Abstract
CYP2J2 epoxygenase is a membrane bound cytochrome P450 that converts omega-3 and omega-6 fatty acids into physiologically active epoxides. In this work, we present a comprehensive comparison of the effects of N-terminal modifications on the properties of CYP2J2 with respect to the activity of the protein in model lipid bilayers using Nanodiscs. We demonstrate that the complete truncation of the N-terminus changes the association of this protein with the E.coli membrane but does not disrupt incorporation in the lipid bilayers of Nanodiscs. Notably, the introduction of silent mutations at the N-terminus was used to express full length CYP2J2 in E. coli while maintaining wild-type functionality. We further show that lipid bilayers are essential for the productive use of NADPH for ebastine hydroxylation by CYP2J2. Taken together, it was determined that the presence of the N-terminus is not as critical as the presence of a membrane environment for efficient electron transfer from cytochrome P450 reductase to CYP2J2 for ebastine hydroxylation in Nanodiscs. This suggests that adopting the native-like conformation of CYP2J2 and cytochrome P450 reductase in lipid bilayers is essential for effective use of reducing equivalents from NADPH for ebastine hydroxylation.
Keywords: eicosanoids, CYP epoxygenases, CYP2J2, cytochrome P450, cytochrome P450 reductase, ebastine hydroxylation, Nanodiscs, lipid bilayers, protein expression, protein purification, N-terminus modifications
Introduction
Eicosanoids are signaling molecules that mediate inflammation in the body and are implicated in many disorders including cancer, arthritis, and cardiovascular disease.1 One class of eicosanoid-synthesizing cytochrome P450s, known as the CYP epoxygenases, catalyzes the epoxidation and/or hydroxylation of various dietary polyunsaturated fatty acids and xenobiotics.2,3 CYP2J and CYP2C are the two primary mammalian subfamilies that comprise the CYP epoxygenases. In humans, CYP2J2 is highly expressed in cardiac myocytes and the endothelium of coronary arteries where it plays significant roles in cardiovascular diseases and inflammation.4–7 This enzyme catalyzes the metabolism of arachidonic acid (AA) into epoxyeicosatrienoic acids (EETs). These metabolites are stereo- and regio-isomers that exert isomer-specific anti-inflammatory, vasodilatory, and proangiogenic effects.8,9 Additionally, CYP2J2 catalyzes the conversion of the omega-3 fatty acids into physiologically active products.5 Both the omega-3 and omega-6 fatty acid metabolites are known dilators of the microvasculature and are essential for heart homeostatic equilibrium.5,10
Recently, CYP2J2 has also been found to be relevant in both cancer and preeclampsia pathophysiology.11–14 EETs are recognized as major promoters of angiogenesis and consequently enhance tumor cell survival.12,13,15,16 In a study by Jiang et al., CYP2J2 was found to be predominantly localized in tumor cells when compared with the adjacent normal tissue samples.16 Additionally, Zeldin and coworkers recently found that EETs stimulate extensive multiorgan metastasis and escape from tumor dormancy in several cancer models.12 In preeclampsia, CYP2J2 was found to be highly upregulated in placental tissues and administration of CYP2J2 inhibitors reversed preeclampsia-associated symptoms in women.11 Hence, the development of drugs inhibiting CYP2J2 has tremendous pharmaceutical potential for the prevention of cancer metastasis and preeclampsia.11,14,17,18
CYP2J2 also plays an important role in the first-pass metabolism of astemizole, terfenadine, and ebastine and is essential for understanding the pharmacokinetic and pharmacodynamic relationships of these drugs in the body.17,19 Thus, to begin to elucidate the structure–function relationships of pharmaceutically relevant CYP2J2, we expressed the protein with high yields in Escherichia coli and focused on understanding the role of the N-terminus of CYP2J2 in association with lipid bilayers and in ebastine hydroxylation activity.
N-terminus role in association with lipid bilayers, activity, and protein–protein interactions
The N-termini of membrane-bound cytochrome P450s have been shown to play an important role in lipid association, protein–protein interactions, and substrate access. There are many reports of the hydrophobic N-terminal domain mediating the P450 membrane-binding association.20–23 Based on this observation, a soluble and monomeric form of CYP2C5 and CYP2C3 was engineered by deleting the N-terminal membrane-spanning region.24,25 Additionally, the N-terminus of CYP1A2 was shown to be involved in membrane anchoring and N-terminal modifications had an effect on the catalytic activity.26 Interestingly, it was also demonstrated that membrane insertion of CYP1A2 was promoted by the presence of anionic phospholipids and this association with membrane is dependent on the N-terminus of CYP1A2.27 In addition to playing a role in association with membrane, the N-terminus can be involved in P450–P450 interactions.28 Tracy and coworkers demonstrated that the hydrophobic N-terminus-binding domains of CYP2C9 and CYP3A4 are involved in heterodimer complex formation and truncation of the N-terminus of CYP2C9 led to the abolition of inhibition by CYP3A4.29 In separate studies, it was hypothesized that the N-terminal domain might be critical for orientation of the protein into the lipid bilayer.30 Thus, the N-terminus affects the localization on the membrane and can influence orientation and activity. Although the N-terminus is considered to be responsible for membrane association, one cannot always obtain a soluble protein by simply truncating the N-terminus. This is likely due to the presence of additional hydrophobic residues, which contribute to membrane association. The other regions thought to be involved in membrane anchoring are the F′ and G′ helices and F-G loop. Hence, it is strongly believed that membrane lipid molecules may play an important role in gating the entrance of substrates and/or release of products, implying that these pathways are likely partially immersed in the membrane.31,32
Model lipid bilayers of Nanodisc for functional interrogation of CYP2J2
Here, we use Nanodiscs for rigorous biochemical interrogation of CYP2J2 and the role of N-terminus in membrane association and activity. Nanodiscs are nanoscale lipid bilayers that are surrounded by a membrane scaffold protein.33–35 Incorporation of a CYP protein into Nanodiscs mimics the endogenous membrane environment of the protein, producing a macromolecule amenable to several important biochemical and biophysical assays hitherto reserved for soluble proteins. Nanodiscs have proven to be excellent for the solubilization and mechanistic investigation of many diverse membrane protein systems. Moreover, this technology has enabled the stabilization of membrane proteins both in solution36–40 and on surfaces.39,41–44
Focus of the article
In this article, we functionally characterize CYP2J2 in model lipid bilayer Nanodiscs. We demonstrate the role of the N-terminus in association with the E. coli membrane and with the lipid bilayers of Nanodiscs. We further evaluate whether lipid bilayers are essential for the productive use of NADPH and product formation by CYP2J2.
Notably, we introduce silent mutations at the N-terminus to express full-length CYP2J2 in E. coli while maintaining wild-type functionality.45 The full-length CYP2J2 provides a nearly identical wild-type construct for comparison to the N-terminus truncated constructs being explored in this work. Although some of the results corroborate more anecdotal results for other P450s, this study presents a more thorough and comprehensive comparison of the effects of N-terminal modifications on the properties of CYP2J2 in native-like lipid bilayers with respect to membrane association and activity.
Results and Discussion
Construction of CYP2J2 N-deletion mutants
The native human CYP2J2 gene was obtained from OriGene. The Kyte-Doolittle hydropathy plot for CYP2J2 (Supporting Information Fig. S1) shows that approximately the first 33 amino acids form the hydrophobic membrane-spanning domain. From literature, it is known that removal of hydrophobic regions from the N-terminus increases the yields of expression of cytochrome P450s.25,46 Therefore, we prepared five different CYP2J2 constructs with varying N-terminal modifications for the purpose of increasing the yield of expression without eliminating their ability to associate with the lipid bilayers. The different N-terminally modified CYP2J2 constructs are listed in Figure 1(A), and the list of primers used for each respective construct are represented in Supporting Information Table S1. The N-terminal modifications made to the CYP2J2 constructs are the following: (1) Replaced the second codon in the gene sequence with Ala (GCT), as many E. coli genes controlled by the lacZ operator have been found to contain this codon.47,48 (2) incorporated silent mutations in the first five to seven codons to render them both AT rich and to minimize the free energy of mRNA secondary structure.48–51 (3) Truncated part or all of the hydrophobic (membrane spanning) region of the N-terminus as done previously.21 Finally, (4) incorporated hydrophilic sequence AKKTSS into the N-terminus.52,53
Figure 1.
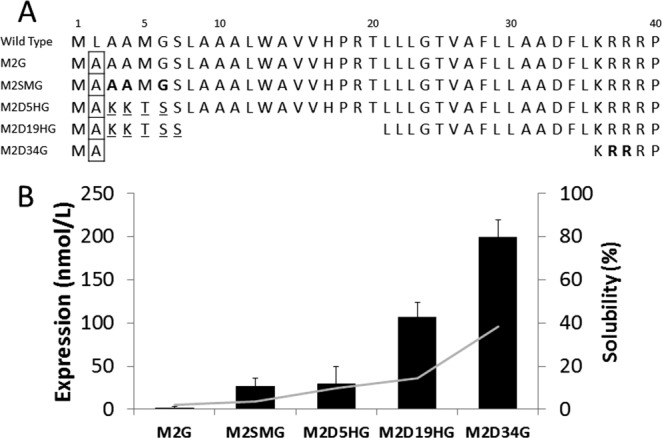
Design and expression of CYP2J2 mutants. (A) CYP2J2 wild-type and design of N-terminal mutant constructs. Boxed residues denote substitution of native Leu with Ala (GCT). Bolded residues represent introduction of AT-rich silent mutations. Underlined residues signify introduction of a hydrophilic motif. Gaps represent deletion of N-terminal amino acids. (B) CYP2J2 N-terminal mutant expression and solubility in DH5alpha cells. The bar graph denotes expression yields that were highly dependent on N-terminal modifications. The line plot signifies percentage of protein found in the soluble fraction during purification, which largely corresponded to N-terminal truncations.
One complication in the molecular cloning of CYP2J2 was the presence of an internal NdeI site in the gene.54 We circumvented this problem by using an easy and elegant method of digesting the CYP2J2 gene with a different enzyme, AseI, which produced an overhang compatible with the NdeI digested fragment and enabled ligation producing the desired constructs.
Heterologous expression of CYP2J2 in E. coli
All the constructs of CYP2J2 were transformed into DH5alpha cells along with the pTGro7 plasmid encoding for the expression of the GroEL-GroES chaperonin system.55 The average yields obtained after Ni-NTA column purification from each of the constructs are shown in Figure 1(B). The overall protein expression yields corresponded to the size of the N-terminal deletion with yields following: M2D34G > M2D19HG > M2D5HG ∼ M2SMG. The protein expression yields of the full-length CYP2J2 significantly varied for constructs M2G and M2SMG with yields of 1–2 and 27 nmol L−1 of purified protein, respectively. Interestingly, both M2G and M2SMG are full-length proteins similar to wild-type CYP2J2 with a single substitution (Leu2Ala) at the second amino acid in the N-terminal domain [Fig. 1(A)]. The difference between these two constructs was the introduction of silent mutations at the third, fourth, and sixth codons of M2SMG that increased expression 20-fold as represented in Figure 1(B). Thus, we were able to greatly enhance the expression of full-length CYP2J2 (M2SMG) with the introduction of silent mutations at the N-terminus, thereby minimizing the free energy of the mRNA secondary structure formation (ΔG of M2G = –3.94 and ΔG of M2SMG = –1.69) as calculated in the experimental procedures section. The expression of full-length CYP2J2 in high yields was essential to fully elucidate the role of the N-terminus of CYP2J2 in membrane binding and association with its redox partner cytochrome P450 reductase.
The complete truncation of the N-terminal hydrophobic region in the M2D34G construct resulted in the protein being located in the soluble fraction at a much higher percentage (∼40%) than the proteins expressed from other constructs [Figure 1(B)]. M2D19HG, which was modified by truncating 19 N-terminal amino acids, was primarily purified from the E. coli membrane fraction (∼85%). This indicated that the additional 14 hydrophobic amino acids of D19HG provided a much stronger membrane anchor for association during purification when compared with M2D34G.
Oligomerization studies
The oligomerization states of the different constructs were estimated by size exclusion chromatography (SEC). In Figure 2(C), we see that the N-terminal hydrophobic regions of M2SMG and M2D5HG induce significantly greater oligomeric states with predominant retention times at 22 min and a much smaller peak at 30 min. The N-terminal truncations of M2D19HG exhibited a different elution profile with two predominant oligomeric states at 22 and 31 min. Most dramatically, the N-terminal deletions of M2D34G produced a large monomeric state at 31 min with a smaller aggregated peak at 19 min. The overall quantity and stokes radius for the larger oligomeric states corresponded closely with the size of the N-terminus, thus following the pattern M2SMG ∼ M2D5HG > M2D19HG > M2D34G.
Figure 2.
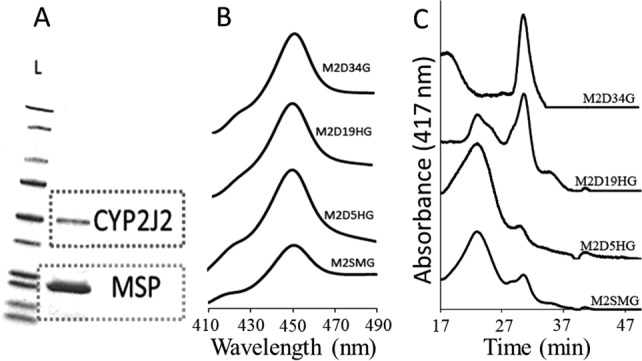
Characterization of CYP2J2 constructs. (A) CYP2J2 mutants were incorporated into Nanodiscs and purified using SEC-HPLC. SDS-PAGE was used to verify successful isolation of each respective CYP2J2-ND. The first column contains a protein ladder standard showing that the molecular weight of M2D19HG is close to the ∼50–60 kDa standard and the scaffolding protein MSP1T2 has molecular weight close to ∼24 kDa. The exact molecular weights were confirmed using ESI-MS. (B) CO-binding spectra were used to characterize each construct and gauge the protein folding after expression and purification. Each construct exhibited ∼100% P450 content, which is indicative of a well-folded and functional CYP. (C) SEC-HPLC was applied to characterize CYP2J2 mutant oligomerization states in phosphate buffer (0.1M). The CYP2J2 elution profile was produced by monitoring the heme absorbance at 417 nm. Using water-soluble protein standards, we observed that the constructs smaller oligomeric state corresponded closely with the size of the N-terminal truncation and followed M2SMG ∼ M2D5HG > M2D19HG > M2D34G.
Spectrophotometric characterization of CYP2J2 N-terminus mutants
The purified oxidized CYP2J2 constructs were analyzed using UV–visible spectra. All the constructs had typical peaks at 417 nm and the α and β bands (Q-bands) at 536 and 570 nm, respectively. The proteins were reduced and the Fe(II) CO spectra were measured for each of the constructs.56 Figure 2(B) shows the Fe(II)-CO spectra of all the different constructs. They all exhibited prominent peak at 450 nm with little or no presence of a peak at 420 nm. This confirmed that the axial-thiolate ligand was retained and suggested maintenance of a typical P450 fold for this motif in the active site.
Spectroscopic substrate-binding assay
Typically for drug-metabolizing and steroid-synthesizing human cytochrome P450s, binding substrate produces a spin state shift accompanied by the shift of Soret from 417 to 393 nm.39,57,58 To probe the active site of a full-length versus truncated CYP2J2, we studied the substrate binding in Nanodiscs. CYP2J2 is primarily expressed in the human heart where its main function is the conversion of AA into EETs by epoxidation. Using this assay, we did not detect an observable spectral spin state change for AA binding. However, we did measure a modest increase in NADPH consumption rates (data not shown) that indicated binding of the substrate. Additionally, Zeldin and coworkers previously determined the in vitro metabolism rate of AA.59 In contrast, the other epoxygenases (CYP2C8 and CYP2C9) directly interact with AA, as evidenced by a 27% spin state change for CYP2C8.60,61 However, this is not unprecedented as there are several examples where substrate binding does not substantially change the spin state for substrates capable of being metabolized. For instance, CYP3A4 metabolizes erythromycin despite a minor spin state shift (5–10%) upon binding.39
CYP2J2 is also involved in the metabolization of certain drugs including ebastine, astemiozole, and terfenadine.20,62 We measured the UV–vis spectra of ebastine binding to CYP2J2-Nanodiscs. As shown in Figure 3, type I substrate-binding spectra are obtained upon titration with ebastine. This is also true for binding of terfenadine to CYP2J2 (data not shown). The spectral dissociation constant of ebastine binding to the two constructs of CYP2J2-Nanodiscs was calculated to be 7.3 ± 1.9 µM for M2SMG and 6.8 ± 0.7 µM for M2D19HG. This reveals that the spectral dissociation constant is likely to be independent of the N-terminus composition of CYP2J2 in Nanodiscs. Unlike other drug-metabolizing P450s, the binding of ebastine fits to a single-substrate binding equation and does not exhibit cooperativity.
Figure 3.
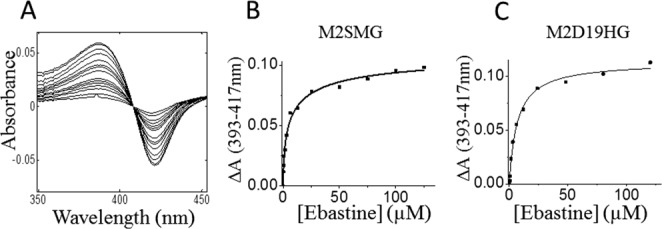
Ebastine-binding spectra were fitted to a single binding isotherm. (A) Difference spectra of D19HGND ebastine titration. Ebastine was added incrementally from 0 to 120 µM. (B, C) The change of absorbance (ΔA) from 393 to 417 nm was calculated for each titration and plotted against the corresponding ebastine concentration (µM). Data were fitted with Origin Lab to the single binding isotherm. A single binding isotherm was applied to calculate the Kd of M2SMG and M2D19HG, which were determined to be 7.3 ± 1.9 and 6.8 ± 0.7 µM, respectively.
Association of CYP2J2 with lipid bilayers
Nanodiscs are native-like lipid bilayer membranes that have been used to stabilize membrane proteins in solution. We assembled the different N-terminal variants of CYP2J2 into Nanodiscs in order to: (a) functionally stabilize the cytochrome P450s, (b) study the stability of the association of different CYP2J2 constructs with the lipid bilayers over time, (c) and assess the role of lipid bilayers in controlling the function of P450s.
The N-terminus of membrane-bound native CYP2J2 is hydrophobic and is thought to be involved in association with the membranes. We analyzed the P450 content of the different N-terminally modified CYP2J2 constructs in both the soluble and membrane fractions of E. coli cells [Figure 1(B)]. We found that M2D34G CYP2J2 (complete N-terminus truncation) was purified from both soluble (40%) and membrane fraction (60%), whereas M2SMG CYP2J2 (full length) was primarily obtained from the membrane fraction (96%) as seen in Figure 1(B). The membrane fraction of M2D34G still contained the majority of protein, indicating that the intrinsic membrane-binding affinity was not completely abolished. These results suggested that the solubility of the protein was increasing owing to the truncation of N-terminal hydrophobic residues similar to other membrane-bound CYPs.24
To evaluate if the N-terminus modifications conferred stable association with lipid bilayers, we measured the stability of the CYP2J2-Nanodisc complex over a period of 7 days at 4°C using SEC. The CYP2J2-Nanodiscs peak eluted at 26 min (flow rate is 0.5 mL min−1) and was measured by monitoring the absorbance at 417 nm. The area under the peak was integrated and used to quantitate percent loss of the CYP2J2-Nanodisc complex. Over 7 days at 4°C there was loss of ∼10–25% of the starting amount of CYP2J2-Nanodiscs [Figure 4(D)]. We did not observe the formation of an empty Nanodisc peak or free CYP2J2 in solution, indicating that the macromolecular complex was likely precipitating upon dissociation and lost in the prefiltration step of this assay.
Figure 4.
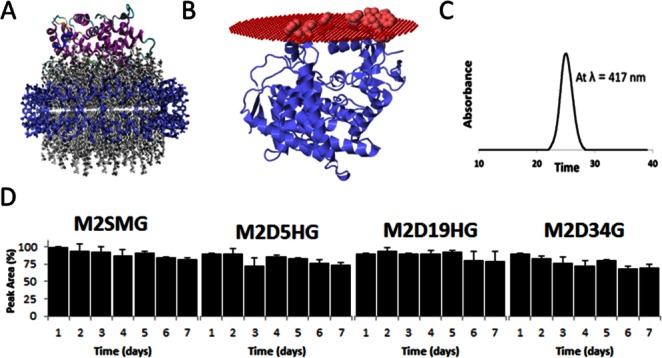
Incorporation of CYP2J2 constructs in Nanodiscs and Nanodisc stability assay. (A) Visual molecular dynamics (VMD) was used to construct a representative model of CYP2J2 incorporated into Nanodiscs (CYP2J2 model was obtained using Phyre). In the model, blue represents the membrane scaffolding protein (MSP1D1), the gray are the phospholipids, and the protein ribbon model is the CYP2J2 enzyme. (B) Membrane association of M2D34HG model. Highlighted residues in the A helical region (Trp48, Arg49, Leu50, Pro51, Phe52, Leu53, Phe57, Leu58, and Phe61), Lys122 in B′ helix and in the FG loop region (Trp235, Phe239, and Leu240) are predicted by OMP software to be associated with the lipid membrane. Predicted Gibbs free energy for this interaction is 13.8 kcal mol−1. (C) CYP2J2 constructs were incorporated into Nanodiscs and monitored at 417 nm SEC-HPLC. The single peak corresponds to homogeneous CYP2J2-Nanodiscs. (D) The relative stability of each CYP2J2 construct incorporated into Nanodiscs was monitored using SEC-HPLC over a 7-day period at 4°C. The percent area of the 417-nm peak corresponding to heme absorption was monitored at the established CYP2J2-Nanodisc elution time (∼26 min). We observed a similar relative stability for each of the constructs with an approximate 10–20% degradation over the time course.
Interestingly, despite the truncation of 34 amino acid residues in the M2D34G construct, we obtained stable Nanodiscs comparable to that of the full-length M2SMG. Surprisingly, M2D19HG demonstrated the greatest stability over the same time course. Thus, the trends observed when purifying proteins from cell lysates do not directly translate to in vitro protein-membrane-associated systems in standard conditions. These observations indicated that additional residues separate from the N-terminus were also responsible for membrane association. To explore this further, we modeled CYP2J2 in membranes using Phyre and OPM software.63,64 We found that similar to other membrane-binding CYPs, the A helix, F-G loop, and F′-G′ helices contain hydrophobic groups that likely aid in the association of this protein with the membrane [Figure 4(B)]. These residues provide interesting targets for additional mutations to further explore the membrane-binding properties of CYP2J2 and engineer a soluble and monomeric CYP2J2 construct.
Additionally, we measured the P450 content by CO binding at the beginning and end of 7 days to gauge CYP2J2 functionality. The Fe(II)-CO P450 formation at the beginning (day 1) and end (day 7) of the time course produced 100% P450 (data not shown), indicating that all the proteins in Nanodiscs retained the typical P450 protein fold with the heme-thiolate motif intact when stored at 4°C.
CYP2J2 activity and role of membranes
We further studied the role of the N-terminus of CYP2J2 in modulating the activity of the protein with respect to the rate of NADPH oxidation and ebastine hydroxylation. The rate of steady-state NADPH consumption was measured for the four constructs in the presence or absence of ebastine in a three different systems—detergent-solubilized system, phospholipid-reconstituted system, and incorporated into Nanodiscs.
Additionally, the enzymatic activities of CYP2J2 constructs were determined in the three systems by measuring the rate of ebastine hydroxylation.65,66 We measured the ebastine hydroxylation activity of the protein to determine whether the protein is using the transferred electrons from NADPH for a productive pathway to generate desired products or using them unproductively in generating unwanted reactive oxygen species.67 We measured the coupling efficiency, which is defined as the ratio of rate of product formation to the rate of NADPH oxidation (Figure 5). These experiments provided insight of the role of N-terminus and that of membranes in controlling the NADPH oxidation and ebastine hydroxylation in CYP2J2-CPR system.
Figure 5.
Schematic of NADPH oxidation and ebastine hydroxylation by a CPR-CYP2J2 Nanodisc complex. Electrons are shuttled from NADPH to CYP2J2 via cytochrome P450 reductase redox centers (step 1). These electrons subsequently drive CYP2J2 P450 catalysis of ebastine to hydroxyebastine (step 2). Coupling efficiency is measured as the ratio of ebastine hydroxylation (step 2) over NADPH oxidation (step 1). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
NADPH oxidation, product formation, and coupling efficiency in CYP2J2:CPR-Nanodiscs
The Nanodisc rates reported here are for a coincorporated 1:1 homogeneous complex of CYP2J2:CPR.58 On average, for the CYP2J2:CPR-Nanodiscs substrate-free form the rate of NADPH consumption was ∼10 nmol min−1 nmol−1 for the four constructs. When a saturating concentration of ebastine was added, a doubling of the NADPH oxidation rate was observed, in the range of 20.3–27.1 nmol min−1 mol−1. Notably, in this defined lipid membrane system, we observed similar NADPH oxidation rates for all the constructs, with M2D19HG:CPR-ND exhibiting a slightly higher consumption rate. Taken together, when the ratio of CYP2J2:CPR was equal and incorporated into membrane bilayers we do not detect a significant difference in the NADPH turnover rates for the constructs.
To gauge functionality, we measured the ebastine hydroxylation activity of this system as given in Table I for M2SMG, M2D5HG, M2D19HG, and M2D34G. Because of the narrow range of product formation it was necessary to calculate the coupling efficiencies to directly compare the four constructs in this system. The coupling efficiencies in Nanodiscs for M2SMG: CPR, M2D5HG:CPR, M2D19HG:CPR, and M2D34G:CPR were determined to be 17.0% ± 0.8%, 17.1% ± 0.7%, 16.5% ± 0.9%, and 13.6% ± 1.3%, respectively. We considered each of these efficiencies to be very similar, with a slight reduction in the efficiency of M2D34G. Overall, these coupling efficiencies were approximately sixfold greater than the detergent-solubilized system and threefold greater than the phospholipid-reconstituted system (Table I). This shows that interaction of CYP2J2 and CPR in a membrane environment plays a more important role in controlling the coupling efficiency when compared with the N-terminus hydrophobic domain of CYP2J2.
Table I.
NADPH oxidation and ebastine hydroxylation rates of CYP2J2 constructs in detergent solubilized and membrane bound systems. Each of the four constructs were incorporated into Nanodiscs, reconstituted with lipids or solubilized in 0.1% CHAPS and used to measure NADPH consumption and hydroxyebastine product formation. The coupling efficiency was calculated by taking the ratio of ebastine hydroxylation measured using LC/MS/MS versus the NADPH Oxidation monitored via UV-Vis spectroscopy at 340 nm
| CYP2J2 Constructs | Substrate-Free NADPH Rate(nmol/min/nmol) | Ebastine NADPH Rate (nmol/min/nmol) | Ebastine Hydroxylation Rate (nmol/min/nmol) | Coupling Efficiency |
|---|---|---|---|---|
| CYP2J2-CPR Nanodiscs | ||||
| M2SMG | 9.2 ± 1.5 | 20.3 ± 0.9 | 3.8 ± 0.7 | 17.0 ± 0.8 |
| M2D5HG | 7.8 ± 0.8 | 21.3 ± 0.4 | 3.7 ± 0.6 | 17.1 ± 0.7 |
| M2D19HG | 12.9 ± 0.7 | 27.1 ± 1.6 | 4.5 ± 0.4 | 16.5 ± 0.9 |
| M2D34G | 12.2 ± 0.3 | 21.2 ± 0.3 | 2.9 ± 0.1 | 13.6 ± 1.3 |
| CYP2J2-POPC Reconstituted | ||||
| M2SMG | 34.0 ± 1.7 | 69 ± 2.0 | 4.3 ± 0.2 | 6.1 ± 0.2 |
| M2D5HG | 31.8 ± 2.8 | 72 ± 6.4 | 4.4 ± 0.3 | 6.1 ± 0.7 |
| M2D19HG | 38.9 ± 3.3 | 88 ± 6.3 | 5.4 ± 0.1 | 6.2 ± 0.4 |
| M2D34G | 65.1 ± 1.3 | 121 ± 8.5 | 3.1 ± 0.7 | 2.5 ± 0.3 |
| CYP2J2-Detegrent | ||||
| M2SMG | 21.5 ± 1.7 | 50.6 ± 3.7 | 1.6 ± 0.2 | 3.2 ± 0.1 |
| M2D5HG | 23.2 ± 1.0 | 44.9 ± 6.1 | 1.4 ± 0.3 | 3.1 ± 0.2 |
| M2D19HG | 27.3 ± 1.5 | 72.2 ± 7.3 | 2.6 ± 0.9 | 3.5 ± 0.9 |
| M2D34G | 28.0 ± 1.4 | 142.5 ± 18.6 | 2.4 ± 0.5 | 1.7 ± 0.1 |
NADPH oxidation, product formation, and coupling efficiency in a phospholipid-reconstituted system
In the absence of substrate the phospholipid-reconstituted system provided a range of NADPH consumption rates (31.8–65.1 nmol min−1 nmol−1) that followed the extent of the N-terminal truncation. Similarly, in the presence of excess substrate a similar trend in NADPH oxidation was observed (69–121 nmol min−1 nmol−1). However, the ebastine hydroxylation rates for all the constructs were similar (Table I). Interestingly, the NADPH oxidation rates and ebastine hydroxylation rates for the M2SMG and M2D5HG constructs with the majority of the N-terminus intact were nearly identical, indicating a similar association with CPR within the reconstituted system. The M2D19HG construct consumed a marginally higher amount of NADPH yet produced the highest amount of product when compared with the other constructs. Strikingly, M2D34G while consuming a relatively high level of NADPH produced the lowest level of product. The coupling efficiencies of this lipid-reconstituted system for M2SMG, M2D5HG, M2D19HG, and M2D34G were calculated to be 6.1% ± 0.2%, 6.1% ± 0.7%, 6.2% ± 0.4% and 2.5% ± 0.3%, respectively. Thus, within this reconstituted system the presence of the full or partially truncated N-terminal domain exhibited similar utilization of NADPH for ebastine hydroxylation. Moreover, it would indicate that despite the different oligomerization states of the different N-terminus constructs, the three constructs' orientation with the exogenous CPR leads to the formation of a productive complex. However, the fully truncated M2D34G construct exhibited a more than two-fold decrease in efficiency, suggesting that the formation of a productive complex between the two proteins is significantly reduced in the absence of the N-terminus. The extent of the analysis is hindered without a comprehensive understanding of the complexes formed within the 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)-reconstituted system. However, sufficient evidence is present to suggest that in the absence of a uniform membrane bilayer environment, the N-terminus is important for productive interaction with CPR.
NADPH consumption, product formation, and coupling efficiency in a CYP2J2-detergent-solubilized system
For detergent-solubilized CYP2J2, there was a dramatic increase of NADPH consumption when ebastine was added and like the phospholipid-reconstituted system the rates ranged from 44.9 nmol min−1 nmol−1 of protein for M2D5HG to 142.5 nmol min−1 nmol−1 for the M2D34G mutant. Interestingly, the differences in the NADPH consumption rates in this system followed the order of the N-terminal truncations: M2D34G > M2D19HG > M2SMG ∼ M2D5HG.
The functional activity of the constructs was measured in the detergent-solubilized system (Table I). We found that the rate of product formation correlated with the rate of NADPH consumption observed. This observation was reinforced upon calculation of the coupling efficiencies for the different constructs. Notably, the coupling efficiencies containing all or part of the N-terminus used NADPH with the same efficiency, whereas the fully truncated construct exhibited ∼50% reduction.
NADPH consumption using AA as a substrate
We also determined that the rate of NADPH oxidation in the presence of excess AA increases marginally (∼1.5-fold) above the substrate-free system (data not shown). Overall, the NADPH oxidation rate with AA and ebastine bound to CYP2J2 was slower when compared with other CYPs bound to substrate.68 In CYP3A4, the presence of a spin state changing substrate can significantly affect NADPH oxidation rates; for instance, the rate in the absence of the substrate is 25 nmol min−1 nmol−1, whereas in the presence of the substrate and a spin state change it can reach 272 nmol min−1 nmol−1. Thus, the modest 1.5-fold increase in NADPH consumption during AA metabolism by CYP2J2 in detergents and Nanodiscs affirms the previous observation that AA does not produce a significant spin state change. Previously, Isin and Guengerich have shown a strong correlation between the steady-state NADPH oxidation rate and the first electron transfer, which can be related to the degree of spin state change.69,70
Comparison of ebastine hydroxylation rate of CYP2J2 isolated from different systems
We compared the ebastine hydroxylation rates of the E. coli-expressed protein to previous recombinant human CYP2J2 expressed in mammalian, yeast, and insect cells. The values obtained had a range of ebastine hydroxylation rates in the different microsomal preparations.19,62,66 Previous studies using mammalian COS-1 and insect baculovirus cells yielded rates (Vmax) of 5.5 and 8.1 nmol min−1 nmol−1, respectively.19,66 The ebastine hydroxylation rates using yeast microsomes exhibited a relatively high Vmax of 40.1 nmol min−1 nmol−1 possibly owing to different post-translational modifications in these systems.62 In this work, the E. coli-expressed CYP2J2 constructs in Nanodiscs exhibited similar catalytic rates to what was reported for microsomal preparations of CYP2J2 from insect and mammalian expression systems. Specifically, the turnover numbers of CYP2J2-CPR-Nanodisc systems were the most similar when compared with the aforementioned systems.
Within the Nanodisc system we were able to control the oligomerization state of CYP2J2:CPR and incorporate these proteins in a 1:1 ratio in a defined lipid membrane environment. This model system enabled us to deconstruct the complex interactions of the N-terminally modified CYPs, which revealed in the membrane lipid bilayer of Nanodiscs, the NADPH consumption rates and coupling ratio were quite similar for the different constructs. This suggests that when the different N-terminal constructs of CYP2J2 are incorporated into lipid bilayers they adopt orientations and conformations that enable similar interactions with CPR. Alternatively, in the detergent- and phospholipid-reconstituted system, the truncation of the N-terminus leads to the increased NADPH oxidation rates and reduced coupling efficiency. It is difficult to delineate the exact factors affecting these rates in detergent- and phospholipid-reconstituted systems in the absence of detailed knowledge of the macromolecular protein complex in these systems.
In Nanodiscs, the dual incorporation of CPR and CYP favors a more productive alignment of the two proteins' redox centers via electrostatic interactions as found in other P450 systems.71–73 Additionally, it is possible that the complementary charge interactions of the incorporated CPR with CYP2J2 could be prompting a more productive orientation in membrane, especially for fully N-terminus truncated M2D34G. Our data show strong evidence that the presence of membrane environment is more critical than the presence of N-terminus for efficient electron transfer from NADPH to CYP2J2 orchestrated by CPR. This suggests the adoption of the native-like conformation in lipid bilayers is essential for effective use of reducing equivalents from NADPH.
Conclusions
In this article, we significantly enhanced the expression of full-length CYP2J2 in E. coli with the introduction of silent mutations at the N-terminus of the protein. The expression of full-length CYP2J2 in high yields was essential to elucidate the role of the N-terminus of CYP2J2 in substrate binding, electron transfer, and association with its redox partner cytochrome P450 reductase. We demonstrated that the use of hydrophilic motifs and truncations of the N-terminal domain dramatically increases CYP2J2 expression in E. coli. All of the constructs expressed exhibited close to 100% P450 content, indicating the maintenance of a typical P450 protein fold. Binding assays revealed that the Kd of binding of ebastine to M2D20HG and M2SMG was similar, thus showing that the N-terminus does not affect the binding of this substrate to the protein.
We further studied the association of CYP2J2 with lipid bilayers. During purification we observed that 40% of the fully truncated M2D34G construct was found in the soluble fraction. However, when the four constructs were incorporated into Nanodiscs, we observed sufficient incorporation and stability over a week at 4°C. Therefore, we concluded that the association of CYP2J2 with lipid bilayers is mediated by other residues besides the N-terminus.
The NADPH consumption and ebastine hydroxylation functionality of each construct was investigated in a detergent-solubilized system, phospholipid-reconstituted system, and CYP2J2-CPR-Nanodiscs. The coupling efficiencies were calculated to compare enzyme productivity, and it was found that the CYP2J2-CPR Nanodisc system was approximately sixfold more efficient than the detergent-solubilized system and threefold more efficient than the phospholipid-reconstituted system.
We concluded that the N-terminus does not interfere with effective coupling of NADPH oxidation with ebastine hydroxylation when the protein is in the membrane environment. However, in a detergent-solubilized system or phospholipid-reconstituted system, an intact N-terminus is required for effective coupling. This suggests that in Nanodiscs, the protein is in a more native-like membrane environment. Additionally, the proteins are monomeric and are effectively interacting with their redox partner CPR. Thus, the membrane environment likely facilitates the proper docking of CPR to CYP2J2.
This study incorporates novel findings with respect to comparing the effects of N-terminal modifications on the properties of CYP2J2 and its ability to interact with its redox partner CPR in the lipid bilayers of Nanodiscs.
Experimental Procedures
Materials
Human CYP2J2 cDNA was obtained from OriGene (Catalog No. SC321730). PCR reagents and other enzymes for molecular biology were purchased from Invitrogen. Qiagen kits were used to purify the plasmid DNA from bacterial cells and from agarose gels. The bacterial strains used were E. coli XL1 Blue obtained from Agilent and DH5α from Invitrogen. Ampicillin, arabinose, chloramphenicol, isopropyl β-d-1-thiogalactopyranoside (IPTG), and Ni-NTA resin were bought from Gold Biotechnology. δ-Aminolevulinic acid was bought from Frontier Scientific. NADPH and NADP were obtained from P212121.com. 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS) was acquired from BioVision. POPC was purchased from Avanti Polar Lipids. Amberlite XAD 2 was acquired from Supelco. Nanosep MF (0.2 µM) and Amicon Ultra (10,000 MWCO) centrifugal filters were procured from Millipore. AA and ebastine were obtained from Cayman Chemical. The protein standards—thyroglobulin, ferritin, bovine serum albumin, and cytochrome c, were purchased from Sigma. All other materials and reagents used were purchased from Sigma and Fisher Scientific.
Design of CYP2J2 constructs and cloning into pCWori+ vector
CYP2J2 cDNA obtained from OriGene was cloned into the pCWori vector. All the constructs were grown in the presence of chaperonin system, hence “G” is used for all constructs. Construct 1-M2G: The amino acid sequence of this construct is similar to the wild-type amino acid sequence except for the substitution of the second amino acid residue leucine (CTC) by alanine (GCT). Construct 2-M2SMG: The sequence of this construct is similar to M2G with the introduction of silent mutations in the third, fourth, and sixth codons to make them AT rich and thereby minimizing the free energy of the mRNA secondary structure formation. The ΔG of first seven codons at N terminus of M2SMG is −1.69, ΔG of WT is −4.27, and ΔG of construct M2G is −3.94. Values were obtained using RNAFold WebServer.49,50 Note that M2G and M2SMG share identical primary sequences but differ in N-terminus codon usage. Construct 3-M2D5HG: This construct was designed not only to increase the yields of expression of human CYP2J2 but also for maintaining the association of this protein with lipid bilayers. Therefore, in this construct, we have replaced the first five amino acids from the N-terminus with a hydrophilic sequence AKKTSS and maintained most of the original hydrophobic sequence.46 Construct 4-M2D19HG: To optimize and enhance the expression while preserving some part of the hydrophobic region to interact with membranes we deleted codons, 2 to 19, from the N-terminus and inserted the hydrophilic sequence AKKTSS. Insertion of a hydrophilic sequence in combination with truncation of a major part of the native hydrophobic sequence has been shown to give higher protein expression. Construct 5-M2D34G: It was engineered with the goal of enhancing yield and making the protein soluble and suitable for crystallization and structural determination by X-ray diffraction studies.74,75 It has been shown previously that deletion of the first 34 amino acids with a hydrophilic region naturally following this deletion increases the yields of CYP2J2 protein in E. coli.76 In all the constructs, we added a penta-His tag to the C-terminus to facilitate purification using metal chelating affinity columns.
The different constructs of the CYP2J2 gene were prepared and amplified by PCR using the appropriate sets of primers listed in Supporting Information Table S1. The PCR conditions are also mentioned. CYP2J2 gene has an internal NdeI site. Hence, an AseI site was introduced within the forward primer. AseI site overhangs are compatible with the overhang produced from digesting NdeI site in the vector. Therefore, all of the constructs had an AseI restriction site in the forward primer along with the various N-terminal modifications. The reverse primer incorporated a penta-His tag and an XbaI restriction site at the C-terminus. The amplified CYP2J2 genes were digested with AseI-XbaI and then ligated with the NdeI-XbaI-digested pCWori vector and cotransformed with chaperonin plasmid pTGro7 into DH5α-competent cells. pTGro7 encodes for the GroEL-GroES gene and is selected via chloramphenicol resistance. Transformed colonies were confirmed for the presence of the respective constructs by DNA sequencing.
Expression and purification of CYP2J2
Double transformant colonies containing the CYP2J2 gene and pTGro7 gene were cultured in 30 mL of Luria Bertani (LB) media with chloramphenicol (20 µg mL−1) and ampicillin (100 µg mL−1) at 37°C and 250 rpm overnight. Five milliliters of this culture was used to inoculate 500 mL of Terrific Broth (TB) media supplemented with trace elements, ampicillin (100 µg mL−1) and chloramphenicol (20 µg mL−1). The culture was grown for 2.5 h at 37°C and 220 rpm. Delta-aminolevulinic acid (500 µL of 0.5 mM) was then added and the culture was grown for another 2 h at 26°C and 160 rpm. After reaching a target absorption (A600 = 1.2), 1 mM of IPTG was added to induce protein expression. In addition, 2 g of arabinose was added to each 500 mL culture.77 The cultures were grown for 44 h and then the cells were harvested by centrifugation. The cell pellet was resuspended in buffer 1 (100 mM potassium phosphate pH 7.4, 20% glycerol, 6 mM magnesium chloride, 0.1 mM dithiothreitol (DTT), and 0.2 mM phenylmethanesulfonylfluoride (PMSF), ∼1 mg each of DNase and RNase). The cells were sonicated on ice for six cycles of 1 min with breaks of 1 min at an output of 4, 40% duty cycle with a Heat Systems Ultrasonics (Qsonica, Newtown, CT) sonicator. This was followed by centrifugation at 35,000 rpm at 4°C for 45 min using a Beckman ultracentrifuge (Beckman Coulter, Brea, CA). The supernatant was collected to quantitate protein in the soluble fraction. The membrane fraction pellet was resuspended in a solubilization buffer 2 (100 mM potassium phosphate pH 7.4, 20% glycerol, 200 mM sodium chloride, 0.1 mM DTT, and 1% CHAPS) for 3–5 h at 4°C. This was further centrifuged at 4°C and 35,000 rpm for 45 min to obtain the supernatant, which contained the target membrane protein. The supernatant was then loaded onto a Ni-NTA column equilibrated with the column buffer 3 (100 mM potassium phosphate pH 7.4, 20% glycerol, and 0.5% CHAPS). The column was washed with five column volumes of the wash buffer (100 mM potassium phosphate pH 7.4, 20% glycerol, 0.5% CHAPS, and 20 mM imidazole). It was further washed with four column volumes of ATP-containing buffer 4 (100 mM potassium phosphate pH 7.4, 20% glycerol, 0.5% CHAPS, 5 mM ATP, 10 mM magnesium chloride, and 150 mM potassium chloride) to remove the GroEL that may be bound to the CYP proteins.78 Finally, the protein was eluted with four column volumes of the elution buffer (100 mM potassium phosphate pH 7.4, 20% glycerol, 0.5% CHAPS, and 200 mM imidazole). The eluted protein was concentrated and buffer exchanged to remove imidazole using Amicon centrifugal filters (10,000 MWCO) and stored at −80°C.
Characterization of CYP2J2
CYP2J2's purity was analyzed using 10% SDS-PAGE where it separated as a single band with an Rz ratio (A417 nm/A280 nm) of 1.3 or higher obtained by measuring the UV–visible spectra [Figure 2(A)]. CYP2J2 concentration was measured by carbon monoxy difference spectra as described previously by Omura and Sato using an extinction coefficient of 91 mM−1 cm−1.56 The protein content from both cytosol and membrane fractions was quantified. For CO analysis we modified the Omura and Sato's method by purging a 500-µL aliquot of the buffer-exchanged sample with nitrogen for 20 min and then adding 10–20 µL of dithionite solution anaerobically. This was then used for CO addition and monitored using a Cary Bio 300 UV–Vis spectrophotometer (Agilent Technologies, Santa Clara, CA). The difference spectra were recorded to quantify the protein as well as to assess the quality of the protein.
Oligomerization states of detergent-solubilized CYP2J2 using SEC
SEC was performed using a Superdex 200 10/300 column (GE Life Sciences, Piscataway, NJ) coupled with a high-performance liquid chromatography (HPLC) system consisting of an Alliance 2695 analytical separation module (Waters, Milford, MA) and a Waters 996 photodiode diode array detector (Waters). The different CYP2J2 constructs were loaded individually at 15 µM on the SEC column and were run with an isocratic mobile phase consisting of 100 mM of potassium phosphate buffer (pH 7.4) containing 150 mM sodium chloride and 0.1 mM EDTA at 0.5 mL min−1 flow rate. The oligomerization states of the CYP2J2 constructs were estimated using a standard curve plotted using thyroglobulin (669 kDa), ferritin (440 kDa), BSA (67 kDa), and cytochrome c (14 kDa) as standards.
Expression and purification of cytochrome p450 reductase
A 10 mL volume of starter culture (grown from a single colony for 18–20 h at 37°C and 220 rpm) was used to inoculate 1 L of LB media supplemented with ampicillin and riboflavin.40 The culture was grown at 37°C and 220 rpm for 4 h (O.D. was 0.8–1). Expression of the reductase was induced by adding IPTG (1 mL of 1 mM) and growing the culture at 30–33°C and 220 rpm for 18–20 h. Cells were harvested by centrifuging at 4000 rpm for 15 min in a JA-10 rotor (Beckman Coulter) at 4°C. The pellet was resuspended in 500 mL of cold lysozyme buffer (75 mM Tris pH 8.0, 0.25 M sucrose, 0.25 mM EDTA, and 0.02 mg mL−1 lysozyme) and allowed to stir at 4°C for 30 min. The spheroplasts were obtained by centrifuging at 4000 rpm for 30 min in the JA-10 rotor at 4°C and then resuspended in 40 mL of lysis buffer (50 mM Tris pH 8.0 and 1 mM PMSF) at 4°C for 30 min. Sonication was carried out on ice with five cycles of 30 s with 1-min breaks. Membranes were isolated by centrifuging at 30,000 rpm for 30 min at 4°C in a Ti-45 rotor. This membrane pellet was then solubilized in 40 mL of column buffer (50 mM Tris pH 7.7, 0.1 mM EDTA, 0.1 mM DTT, and 20% glycerol) containing 0.2% Triton for an hour at 4°C. Subsequent centrifugation at 30,000 rpm for 30 min at 4°C in a Ti-45 rotor (Beckman Coulter) gave the protein in the supernatant fraction, which was then loaded onto a 2′5′ADP Agarose column equilibrated with the column buffer. The column was then washed with 10 column volumes (c.v.) of the wash buffer (100 mL of column buffer containing 2 mM adenosine) and eluted with 5 c.v. of the elution buffer (50 mL of column buffer containing 2 mM NADP or 2 mM AMP). To remove the detergent and obtain high purity of protein, the eluate was passed through a DEAE column equilibrated with the wash buffer, washed with 10 column volumes of the wash buffer, and eluted with 5 column volumes of the elution buffer. The final eluate was concentrated and the CPR concentration was measured with a Cary Bio 300 UV–Vis spectrophotometer (Agilent Technologies) at 456 nm. Purified CPR was characterized by measuring the cytochrome c reduction for 10 min at 550 nm in a solution of 1 mL of 0.3M potassium phosphate buffer pH 7.4 containing 50 µM cytochrome c, 50 µM NADPH, and 2 µM of the purified CPR. The purified reductase was stored at −80°C.
Assembly of CYP2J2-Nanodiscs
The lipid, POPC was solubilized using 100 mM cholate and added to the membrane scaffold protein MSP1D1 (65:1 ratio) and allowed to mix at 4°C for 1 h. To assemble the Nanodiscs with the protein, CYP2J2 in 100 mM potassium phosphate pH 7.4, 20% glycerol, and 0.5% CHAPS was added to the mixture at a CYP:MSP ratio of 1:15 and allowed to mix for an additional hour. Amberlite beads were then added to the final mixture to remove the detergent overnight at 4°C. The mixture was separated from the beads by filtration. The resulting solution was concentrated, loaded onto a SEC column, and the protein-incorporated Nanodiscs peaks were collected and concentrated. The concentrated CYP2J2-ND complex was stored at −80°C in the presence of 10% glycerol.
Assembly of CYP2J2-CPR Nanodisc complex for activity assay
The assembly of the CYP2J2 and CPR into a single macromolecular complex was achieved following previously described methods.58 Briefly, CYP2J2 and CPR were incubated in a 1:1.5 ratio in 0.1% (v/v) cholate for 1 h at 4°C before being added to the detergent-solubilized MSP1E3D1- and POPC mixture described above. The final molar ratio of the mixture contained a CYP2J2/CPR/MSP1E3D1(−)/POPC ratio of 0.1:0.15:1:130. Amberlite beads were then added to the final mixture to remove the detergent overnight at 4°C. For the isolation of Nanodiscs containing both CPR and CYP2J2 the filtered solution was passed through an ADP agarose column to selectively bind CPR and eluted with 2.5 mM NADP phosphate buffer (7.4). The eluent was then passed through a Ni-NTA column to selectively bind the His-tagged CYP2J2 protein and eluted phosphate buffer (7.4) containing 200 mM imidazole. The eluent was concentrated and then loaded on a SEC column where the relatively large Nanodisc containing both CPR and CYP was confirmed by the elution time at ∼21.5 min. The final reconstitution molar ratio of all components was 0.1:0.2:240:120:1 CYP2J2/CPR/cholate/POPC/MSP1E3D1(−). The 1:1 ratio of CYP2J2 and CPR was confirmed with a UV–Vis scan using the respective absorbance of CYP2J2 at 417 nm (ε = 110 mM−1 cm−1) and CPR at 456 nm (ε = 24 mM−1 cm−1).
Preparation of CYP2J2 and CPR in a POPC-reconstituted system
The in vitro activity of CYP2J2 and the redox partner CPR was assessed in a standard phospholipid-reconstituted system following previously described methodology.79 The lipids were prepared by reconstituting POPC in 100 mM phosphate buffer (pH 7.4) containing 100 mM sodium cholate to a final lipid stock concentration of 10 mM. The solution was then dispersed by sonication until clarity was achieved before mixing with the proteins. The detergent-solubilized POPC stock was then added at a final concentration of 50 µM into a cuvette containing 0.6 µM CPR and 0.2 µM CYP2J2 and incubated at 37°C for 5 min before use in the NADPH oxidation and ebastine hydroxylation assay.
Stability of CYP2J2-Nanodiscs
The effect of N-terminal modifications on association of CYP2J2 with lipid bilayers of Nanodiscs was investigated by monitoring the stability of CYP2J2-Nanodiscs at 4°C using SEC-HPLC. Samples containing CYP2J2 N-terminal mutants incorporated into Nanodiscs were prepared with 50 mM POPC, 290 nmol of MSP1T2(−) (MSP1T2 His-tag was cleaved using TEV protease),80 and 29 nmol of CYP2J2 as described above. Ni-NTA affinity column was used to remove empty Nanodiscs from CYP2J2-Nanodiscs mixture because the CYP2J2 constructs contained a His-tag but the MSP1T2(−) did not. Samples were aliquoted and stored in 100 mM phosphate buffer (pH 7.4) at 4°C and analyzed over a period of 7 days with an isocratic gradient consisting of a 100 mM phosphate buffer (pH 7.4) mobile phase over a 60-min period. The CYP2J2-ND assembly was monitored at 417 and 280 nm. Overall stability of M2SMG-ND, M2D5HG-ND, M2D19HG-ND, and M2D34G-ND was calculated from the integrated areas of each time point relative to the initial injection for each respective construct. The day 0 and day 7 samples were also evaluated for stability using a CO binding assay as described above.
Binding titration of CYP2J2-Nanodiscs with ebastine
The binding of ebastine to CYP2J2 constructs was calculated by monitoring the spin state shift using a Cary Bio 300 UV–Vis spectrophotometer (Agilent Technologies). Two constructs, M2SMG and M2D19HG, were incorporated into Nanodiscs and purified using the methods listed above. Samples contained 2–5 µM of CYP2J2-ND, 100 mM phosphate buffer (pH 7.4), and ebastine. Ebastine was added incrementally from a 10 mM ethanol stock with the final concentration of ethanol not exceeding 1%. The absorption spectra were measured between 800 and 200 nm after each ebastine titration. Spectra were plotted and analyzed in Origin Lab (Origin Lab, Northampton, MA). For each titration the characteristic type I spectral shift difference, from a low spin to a high spin state, was calculated (by measuring difference A386 − A417) and plotted against the ebastine concentration. The data were then fitted using the Hill equation [Eq. (1)]. Additionally, the percent of high spin was calculated using a standard MATLAB subroutine.
| (1) |
where ΔA is the absorbance difference at 386 and 417 nm, Amax is the amplitude corresponding to maximal spin shift, Ks is the spectral dissociation constant, S is the substrate concentration, and n is the Hill coefficient. The ebastine-binding data fit n = 1 better than n = 2; therefore, for final fitting a single binding isotherm was used. The calculated spectral binding constants are averages of three independent experiments.
NADPH activity assays
CYP2J2 NADPH oxidation rates were examined native-like in membrane-bound systems (Nanodiscs) and detergent-solubilized systems (0.1% CHAPS). The NADPH oxidation rates were measured using kinetics mode in a Cary 300 UV–Vis spectrometer (Agilent Technologies). Incubation mixtures were calculated in the presence of saturating concentrations of substrate (AA or ebastine) and electron donor (NADPH). The incubation mixtures contained 100 mM phosphate buffer (pH 7.4), 150 pmol CPR, 50 pmol CYP2J2 N-terminal mutant, and either AA (70 µM) or ebastine (50 µM). Samples were equilibrated at 37°C for 5 min before reaction initiation with 200 µM NADPH (total volume 250 µL). NADPH oxidation was monitored for 10 min at 340 nm and the slope was used to determine the absorption change over time. The NADPH consumption rate was calculated using an extinction coefficient of 6.22 mM−1 cm−1. Reactions were quenched and products of the reaction were further analyzed as given in the next section.65
Ebastine hydroxylation assay
CYP2J2 N-terminal mutant ebastine metabolism rates were examined in membrane-bound systems (Nanodiscs) and detergent-solubilized systems (0.1% CHAPS). Saturating concentrations of the substrate (ebastine) and electron donor (NADPH) were used to calculate the reaction Vmax (nmol min−1 nmol−1 of CYP) for each respective CYP2J2 N-terminus mutant. The incubation mixture for ebastine hydroxylation was the same as above. After a 10-min incubation period, the reaction was terminated with 100 µL of ice-cold acetonitrile (ACN) and then spiked with the internal standard terfenadine (100 ng mL−1). The reactions were stopped at 10 min to minimize secondary metabolite formation. Quenched samples were centrifuged at 10,000 rpm for 15 min on a desktop minicentrifuge (Eppendorf AG Hamburg, Germany). A 100-µL sample of supernatant was flash frozen for hydroxyebastine concentration determination via LC/MS/MS analysis. Each construct assay was completed in duplicate or greater. Rates and coupling efficiency (%) are reported as the mean ± SE.
Liquid chromatography-tandem mass spectrometry for quantitation of ebastine and hydroxyebastine
Liquid chromatography-tandem mass spectrometry (LC/MS/MS) was used for quantitation of ebastine, hydroxyebastine, and terfenadine parent ions [M + H]+ and their corresponding fragment ions. The LC/MS/MS system consisted of a Waters Alliance 2795 analytical HPLC separation module (Waters) coupled with an electrospray ionization mass spectrometer (Waters QuattroUltima, Waters) operated in positive mode. Samples were analyzed using a reversed-phase C18, 1.3 Å, 2.1 mm × 20 mm, and 2.5 µm pore size column (Waters). The solvent system was composed of two solutions: solvent A [95% H2O, 5% ACN, and 0.1% formic acid (FA)] and solvent B (5% H2O, 95% ACN, and 0.1% FA). The 10-min gradient LC separation included eight steps: (1) 90–80% solvent A in 0–1 min (linear); (2) 80–65% solvent A for 1–4 min (linear); (3) 65% solvent A for 4–5 min (isocratic); (5) 65–50% solvent A for 5–7 min (linear); (6) 50% solvent A for 7–8 min (isocratic); (7) 50–90% solvent A for 8–9 min (linear); and (8) 90% solvent A for 9–10 min (isocratic).
The MRM data acquisition consisted of monitoring the following analytes in positive mode using these transitions (parent ion → fragment ion, cone voltage, collision voltage): ebastine (470.4 → 167.0, 30 eV, 30 eV and 470.4 → 203.1, 30 eV, 30 eV), terfenadine (472.4 → 57.2, 30 eV, 35 eV and 472.4 → 436.3, 30 eV, 25 eV), and hydroxyl-ebastine (486.4 → 167.0, 30 eV, 30 eV and 486.4 → 219.1, 30 eV, 30 eV) all with 100-ms dwell time. Quantitation of ebastine and hydroxyebastine was determined using the peak areas normalized to the internal standard (terfenadine 100 ng mL−1) from a previously calculated standard curve. Analytical data were processed using Waters Mass Lynx software (version 4.1).
Modeling of CYP2J2
The primary amino acid sequence of M2D34G was used as input to generate a three-dimensional model of CYP2J2 created using the Phyre Web server.63 The resulting pdb file was used as input for the Orientations of Proteins in Membranes (OPM) database64 to generate a model of M2D34 binding with a membrane [Figure 4(B)].
Acknowledgments
The authors thank Ms. Snehita Sri Varma, Dr. Mary Schuler, Dr. Ilia Denisov, and Yelena Grinkova at the University of Illinois for helpful discussions. LC/MS/MS analyses were performed at the University of Illinois—School of Chemical Sciences Mass Spectrometry Laboratory. They thank Dr. Kevin Tucker and Dr. Furong Sun for help with the optimization of the LC/MS/MS method and analysis of the samples. They also thank Dr. Ferguson, Dr. Bagchi, and Dr. Bunick for use of their equipment. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. A.D. and A.P. designed research; A.P. and D.R.M. performed research; E.M. expressed proteins for the research; A.D., A.P., D.R.M., and D.D.M. analyzed data; and A.D., A.P., and D.R.M. wrote the article.
Glossary
- CPR
cytochrome P450 reductase
- CYP2J2
cytochrome P450 2J2
- ND
Nanodiscs
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPS
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Samuelsson B. Role of basic science in the development of new medicines: examples from the eicosanoid field. J Biol Chem. 2012;287:10070–10080. doi: 10.1074/jbc.X112.351437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hannemann F, Bichet A, Ewen KM, Bernhardt R. Cytochrome P450 systems—biological variations of electron transport chains. Biochim Biophys Acta. 2007;1770:330–344. doi: 10.1016/j.bbagen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 3.Bernhardt R. Cytochromes P450 as versatile biocatalysts. J Biotechnol. 2006;124:128–145. doi: 10.1016/j.jbiotec.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 4.Delozier TC, Kissling GE, Coulter SJ, Dai D, Foley JF, Bradbury JA, Murphy E, Steenbergen C, Zeldin DC, Goldstein J. Detection of human CYP2C8, CYP2C9, and CYP2J2 in cardiovascular tissues. Drug Metab Dispos. 2007;35:682–688. doi: 10.1124/dmd.106.012823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westphal C, Konkel A, Schunck WH. CYP-eicosanoids—a new link between omega-3 fatty acids and cardiac disease? Prostaglandins Other Lipid Mediat 9. 2011;6:99–108. doi: 10.1016/j.prostaglandins.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Jiang Y, Liu Y, Lin C, Cheng G, Chen X, Hao B, Tan W, Lin D, He F. CYP2J2*7 single nucleotide polymorphism in a Chinese population. Clin Chim Acta. 2006;365:125–128. doi: 10.1016/j.cca.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Xiao B, Li X, Yan J, Yu X, Yang G, Xiao X, Voltz JW, Zeldin DC, Wang DW. Overexpression of cytochrome P450 epoxygenases prevents development of hypertension in spontaneously hypertensive rats by enhancing atrial natriuretic peptide. J Pharmacol Exp Ther. 2010;334:784–794. doi: 10.1124/jpet.110.167510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imig JD. Epoxides and soluble epoxide hydrolase in cardiovascular physiology. Physiol Rev. 2012;92:101–130. doi: 10.1152/physrev.00021.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imig JD, Hammock BD. Soluble epoxide hydrolase as a therapeutic target for cardiovascular diseases. Nat Rev Drug Discov. 2009;8:794–805. doi: 10.1038/nrd2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, El-Sikhry H, Chaudhary KR, Batchu SN, Shayeganpour A, Jukar TO, Bradbury JA, Graves JP, DeGraff LM, Myers P, Rouse DC, Foley J, Nyska A, Zeldin DC, Seubert JM. Overexpression of CYP2J2 provides protection against doxorubicin-induced cardiotoxicity. Am J Physiol Heart Circ Physiol. 2009;297:H37–H46. doi: 10.1152/ajpheart.00983.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herse F, Lamarca B, Hubel CA, Kaartokallio T, Lokki AI, Ekholm E, Laivuori H, Gauster M, Huppertz B, Sugulle M, Ryan MJ, Novotny S, Brewer J, Park JK, Kacik M, Hoyer J, Verlohren S, Wallukat G, Rothe M, Luft FC, Muller DN, Schunck WH, Staff AC, Dechend R. Cytochrome P450 subfamily 2J polypeptide 2 expression and circulating epoxyeicosatrienoic metabolites in preeclampsia. Circulation. 2012;126:2990–2999. doi: 10.1161/CIRCULATIONAHA.112.127340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panigrahy D, Edin ML, Lee CR, Huang S, Bielenberg DR, Butterfield CE, Barnes CM, Mammoto A, Mammoto T, Luria A, Benny O, Chaponis DM, Dudley AC, Greene ER, Vergilio JA, Pietramaggiori G, Scherer-Pietramaggiori SS, Short SM, Seth M, Lih FB, Tomer KB, Yang J, Schwendener RA, Hammock BD, Falck JR, Manthati VL, Ingber DE, Kaipainen A, D'Amore PA, Kieran MW, Zeldin DC. Epoxyeicosanoids stimulate multiorgan metastasis and tumor dormancy escape in mice. J Clin Invest. 2012;122:178–191. doi: 10.1172/JCI58128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panigrahy D, Greene ER, Pozzi A, Wang DW, Zeldin DC. EET signaling in cancer. Cancer Metastasis Rev. 2011;30:525–540. doi: 10.1007/s10555-011-9315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panigrahy D, Kaipainen A, Greene ER, Huang S. Cytochrome P450-derived eicosanoids: the neglected pathway in cancer. Cancer Metastasis Rev. 2010;29:723–735. doi: 10.1007/s10555-010-9264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang JG, Chen CL, Card JW, Yang S, Chen JX, Fu XN, Ning YG, Xiao X, Zeldin DC, Wang DW. Cytochrome P450 2J2 promotes the neoplastic phenotype of carcinoma cells and is up-regulated in human tumors. Cancer Res. 2005;65:4707–4715. doi: 10.1158/0008-5472.CAN-04-4173. [DOI] [PubMed] [Google Scholar]
- 16.Jiang JG, Ning YG, Chen C, Ma D, Liu ZJ, Yang S, Zhou J, Xiao X, Zhang XA, Edin ML, Card JW, Wang J, Zeldin DC, Wang DW. Cytochrome p450 epoxygenase promotes human cancer metastasis. Cancer Res. 2007;67:6665–6674. doi: 10.1158/0008-5472.CAN-06-3643. [DOI] [PubMed] [Google Scholar]
- 17.Lee CA, Jones JPIII, Katayama J, Kaspera R, Jiang Y, Freiwald S, Smith E, Walker GS, Totah RA. Identifying a selective substrate and inhibitor pair for the evaluation of CYP2J2 activity. Drug Metab Dispos. 2012;40:943–951. doi: 10.1124/dmd.111.043505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, Li G, Liao W, Wu J, Liu L, Ma D, Zhou J, Elbekai RH, Edin ML, Zeldin DC, Wang DW. Selective inhibitors of CYP2J2 related to terfenadine exhibit strong activity against human cancers in vitro and in vivo. J Pharmacol Exp Ther. 2009;329:908–918. doi: 10.1124/jpet.109.152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto S, Hirama T, Matsubara T, Nagata K, Yamazoe Y. Involvement of CYP2J2 on the intestinal first-pass metabolism of antihistamine drug, astemizole. Drug Metab Dispos. 2002;30:1240–1245. doi: 10.1124/dmd.30.11.1240. [DOI] [PubMed] [Google Scholar]
- 20.Skorupa E, Kemper B. Signals for retention of cytochrome P450 in the endoplasmic reticulum. Methods Mol Biol. 1998;107:251–266. doi: 10.1385/0-89603-519-0:251. [DOI] [PubMed] [Google Scholar]
- 21.Cosme J, Johnson EF. Analyzing binding of N-terminal truncated, microsomal cytochrome P450s to membranes. Methods Enzymol. 2002;357:116–120. doi: 10.1016/s0076-6879(02)57671-0. [DOI] [PubMed] [Google Scholar]
- 22.Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Microsomal cytochrome P450 2C5: comparison to microbial P450s and unique features. J Inorg Biochem. 2000;81:183–190. doi: 10.1016/s0162-0134(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 23.Hsu PY, Wang LH. Protein engineering of thromboxane synthase: conversion of membrane-bound to soluble form. Arch Biochem Biophys. 2003;416:38–46. doi: 10.1016/s0003-9861(03)00287-x. [DOI] [PubMed] [Google Scholar]
- 24.Cosme J, Johnson EF. Engineering microsomal cytochrome P450 2C5 to be a soluble, monomeric enzyme. Mutations that alter aggregation, phospholipid dependence of catalysis, and membrane binding. J Biol Chem. 2000;275:2545–2553. doi: 10.1074/jbc.275.4.2545. [DOI] [PubMed] [Google Scholar]
- 25.von Wachenfeldt C, Richardson TH, Cosme J, Johnson EF. Microsomal P450 2C3 is expressed as a soluble dimer in Escherichia coli following modification of its N-terminus. Arch Biochem Biophys. 1997;339:107–114. doi: 10.1006/abbi.1996.9859. [DOI] [PubMed] [Google Scholar]
- 26.Kim HJ, Lee SB, Guengerich FP, Park YI, Dong MS. Effects of N-terminal modification of recombinant human cytochrome P450 1A2 on catalytic activity. Xenobiotica. 2007;37:356–365. doi: 10.1080/00498250601178189. [DOI] [PubMed] [Google Scholar]
- 27.Ahn T, Guengerich FP, Yun CH. Membrane insertion of cytochrome P450 1A2 promoted by anionic phospholipids. Biochemistry. 1998;37:12860–12866. doi: 10.1021/bi980804f. [DOI] [PubMed] [Google Scholar]
- 28.Davydov DR. Microsomal monooxygenase as a multienzyme system: the role of P450-P450 interactions. Expert Opin Drug Metab Toxicol. 2011;7:543–558. doi: 10.1517/17425255.2011.562194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian M, Tam H, Zheng H, Tracy TS. CYP2C9-CYP3A4 protein-protein interactions: role of the hydrophobic N terminus. Drug Metab Dispos. 2010;38:1003–1009. doi: 10.1124/dmd.109.030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pernecky SJ, Larson JR, Philpot RM, Coon MJ. Expression of truncated forms of liver microsomal P450 cytochromes 2B4 and 2E1 in Escherichia coli: influence of NH2-terminal region on localization in cytosol and membranes. Proc Natl Acad Sci USA. 1993;90:2651–2655. doi: 10.1073/pnas.90.7.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berka K, Hendrychová T, Anzenbacher P, Otyepka M. Membrane position of ibuprofen agrees with suggested access path entrance to cytochrome P450 2C9 active site. J Phys Chem A. 2011;115:11248–11255. doi: 10.1021/jp204488j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conner KP, Woods CM, Atkins WM. Interactions of cytochrome P450s with their ligands. Arch Biochem Biophys. 2011;50:56–65. doi: 10.1016/j.abb.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denisov IG, Sligar SG. Cytochromes P450 in nanodiscs. Biochim Biophys Acta. 2011;1814:223–229. doi: 10.1016/j.bbapap.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Chapter 11—Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leitz AJ, Bayburt TH, Barnakov AN, Springer BA, Sligar SG. Functional reconstitution of Beta2-adrenergic receptors utilizing self-assembling Nanodisc technology. Biotechniques. 2006;40:601–612. doi: 10.2144/000112169. [DOI] [PubMed] [Google Scholar]
- 37.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci USA. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 39.Das A, Grinkova YV, Sligar SG. Redox potential control by drug binding to cytochrome P450 3A4. J Am Chem Soc. 2007;129:13778–13779. doi: 10.1021/ja074864x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das A, Sligar SG. Modulation of the cytochrome P450 reductase redox potential by the phospholipid bilayer. Biochemistry. 2009;48:12104–12112. doi: 10.1021/bi9011435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tark SH, Das A, Sligar S, Dravid VP. Nanomechanical detection of cholera toxin using microcantilevers functionalized with ganglioside nanodiscs. Nanotechnology. 2010;21:435502. doi: 10.1088/0957-4484/21/43/435502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marty MT, Das A, Sligar SG. Ultra-thin layer MALDI mass spectrometry of membrane proteins in nanodiscs. Anal Bioanal Chem. 2012;402:721–729. doi: 10.1007/s00216-011-5512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das A, Zhao J, Schatz GC, Sligar SG, Van Duyne RP. Screening of type I and II drug binding to human cytochrome P450-3A4 in nanodiscs by localized surface plasmon resonance spectroscopy. Anal Chem. 2009;81:3754–3759. doi: 10.1021/ac802612z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao J, Das A, Zhang X, Schatz GC, Sligar SG, Van Duyne RP. Resonance surface plasmon spectroscopy: low molecular weight substrate binding to cytochrome p450. J Am Chem Soc. 2006;128:11004–11005. doi: 10.1021/ja0636082. [DOI] [PubMed] [Google Scholar]
- 45.Kim DH, Kim KH, Isin EM, Guengerich FP, Chae HZ, Ahn T, Yun CH. Heterologous expression and characterization of wild-type human cytochrome P450 1A2 without conventional N-terminal modification in Escherichia coli. Protein Expr Purif. 2008;57:188–200. doi: 10.1016/j.pep.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 46.Richardson TH, Jung F, Griffin KJ, Wester M, Raucy JL, Kemper B, Bornheim LM, Hassett C, Omiecinski CJ, Johnson EF. Universal approach to the expression of human and rabbit cytochrome P450s of the 2c subfamily in Escherichia coli. Arch Biochem Biophys. 1995;323:87–96. doi: 10.1006/abbi.1995.0013. [DOI] [PubMed] [Google Scholar]
- 47.Looman AC, Bodlaender J, Comstock LJ, Eaton D, Jhurani P, Deboer HA, Vanknippenberg PH. Influence of the codon following the aug initiation codon on the expression of a modified Lacz gene in Escherichia coli. EMBO J. 1987;6:2489–2492. doi: 10.1002/j.1460-2075.1987.tb02530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barnes HJ, Arlotto MP, Waterman MR. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc Natl Acad Sci USA. 1991;88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stormo GD, Schneider TD, Gold LM. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982;10:2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schauder B, McCarthy JE. The role of bases upstream of the Shine-Dalgarno region and in the coding sequence in the control of gene expression in Escherichia coli: translation and stability of mRNAs in vivo. Gene. 1989;78:59–72. doi: 10.1016/0378-1119(89)90314-4. [DOI] [PubMed] [Google Scholar]
- 51.Yamasaki T, Izumi S, Ide H, Ohyama Y. Identification of a novel rat microsomal vitamin D3 25-hydroxylase. J Biol Chem. 2004;279:22848–22856. doi: 10.1074/jbc.M311346200. [DOI] [PubMed] [Google Scholar]
- 52.DeVore NM, Scott E. Structures of cytochrome P450 17A1 with prostate cancer drugs abiraterone and TOK-001. Nature. 2012;482:116–119. doi: 10.1038/nature10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fisher CW, Caudle DL, Martin-Wixtrom C, Quattrochi LC, Tukey RH, Waterman MR, Estabrook R. High-level expression of functional human cytochrome P450 1A2 in Escherichia coli. FASEB J. 1992;6:759–764. doi: 10.1096/fasebj.6.2.1537466. [DOI] [PubMed] [Google Scholar]
- 54.Messina A, Nencioni S, Gervasi PG, Gotlinger KH, Schwartzman ML, Longo V. Molecular cloning and enzymatic characterization of sheep CYP2J. Xenobiotica. 2010;40:109–118. doi: 10.3109/00498250903410590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Z, Sigler P. GroEL/GroES: structure and function of a two-stroke folding machine. J Struct Biol. 1998;124:129–141. doi: 10.1006/jsbi.1998.4060. [DOI] [PubMed] [Google Scholar]
- 56.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J Biol Chem. 1964;239:2379–2385. [PubMed] [Google Scholar]
- 57.Sligar SG. Coupling of spin, substrate, and redox equilibria in cytochrome P450. Biochemistry. 1976;15:5399–5406. doi: 10.1021/bi00669a029. [DOI] [PubMed] [Google Scholar]
- 58.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. Cooperativity in cytochrome P450 3A4: linkages in substrate binding, spin state, uncoupling, and product formation. J Biol Chem. 2007;282:7066–7076. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 59.Wu S, Chen W, Murphy E, Gabel S, Tomer KB, Foley J, Steenbergen C, Falck JR, Moomaw CR, Zeldin DC. Molecular cloning, expression, and functional significance of a cytochrome P450 highly expressed in rat heart myocytes. J Biol Chem. 1997;272:12551–12559. doi: 10.1074/jbc.272.19.12551. [DOI] [PubMed] [Google Scholar]
- 60.Schoch GA, Yano JK, Sansen S, Dansette PM, Stout CD, Johnson EF. Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid. J Biol Chem. 2008;283:17227–17237. doi: 10.1074/jbc.M802180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF. Structure of human microsomal cytochrome P450 2C8. Evidence for a peripheral fatty acid binding site. J Biol Chem. 2004;279:9497–9503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]
- 62.Hashizume T, Imaoka S, Mise M, Terauchi Y, Fujii T, Miyazaki H, Kamataki T, Funae Y. Involvement of CYP2J2 and CYP4F12 in the metabolism of ebastine in human intestinal microsomes. J Pharmacol Exp Ther. 2002;300:298–304. doi: 10.1124/jpet.300.1.298. [DOI] [PubMed] [Google Scholar]
- 63.Kelley LA, Sternberg M. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 64.Lomize MA, Lomize AL, Pogozheva ID, Mosberg H. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 65.Kang W, Liu KH, Ryu JY, Shin J. Simultaneous determination of ebastine and its three metabolites in plasma using liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;813:75–80. doi: 10.1016/j.jchromb.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 66.Liu KH, Kim MG, Lee DJ, Yoon YJ, Kim MJ, Shon JH, Choi CS, Choi YK, Desta Z, Shin JG. Characterization of ebastine, hydroxyebastine, and carebastine metabolism by human liver microsomes and expressed cytochrome P450 enzymes: major roles for CYP2J2 and CYP3A. Drug Metab Dispos. 2006;34:1793–1797. doi: 10.1124/dmd.106.010488. [DOI] [PubMed] [Google Scholar]
- 67.Denisov IG, Makris TM, Sligar SG, Schlichting I. Structure and chemistry of cytochrome P450. Chem Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 68.Sulistyaningdyah WT, Ogawa J, Li QS, Maeda C, Yano Y, Schmid RD, Shimizu S. Hydroxylation activity of P450 BM-3 mutant F87V towards aromatic compounds and its application to the synthesis of hydroquinone derivatives from phenolic compounds. Appl Microbiol Biotechnol. 2005;67:556–562. doi: 10.1007/s00253-004-1761-9. [DOI] [PubMed] [Google Scholar]
- 69.Isin EM, Guengerich FP. Substrate binding to cytochromes P450. Anal Bioanal Chem. 2008;392:1019–1030. doi: 10.1007/s00216-008-2244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fisher MT, Sligar SG. Control of heme protein redox potential and reduction rate—linear free-energy relation between potential and ferric spin state equilibrium. J Am Chem Soc. 1985;107:5018–5019. [Google Scholar]
- 71.Bridges A, Gruenke L, Chang YT, Vakser IA, Loew G, Waskell L. Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J Biol Chem. 1998;273:17036–17049. doi: 10.1074/jbc.273.27.17036. [DOI] [PubMed] [Google Scholar]
- 72.Shen S, Strobel HW. Role of lysine and arginine residues of cytochrome P450 in the interaction between cytochrome P4502B1 and NADPH-cytochrome P450 reductase. Arch Biochem Biophys. 1993;304:257–265. doi: 10.1006/abbi.1993.1347. [DOI] [PubMed] [Google Scholar]
- 73.Shimizu T, Tateishi T, Hatano M, Fujii-Kuriyama Y. Probing the role of lysines and arginines in the catalytic function of cytochrome P450d by site-directed mutagenesis. Interaction with NADPH-cytochrome P450 reductase. J Biol Chem. 1991;266:3372–3375. [PubMed] [Google Scholar]
- 74.Poulos TL. Approaches to crystallizing P450s. Methods Enzymol. 1996;272:358–368. doi: 10.1016/s0076-6879(96)72041-4. [DOI] [PubMed] [Google Scholar]
- 75.Poulos TL. Cytochrome P450. Curr Opin Struct Biol. 1995;5:767–774. doi: 10.1016/0959-440x(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 76.Aiba I, Yamasaki T, Shinki T, Izumi S, Yamamoto K, Yamada S, Terato H, Ide H, Ohyama Y. Characterization of rat and human CYP2J enzymes as vitamin D 25-hydroxylases. Steroids. 2006;71:849–856. doi: 10.1016/j.steroids.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 77.Kagawa N, Cao Q. Osmotic stress induced by carbohydrates enhances expression of foreign proteins in Escherichia coli. Arch Biochem Biophys. 2001;393:290–296. doi: 10.1006/abbi.2001.2516. [DOI] [PubMed] [Google Scholar]
- 78.Joseph RE, Andreotti AH. Bacterial expression and purification of interleukin-2 tyrosine kinase: single step separation of the chaperonin impurity. Protein Expr Purif. 2008;60:194–197. doi: 10.1016/j.pep.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Causey KM, Eyer CS, Backes WL. Dual role of phospholipid in the reconstitution of cytochrome P-450 LM2-dependent activities. Mol Pharmacol. 1990;38:134–142. [PubMed] [Google Scholar]
- 80.Kapust RB, Tözsér J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh D. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


