added and incubated at room temperature for 20 min. Wells were then washed several times under running water and air-dried. Spots were counted using an automated ELISPOT reader system (A-EL-Vis, Hannover, Germany).
Depletion of CD8+ or CD4+ cells from PBMC
Before starting the 10-day culture period, assays were also performed after depleting CD8+ or CD4+ cells from PBMC by using CD8 MicroBeads or CD4 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) specific for human cells following the manufacturer's instructions. Magnetic separations were performed using MS columns (Miltenyi Biotec). The depleted fractions contained < 5% of target cells, as determined by flow cytometry.
Data analysis
The mean number of spots from duplicate wells was adjusted to 1 × 106 PBMC. Results are presented as net spots per million PBMC calculated as follows: mean number of spots per million PBMC in wells from each EBV peptide pool minus the mean number of spots per million PBMC in wells with culture medium only. Results from the cultured ELISPOT were adjusted per proliferation index (number of antigen-stimulated cells after 10 days of culture divided by the number of medium-stimulated cells after 10 days of culture). For analysis of intra-assay cultured ELISPOT variability, the mean and SD were calculated for each individual set of duplicate wells and each coefficient of variation (%CV) was determined (SD divided by the mean and converted to a percentage). The inter-assay cultured ELISPOT variability was calculated as the %CV of the mean from duplicate wells from each assay performed on different days. Statistical analyses were performed using GraphPad Prism (version 5; GraphPad Software Inc., La Jolla, CA). The Wilcoxon signed rank test was used to compare medians and the Spearman's test was used for correlation analysis. All tests were two-tailed. A P-value < 0·05 was considered statistically significant.
Results
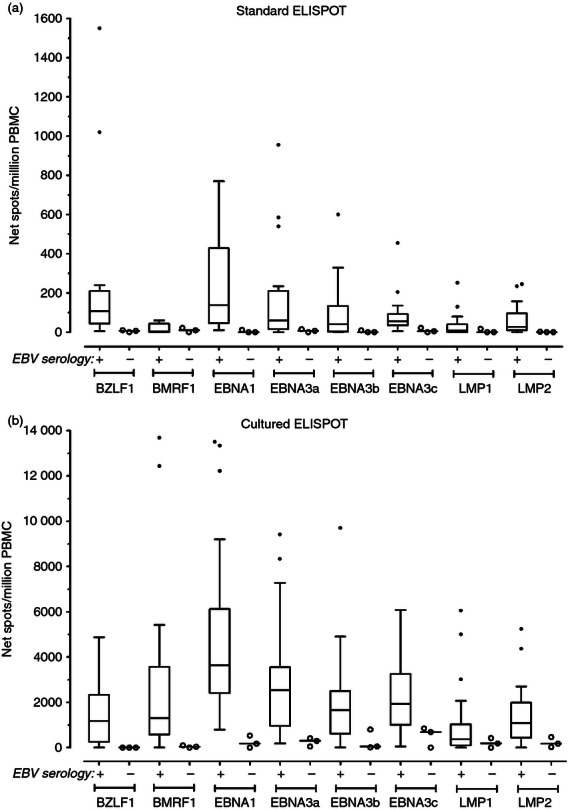
EBV-specific T-cell responses in healthy subjects as measured by standard ELISPOT
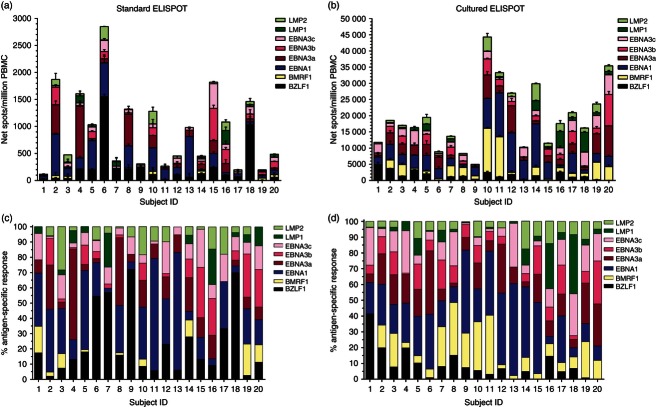
The 23 healthy subjects evaluated in the study had a mean CD4 T-cell count of 984·7 (SD 357) cells/µl and a mean CD8 T-cell count of 557·3 (SD 232·3) cells/µl. We evaluated, by standard ELISPOT assay, antigen-specific IFN-γ production in PBMC from the 23 healthy subjects in response to overlapping peptide pools spanning full-length EBV lytic BZLF1 and BMRF1 proteins as well as full-length EBV latent EBNA1, EBNA3a, EBNA3b, EBNA3c, LMP1 and LMP2 proteins. EBV-specific T-cell responses are presented in Fig. 1(a). In EBV-seropositive subjects (n = 20), the median number of net spots/million PBMC in response to lytic BZLF1 and BMRF1 proteins was 107·5 [interquartile range (IQR) 43·8–210] and 3·5 (IQR 0–43·8), respectively, whereas in response to latent EBNA1, EBNA3a, EBNA3b, EBNA3c, LMP1 and LMP2 proteins it was 137·5 (IQR 45·5–428·8), 60 (IQR 16·3–210·5), 40 (IQR 2·5–133·8), 55 (IQR 35–92·5), 7·5 (IQR 0–40) and 27·5 (IQR 10–96·3), respectively. In three EBV-seronegative healthy subjects, the overall median EBV-specific T-cell response detected by standard ELISPOT was 1 (IQR 0–10) net spots/million PBMC, consistent with non-existent EBV immunity and demonstrating assay specificity. For all 23 subjects, incubation of PBMC with culture medium only resulted in a mean number of spots per well of 3·4 (SD 2·2), whereas the mean number of spots per well in response to PHA was 695·3 (SD 158·6) (not shown).
Figure 1.
Epstein–Barr virus (EBV)-specific T-cell responses determined by standard and cultured ELISPOT assays in healthy subjects. Peripheral blood mononuclear cells (PBMC) from EBV-seropositive (n = 20) and EBV-seronegative (n = 3) healthy subjects were evaluated in response to peptide pools (15 amino acids in length with an 11-amino-acid overlap) representing the full-length lytic (BZLF1 and BMRF1) and latent (EBNA1, EBNA3a, EBNA3b, EBNA3c, LMP1 and LMP2) EBV proteins. Results are shown as net spots/million PBMC for standard (a) and cultured (b) ELISPOT responses. EBV-specific T-cell responses determined by the cultured ELISPOT assay are significantly higher than those detected by the standard ELISPOT assay (P ≤ 0·002, two-tailed Wilcoxon signed rank test). Box and whisker plots (Tukey) indicate median (middle line in the box) EBV-specific T-cell responses detected in EBV-seropositive individuals. Data from each EBV-seronegative subject were plotted individually and the horizontal line marks the respective median number.
EBV-specific T-cell responses in healthy subjects as measured by cultured ELISPOT
We simultaneously evaluated EBV-specific memory T-cell responses in PBMC from the 23 healthy subjects by cultured ELISPOT assay (Fig. 1b). In EBV-seropositive healthy individuals, EBV-specific cultured T-cell responses were significantly higher than T-cell responses quantified by standard ELISPOT (P ≤ 0·002, two-tailed Wilcoxon signed rank test). The median numbers of net spots/million PBMC obtained by cultured ELISPOT in response to BZLF1 (1182, IQR 250·5–2331) and BMRF1 (1301, IQR 582·3–3572) lytic proteins were 11-fold and 372-fold higher than those detected by standard ELISPOT. The median EBNA1-specific T-cell-cultured ELISPOT response was the highest (3632, IQR 2409–6118 net spots/million PBMC) and it was 26-fold higher than that detected by the standard assay. The median cultured ELISPOT responses (net spots/million PBMC) to EBNA3a (2539, IQR 964–3555), EBNA3b (1654, IQR 617–2498), EBNA3c (1928, IQR 999–3258), LMP1 (367, IQR 105·5–1033) and LMP2 (1084, IQR 444–1989) were 42-fold, 41-fold, 36-fold, 49-fold and 39-fold higher than those detected by standard ELISPOT. In three EBV-seronegative healthy subjects, the overall median EBV-specific T-cell response detected by cultured ELISPOT was 75 (IQR 0–422) net spots/million PBMC, which was 21-fold lower than that detected in EBV-seropositive healthy individuals. For all 23 subjects, incubation of PBMC with culture medium only resulted in a mean number of spots per well of 8·5 (SD 16·4), whereas a mean number of 167·1 (SD 83·6) spots per well in response to SEB stimulation was detected (not shown).
We further compared the antigen-specific T-cell responses measured by standard and cultured ELISPOT assays. No correlation was found between the two ELISPOT assays in response to EBV antigens (P > 0·05, Spearman's test, not shown), supporting the concept that different antigen-specific T-cell populations are being measured.
Reproducibility of the cultured ELISPOT assay
Experiments were conducted to assess quantitative reproducibility of the cultured ELISPOT in PBMC from three EBV-seropositive healthy donors in response to EBV antigens. Three aliquots of the same PBMC sample were examined on three different days by a single operator. As shown in Fig. 2(a), the mean intra-assay CV, describing the variation among duplicate wells on the same plate from the same sample for each stimulation condition, ranged from 3·6% to 12·3%. The mean inter-assay CV, describing the variation among three assays performed on different days, ranged from 8·8% to 49·3% (Fig. 2b).
Figure 2.
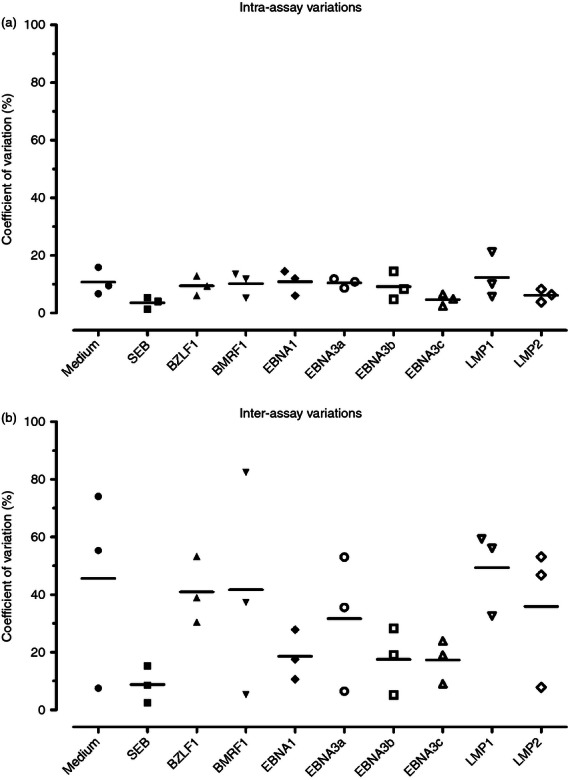
Reproducibility of the cultured ELISPOT assay. Intra-assay (a) and inter-assay (b) variations were evaluated in peripheral blood mononuclear cells (PBMC) from three Epstein–Barr virus (EBV)-seropositive healthy donors to peptide pools (15 amino acids in length with an 11-amino-acid overlap) representing the full-length lytic (BZLF1 and BMRF1) and latent (EBNA1, EBNA3a, EBNA3b, EBNA3c, LMP1 and LMP2) EBV proteins as well as medium alone and staphylococcal enterotoxin B (SEB). For the intra-assay variability, each symbol represents the coefficient of variation (expressed as a percentage, %CV) calculated from duplicate ELISPOT wells of each sample to each stimulation condition and the horizontal line represents the respective mean %CV. For the inter-assay variability, each symbol represents the %CV of the mean from duplicate wells from each assay performed in three different days and the horizontal line represents the respective mean %CV.
Magnitude and distribution of antigen-specific T-cell responses in EBV-seropositive healthy subjects
Figure 3 separates standard and cultured EBV-specific T-cell responses in the 20 EBV-seropositive healthy subjects and shows that responses are different not only in magnitude but also in distribution. Taking into account the cumulative magnitude of the EBV-specific response for each individual, the median EBV-specific standard ELISPOT response was 732·5 (IQR 318·8–1425) net spots/million PBMC (Fig. 3a), whereas the median cultured ELISPOT response was 17 290 (IQR 11 561–26 103) net spots/million PBMC (Fig. 3b). When analysing the specific response to each antigen as a percentage of the total EBV-specific response, we observed that the distribution of antigen-specific T-cell responses to lytic and latent EBV antigens detected by standard and cultured ELISPOT assays was distinct (Fig. 3c,d). Antigen-specific cultured ELISPOT responses were more consistently detected compared with standard ELISPOT responses. Most of the subjects (15/20, 75%) had a detectable cultured ELISPOT response to all eight EBV proteins, whereas only 30% (6/20) had a detectable standard ELISPOT response to the eight EBV antigens. Differences in T-cell response to lytic antigens were clearly evident, the median percentages of standard ELISPOT responses to BZLF1 and BMRF1 lytic proteins were 14·6 (IQR 7·7–31·9) and 0·3 (IQR 0–8·3), respectively, whereas the median percentages of cultured ELISPOT responses were 6·8 (IQR 1·6–13·5) and 8·4 (IQR 3·8–22·7), respectively. The analysis considering the proportion of T-cell responses to latent antigens further showed that cultured ELISPOT responses were less variable than standard ELISPOT responses. While the median percentages of standard ELISPOT responses to EBNA1, EBNA3a, EBNA3b, EBNA3c, LMP1 and LMP2 were 26·3 (IQR 7·4–34·6), 9·1 (IQR 3·9–17·1), 3·8 (IQR 0·2–16·4), 9·1 (IQR 5·2–15·7), 1·6 (IQR 0–5·5) and 5·4 (IQR 1–9·8), respectively, the median percentages of cultured ELISPOT responses were 25 (IQR 12·7–43·7), 15·2 (IQR 6·7–19·1), 8·6 (IQR 5·3–15·7), 11·6 (IQR 5·3–20·5), 2·6 (IQR 0·7–7) and 5·3 (IQR 3·7–9·8), respectively.
Figure 3.
Magnitude and distribution of Epstein–Barr virus (EBV)-specific T-cell responses in healthy subjects. The magnitude of standard (a) and cultured (b) ELISPOT responses in peripheral blood mononuclear cells (PBMC) from each EBV-seropositive healthy subject to peptide pools (15 amino acids in length with an 11-amino-acid overlap) representing the full-length lytic (BZLF1 and BMRF1) and latent (EBNA1, EBNA3a, EBNA3b, EBNA3c, LMP1 and LMP2) EBV proteins are shown. Results are shown as net spots/million PBMC. Error bars represent the SD of the mean. The specific response to each EBV antigen as a percentage of the total summed EBV-specific standard (c) and cultured (d) ELISPOT responses are shown.
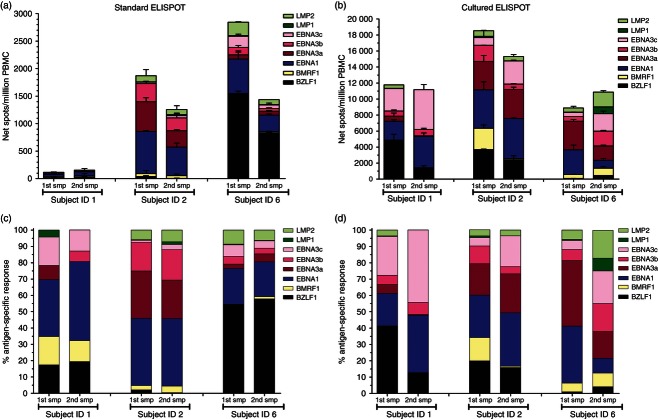
EBV-specific T-cell responses in a subsequent PBMC sample
To determine whether the magnitude and distribution of EBV-specific T-cell responses detected by standard and cultured ELISPOT assays were stable over time, PBMC from three EBV-seropositive healthy subjects were subsequently analysed 4–5 months apart. A wide divergence in the cumulative magnitude of the EBV-specific standard ELISPOT response between subjects was observed and it also displayed a certain degree of variability within the same individual (Fig. 4a); however, the cumulative magnitude of the EBV-specific cultured ELISPOT response was quite comparable between subjects and remained relatively stable in the same individual within this period of time (Fig. 4b). A distinct distribution of antigen-specific T-cell responses to lytic and latent EBV antigens detected by standard (Fig. 4c) and cultured ELISPOT (Fig. 4d) assays was also observed, with the cultured ELISPOT responses being more stable over time.
Figure 4.
Analysis of Epstein–Barr virus (EBV)-specific T-cell responses by ELISPOT assays in a subsequent peripheral blood mononuclear cell (PBMC) sample. Standard (a) and cultured (b) ELISPOT responses to peptide pools (15 amino acids in length with an 11-amino-acid overlap) representing the full-length lytic (BZLF1 and BMRF1) and latent (EBNA1, EBNA3a, EBNA3b, EBNA3c, LMP1 and LMP2) EBV proteins were analysed in a subsequent PBMC sample (4–5 months apart) obtained from three EBV-seropositive healthy subjects. Results are shown as net spots/million PBMC. Error bars represent the SD of the mean. The specific response to each EBV antigen as a percentage of the total summed EBV-specific standard (c) and cultured (d) ELISPOT responses are shown. smp, sample.
EBV-specific cultured ELISPOT responses following T-cell subset depletion
Studies have documented the predominance and immunodominance of EBV-specific CD8+ T-cell responses versus CD4+ T-cell responses in healthy virus carriers.3 Therefore, cultured ELISPOT assays were performed using PBMC, CD8-depleted PBMC and CD4-depleted PBMC from four healthy donors in response to overlapping peptide pools (15 amino acids in length with an 11-amino-acid overlap) representing the full-length lytic (BZLF1 and BMRF1) and latent (EBNA1, EBNA3a, EBNA3b, EBNA3c, LMP1 and LMP2) EBV proteins. The results presented in Fig. 5 consistently show that CD8+ T-cell depletion had no effect on the cultured ELISPOT responses compared with results from PBMC. In contrast, CD4+ T-cell depletion decreased, or completely reduced, EBV-specific cultured ELISPOT responses, indicating that CD4+ T cells are the source of these responses. To show that the healthy EBV carriers had representative EBV-specific CD8+ T-cell responses, cultured ELISPOT assays were also performed in response to an EBV peptide pool (15 peptides, 8–10 amino acids in length) representing defined EBV-derived HLA class I-restricted T-cell epitopes. As also shown in Fig. 5, the median number of net spots/million PBMC in response to EBV peptide pool (class I) was 3938 (IQR 2941–5797) and was similar to that obtained in CD4-depleted PBMC (median 3754, IQR 2325–4446, net spots/million PBMC), whereas CD8+ T-cell depletion almost completely abolished the cultured ELISPOT response, indicating that CD8+ T cells are the source of these responses.
Figure 5.
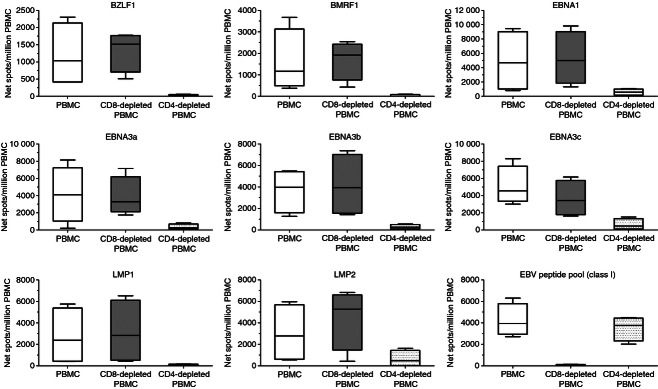
Epstein–Barr virus (EBV)-specific cultured ELISPOT responses following T-cell subset depletion. Peripheral blood mononuclear cells (PBMC), CD8-depleted PBMC and CD4-depleted PBMC from four EBV-seropositive healthy subjects were evaluated by cultured ELISPOT in response to peptide pools (15 amino acids in length with an 11-amino-acid overlap) representing the full-length lytic (BZLF1 and BMRF1) and latent (EBNA1, EBNA3a, EBNA3b, EBNA3c, LMP1 and LMP2) EBV proteins as well as to an EBV peptide pool (15 peptides, 8–10 amino acids in length) representing defined EBV-derived HLA class I-restricted T-cell epitopes. Results are shown as net spots/million PBMC. Box and whisker plots (Tukey) indicate median (middle line in the box) EBV-specific T-cell responses.
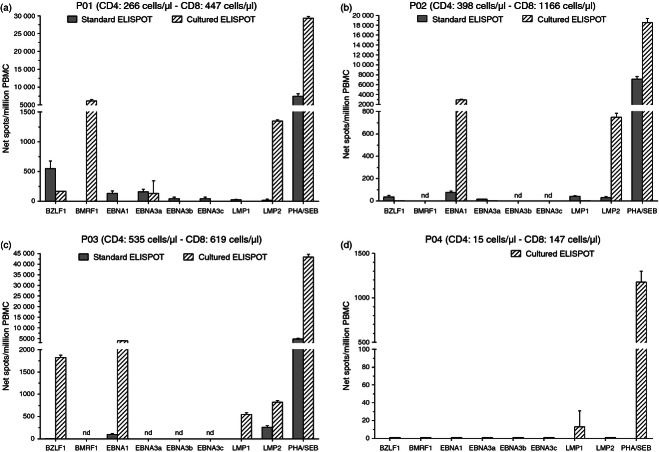
EBV-specific T-cell responses in solid organ transplant recipients
Epstein–Barr virus-specific T-cell responses were characterized by standard and cultured ELISPOT assays in four solid organ transplant recipients. Due to limited PBMC availability from each patient, evaluation of T-cell responses could not be assessed against all eight EBV antigens evaluated in healthy subjects and for one patient (P04) T-cell responses were only assessed by cultured ELISPOT. Responses to non-specific stimulation (PHA or SEB) in three of the four solid organ transplant recipients (P01–P03) were comparable to those obtained in healthy subjects (Fig. 6a–c), whereas the magnitude of response to SEB was low in P04 (who had a low number of both T-cell subsets) (Fig. 6d). EBV-specific standard and cultured ELISPOT responses in three patients differed in both magnitude and distribution. In general, the magnitude of EBV-specific T-cell responses detected by standard and cultured ELISPOT assays in three heart transplant recipients without EBV-associated diseases were comparable to the corresponding median response in EBV-seropositive healthy subjects (Fig. 6a–c), only a few EBV-specific immune responses were of a lower magnitude compared with healthy carriers. In contrast, almost no EBV-specific T-cell responses were detected in the patient with EBV-associated diseases (P04) (Fig. 6d).
Figure 6.
Epstein–Barr virus (EBV)-specific T-cell responses determined by standard and cultured ELISPOT assays in solid organ transplant recipients. Peripheral blood mononuclear cells (PBMC) from three heart transplant recipients (a–c) and a lung transplant recipient (d) were evaluated by standard and/or cultured ELISPOT assays in response to peptide pools (15 amino acids in length with an 11-amino-acid overlap) representing the full-length lytic (BZLF1 and BMRF1) and latent (EBNA1, EBNA3a, EBNA3b, EBNA3c, LMP1 and LMP2) EBV proteins. Results are shown as net spots/million PBMC. Error bars represent the SD of the mean. Blood was obtained at month 6·1 (patient P01), 9·7 (patient P02), 6·7 (patient P03) and 62·8 (patient P04), respectively, after transplantation. T-cell subsets indicated are at the time of ELISPOT analyses. nd, not done.
Discussion
In the present study, we simultaneously analysed EBV-specific T-cell responses to sets of peptides that were 15 amino acids in length with 11-amino-acid overlap by standard and cultured ELISPOT assays. In EBV-seropositive healthy subjects (i) EBV-specific cultured ELISPOT responses were significantly greater than standard ELISPOT responses; (ii) the distribution of antigen-specific T-cell responses to lytic and latent EBV proteins showed that antigen-specific cultured ELISPOT responses were more consistently detectable compared with standard ELISPOT responses; (iii) T-cell responses to EBV antigens detected by cultured ELISPOT were mainly mediated by CD4+ T cells. Furthermore, in general, the magnitude of EBV-specific T-cell responses detected by standard and cultured ELISPOT assays in three heart transplant recipients without EBV-associated diseases were comparable to the corresponding median response in healthy subjects. In contrast, EBV-specific cultured ELISPOT responses were almost undetectable in a lung transplant patient with PTLD.
Although EBV-specific CD8+ T-cell responses have been studied in detail and their importance has been clearly demonstrated,3 the corresponding EBV-specific CD4+ T-cell response is still poorly defined, although antigen-specific CD4+ T-cell help is necessary to generate an effective memory CD8+ T-cell response17 and to maintain CD8+ T-cell function during chronic infection.18 In this study, we used overlapping peptide pools representing both lytic and latent EBV cycle proteins for in vitro stimulation of T lymphocytes, allowing the evaluation of T-cell responses independent of the subject's HLA and EBV antigenic peptides. Peptides were 15 amino acids in length with an 11-amino-acid overlap. Sets of such peptides have been shown to represent a good compromise for stimulating both CD8+ and CD4+ T-cell responses in a number of applications.19,20 However, our data indicate that the stimulation conditions used in our assay efficiently expand CD4+ rather than CD8+ T-cell responses. Additionally, peptides that are 8–10 amino acids in length specifically stimulated EBV-specific CD8+ T-cell responses. It is important to note that our study does not argue against the predominance of EBV-specific CD8+ over CD4+ T-cell responses, nor does it make a comparison of EBV-specific CD4+ versus CD8+ T-cell responses. Our experiments were aimed at determining the frequency and distribution of EBV-specific T-cell responses simultaneously analysed by standard and cultured ELISPOT assays. Based on our findings, the cultured ELISPOT presents important advantages because of the possibility of expanding EBV-specific CD4+ T-cell responses as few studies have been able to detect EBV-specific memory CD4+ T cells capable of both expansion and IFN-γ production upon antigenic challenge.21–23
Studies evaluating the relationship between memory T-cell phenotype and the ability to secrete IFN-γ in response to short-term peptide stimulation, as measured by intracellular cytokine staining, have shown that this cytokine can be detected in CD45RO+ and CD45RA+ CD8+ T-cell subsets24 as well as CD4+ T-cell subsets25 and, when analysed for CCR7 expression, IFN-γ production was mainly detected in CCR7− cells rather than CCR7+ cells, indicating that IFN-γ is mainly produced by the effector memory (CD45RO+ CCR7−) rather than central memory (CD45RO+ CCR7+) T-cell subset.13 However, recent studies indicate that depletion of CCR7+ or CD62L+ cells reduced CD4 and CD8 T-cell responses detected by standard ELISPOT but the cultured ELISPOT responses were reduced more dramatically, indicating the predominant role of central memory T cells in the cultured ELISPOT response and suggesting a role for this T-cell subset in the standard ELISPOT response.15 More recent studies further indicate that expandable antigen-specific T cells, as measured by cultured ELISPOT, predominantly display a central memory phenotype.26 In the present study, the role of memory T-cell phenotypes in the ELISPOT assays was not analysed. However, our results suggest that the cultured ELISPOT not only increases the frequency of the antigen-specific T-cell responses but did not correlate with the standard ELISPOT responses, indicating that different IFN-γ producing cell populations are being quantified.
Antigen-specific T-cell responses measured by cultured cellular assays, but not by standard ELISPOT, have been shown to correlate with slow disease progression in HIV-1-infected individuals naive to antiretroviral therapy,14 protection against malaria,27 suppression of viral rebound in chronic hepatitis B carriers,28 and a favourable outcome in tuberculosis.29 Longitudinal studies are warranted to evaluate whether EBV-specific memory T-cell responses measured by the cultured ELISPOT assay could represent a relevant approach in the clinical setting.
Among the lytic proteins, BZLF1 (immediate early) and BMRF1 (early) were chosen because they are reported to be the most frequently recognized by CD8+ T cells in healthy EBV carriers.3 We detected a modest BZLF1-specific standard ELISPOT T-cell response in all EBV-seropositive healthy subjects, whereas the response to BMRF1 was low (if any). A recent study indicates that BZLF1 is the most frequently recognized by CD4+ T cells from healthy virus-immune donors compared with BMRF1, as determined by standard IFN-γ ELISPOT assay using 15-mer peptides overlapping by 11 amino acids.30 In our study, BZLF1- and BMRF1-specific cultured T-cell responses were significantly higher than T-cell responses detected by standard ELISPOT and CD4+ T cells mainly mediated cultured ELISPOT responses to both lytic antigens. Piriou et al.21 combined in vitro expanded (12 days) specific T cells with flow cytometric analysis of intracellular IFN-γ production and reported that the frequency of BZLF1-specific CD4+ T-cell responses in healthy EBV carriers was higher than that measured directly after short-term stimulation (18 hr) with a BZLF1 peptide pool (15-mers with 11-amino-acid overlap). Remarkably, we found that nearly all EBV-seropositive healthy subjects mounted a significant BMRF1-specific cultured ELISPOT response. These observations, together with the results presented here, indicate that T-cell responses to lytic EBV antigens persist in healthy virus carriers and may have a role in controlling viral reactivation in the carrier state.
Although no EBV-specific T-cell responses were detected by standard ELISPOT in PBMC from EBV-seronegative healthy subjects, consistent with non-existent EBV immunity and confirming the assay specificity, a modest response to EBNA3b and EBNA3c peptide pools was detected after stimulation in vitro for 10 days. It is possible that cross-reactivity to self or non-EBV antigens occurred. In future studies, T-cell responses will be mapped using EBV sub-pools and single peptides in an attempt to identify the trigger of this cross-reactivity. An alternative explanation could be that our in vitro stimulation protocol has led to in vitro priming of naive T cells. However, we consider that it is unlikely to see significant de novo priming in EBV naive healthy controls because the assay is of short duration (10 days), with no addition of peptide antigens other than the initial set-up, no addition of other exogenous cytokines apart from IL-2, and no enrichment of dendritic cells, all of which are necessary for in vitro primary responses.31
Antigen-specific memory T-cell responses to latent EBV proteins, as measured by cultured ELISPOT, were significantly higher than those detected by the standard assay. Interestingly, we found that median EBNA1-specific T-cell responses detected by both standard and cultured ELISPOT assays in EBV-seropositive healthy subjects were the highest compared with the other latent EBV antigens evaluated. EBNA1 is present in all three EBV latency programmes.32 The pattern of immunodominance, which was more evident for T-cell responses detected by cultured ELISPOT, can be summarized as EBNA1 > EBNA3 family antigens > LMP2 > LMP1. Early studies indicate that EBNA1 evades T-cell recognition.33,34 However, other studies have shown that EBNA1 is an important target for EBV-specific T-cell responses. In healthy long-term carriers, CD4+ T cells showed a stronger response to EBNA1 than other EBV proteins.35,36 A more recent study demonstrated that it is possible to expand EBNA1-specific T cells, indicating that EBNA1-specific T cells might be included in adoptive immunotherapy, for instance in PTLD.37 We demonstrated that CD4+ T cells mainly mediated the EBV-specific cultured T-cell responses in healthy subjects. Studies have shown that EBV-specific CD4+ T cells could control proliferation and outgrowth of EBV-infected cells in vitro.38,39 Put together, these results suggest an important role of memory CD4+ T cells in EBV-specific immunity.
Epstein–Barr virus is associated with a wide range of malignancies such as lymphomas, sarcomas and nasopharyngeal and gastric carcinomas. It is assumed that impairment of EBV-specific immunity due to immunosuppression may allow the outgrowth of EBV-transformed B cells, giving rise to PTLD, a life-threatening complication after solid organ40 or haematopoietic stem cell transplantation.41 We demonstrated that EBV-specific standard and cultured T-cell responses also differed in solid organ transplant recipients, the magnitude of EBV-specific T-cell responses detected by standard and cultured ELISPOT assays in three heart transplant recipients were generally comparable to the corresponding median response in EBV-seropositive healthy individuals, whereas no EBV-specific memory T-cell responses were detected in a patient with PTLD. Macedo et al.,42 using standard IFN-γ ELISPOT and short HLA-A2-restricted EBV peptides derived from BMLF1, LMP2 and EBNA3a, reported that EBV-specific CD8+ T cells from solid organ transplant recipients showed decreased IFN-γ production when compared with healthy donors. Further longitudinal studies in large patient cohorts are needed to correlate the magnitude and distribution of EBV-specific standard and cultured T-cell responses with the risk of developing EBV-related malignancies in immunocompromised patients, such as solid organ transplant recipients.
In summary, this study represents the first evaluation of EBV-specific T-cell responses quantified simultaneously by standard and cultured ELISPOT assays. Additionally, our results indicate that the cultured ELISPOT assay is reproducible; low variation across replicate wells was found and the mean inter-assay CV was below 50%, which is acceptable for a biological assay.43 Our study demonstrated that both the magnitude and distribution of memory T-cell responses, quantified by a cultured ELISPOT assay, are relatively stable in healthy EBV carriers, suggesting that the assay can be reliably used to measure EBV-specific T-cell responses. Additionally, by using the cultured ELISPOT it will not only be possible to investigate EBV-specific CD4+ T-cell responses in EBV-related disease but also it would be useful to differentiate between T-cell responses to antigens expressed in the different latency programmes associated with the malignancy in question. Therefore, analysis of EBV-specific T-cell responses by ELISPOT assays might provide new insights in the pathogenesis of EBV-associated diseases and new tools in the monitoring of EBV infection in immunocompromised patients.
Acknowledgments
We thank Daniela Sartori for careful preparation of the manuscript and Laurene Kelly for revision of the English. This work was supported by the Ministero della Salute, Fondazione IRCCS Policlinico San Matteo Ricerca Corrente grant no. 80207 to FB.
Disclosures
The authors declare no conflicts of interest.
References
- 1.Vetsika EK, Callan M. Infectious mononucleosis and Epstein–Barr virus. Expert Rev Mol Med. 2004;6:1–16. doi: 10.1017/S1462399404008440. [DOI] [PubMed] [Google Scholar]
- 2.Callan MF. The evolution of antigen-specific CD8+ T cell responses after natural primary infection of humans with Epstein–Barr virus. Viral Immunol. 2003;16:3–16. doi: 10.1089/088282403763635401. [DOI] [PubMed] [Google Scholar]
- 3.Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: lessons from Epstein–Barr virus. Annu Rev Immunol. 2007;25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- 4.Thompson MP, Kurzrock R. Epstein–Barr virus and cancer. Clin Cancer Res. 2004;10:803–21. doi: 10.1158/1078-0432.ccr-0670-3. [DOI] [PubMed] [Google Scholar]
- 5.Sugaya N, Kimura H, Hara S, Hoshino Y, Kojima S, Morishima T, Tsurumi T, Kuzushima K. Quantitative analysis of Epstein–Barr virus (EBV)-specific CD8+ T cells in patients with chronic active EBV infection. J Infect Dis. 2004;190:985–8. doi: 10.1086/423285. [DOI] [PubMed] [Google Scholar]
- 6.Guppy AE, Rawlings E, Madrigal JA, Amlot PL, Barber LD. A quantitative assay for Epstein–Barr virus-specific immunity shows interferon-γ producing CD8+ T cells increase during immunosuppression reduction to treat posttransplant lymphoproliferative disease. Transplantation. 2007;84:1534–9. doi: 10.1097/01.tp.0000290232.65830.e7. [DOI] [PubMed] [Google Scholar]
- 7.Bhaduri-McIntosh S, Rotenberg MJ, Gardner B, Robert M, Miller G. Repertoire and frequency of immune cells reactive to Epstein–Barr virus-derived autologous lymphoblastoid cell lines. Blood. 2008;111:1334–43. doi: 10.1182/blood-2007-07-101907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Lemas VM, Flinn IW, Krone C, Ambinder RF. Application of the ELISPOT assay to the characterization of CD8+ responses to Epstein–Barr virus antigens. Blood. 2000;95:241–8. [PubMed] [Google Scholar]
- 9.Smets F, Latinne D, Bazin H, Reding R, Otte JB, Buts JP, Sokal EM. Ratio between Epstein–Barr viral load and anti-Epstein–Barr virus specific T-cell response as a predictive marker of posttransplant lymphoproliferative disease. Transplantation. 2002;73:1603–10. doi: 10.1097/00007890-200205270-00014. [DOI] [PubMed] [Google Scholar]
- 10.Kieff E, Rickinson AB. Epstein–Barr virus and its replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th edn. Philadelphia,PA: Lippincott Williams and Wilkins; 2007. pp. 2603–54. [Google Scholar]
- 11.McAulay KA, Haque T, Urquhart G, Bellamy C, Guiretti D, Crawford DH. Epitope specificity and clonality of EBV-specific CTLs used to treat posttransplant lymphoproliferative disease. J Immunol. 2009;182:3892–901. doi: 10.4049/jimmunol.0803572. [DOI] [PubMed] [Google Scholar]
- 12.Adhikary D, Behrends U, Boerschmann H, et al. Immunodominance of lytic cycle antigens in Epstein–Barr virus-specific CD4+ T cell preparations for therapy. PLoS ONE. 2007;2:e583. doi: 10.1371/journal.pone.0000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 14.Calarota SA, Foli A, Maserati R, et al. HIV-1-specific T cell precursors with high proliferative capacity correlate with low viremia and high CD4 counts in untreated individuals. J Immunol. 2008;180:5907–15. doi: 10.4049/jimmunol.180.9.5907. [DOI] [PubMed] [Google Scholar]
- 15.Todryk SM, Pathan AA, Keating S, Porter DW, Berthoud T, Thompson F, Klenerman P, Hill AV. The relationship between human effector and memory T cells measured by ex vivo and cultured ELISPOT following recent and distal priming. Immunology. 2009;128:83–91. doi: 10.1111/j.1365-2567.2009.03073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Currier JR, Kuta EG, Turk E, et al. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J Immunol Methods. 2002;260:157–72. doi: 10.1016/s0022-1759(01)00535-x. [DOI] [PubMed] [Google Scholar]
- 17.Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–6. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 18.Cardin RD, Brooks JW, Sarawar SR, Doherty PC. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J Exp Med. 1996;184:863–71. doi: 10.1084/jem.184.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maecker HT, Dunn HS, Suni MA, et al. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J Immunol Methods. 2001;255:27–40. doi: 10.1016/s0022-1759(01)00416-1. [DOI] [PubMed] [Google Scholar]
- 20.Kiecker F, Streitz M, Ay B, Cherepnev G, Volk HD, Volkmer-Engert R, Kern F. Analysis of antigen-specific T-cell responses with synthetic peptides – what kind of peptide for which purpose? Hum Immunol. 2004;65:523–36. doi: 10.1016/j.humimm.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 21.Piriou ER, van Dort K, Nanlohy NM, van Oers MH, Miedema F, van Baarle D. Novel method for detection of virus-specific CD4+ T cells indicates a decreased EBV-specific CD4+ T cell response in untreated HIV-infected subjects. Eur J Immunol. 2005;35:796–805. doi: 10.1002/eji.200425792. [DOI] [PubMed] [Google Scholar]
- 22.Piriou E, van Dort K, Nanlohy NM, van Oers MH, Miedema F, van Baarle D. Loss of EBNA1-specific memory CD4+ and CD8+ T cells in HIV-infected patients progressing to AIDS-related non-Hodgkin lymphoma. Blood. 2005;106:3166–74. doi: 10.1182/blood-2005-01-0432. [DOI] [PubMed] [Google Scholar]
- 23.Heller KN, Upshaw J, Seyoum B, Zebroski H, Münz C. Distinct memory CD4+ T-cell subsets mediate immune recognition of Epstein–Barr virus nuclear antigen 1 in healthy virus carriers. Blood. 2007;109:1138–46. doi: 10.1182/blood-2006-05-023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hislop AD, Gudgeon NH, Callan MF, Fazou C, Hasegawa H, Salmon M, Rickinson AB. EBV-specific CD8+ T cell memory: relationships between epitope specificity, cell phenotype, and immediate effector function. J Immunol. 2001;167:2019–29. doi: 10.4049/jimmunol.167.4.2019. [DOI] [PubMed] [Google Scholar]
- 25.Amyes E, McMichael AJ, Callan MF. Human CD4+ T cells are predominantly distributed among six phenotypically and functionally distinct subsets. J Immunol. 2005;175:5765–73. doi: 10.4049/jimmunol.175.9.5765. [DOI] [PubMed] [Google Scholar]
- 26.Ndhlovu ZM, Proudfoot J, Cesa K, et al. Elite controllers with low to absent effector CD8+ T cell responses maintain highly functional, broadly directed central memory responses. J Virol. 2012;86:6959–69. doi: 10.1128/JVI.00531-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keating SM, Bejon P, Berthoud T, et al. Durable human memory T cells quantifiable by cultured enzyme-linked immunospot assays are induced by heterologous prime boost immunization and correlate with protection against malaria. J Immunol. 2005;175:5675–80. doi: 10.4049/jimmunol.175.9.5675. [DOI] [PubMed] [Google Scholar]
- 28.Yang SH, Lee CG, Park SH, et al. Correlation of antiviral T-cell responses with suppression of viral rebound in chronic hepatitis B carriers: a proof-of-concept study. Gene Ther. 2006;13:1110–7. doi: 10.1038/sj.gt.3302751. [DOI] [PubMed] [Google Scholar]
- 29.Goletti D, Butera O, Bizzoni F, Casetti R, Girardi E, Poccia F. Region of difference 1 antigen-specific CD4+ memory T cells correlate with a favorable outcome of tuberculosis. J Infect Dis. 2006;194:984–92. doi: 10.1086/507427. [DOI] [PubMed] [Google Scholar]
- 30.Long HM, Leese AM, Chagoury OL, Connerty SR, Quarcoopome J, Quinn LL, Shannon-Lowe C, Rickinson AB. Cytotoxic CD4+ T cell responses to EBV contrast with CD8 responses in breadth of lytic cycle antigen choice and in lytic cycle recognition. J Immunol. 2011;187:92–101. doi: 10.4049/jimmunol.1100590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moser JM, Sassano ER, Leistritz DC, Eatrides JM, Phogat S, Koff W, Drake DR., 3rd Optimization of a dendritic cell-based assay for the in vitro priming of naïve human CD4+ T cells. J Immunol Methods. 2010;353:8–19. doi: 10.1016/j.jim.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Thorley-Lawson DA. Epstein–Barr virus: exploiting the immune system. Nat Rev Immunol. 2001;1:75–82. doi: 10.1038/35095584. [DOI] [PubMed] [Google Scholar]
- 33.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen PM, Klein G, Kurilla MG, Masucci MG. Inhibition of antigen processing by the internal repeat region of the Epstein–Barr virus nuclear antigen-1. Nature. 1995;375:685–8. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 34.Yin Y, Manoury B, Fåhraeus R. Self-inhibition of synthesis and antigen presentation by Epstein–Barr virus-encoded EBNA1. Science. 2003;301:1371–4. doi: 10.1126/science.1088902. [DOI] [PubMed] [Google Scholar]
- 35.Münz C, Bickham KL, Subklewe M, et al. Human CD4+ T lymphocytes consistently respond to the latent Epstein–Barr virus nuclear antigen EBNA1. J Exp Med. 2000;191:1649–60. doi: 10.1084/jem.191.10.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leen A, Meij P, Redchenko I, Middeldorp J, Bloemena E, Rickinson A, Blake N. Differential immunogenicity of Epstein–Barr virus latent-cycle proteins for human CD4+ T-helper 1 responses. J Virol. 2001;75:8649–59. doi: 10.1128/JVI.75.18.8649-8659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones K, Nourse JP, Morrison L, Nguyen-Van D, Moss DJ, Burrows SR, Gandhi MK. Expansion of EBNA1-specific effector T cells in posttransplantation lymphoproliferative disorders. Blood. 2010;116:2245–52. doi: 10.1182/blood-2010-03-274076. [DOI] [PubMed] [Google Scholar]
- 38.Omiya R, Buteau C, Kobayashi H, Paya CV, Celis E. Inhibition of EBV-induced lymphoproliferation by CD4+ T cells specific for an MHC class II promiscuous epitope. J Immunol. 2002;169:2172–9. doi: 10.4049/jimmunol.169.4.2172. [DOI] [PubMed] [Google Scholar]
- 39.Nikiforow S, Bottomly K, Miller G, Münz C. Cytolytic CD4+-T-cell clones reactive to EBNA1 inhibit Epstein–Barr virus-induced B-cell proliferation. J Virol. 2003;77:12088–104. doi: 10.1128/JVI.77.22.12088-12104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baldanti F, Rognoni V, Cascina A, Oggionni T, Tinelli C, Meloni F. Post-transplant lymphoproliferative disorders and Epstein–Barr virus DNAemia in a cohort of lung transplant recipients. Virol J. 2011;8:421. doi: 10.1186/1743-422X-8-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baldanti F, Gatti M, Furione M, Paolucci S, Tinelli C, Comoli P, Merli P, Locatelli F. Kinetics of Epstein–Barr virus DNA load in different blood compartments of pediatric recipients of T-cell-depleted HLA-haploidentical stem cell transplantation. J Clin Microbiol. 2008;46:3672–7. doi: 10.1128/JCM.00913-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macedo C, Donnenberg A, Popescu I, et al. EBV-specific memory CD8+ T cell phenotype and function in stable solid organ transplant patients. Transpl Immunol. 2005;14:109–16. doi: 10.1016/j.trim.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Maecker HT, Hassler J, Payne JK, et al. Precision and linearity targets for validation of an IFN-γ ELISPOT, cytokine flow cytometry, and tetramer assay using CMV peptides. BMC Immunol. 2008;9:9. doi: 10.1186/1471-2172-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]