Abstract
A simple, sensitive, precise and specific method for the determination of curcuminoids and curcuminoid-loaded liposome formulation was developed using reverse-phase high-performance liquid chromatography method. The analysis was performed isocratically on Zorbax Eclipse XDB-C18 column (150×4 mm, 5 μm), analytical column using UV detector and mobile phase consisting of 0.1% orthophosphoric acid and acetonitrile. The proposed method for curcuminoids was validated for linearity in the range from 50 to 300 ΅g/ml with correlation coefficient above 0.997. Intraday and interday precision studies showed the relative standard deviation less than 2%. The limit of detection and limit of quantitation values were 2.5 and 8.25 ΅g/ml, respectively. Forced degradation study for curcuminoids and liposomal curcuminoids sample was carried out and observed that proposed method was also suitable for finding degradation products in the sample. Proposed method was successfully applied to estimate curcuminoids content without any interference of other excipients from liposomal formulation. Therefore, the method developed is well suited for curcuminoids and its liposome estimation.
Keywords: Curcuminoids, degradation study, liposome, reversed phase-high performance liquid chromatography
The growing public interest in traditional medicine, particularly plants-based medicine, has led to extensive research on the potential of natural origin substances. This interest primarily stems from the belief that green medicine is safe and dependable, compared with costly synthetic drugs that have adverse side-effects. Presently curcumin enjoys the first place among all medicinal molecules derived from plant origin, which was and still being studied very extensively for its extraction, analytical, pharmacological and clinical features.
Turmeric, dried rhizomes of Curcuma longa L. (Zingiberaceae), contains 5% curcuminoids, which is a group of phenolic compounds and is a mixture of three principal compounds namely curcumin (77%), demethoxycurcumin (17%), and bisdemethoxycurcumin (3%).[1,2] These compounds are practically insoluble in water at acidic and neutral pH, and soluble in methanol, ethanol, dimethylsulfoxide and acetone. The maximum absorption (λ max) of curcuminoids in methanol occurs at 425 nm. All the three compounds contribute to potency and safety when used as medicinal product. Structure of curcuminoids is shown in fig. 1.
Fig. 1.
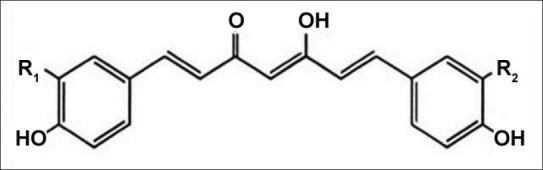
Structure of curcuminoids.
Curcumin R1=OCH3, R2=OCH3; demethoxycurcumin R1=OCH3, R2=H; bisdemethoxycurcumin R1=H, R2=H.
Literature on analysis of curcuminoids revealed several methods based on different techniques namely high-performance liquid chromatography (HPLC),[3–6] high-performance thin layer chromatography (HPTLC) ,[7,8] dihydrogen phosphate impregnated silica gel TLC plates,[9] UV fluorescence detection,[10] capillary electrophoresis[11,12] and liquid chromatography-mass spectroscopy (LC/MS) analysis in food products.[13] The simple analysis of individual curcuminoids is possible by HPLC on normal phase or on reversed phase (RP) C18 columns.[14] Due to the very labile characteristics of curcuminoids C18 columns are preferred for HPLC analysis.[15] Moving to the mobile phase, commonly used RP solvents; methanol, acetonitrile, and tetrahydrofuran (THF) of which methanol does not provide the necessary resolution/selectivity for the separation of curcuminoids. Using THF instead of acetonitrile as the organic modifier reverses the elution order of the curcuminoids.[14] Literature reports pH variation in the mobile phase for curcuminoids analysis.[16]
Curcuminoids, a natural polyphenol, exhibits wide range of pharmacological actions such as antiinflammatory, antineoplastic, antioxidant, and chemopreventive activity.[17–22] Several clinical trials dealing with cancers have addressed the pharmacokinetics, safety, and efficacy of curcuminoids in humans.[19] Despite extensive research and development, poor solubility of this molecule, due to its hydrophobic property and preferential interaction with lipid membranes, remains a major barrier in its bioavailability and clinical efficacy.[23] To increase its solubility and bioavailability, attempts have been made through encapsulation in liposomes, polymeric nanoparticles, biodegradable microspheres, cyclodextrin, and hydrogels.[24–32] Degradation study of curcumin has been done in the literature.[33,34] Liposome containing curcuminoids formulation degradation study has not been previously reported so far, to our present knowledge.
The aim of the present work was to develop a simple, precise and rapid RP-HPLC method for the quantitative analysis of curcuminoids. The proposed method was studied for its linearity, precision, accuracy, detection and quantitation limits (limit of detection (LOD) and limit of quantification (LOQ)) as well as system performance parameters such as theoretical plates, capacity factor, tailing factor, separation factor and peak resolution. The developed method was validated as per International Conference on Harmonization (ICH) guidelines.[35] At the end, forced degradation study was also performed for both the curcuminoids and liposomal curcuminoids samples. Furthermore, developed method was used for the quantification of unknown drug sample in the curcuminoid-loaded liposome formulation and freeze dried formulation.
MATERIALS AND METHODS
Curcuminoids sample (purity 99%) was received as a gift sample from Konark Herbals and Healthcare, Mumbai, India. Soy Phosphatidylcholine 70% (PC) (Sonic Biochem, Indore, India); cholesterol (Chol), sucrose, orthophosphoric acid (OPA) and Triton X-100 were used of extra pure grade from SD Fine Chemicals, Mumbai, India. Methanol, acetonitrile used were of HPLC grade purchased from SD Fine Chemicals, Mumbai, India. All the other reagents and chemicals used were of analytical grade or pharmaceutical grade.
Mobile phase preparation:
The mobile phase previously optimized consisted of acetonitrile and 0.1% OPA in the ratio of 50:50 v/v. Both the solvents were filtered through 0.2 μm Pall Ultipor N66 membrane. The isocratic elution was carried out with the flow rate of 1 ml/min at ambient temperature and a wavelength of 425 nm was used for detection.
Instrument details and optimization:
HPLC system used was (Agilent 1200 Series, Agilent Technologies India Pvt. Ltd., India) equipped with ChemStation software, a model G1322A degasser, a model G1311A quaternary pump, auto sampler, a model G1316A column oven and variable wavelength UV/Vis detector. The separation was performed on ZORBAX Eclipse XDB-C18 column (150×4 mm, 5 μm).
Any solvent remaining in the system from the previous analysis was washed out by passing methanol:water (50:50) for 10 min at 1 ml/min flow rate with purge valve open. After closing the purge valve, 100% methanol was passed for 30 min through column for proper column washing. After this step, for column conditioning and base line stability, acetonitrile:OPA (0.1%) mobile phase in the ratio of 50:50 v/v was passed through HPLC system for 1 h at 1 ml/min flow rate. The total injection volume was set to 5 μl.
Standard solution preparation:
Standard curcuminoid 50 mg was accurately weighed and transferred to a 50 ml volumetric flask and the volume was made with methanol. Solutions of 50, 100, 150, 200, 250 and 300 μg/ml were made by transferring the aliquot from stock solution and the volume was made with methanol in each case. Further standard solutions were prepared freshly each day by appropriate dilution of stock solution with methanol for intraday as well as interday analysis. The solution was filtered through a 0.2 μm Ultipor N66 membrane syringe filter, before injecting it into the chromatographic system.
Validation of the method:
Validation of the analytical method was done according to the ICH guidelines. The method was validated for linearity, precision, accuracy, LOD and LOQ.
Linearity:
The linearity of measurement was evaluated by analysing different concentrations (50-300 μg/ml) of the standard solutions. Calibration curve was constructed for curcuminoids by plotting average peak area against concentration and regression equation. The correlation coefficient and the slope of the peak were also computed. All the samples were analysed in triplicate.
Precision:
The precision of the method was investigated with respect to repeatability and intermediate precision. The repeatability (intraday precision) of the method was evaluated by assaying three replicate injections of the curcuminoids standard at concentration of 100, 200 and 300 μg/ml on the same day at different times. The percentage relative standard deviation (RSD) of the peak area was calculated. The intermediate precision (interday precision) was demonstrated by evaluating the relative peak area at three different concentration levels as taken in intraday study that cover the assay method. The precision was expressed as % RSD of the system and the samples analysed in triplicate.
Accuracy study:
The accuracy of the method was tested by performing recovery studies at 3 levels of curcuminoids reference standards added to the samples. Three different volumes (0.5, 1 and 1.5 ml) of the standard solution (containing 100 μg/ml of curcuminoids in methanol) were added to the sample solution (60.3 μg/ml) and analysed by the proposed HPLC method. All the samples were determined in triplicate. The % RSD was calculated by using the formula, % recovery=[(b-a)/c]×100…(1), where, a is the amount of drug found in the sample before addition of standard drug, b is the amount of drug found after addition of standard drug, c is the amount of standard drug added.
Limit of detection and limit of quantitation:
LOD and LOQ of the developed method were determined by injecting progressively low concentrations of the standard solutions using the developed RP-HPLC method. The LOD is the smallest concentration of the analyte that gives a measurable response (signal to noise ratio of 3). The LOQ is the smallest concentration of the analyte, which gives response that can be accurately quantified (signal to noise ratio of 10).
Preparation and analysis of curcuminoid-loaded liposomes:
The phospholipids curcuminoids (PC), and cholesterol were mixed together at previously optimized ratio in chloroform solvent. Amber coloured 250 ml round bottom flask was used to get the PCs film with the help of rotary evaporator (Rotavapor RII, BÜCHI, Switzerland). Any residual chloroform that remained in the flask was removed by placing the film overnight in a vacuum desiccator. Multilamellar vesicles were formed by reconstituting the lipid film with Milli-Q water under vigorous shaking at 35°. Unilamellar vesicles were then formed using a probe sonicator (Sonics Vibra-cell, Make: Ace, 750 W) for 150 s on ice bath (5 s pulse with 5 s interval between each pulse). After sonication, the liposome dispersion was centrifuged at 4000×g for 90 s to pellet form. The nonintercalated curcuminoids remained in the solution.
Approximately 1 ml of the liposomal dispersion containing PC, cholesterol and curcuminoids in the ratio of 20:2:1 was dissolved in 10% Triton X-100 of 5 ml methanol and allows standing for 15 min. 5 μl aliquot of the solution was injected into a HPLC system. All the analytical conditions were kept constant as used for the standard curve preparation. The percent entrapment efficiency (EE) was calculated by the standard equation, EE (%)=(encapsulated drug/[encapsulated drug + free drug])×100…(2)
The curcuminoids retention (CR) after freeze-drying was calculated using the formula, CR(%)=(EE2/EE1)×100…(3), where, EE1 and EE2 are EE before and after freeze-rying respectively.
Degradation studies of curcuminoids and liposomal curcuminoids:
For acid and base degradation study, 1 ml of 1 N aqueous hydrochloric acid solution (HCl) and 1 ml of 1 N aqueous NaOH solutions were added in each 1 ml of methanol solution of curcuminoids (100 μg/ml) in 10 ml amber coloured volumetric flask, respectively. Both the flasks were sealed and placed at 40° for 2 h. Thereafter the solutions were cooled and neutralized with 1 ml of 1 N aqueous NaOH and 1 N aqueous HCl for acid and base degradation, respectively. Finally, volume was made up with methanol to 10 ml and solutions were filtered through syringe filter of 0.2 μm and subjected to HPLC analysis. In case of liposome sample, after neutralisation, 0.5 ml of Triton X-100 was added for breaking the liposomes and the volume was made up with methanol and the solution was filtered through a 0.2 μm syringe filter, before injecting the solution into the chromatographic system. The study was performed in triplicate.
The oxidative degradation was performed by transferring 1 ml of methanol solution of curcuminoids (100 μg/ml) to a 10 ml amber volumetric flask to which 1 ml of hydrogen peroxide solution (H2O2; 30% v/v) was added. The flask was sealed and placed at 40° for 2 h. Thereafter, the volume was made up with methanol and the solution was filtered through a 0.2 μm syringe filter, before injecting the solution into the chromatographic system. For liposome sample, additionally 0.5 ml Triton X-100 was added and volume was made with methanol. The solution filtered through 0.2 μm syringe filter and was subjected to HPLC analysis. The study was performed in triplicate.
For thermal degradation studies, 1 ml of methanol solution of curcuminoids (100 μg/ml) was transferred to a 100 ml amber volumetric flask to which 2 mL of methanol was added. The flask was sealed and placed at 80° for 2 h. Thereafter, the volume was made up with methanol and the solution was filtered through a 0.2 μm syringe filter, before injecting the solution into the chromatographic system. Thermal degradation of liposomal sample was performed in the same way for curcuminoids sample and by adding Triton X-100 before volume was made up with methanol. The solution was filtered through 0.2 μm syringe filter and subjected to HPLC analysis. The study was performed in triplicate.
Photo degradation study was carried out by transferring curcuminoids solution (10 μg/ml) to 10 ml transparent volumetric flask and flask was sealed and exposed to direct sunlight for a period of 6 h. Thereafter, the solution was filtered through a 0.2 μm syringe filter, before injecting the solution into the chromatographic system. Liposomal sample (0.2 mg which corresponds to 10 μg/ml of curcuminoids) was transferred to 10 ml volumetric flask and volume was made to 9.5 ml with methanol. Flask was sealed and exposed to direct sunlight for a period of 6 h. Triton X-100 was added to the final sample and filtered through 0.2 μm syringe filter before subjecting to the HPLC analysis. The study was performed in triplicate.
RESULT AND DISCUSSION
HPLC separation of curcuminoids was carried out on C18 column by an isocratic elution with 0.1% OPA-acetonitrile (50:50 v/v) solution. The flow rate was constant at 1 ml/min and the column temperature was at room temperature (25±1°). The UV wavelength was set at 425 nm. No interference from diluents, impurities, or excipients present in the curcuminoids sample was observed at this detection wavelength. Sharp and symmetrical peaks were obtained for curcuminoids when analysed under these conditions.
Chromatogram of curcuminoids peaks are shown in fig. 2. Commercially available curcumin consists of a mixture of three naturally occurring curcuminoids with curcumin as the main (≈77%) constituent. Curcuminoids are recognized for their broad spectrum of biological activities and safety in foods or pharmaceuticals. The validation parameters for the curcuminoids sample were studied. The retention time for bisdemethoxycurcumin, demethoxycurcumin and curcumin were found to be 4.97, 5.62 and 6.36 respectively, as shown in fig. 2.
Fig. 2.
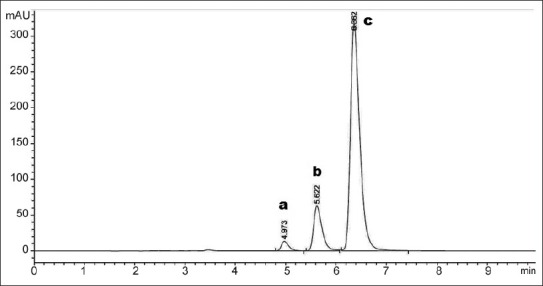
Chromatogram of standard curcuminoids at 425 nm. a. bisdemethoxycurcumin; b. demethoxycurcumin; c. curcumin
The method was validated for linearity, precision, accuracy, LOD and LOQ. Curcuminoids showed three peaks namely bisdemethoxycurcumin, demethoxycurcumin and curcumin as discussed earlier. The standard curve for curcuminoid was prepared, which shows good linearity in the range of 50-300 μg/ml with correlation coefficient (R2) of 0.9977 as this was the working concentration range for the sample formulations. The % RSD for intraday and interday precisions of curcuminoids was 0.2219 and 0.4042, respectively. The results showed acceptable precision of the method, with RSD values much lower than 2%. The results showed that an excellent correlation exists between the peak area and the concentration of drugs within the concentration range. The accuracy of the method was evaluated by adding the standard solution of 50, 100 and 150 (μg/ml) to known sample solutions. The % recovery of curcuminoids was obtained with an average of 98.13%, which was well within the range of 98-102%. The LOD and LOQ for curcuminoids sample were 0.124 μg/ml and 0.375 μg/ml, respectively. The high sensitivity of the proposed method is reflected by the signal to noise ratio of 3.16 and 10.43, for LOD and LOQ, respectively.
Curcuminoid-loaded liposomes were successfully analysed by the proposed HPLC method with no interference of other excipients present in the liposome formulation. The results showed 99% EE of the formulated liposome containing curcuminoids with 97% retention after freeze drying. These results suggest the suitability of HPLC for quantitative analysis of curcuminoids.
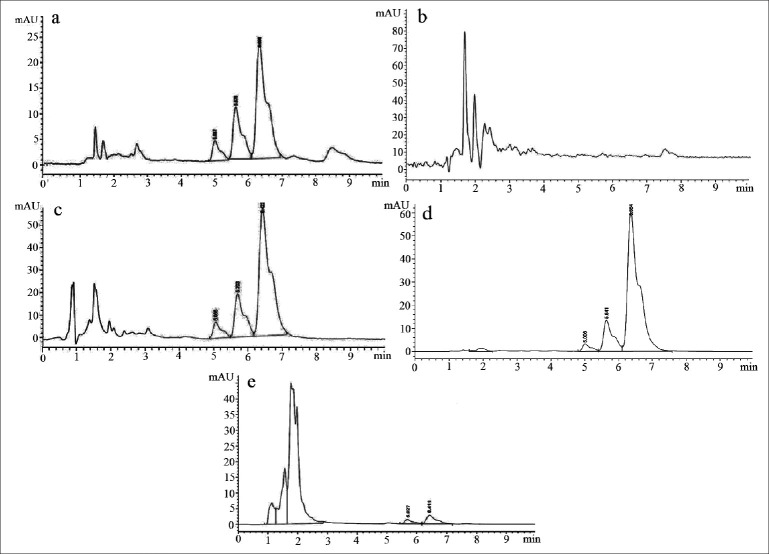
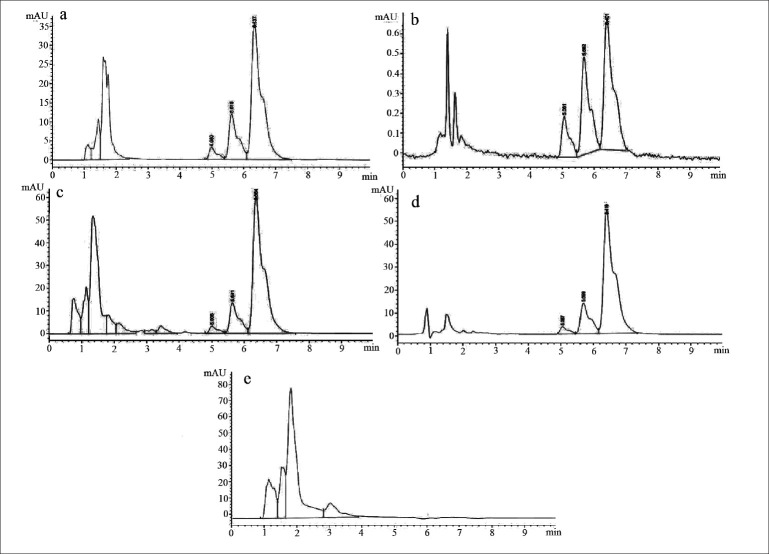
Curcuminoids sample can be detected at 425 nm wavelength whereas its degradation products can be detected at 280 nm.[33] Chromatogram of curcuminoids sample at 280 nm was shown in fig. 3. The chromatograms of the samples degraded with acid, base, hydrogen peroxide and direct sunlight showed major degradation peaks of pure curcuminoids sample whereas thermal conditions showed good stability of curcuminoids sample. Curcuminoids were completely degraded in alkaline conditions as compared to acid and oxidative conditions. Liposome containing curcuminoids showed better stability as compared to pure curcuminoids sample. Percent degradation of curcuminoids and liposomal curcuminoids using various conditions was shown in Table 1. Chromatograms of the degradation products were shown in fig. 4 and 5.
Fig. 3.
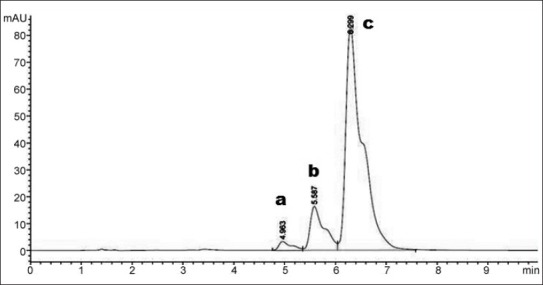
Chromatogram of standard curcuminoids at 280 nm.
a. bisdemethoxycurcumin; b. demethoxycurcumin; c. curcumin
Table 1.
FORCED DEGRADATION OF CURCUMINOIDS AND LIPOSOMAL CURCUMINOIDS (N=3)
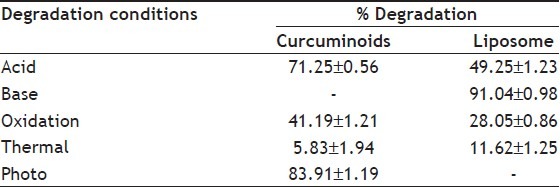
Fig. 4.
HPLC chromatograms of stress degradation studies of curcuminoids
Chromatograms of curcuminoid analytes and degradation products under various stress conditions such as a. acid degradation; b. base degradation; c. oxidation degradation; d. thermal degradation and e. photodegradation.
Fig. 5.
HPLC chromatograms of stress degradation studies of liposomal curcuminoids
Chromatograms of curcuminoid analytes from liposomal formulation and degradation products under various stress conditions such as a. acid degradation; b. base degradation; c. oxidation degradation; d. thermal degradation and e. photodegradation
A convenient, rapid, accurate and precise RP-HPLC method was developed for the estimation of curcuminoids as well as liposome containing curcuminoids formulation. The assay provides a linear response across a wide range of concentrations. The proposed method assures the prolong life of column and the system due to lower concentration of acid in the mobile phase and the use of two solvents as mobile phase. Forced degradation study of pure and liposome containing curcuminoids samples showed better sensitivity to degraded products. This method can be said to be more economical as compared to other methods reported in literature. The proposed RP-HPLC method was successfully implemented for curcuminoid-loaded liposome formulation without any interference of other formulation additives.
ACKNOWLEDGEMENTS
The authors are thankful to University Grant Commission Government of India, for providing funding for this research work.
Footnotes
Jangle and Thorat: RP-HPLC Method for Curcuminoids and Its Liposome
REFERENCES
- 1.Strimpakos AS, Sharma RA. Curcumin: Preventive and therapeutic properties in laboratory studies and clinical trials. Antioxid Redox Signal. 2008;10:511–45. doi: 10.1089/ars.2007.1769. [DOI] [PubMed] [Google Scholar]
- 2.Goel A, Kunnumakkara AB, Aggarwal BB. Curcumin as Curecumin: From kitchen to clinic. Biochem Pharmacol. 2008;75:787–809. doi: 10.1016/j.bcp.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Antony B, Merina B, Iyer VS, Judy N, Lennertz K, Joyal S. A pilot cross-over study to evaluate human oral bioavailability of BCM-95CG (Biocurcumax), a novel bioenhanced preparation of Curcumin. Indian J Pharm Sci. 2008;70:445–9. doi: 10.4103/0250-474X.44591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chauhan S, Singh B, Agrawala S. Estimation of curcuminoids in Curcuma longa by HPLC and spectrophotometric methods. Indian J Pharm Sci. 1999;61:58–60. [Google Scholar]
- 5.Pak Y, Patek R, Mayersohn M. Sensitive and rapid isocratic liquid chromatography method for the quantitation of curcumin in plasma. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;796:339–46. doi: 10.1016/j.jchromb.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 6.Jayaprakasha GK, Jagan Mohan Rao L, Sakariah KK. Improved HPLC method for the determination of curcumin, demethoxycurcumin, and bisdemethoxycurcumin. J Agric Food Chem. 2002;50:3668–72. doi: 10.1021/jf025506a. [DOI] [PubMed] [Google Scholar]
- 7.Guptha A, Gupta M, Kumar S. Simultaneous determination of curcuminoids in Curcuma samples using high performance thin layer chromatography. J Liq Chromatogr Relat Technol. 1999;22:1561–9. [Google Scholar]
- 8.Pathania V, Gupta A, Singh B. Improved HPTLC method for determination of curcuminoids from Curcuma longa. J Liq Chromatogr Relat Technol. 2006;29:877–87. [Google Scholar]
- 9.Rasmussen HB, Christensen SB, Kvist LP, Karazmi A. A simple and efficient separation of the curcumins, the antiprotozoal constituents of Curcuma longa. Planta Med. 2000;66:396–8. doi: 10.1055/s-2000-8533. [DOI] [PubMed] [Google Scholar]
- 10.Inoue K, Yoshimura Y, Nakazawa H. Evaluation of the turmeric (Curcuma longa L) based on flow injection analysis with ultraviolet and fluorometric detections. Anal Lett. 2001;34:1711–8. [Google Scholar]
- 11.Yuan K, Weng Q, Zhang H, Xiong J, Xu G. Application of capillary zone electrophoresis in the separation and determination of the curcuminoids in urine. J Pharm Biomed Anal. 2005;38:133–8. doi: 10.1016/j.jpba.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Sun X, Gao C, Cao W, Yang X, Wang E. Capillary electrophoresis with amperometric detection of curcumin in Chinese herbal medicine pretreated by solid-phase extraction. J Chromatogr A. 2002;962:117–25. doi: 10.1016/s0021-9673(02)00509-5. [DOI] [PubMed] [Google Scholar]
- 13.Inoue K, Hamasaki S, Yoshimura Y, Yamada N, Nakamura M, Ito Y, Nakazawa H. Validation of LC/electrospray-MS for the determination of major curcuminoids in foods. J Liq Chromatogr Relat Technol. 2003;26:53–62. [Google Scholar]
- 14.Tonnesen H, Karlsen J. High-performance liquid chromatography of curcumin and related compounds. J Chromatogr. 1983;259:367–71. [Google Scholar]
- 15.Smith R, Witowska B. Comparison of detectors for the determination of curcumin in turmeric by high performance liquid chromatography. Analyst. 1984;109:259–61. [Google Scholar]
- 16.Pan MH, Huang TM, Lin JK. Biotransformation of curcumin through reduction and glucuronidation in mice. Drug Metab Dispos. 1999;27:486–94. [PubMed] [Google Scholar]
- 17.Ravindran J, Prasad S, Aggarwal BB. Curcumin and cancer cells: How many ways can curry kill tumor cells selectively? AAPS J. 2009;11:495–510. doi: 10.1208/s12248-009-9128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anand P, Thomas SG, Kunnumakkara AB, Sundaram C, Harikumar KB, Sung B, et al. Biological activities of curcumin and its analogues (Congeners) made by man and Mother Nature. Biochem Pharmacol. 2008;76:1590–611. doi: 10.1016/j.bcp.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Dhillon N, Aggarwal BB, Newman RA, Wolff RA, Kunnumakkara AB, Abbruzzese JL, et al. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin Cancer Res. 2008;14:4491–9. doi: 10.1158/1078-0432.CCR-08-0024. [DOI] [PubMed] [Google Scholar]
- 20.Anand P, Sundaram C, Jhurani S, Kunnumakkara AB, Aggarwal BB. Curcumin and cancer: An old-age disease with an age-old solution. Cancer Lett. 2008;267:133–64. doi: 10.1016/j.canlet.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 21.Aggarwal BB, Harikumar KB. Potential therapeutic effects of curcumin, the antiinflammatory agent, against neurodegenerative, cardiovascular, pulmonary, metabolic, autoimmune and neoplastic diseases. Int J Biochem Cell Biol. 2009;41:40–59. doi: 10.1016/j.biocel.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulkarni SK, Dhir A. An overview of curcumin in neurological disorders. Indian J Pharm Sci. 2010;72:149–54. doi: 10.4103/0250-474X.65012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: Problems and promises. Mol Pharm. 2007;4:807–18. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- 24.Mulik R, Mahadik K, Paradkar A. Development of curcuminoids loaded poly (butyl) cyanoacrylate nanoparticles: Physicochemical characterization and stability study. Eur J Pharm Sci. 2009;37:395–404. doi: 10.1016/j.ejps.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Shaikh J, Ankola DD, Beniwal V, Singh D, Kumar MN. Nanoparticle encapsulation improves oral bioavailability of curcumin by at least 9-fold when compared to curcumin administered with piperine as absorption enhancer. Eur J Pharm Sci. 2009;37:223–30. doi: 10.1016/j.ejps.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 26.Narayanan NK, Nargi D, Randolph C, Narayanan BA. Liposome encapsulation of curcumin and resveratrol in combination reduces prostate cancer incidence in PTEN knockout mice. Int J Cancer. 2009;125:1–8. doi: 10.1002/ijc.24336. [DOI] [PubMed] [Google Scholar]
- 27.Yadav VR, Suresh S, Devi K, Yadav S. Novel formulation of solid lipid microparticles of curcumin for antiangiogenic and antiinflammatory activity for optimization of therapy of inflammatory bowel disease. J Pharm Pharmacol. 2009;61:311–21. doi: 10.1211/jpp/61.03.0005. [DOI] [PubMed] [Google Scholar]
- 28.Cui J, Yu B, Zhao Y, Zhu W, Li H, Lou H, et al. Enhancement of oral absorption of curcumin by self-microemulsifying drug delivery systems. Int J Pharm. 2009;371:148–55. doi: 10.1016/j.ijpharm.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 29.Chen C, Johnston TD, Jeon H, Gedaly R, McHugh PP, Burke TG, et al. An in vitro study of liposomal curcumin: Stability, toxicity and biological activity in human lymphocytes and Epstein-Barr virus-transformed human B-cells. Int J Pharm. 2009;366:133–9. doi: 10.1016/j.ijpharm.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Bisht S, Feldmann G, Soni S, Ravi R, Karikar C, Maitra A, et al. Polymeric nanoparticle-encapsulated curcumin (nanocurcumin): A novel strategy for human cancer therapy. J Nanobiotechnology. 2007;5:3. doi: 10.1186/1477-3155-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Braiteh FS, Kurzrock R. Liposome-encapsulated curcumin: in vitro and in vivo effects on proliferation, apoptosis, signalling, and angiogenesis. Cancer. 2005;104:1322–31. doi: 10.1002/cncr.21300. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Jiang Y, Wen J, Fan G, Wu Y, Zhang C. A rapid and simple HPLC method for the determination of curcumin in rat plasma: Assay development, validation and application to a pharmacokinetic study of curcumin liposome. Biomed Chromatogr. 2009;23:1201–7. doi: 10.1002/bmc.1244. [DOI] [PubMed] [Google Scholar]
- 33.Dandekar P, Patravale V. Development and validation of a stability-indicating LC method for curcumin. Chromatographia. 2009;69:871–7. [Google Scholar]
- 34.Ansari MJ, Ahmad S, Kohli K, Ali J, Khar RK. Stability-indicating HPTLC determination of curcumin in bulk drug and pharmaceutical formulations. J Pharm Biomed Anal. 2005;39:132–8. doi: 10.1016/j.jpba.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 35.ICH (Q2B) Note for Guidance on Validation of Analytical Procedures: Methodology. International Conference on Harmonization; Geneva, Switzerland: IFPMA. 1996. [Google Scholar]