Summary
The stringent response to amino acid starvation, whereby stable RNA synthesis is curtailed in favor of transcription of amino acid biosynthetic genes, is controlled by the alarmone ppGpp. To elucidate the extent of gene expression effected by ppGpp, we designed an experimental system based on starvation for isoleucine, which could be applied to both wildtype Escherichia coli and the multi-auxotrophic relA spoT mutant (ppGpp0). We used microarrays to profile the response to amino acid starvation in both strains. The wildtype response included induction of the general stress response, down regulation of genes involved in production of macromolecular structures, and comprehensive restructuring of metabolic gene expression, but not induction of amino acid biosynthesis genes en masse. This restructuring of metabolism was confirmed using kinetic Biolog assays. These responses were profoundly altered in the ppGpp0 strain. Furthermore, upon isoleucine starvation, the ppGpp0 strain exhibited a larger cell size and continued growth, ultimately producing 50% more biomass than the wildtype, despite producing a similar amount of protein. This mutant phenotype correlated with aberrant gene expression in diverse processes including DNA replication, cell division, and fatty acid and membrane biosynthesis. We present a model that expands and functionally integrates the ppGpp-mediated stringent response to include control of virtually all macromolecular synthesis and intermediary metabolism.
Introduction
When nutrients become limiting for growth, E. coli cells adjust their gene expression program from one that supports growth to one that allows for prolonged survival in stationary phase. In many bacteria, a key potentiator of this physiological switch is the accumulation of the alarmones guanosine 5′,3′ bispyrophosphate and guanosine pentaphosphate (ppGpp and pppGpp: collectively referred to here as ppGpp) (Cashel et al., 1996). When amino acids become limiting, uncharged tRNAs bind to the ribosomal A site, signaling ribosome-associated RelA to synthesize ppGpp (Wendrich et al., 2002). Aided by DksA, ppGpp binds in the secondary channel of RNA polymerase (RNAP) near the active site (Artsimovitch et al., 2004); the immediate consequence is the cessation of transcription of stable RNAs (ribosomal and transfer RNAs), termed the stringent response(Maaloe & Kjeldgaard, 1966, Cashel et al., 1996). ppGpp decreases the half-life of the open complex at most promoters tested thus far; the physiological result is the strong down regulation of promoters with intrinsically short half-lives, such as those of stable RNA genes (Barker et al., 2001b). Since expression of ribosomal protein genes is controlled by rRNA levels, the stringent response includes a large-scale down regulation of the translation apparatus (Paul et al., 2004). As transcription of the translation apparatus genes and stable RNAs can account for a large percentage (60-80%) of the transcription occurring in rapidly growing cells, the liberation of RNAP from these genes is thought to passively allow up regulation of diverse promoters activated at the onset of stationary phase (Barker et al., 2001b, Barker et al., 2001a). Also, ppGpp in concert with DksA has been shown to directly stimulate transcription from promoters of several amino acid biosynthesis genes (Paul et al., 2005). In support of the ‘passive mechanism’, we recently showed that a ΔrelA mutant is constrained in its ability to up regulate genes in diverse regulatory networks during carbon starvation (Traxler et al., 2006).
In addition to RelA, ppGpp is also produced by SpoT, apparently in response to diverse signals including carbon (Xiao et al., 1991), iron (Vinella et al., 2005), and fatty acid starvation (Battesti & Bouveret, 2006). The synthase activity of SpoT is not as robust as that of RelA, and SpoT also contains ppGpp hydrolase activity, which mediates ppGpp turnover and thus is important for determining the intracellular ppGpp concentration (Xiao et al., 1991). Mutants lacking relA and spoT are completely devoid of ppGpp (ppGpp0), a state that results in a pleiotropic phenotype (Xiao et al., 1991). Most notably, ppGpp0 strains exhibit a ‘relaxed’ phenotype, i.e., stable RNA synthesis continues after exhaustion of amino acids (Stent & Brenner, 1961). ppGpp0 strains are also auxotrophic for eleven amino acids, apparently because ppGpp is required for effective transcription of amino acid biosynthetic genes (Xiao et al., 1991). Additionally, relaxed strains exhibit a prolonged period of growth arrest after amino acid starvation has been relieved (Uzan & Danchin, 1978). Years of experimentation have linked ppGpp to a wide variety of physiological processes beyond translation and amino acid biosynthesis, including catabolite (de)repression (Johansson et al., 2000, Traxler et al., 2006), DNA synthesis (Hernandez & Bremer, 1993, Chiaramello & Zyskind, 1990), fatty acid metabolism (Heath et al., 1994, Eichel et al., 1999), general stress response (Gentry et al., 1993), surface organelle production (fimbriae and flagella) (Magnusson et al., 2007, Aberg et al., 2006), and virulence (Magnusson et al., 2005). Such wide-ranging regulation suggests ppGpp is a critical element of the response network that allows cells to adapt their physiology to their surroundings; however, the manner in which these processes are integrated within the stringent response remains unclear. Furthermore, the role of ppGpp in controlling intermediary metabolism beyond that of amino acid biosynthesis has not been experimentally defined.
To address these questions, we sought to examine the extent of the stringent response under conditions of amino acid starvation. To do so, we exploited a long-known metabolic anomaly characteristic of E. coli K12 strains, whereby starvation for isoleucine is caused by excess valine (Leavitt & Umbarger, 1962).
To determine the extent of regulation by ppGpp we obtained transcription profiles of the wildtype (WT) and ppGpp0 strains, using whole genome microarrays. In comparison to the WT, the strain lacking ppGpp was crippled in its ability to regulate genes involved in diverse areas of metabolism, including central metabolism, amino acid biosynthesis/degradation, and nucleotide biosynthesis. We also conducted a number of experiments that provide a context for interpreting these transcription profiles, including measurements of metabolites in the culture medium, changes in the metabolic proteome, viability assays, and measurements of total protein, RNA, and biomass. Based on these data, we present a model that expands and integrates our view of the metabolic and structural rearrangements that accompany the stringent response, which is at once more massive and finely tuned than previously appreciated.
Results
Experimental system for eliciting the stringent response to amino acid starvation
We were severely constrained in our choice of experimental systems because ppGpp0 strains are multiply auxotrophic and because the stringent response is thought to broadly impact amino acid biosynthesis (Cashel et al., 1996). To elicit the stringent response to amino acid starvation, many studies have utilized serine hydroxamate, which binds to and interferes with seryl-tRNA synthetases. While this strategy is excellent for inducing ppGpp accumulation, it falls short of modeling the concerted response to depletion of the intracellular amino acid pool because little or no new protein can be produced, post-treatment (Tosa & Pizer, 1971). Thus, after serine hydroxamate treatment, no reorganization of the proteome can occur.
Another widely used experimental system is based on the vulnerability of K-12 strains to valine toxicity (Leavitt & Umbarger, 1962). The first dedicated reaction in branched chain amino acid biosynthesis is catalyzed by acetohydroxy acid synthase (AHAS), which forms α-acetolactate from two molecules of pyruvate during the synthesis of valine or the formation of α-acetohydroxybutyrate from one molecule of pyruvate and one molecule of α-ketobutyrate during the synthesis of isoleucine, (for review, see (Umbarger, 1996). E. coli has three different AHAS enzymes: AHAS I (ilvBN) uses pyruvate exclusively for the production of valine, AHAS II (ilvGM) uses pyruvate and α-ketobutyrate for the production of isoleucine, and AHAS III (ilvIH) catalyzes both reactions but favors pyruvate and α-ketobutyrate as substrates. Both AHAS I and III are feedback inhibited by valine. K-12 strains of E. coli harbor a frame-shift mutation in the ilvG gene, which renders the AHAS II enzyme inactive. Thus, when isoleucine is limiting and valine is in excess, AHAS I and III are inhibited, resulting in an inability to biosynthesize isoleucine. While starvation for isoleucine can be induced by dosing cells with valine in minimal medium (i.e., isoleucine absent) (Leavitt & Umbarger, 1962), this strategy was not available to us because of the limitations imposed by the multiple amino acid auxotrophy associated with lack of ppGpp in E. coli MG1655 ΔrelA ΔspoT (ppGpp0).
To circumvent this problem, we retroverted the valine-dosing strategy by imposing isoleucine starvation in the presence of the other 19 amino acids, including valine, which we reasoned would inhibit isoleucine biosynthesis, as described above. We grew the cells in MOPS medium containing glucose (0.2%) plus all 20 amino acids (Wanner et al., 1977), except that isoleucine was provided at 60 μM instead of 400 μM, which allowed WT cells to reach an OD of 0.6-0.7 (Fig. 1A). At this OD, isoleucine was exhausted, but valine was still in excess (Fig. 1), and as expected, growth was arrested; this could be alleviated by re-addition of isoleucine (data not shown).
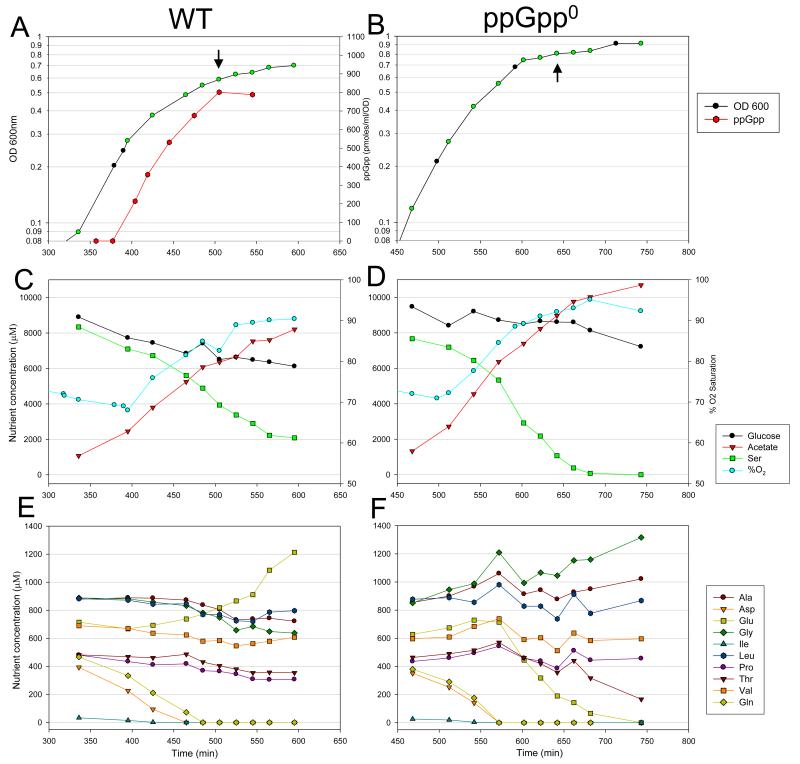
Fig. 1.
Growth curves and selected medium component concentrations for WT and ppGpp0 strains.
A. and B. Growth curves and ppGpp accumulation for WT and ppGpp0 strains, respectively, grown at 37° C in MOPS medium with glucose plus all 20 amino acids, with a limiting amount of isoleucine. ppGpp was undetectable in the mutant strain. Time points marked in green indicate times of sampling for medium component analysis presented in C-F. Black arrows indicate sampling times for microarray analysis. Note the higher OD reached by the ppGpp0 strain despite being grown in identical medium.
C. and D. Concentrations of carbon sources and acetate, and % O2 saturation for WT and ppGpp0 strains, respectively.
E. and F. Concentrations of selected amino acids. Concentrations of amino acids not shown did not change appreciably over the course of the experiments.
While we favored isoleucine starvation over serine hydroxamate treatment, this system is not without potential confounding factors. The growth arrest caused by excess valine has been attributed to metabolic consequences other than isoleucine starvation, namely α-ketobutyrate accumulation. This conclusion is based on valine inhibited cells grown on glucose minimal medium having a slightly above normal (144%) physiological intracellular level of isoleucine (Herring et al., 1995). Mathematical modeling predicted an accumulation of α-ketobutyrate (Yang et al., 2005), which is known to inhibit the glucose PTS transporter (Danchin et al., 1984), leading to the idea that valine toxicity should be attributed to carbon starvation-induced growth arrest (Yang et al., 2005). To test the possibility that glucose transport was inhibited in our experiments, we examined the array data, but could find no evidence for glucose starvation, i.e., catabolite derepression (discussed in detail below), as we previously observed for carbon starved cells (Chang et al., 2002, Traxler et al., 2006). Moreover, a microarray time course showed that the first genes induced in this experimental setup are those of the branched chain amino acid biosynthetic pathways (data not shown), implying that from a physiological stand point, the cells acutely sensed the depletion of isoleucine. A second potential complication was also considered: α-ketobutyrate accumulation, concomitant with high expression of the leucine biosynthetic pathway, leads to production of norleucine, a methionine analog (Bogosian et al., 1989). Norleucine can be incorporated into protein in place of methionine (Cohen & Munier, 1956). However, the incorporation of norleucine into protein was found to be completely prevented by supplementation with exogenous methionine (Bogosian et al., 1989). Since our medium contained exogenous methionine, we expect any negative effects of norleucine accumulation to be minimized. Thus, we interpret our results in the context of isoleucine starvation.
Growth and pattern of ppGpp accumulation in isoleucine starved cultures
To maximize reproducibility, cultures were grown in 1 L volumes in a fermenter under steady pH and O2 saturation levels. To examine the response to isoleucine starvation, isogenic E. coli MG1655 wild type (WT) and ppGpp0 strains were grown in isoleucine-limited medium and samples were taken in log phase and following growth arrest. The WT and ppGpp0 strains grew at similar rates in logarithmic phase. However, the ppGpp0 strain exhibited a prolonged lag phase before achieving robust growth; this accounts for the differences in the two growth curves (Fig 1). The response to limiting isoleucine was evident by an initial slowing of growth followed by growth arrest. Growth began to slow in the WT at OD of ~0.4 and growth arrested at OD of ~0.6. In the ppGpp0 strain growth began to slow at OD ~0.5 and arrested at OD ~0.8 ppGpp began to accumulate in the WT before growth slowed, and was first detectable at OD ~0.3. The ppGpp level increased over the next 100 minutes, leveling off in growth arrested cells (OD ~0.6 at about 500 min in Fig. 1). ppGpp was undetectable in the ΔrelA ΔspoT mutant. In our assays, the level of ppGpp that ultimately accumulated in the WT was ~800 pmoles/ml/OD, which equates to an intracellular concentration of ~0.9 mM. A similar intracellular concentration was observed after isoleucine starvation was provoked by addition of valine to cells growing in minimal media (VanBogelen et al., 1987) .
Altered flux through amino acid degradative and biosynthetic pathways in response to isoleucine starvation is ppGpp-dependent
We sought to characterize the pattern of amino acid utilization under the experimental conditions described above to allow for better interpretation of the high-throughput data described in subsequent sections. During the course of growth and entry into growth arrest, culture samples were harvested, filtered, and the filtrates were immediately frozen. Samples were then analyzed by capillary electrophoresis-mass spectrometry (CE-MS) to measure the concentrations of 19 amino acids at each time point (cysteine could not be measured). The results for selected amino acids are shown in Fig 1. Concentrations of the 8 amino acids not shown in Fig. 1 did not change significantly over the course of the experiment. At an OD of ~0.4 in both cultures, isoleucine levels dropped below detectable limits, leading to growth arrest of the WT at OD ~0.6 and in the ppGpp0 strain at OD ~0.8 (Fig. 1B). Starved WT cultures immediately resumed growth upon addition of isoleucine, accompanied by depletion of oxygen; these responses were significantly delayed in the ppGpp0 strain (the period of delay depended on the length of growth arrest; data not shown).
The next three amino acids consumed from the medium were aspartate, followed by glutamine, followed by serine. Serine was included in the medium at a much higher level (10 mM) than the other amino acids since it also serves as a carbon source that is co-metabolized with glucose (Wanner et al., 1977). In both strains, the usage of serine in the medium accelerated approximately 2-fold at the onset of growth arrest concomitant with a 2.8-fold decrease in the rate of glucose consumption in the WT (discussed below) (Fig. 1C-D). As the cells transitioned into growth arrest, we observed that the saturation of O2 in the medium increased by approximately 20%, implying that the cells consumed less oxygen (Fig. 1C-D). Acetate accumulated steadily over the entire time-course. Taken together, these results imply that amino acid starvation causes aerobic metabolism of glucose to be curtailed in favor of fermentation of serine to acetate via pyruvate.
We observed a large difference in the pattern of glutamate production/consumption between the WT and ppGpp0 strains (Fig. 1E-F). In the WT, the level of glutamate in the medium increased across the time series, a trend that accelerated noticeably at the onset of growth arrest. In contrast, the level of glutamate in the ppGpp0 culture decreased significantly over time, starting at the onset of growth arrest and dropping below detection by the conclusion of the experiment. Another observable trend was seen in the utilization of glycine, alanine, and leucine. In the WT culture, the levels of these three amino acids remained tightly associated, showing a modest decrease by the end of the experiment. In the ppGpp0 culture, the levels of glycine, alanine, and leucine diverged at the onset of growth arrest, with the glycine level increasing to a much higher level than in the WT culture.
Overall, these results suggest that the metabolic response to isoleucine starvation is complex and involves the rerouting of flux through multiple pathways. Most notably these adjustments entailed the apparent conversion of serine to acetate as the primary energy-generating process and diminished metabolism of glucose. Differences in the patterns of amino acid utilization/formation between the WT and ppGpp0 cultures suggest that the mutant strain was defective in its ability to restructure its metabolism. Specifically, the aberrant production of glycine by the ppGpp0 strain suggests heteroclitic metabolism of serine, the precursor of glycine (Stauffer & Brenchley, 1974). Finally, the depletion of glutamate observed in the ppGpp0 culture suggests a potential problem in maintaining the balance of glutamate and α-ketoglutarate, and thus has profound implications for the ability of the ppGpp0 strain to fulfill the many needs met by glutamate in redox homeostasis, as a metabolic precursor, and as an amine donor.
Overview of microarray datasets
To test the transcriptional response to isoleucine starvation, we extracted RNA from growth arrested WT and isogenic mutants (OD ~0.6 at 500 min for the WT and OD ~0.8 at 640 min for the ppGpp0 strain). In the WT, this corresponded to the time of maximum ppGpp accumulation (Fig. 1A). The control RNA was extracted from exponentially growing WT cells in identical medium replete with isoleucine. Since the stringent response is known to inhibit stable RNA synthesis, it is likely that the proportion of mRNA to stable RNA is different in the WT compared to the ppGpp0 strain. However, it is not possible to estimate these differences using the microarrays employed in this study because they do not contain probes for stable RNA transcripts. Moreover, the normalization strategy (RMA) used for the data processing offsets differences in total RNA and/or the relative proportion of mRNA and stable RNA. Hence, the data allow direct comparison of mRNA levels between strains and growth conditions, but do not compensate for gross differences in RNA content. The WT transcriptional response to isoleucine starvation was extensive, with 1024 genes differentially regulated >2-fold (log2 = 1). A list of these genes is shown in Supplemental table T1. Of the 532 genes that were induced >2-fold in the WT strain, about one-third are involved in metabolism (174 genes), 139 are involved in the RpoS-dependent general stress response, and >200 have unknown functions. The WT down regulated 492 genes, including many genes associated with the translation apparatus (>40 ribosomal protein genes and 19 accessory translation genes), a hallmark of the stringent response.
Global comparison of the WT and isogenic ppGpp0 mutant transcriptional responses to isoleucine starvation revealed profound differences between them (Fig. 2A). In fact, when the transcription profiles of the two strains were compared directly, 1427 genes (>30% of the genome) showed expression levels that deviated 2-fold or more in the ppGpp0 strain. A list of these genes is available in Supplemental table T2. While both the WT and ppGpp0 strain induced over 500 genes in response to isoleucine starvation, only 133 genes were commonly induced in both strains (Fig. 2B). Moreover, both strains down regulated over 450 genes >2-fold, but only 198 of these genes were down regulated in both. The comprehensive down regulation of translation apparatus genes observed in the WT was essentially absent in the ppGpp0 strain, and induction of the RpoS-dependent general stress response was severely diminished (Fig. 2C). Overall, these results suggest that the response to isoleucine starvation is fundamentally altered in the ppGpp0 strain.
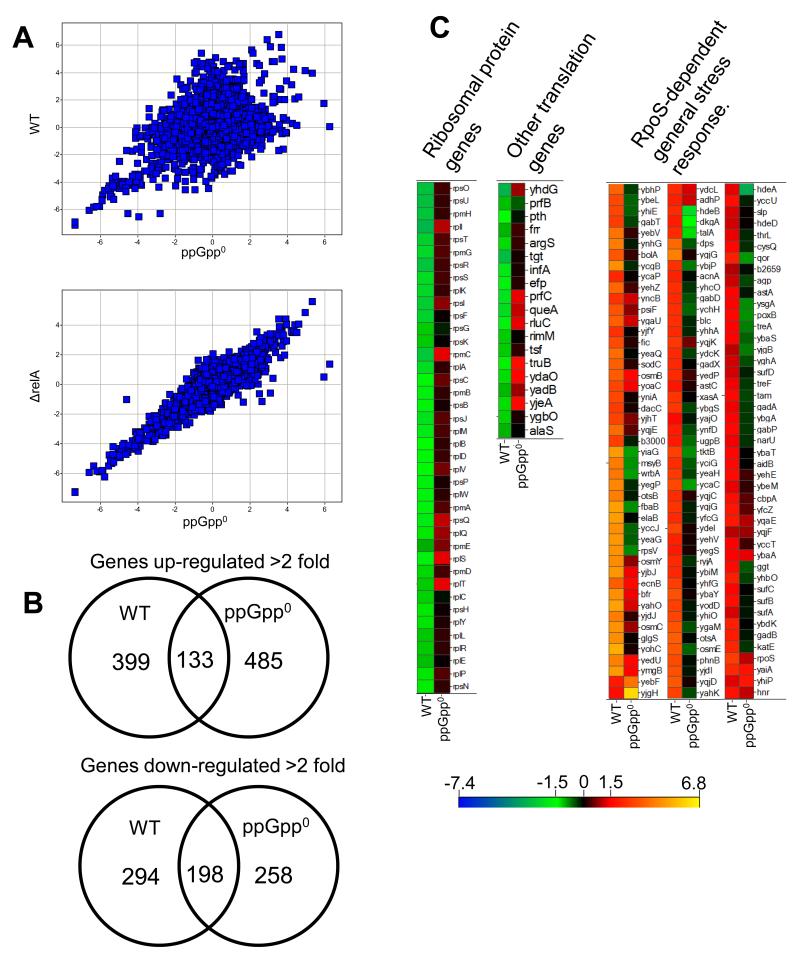
Fig. 2.
Comparisons of microarray results for WT and ppGpp0 strains. Control RNA was extracted from log phase WT cells at an OD of ~0.4 grown in medium with replete isoleucine. Test RNA was harvested from WT, ΔrelA and ppGpp0 strains ~40 min after onset of growth arrest. All test array data were normalized to control array data before comparative analysis of strain-specific responses to isoleucine starvation. Array data presented in all figures are log2 expression ratios (test:control).
A. Upper plot: comparison of isoleucine-starved WT and ppGpp0 strain transcriptome profiles. Lower plot: comparison of isoleucine-starved ΔrelA and ppGpp0 strain transcriptome profiles.
B. Upper diagram: Venn diagram comparing up regulated genes between the WT and ppGpp0 strain. Lower diagram: Venn diagram comparing down regulated genes between the WT and ppGpp0 strain.
C. Heat maps of log2 expression ratios for the WT and ppGpp0 strain for ribosomal protein genes, other genes involved in translation, and the general stress response. All genes shown in C differed in their expression >2-fold between the WT and ppGpp0 strain. Ribosomal protein and translation genes shown were all down regulated >2-fold in the WT, while all the general stress response genes shown were induced >2-fold in the WT.
We also obtained a transcription profile of an isogenic ΔrelA strain starved for isoleucine and found that its response was strikingly similar to the ppGpp0 strain, indicating a minimal role for SpoT in the stringent response to isoleucine starvation (Fig 2A lower panel). Genes which were expressed differently in the ΔrelA and ppGpp0 strain are listed in Supplementary table T3. Based on the similarity of the transcription profiles of the ΔrelA and ppGpp0 strains, it is reasonable to assume that the differences observed between the WT and ppGpp0 strain qualitatively hold true also for the ΔrelA strain. Thus, we focused our analysis on the ppGpp0 strain, so the results could be interpreted in the complete absence of ppGpp.
The WT response to isoleucine starvation involves regulation of diverse metabolic pathways at the transcriptional level
To interpret microarray data in a metabolic context, log2 gene expression ratios were overlaid onto metabolic maps (Figs. 3 and 4). This analysis shows a large number of genes in multiple pathways were differentially regulated in response to isoleucine starvation. We interpret these data with the caveat that the relationship between simple induction or repression of a pathway at the transcriptional level does not necessarily reflect the level of flux or active enzymes in these pathways. However, general correlations between gene expression and metabolic activity have been observed (Oh et al., 2002).
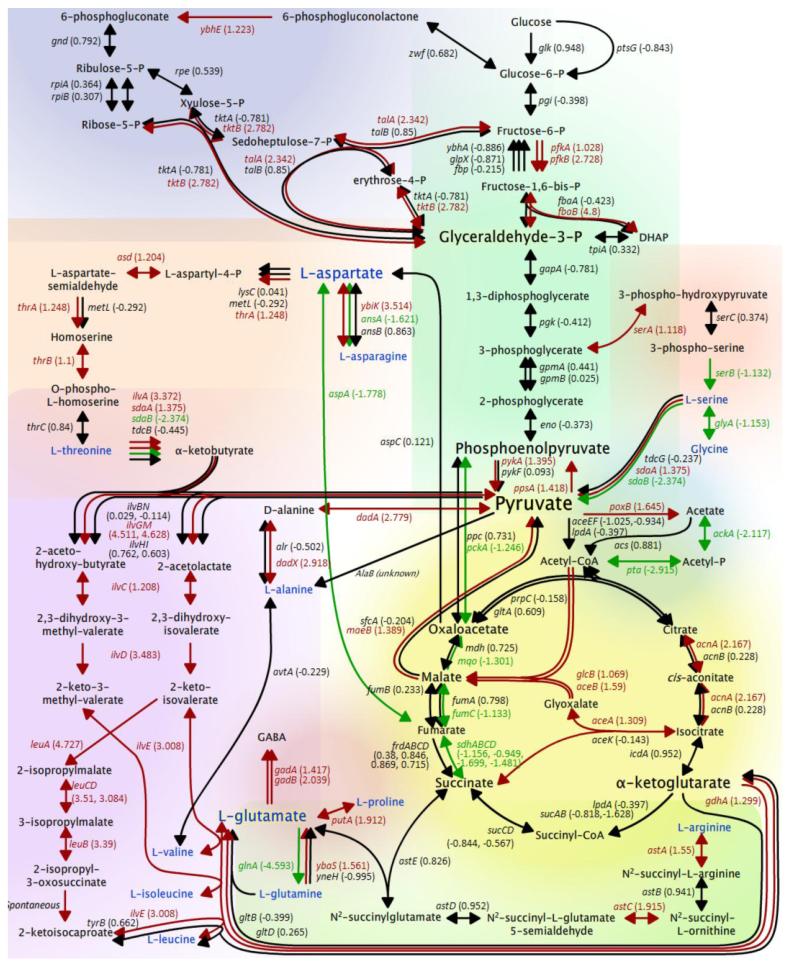
Fig. 3.
WT transcriptome data overlaid on selected metabolic pathways. Genes up regulated >2-fold are shown in red, while genes down regulated >2-fold are shown in green. Genes whose expression did not change >2-fold are shown in black. Where multiple gene products are required for a single conversion, the corresponding arrow is colored according to the average expression value of the corresponding genes. Amino acids are in blue font. Background colors are intended to delineate various pathways as follows: glycolysis: green (upper center), pentose phosphate pathway: dark blue (upper left), TCA cycle: yellow (lower right), threonine biosynthesis: light orange (middle left), branched chain amino acid biosynthesis: purple (lower left), serine metabolism: dark orange (middle right), acetate metabolism: light blue (middle right), glutamate biosynthesis: red (lower center), arginine degradation: light green (bottom right).
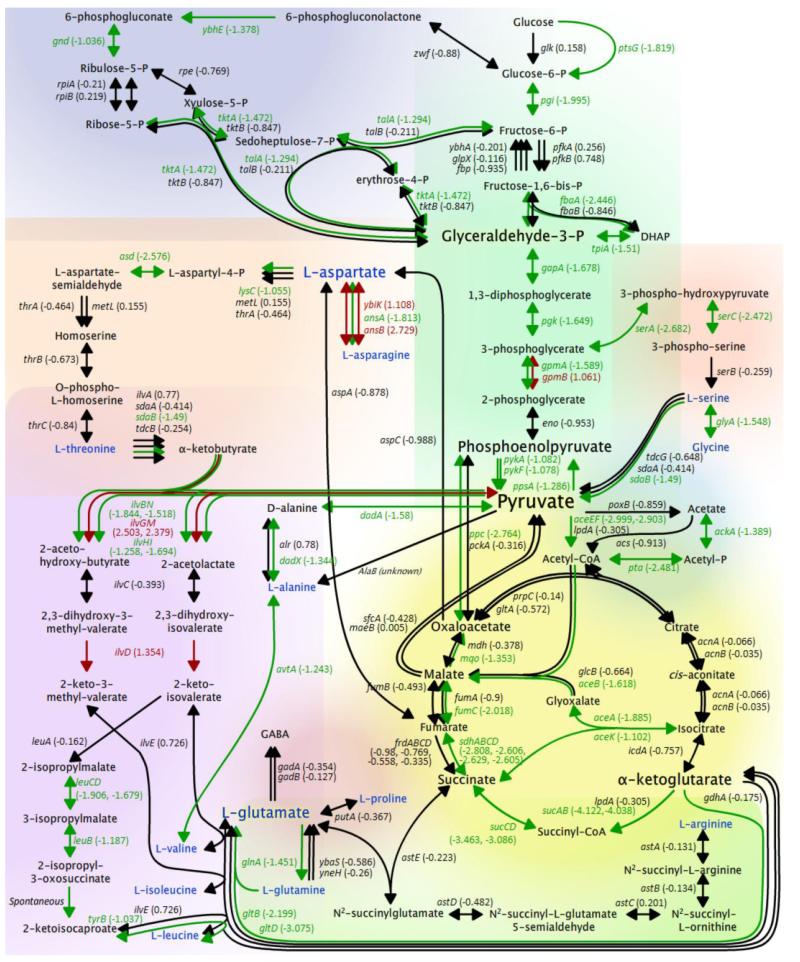
Fig. 4.
ppGpp0 strain transcriptome data overlaid on selected metabolic pathways. Color coding is identical to that for Fig. 3.
The WT induced genes in all branches of central metabolism (Fig. 3), including the pentose phosphate pathway, glycolysis, TCA cycle, and the glyoxylate shunt. Within the pentose phosphate pathway three genes were induced, including two in the non-oxidative branch, tktB and talA, which are known members of the RpoS regulon. Among glycolytic genes, three were induced, including two whose products catalyze the formation of metabolic intermediates known to exert feedback control of glycolytic flux (fbaB and pykA). Many genes encoding enzymes involved in pyruvate metabolism were induced in the WT, suggesting that pyruvate is a critical nexus in the metabolic adjustment to isoleucine starvation. sdaA, which encodes the major serine deaminase, was induced 2.6-fold. This induction correlates well with increased serine uptake from the medium (Fig 1). When considered together, these observations are consistent with increased flux from glycolytic intermediates and serine to pyruvate. Expression of genes involved in the TCA cycle was complex with the induction of acnA, genes of the glyoxylate shunt, and the gene encoding malate synthase G, maeB and the down regulation of genes involved in the conversion of succinate to oxaloacetate. Together these alterations in gene expression suggest that carbon entering the TCA cycle from acetate or β-oxidation of fatty acids may be channeled to the production of pyruvate, as well as α-ketoglutarate, the precursor of glutamate, which accumulated in the medium under these conditions (Fig. 1).
Isoleucine starvation triggered induction of genes directly involved in the branched chain amino acid pathways as well as genes in pathways which generate precursors for branched chain amino acid biosynthesis. E. coli synthesizes isoleucine from two precursor metabolites: pyruvate and oxaloacetate, (for review, see (Patte, 1996)). Oxaloacetate from the TCA cycle is converted in one step to aspartate. Aspartate can also be made from asparagine in a single reaction. Aspartate is converted to threonine in five steps (Fig. 3). Threonine is then deaminated to form α-ketobutyrate, which is a substrate for AHAS II and III. Thus, from a physiological standpoint, the cell can convert asparagine→aspartate→threonine→isoleucine in 11 steps, 10 of which were up regulated under the experimental conditions examined here. IlvGM (AHAS II) was the only one of the three AHAS enzymes whose genes were induced by isoleucine starvation in the WT (ilvG 22-fold and ilvM 25-fold). While not enzymatically effective, the up regulation of ilvG and ilvM likely represents the cells’ attempt to induce valine insensitive AHAS II. In addition to ilvG and ilvM, all of the genes for enzymes involved in synthesis of the branched chain amino acids valine and isoleucine were strongly induced: ilvC (2.3-fold), ilvE (8.0-fold), and ilvD (11-fold). The genes of the leuABCD operon, which are responsible for leucine biosynthesis, were up regulated (8.5- to 26-fold). Collectively, this strong, comprehensive induction constitutes a direct, albeit impotent, response to isoleucine starvation.
The ppGpp0 metabolic response to isoleucine starvation deviates from WT at the transcriptional level
The transcriptional differences observed in the profiles of the WT and ppGpp0 strains in response to isoleucine starvation are far-reaching. It is long known that ppGpp is required for stringent induction of genes involved in amino acid biosynthesis (Cashel et al., 1996). This trend is observed in the data presented here, wherein the ppGpp0 strain failed to induce genes associated with biosynthesis of the branched chain amino acids, as well as threonine and glutamate (Fig. 4). The transcription profiles show that induction of multiple genes involved in the metabolism of arginine, alanine, serine, and glutamate is contingent upon ppGpp. Most notably, the data presented here also establish that the expression of many metabolic genes beyond the scope of amino acid biosynthesis is also ppGpp-dependent. These include genes of glycolysis, the pentose phosphate pathway, the TCA cycle and the glyoxylate shunt. The strong repression of numerous central metabolism genes observed in the ppGpp0 strain implies a comprehensive down-shift in metabolic potential. Diminished carbon flux through glycolysis would be expected to impact production of precursor metabolites, and ultimately, the flux of carbon into other central metabolic pathways. Additionally, normal regulation of genes involved in pyruvate metabolism, a metabolic focal point, also required ppGpp. While the regulation of many of the metabolic genes discussed here may be indirectly regulated by ppGpp, taken together these results suggest that ppGpp plays a larger role in regulating intermediary metabolism than previously recognized.
Biolog analysis shows diversification of carbon source utilization in response to isoleucine limitation
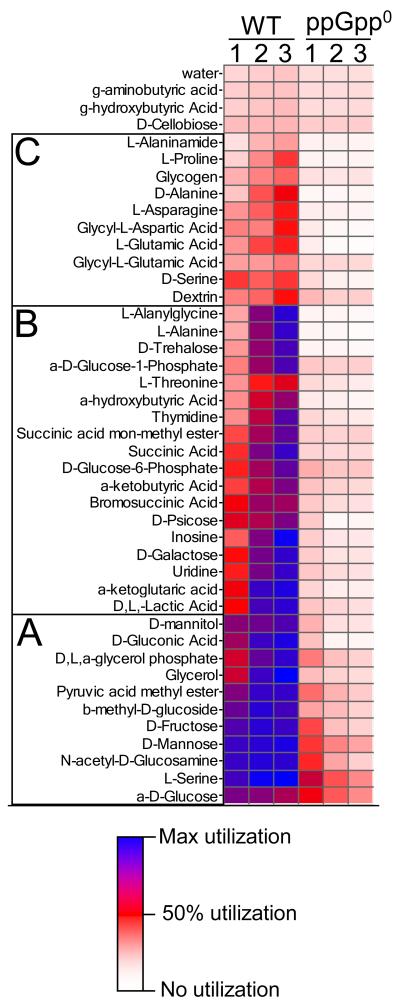
In light of the metabolic rearrangements shown in the transcription profiles, we sought a strategy to measure the response to isoleucine starvation with respect to changes in the overall metabolic capacity of the cells. To do this, we harvested cells for Biolog GN2 microplate analysis from the isoleucine-starved cultures and immediately added chloramphenicol to inhibit protein synthesis and preserve their metabolic capacity, as described elsewhere (Ihssen & Egli, 2005). Thus, the pattern of carbon sources utilized represents a metabolic snapshot of the enzymes present in the cells at the time of sampling. It should also be noted that because the cells are washed and incubated in basal, minimal medium, the allosteric constraints applied by the components of the growth medium were relieved, theoretically allowing for an uninhibited display of metabolic capacity. Cells where harvested for Biolog analysis at times corresponding to active growth (OD 0.3), the onset of growth arrest, and 1.5 hours after the onset of growth arrest (Fig 5). The cells were incubated in Biolog plates for 24 hours in an Omnilog system, and the amount of reduced tetrazolium violet dye in each well was quantified every 15 min. The amount of dye reduced corresponds to the extent of carbon source oxidized. Dye reduction was plotted vs. time, and the area under each resulting kinetic curve was used as an overall measure of the cells’ ability to utilize each carbon source.
Fig. 5.
Cluster analysis of carbon sources utilized in kinetic Biolog assays for WT and ppGpp0 strains. White represents negligible utilization, dark blue represents maximal utilization, and bright red represents half-maximal utilization, as shown by key. Units are arbitrary. Column 1 contains data for both strains at OD ~0.3, during rapid growth before isoleucine starvation. Column 2 contains data for cells harvested at the onset of isoleucine starvation. Column 3 contains data for cells harvested 1.5 hours into growth arrest. These sampling times correspond to time points T2, T3 and T5 in Fig 6A and 6B. Clusters A, B, and C are described in the text.
Hierarchical cluster analysis of the WT Biolog data indicated that the number of carbon sources used increased as the cells progressed into growth arrest. Distinct groups of carbon sources were utilized at each of the three time points. The first group of carbon sources was utilized strongly at all time points and consisted mainly of carbohydrates that are assimilated directly into glycolysis. These included mannitol, gluconate, glycerol, fructose, mannose, N-acetyl-glucosamine, and glucose. Serine was the only amino acid that fell into this group. This is not surprising given the concomitant consumption of serine and glucose observed in Fig. 1C. The next cluster of carbon sources represents those that were utilized in the second and third time points, but not the first. Compounds in this category were diverse and included TCA cycle intermediates (α-ketoglutarate, succinate, and derivatives thereof), nucleotides (uridine, inosine, and thymidine), and others (L-alanine, lactate, galactose, trehalose, psicose, etc.) Several intermediates of branched chain amino acid biosynthesis (L-threonine, α-ketobutyrate, α-hydroxybutyrate) were also utilized at the second time point, reflecting the specific induction of this pathway in response to isoleucine starvation. The third cluster of substrates, which was only utilized after 1.5 hours of growth arrest, was comprised of mainly amino acids and their derivatives including L-alaninamide, L-proline, D-alanine, L-asparagine, Glycyl-L-asparate, L-glutamate, Glycyl-L-glutamate, D-serine and also dextrin and glycogen. The induction of pathways indicated by these Biolog assays correlated well with the pattern of metabolic gene induction noted in the transcriptome profiles.
We considered whether the pattern of carbon source utilization in the WT reflected a foraging strategy, the induction of pathways to re-route internal flux, or both. The expanded metabolic capacity of the WT did not resemble Biolog results from carbon starved cells described elsewhere (Ihssen & Egli, 2005). Nor did we observe in our microarray experiments expression patterns of induced transporter genes consistent with known patterns of carbon or nitrogen foraging (Chang et al., 2002, Gyaneshwar et al., 2005, Liu et al., 2005). The transporter expression pattern was not directly predictive of carbon sources readily utilized in the Biolog assays (data not shown). For these reasons, we think the WT Biolog assay results probably do not represent a foraging response alone, but are also indicative of a re-routing of intracellular flux through newly induced pathways. Thus, we interpret the expansion of the WT metabolic capacity as evidence for a large-scale reorientation of metabolism from anabolism and macromolecular synthesis to reassimilation of carbon back into central metabolism and amino acid biosynthesis pathways. The observed metabolic rearrangement is a ppGpp-dependent process, as described below.
The ppGpp0 strain was radically impacted, by comparison to the WT, in substrate utilization (Fig. 5). The only substrates readily metabolized by the ppGpp0 strain were a subset of those found in the first cluster of carbon sources used by the WT, namely, L-serine, glucose, and a few other substrates that are directly assimilated into glycolysis. Furthermore, the number of carbon substrates utilized by the ppGpp0 strain did not increase in response to growth arrest, rather, the ability of the cells to metabolize those substrates that were used during active growth actually declined. This result suggests that the metabolic capacity of the ppGpp0 strain diminished in response to isoleucine starvation. The extreme nature of the ppGpp0 Biolog phenotype prompted us to consider whether or not these results were artifactual. However, we think that the Biolog phenotype observed here is accurate because the transcriptome of the ppGpp0 strain is consistent with a large-scale metabolic shut-down and the ΔrelA strain had a nearly identical transcription profile and Biolog phenotype (data not shown). These data demonstrate that the transcriptional impairment observed in the ppGpp0 strain leads to compromised ability to expand metabolic potential at the proteomic level.
The ppGpp0 cells are viable and enlarged during isoleucine starvation
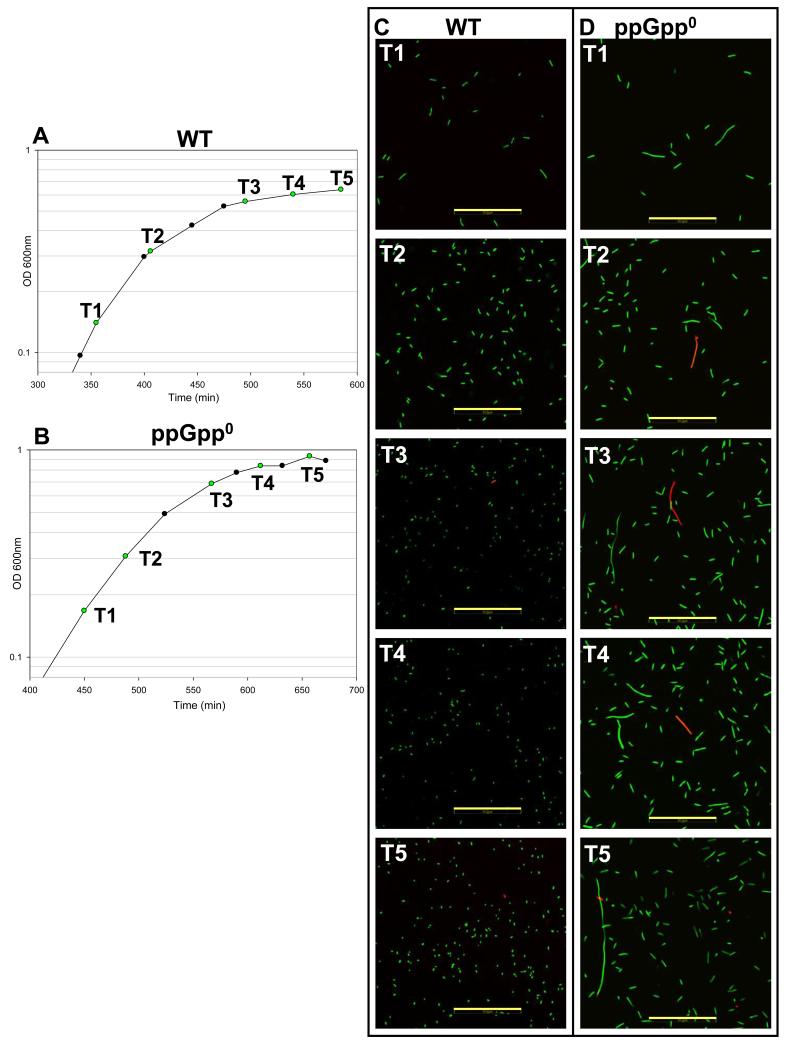
One possible explanation for the decreased metabolic activity exhibited by the ppGpp0 strain in response to isoleucine starvation is simply that the cells had died. Plate counts of isoleucine starved ppGpp0 strain indicated a ~1 log lower number of colony forming units across the entire time course (even during rapid growth) by comparison to the WT (data not shown). A several-fold lower plating efficiency (Xiao et al., 1991) and filamentous morphology (Xiao et al., 1991, Magnusson et al., 2007) have been demonstrated previously for strains lacking ppGpp. To directly check whether or not the ppGpp0 strain suffered a decrease in viability due to amino acid starvation, we stained culture samples with a live-dead stain and observed them using confocal microscopy (Fig. 6). This technique stains cells with compromised membrane integrity red (via propidium iodide) while intact (viable) cells are stained green (SYTO-9) (Stocks, 2004). Samples were checked across the entire time-course. WT cells were large and rod-shaped during rapid growth and became coccoid as growth ceased (Fig. 6D). No increase in non-viable cells was observed during isoleucine starvation, with non-viable cells making up a negligible portion of the population. The ppGpp0 cells were observed to be as long as or longer than the WT during rapid growth, with occasional filamentation (Fig. 6E). There was no decline in viability of the ppGpp0 culture upon isoleucine starvation (< 1% die-off). However, we observed that the ppGpp0 cells remained large and rod shaped, even 1.5 hours after growth had stopped. Stationary phase ppGpp0 cells routinely ranged from 4- to 10-fold longer than WT stationary phase cells and in the most extreme cases, filaments up to ~200 μm long were observed (data not shown). These results point to a larger role for ppGpp in modulating cell division, as has been suggested (Schreiber et al., 1995, Vinella & D’Ari, 1995).
Fig. 6.
Growth curves and viability staining of WT and ppGpp0 strains.
A. Growth curve of WT for viability staining. Time points where viability was tested are marked in green and labeled T1-T5.
B. Growth curve of ppGpp0 strain for viability staining. Labels are as in (A).
C. Micrographs of differentially stained WT cells harvested at time points T1-T5 as labeled in (A). Cells stained green are viable, while cells stained red have compromised membranes (non-viable). Gold scale bar = 50 μm.
D. Micrographs of differentially stained ppGpp0 cells harvested at time points T1-T5 as labeled in (B). Staining and scale bars are identical to (C).
The ppGpp0 strain has altered macromolecular composition
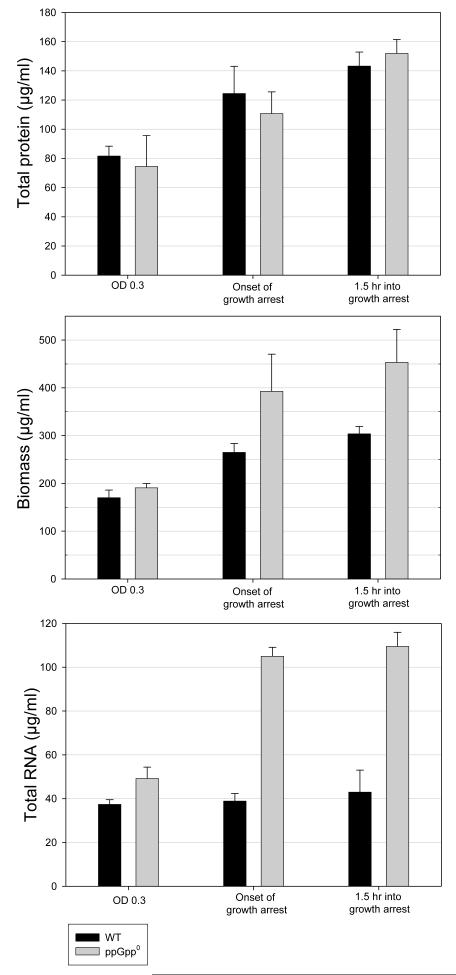
We also observed that the WT isoleucine-limited cultures routinely reached an OD of ~0.7, whereas the ppGpp0 strain reproducibly reached a higher OD of ~0.9 (Fig. 6A vs.6B). Theoretically, the total amount of protein produced by both cultures is dictated by the amount of isoleucine included in the growth medium. However, precursors of other major cell components that contribute to biomass might still be formed from glucose, serine, etc., which were not limiting. Keeping in mind these possibilities, we measured total protein, biomass, and RNA production for both the WT and ppGpp0 strain (Fig. 7). Both strains produced a comparable amount of protein (~150 μg/ml culture by 1.5 hours into stationary phase). In contrast, the ppGpp0 strain produced an average of ~50% more biomass than the WT under identical conditions. From these findings, we conclude that ppGpp is required to maintain a normal protein:biomass ratio, and that during times of amino acid starvation, ppGpp plays a critical role in keeping the production of macromolecular components in line with the translational capacity of the cell. Since the defining phenotype of relaxed strains is continued stable RNA synthesis in times of starvation, we determined the RNA content of the WT and ppGpp0 strains (Fig. 7). The WT RNA level did not increase significantly after an OD of 0.3, which correlates with the onset of ppGpp accumulation. As expected, the ppGpp0 strain did not curtail RNA synthesis, ultimately producing ~2.5-fold more RNA compared to the WT 1.5 hours after growth arrest. This increase in RNA accounts for ~45% of the mutant’s extra biomass. Since only about one-half of the excess biomass produced by the ppGpp0 strain was RNA, we conclude that ppGpp accumulation in the WT also limits production of other macromolecular cell components, i.e., cell membranes, cell wall, and DNA.
Fig. 7.
Total protein, biomass, and RNA produced by WT (black bars) and ppGpp0 strains (grey bars) during isoleucine starvation. The first time point (rapid growth) corresponds to T2 in (Fig 6A and 6B). Onset of growth arrest corresponds to (T3) in (Fig 6A and 6B). Final time point (1.5 hrs into growth arrest) corresponds to (T5) in (Fig 6A and 6B). Error bars indicate standard deviations. Both strains produce a similar amount of protein, however, the ppGpp0 strain produces 50% more biomass and >150% more RNA than the WT.
The ppGpp0 strain shows deviations in gene expression for major physiological processes
Prompted by observations that cells lacking ppGpp are much larger than WT stationary phase cells, and that this likely contributes to a larger biomass yield, we interrogated the microarray data to look for abnormal expression of genes that could account for this phenotype. Accordingly, we considered genes in various functional categories whose difference in expression between the WT and the ppGpp0 strain was ≥2-fold. These differences are summarized in Table 1. Heat maps, comparative expression values, and gene functions are available for these functional groups in supplementary Fig 1 with an accompanying note.
Table 1.
Number of genes in various functional groups differentially expressed in relA spoT- strain
| Functional group | Total number differentially expressed |
Genes expressed >2 fold higher |
Genes expressed >2 fold lower |
|---|---|---|---|
| Cell Division | 15 | 9 | 6 |
| DNA replication | 16 | 15 | 1 |
| DNA repair | 23 | 20 | 3 |
| Nucleotide Biosynthesis | 27 | 24 | 3 |
| Nucleotide Catabolism | 6 | 0 | 6 |
| Nucleotide Salvage | 6 | 6 | 0 |
| Fatty acid/Phospholipid Biosynthesis | 17 | 12 | 5 |
| Fatty acid B-oxidation | 6 | 1 | 5 |
| Peptidoglycan Biosynthesis | 13 | 9 | 4 |
| LPS/Outer membrane Biosynthesis | 24 | 23 | 1 |
| Glycogen metabolism | 7 | 0 | 7 |
In general, the data presented in Table 1 show a trend in which genes involved in macromolecular synthesis/biomass production were consistently expressed at higher levels in the ppGpp0 strain than the WT. This included some 92 genes involved in cell division, DNA replication, and the biosynthesis of nucleotides, fatty acids, cell wall, and LPS/outer membrane. Interestingly, the WT strain did not exhibit increased expression of genes associated with the SOS response to DNA damage, however, 20 genes involved in DNA repair were expressed higher in the ppGpp0 strain. Taken together with the higher expression of genes involved in DNA replication, this result suggests that chromosome replication continued abnormally in the ppGpp0 strain, ultimately resulting in DNA damage. Another trend evident in Table 1 is that whereas the WT induced genes involved in catabolism of macromolecular precursors, the ppGpp0 strain did not. This included the lower expression in ppGpp0 strain of 11 genes involved in nucleotide catabolism and fatty acid β-oxidation. The ppGpp0 strain also expressed six genes more highly that are involved in the salvage of endogenous nucleotide precursors for the production of new nucleotides. This trend suggests that the ppGpp0 strain actively attempted to salvage nucleotides in keeping with continued synthesis of nucleic acid. Finally, the ppGpp0 strain failed to induce genes involved in glycogen metabolism as observed in the WT, suggesting that glycogen probably does not contribute to the higher biomass produced by the ppGpp0 strain.
Discussion
Summary of results
We sought to examine the extent of the stringent response to amino acid starvation. To do so, we used isoleucine starvation as a model system. Global transcriptome profiling showed that isoleucine starvation of the WT resulted in changes in expression of genes in many metabolic pathways, curtailed expression of genes involved in macromolecular synthesis, and initiated the general stress response. In stark contrast, the ppGpp0 strain failed to make these changes. To further examine the transcriptional response to isoleucine starvation, snapshots of the metabolic capacities of the WT and ppGpp0 mutant were determined using Biolog GN2 microplates in a protocol that prevented changes to the functional metabolic proteome. The results showed that the WT greatly expanded its repertoire of active metabolic pathways, while the ppGpp0 strain failed to diversify its metabolic capacity and showed diminished ability to utilize pathways that were active before the onset of isoleucine starvation.
Noting that the ppGpp0 strain showed greatly diminished metabolic activity, we also checked for a possible decrease in viability as a result of amino acid starvation through differential staining/confocal microscopy. Although we noted no decrease in cell viability based on membrane integrity, we did observe that ppGpp0 mutant cells were considerably longer than WT isoleucine-starved cells at all time points and ppGpp0 cultures routinely reached a higher density. We found that although the two strains produced similar amounts of protein, the ppGpp0 strain continued to grow unchecked, producing 50% more biomass than the WT. Nearly one-half of this extra biomass was composed of RNA. Furthermore, we found that this mutant phenotype correlates with aberrant gene expression in diverse cellular processes including cell division, DNA replication, and nucleotide, fatty acid, cell wall, and LPS/outer membrane biosynthesis. The comprehensive deficiencies of the ppGpp0 strain, as evidenced by the global measurements presented here, imply that ppGpp is the pivotal signal required to successfully develop virtually all physiological responses to amino acid starvation.
A recent study described the transcriptional changes resulting from serine hydroxamate treatment (Durfee et al., 2008). This report compared the WT transcriptional response to that of a relAΔ251 strain. Though the strains and conditions differ from ours, several overlapping trends are evident. These include down regulation of genes involved in translation and induction of the RpoS-dependent general stress response. However, we note that in these cases the response to isoleucine starvation was more robust, with >40 ribosomal protein genes down regulated compared to only 5 genes down regulated in response to serine hydroxamate treatment. Also, >130 RpoS regulon members were induced in isoleucine starved cells compared to ~20 RpoS regulon members induced by serine hydroxamate. The latter difference may be attributable to inhibited translation caused by serine hydroxamate and therefore decreased accumulation of RpoS. Global defects in regulation were observed for the relAΔ251 strain in response to serine hydroxamate treatment, but the defects were far less comprehensive than those observed here for the ppGpp0 and ΔrelA strains starved for isoleucine. Differences in the transcriptomes of isoleucine starved ppGpp0 cultures and the serine hydroxamate treated relAΔ251 strain likely result from the residual ppGpp present as result of having an intact spoT allele in the latter strain, as well as physiological differences between isoleucine starvation and serine hydroxamate induced translation inhibition.
Metabolic restructuring during the stringent response to isoleucine starvation
Often the stringent response to amino acid starvation is thought of as a general reorganization of transcription involving down regulation of stable RNA synthesis and large-scale induction of amino acid biosynthetic operons. While the basic elements of this paradigm are clearly correct, the data presented in this report suggest that the response to starvation for a given amino acid leads to a more complex pattern of transcription than previously thought, especially with respect to metabolic genes. Instead of a general stimulation of amino acid biosynthetic genes, we observed induction of a range of amino acid biosynthetic and catabolic pathways, which, given the composition of the growth medium, would best allow the cell to route metabolic flux into formation of the limiting amino acid. The transcriptome data obtained for the mutant lacking ppGpp showed radically impacted ability to appropriately regulate expression of genes involved in almost all areas of metabolism, from central metabolism to disparate biosynthetic pathways. We therefore suggest that the transcriptional program initiated by ppGpp accumulation is a larger framework within which the pathways specific for the limiting amino acid are readily induced, accompanied by changes in expression of central metabolic genes as necessary to maximize the production of the required precursor metabolites. While it remains to be tested whether alternative metabolic rearrangements occur when cultures are starved for other amino acids, we envision the stringent response to be a global response to nutritional stress that halts growth processes and makes those resources available for the cell to specifically remediate the offending stress, as described in the model outlined below.
The temporal unfolding of a complex global response likely requires continued feedback between the changing transcriptome and the resulting proteome for its appropriate development. Under the conditions employed here, since isoleucine is not replenished once it is depleted, translation which occurs after the exhaustion of exogenous isoleucine is likely made possible through the recycling of amino acids liberated from turnover of existing cellular proteins. Indeed, significant reorganization of the proteome was found to occur in the WT after the exhaustion of isoleucine, as evidenced by the cells’ changing metabolic capacities measured by kinetic Biolog assays. This did not happen in the ppGpp0 strain. In keeping with the constrained expression of metabolic genes observed in our array experiments, the ppGpp0 strain displayed no ability to diversify its repertoire of readily metabolized carbon sources in our Biolog assays, further implicating ppGpp in the large-scale restructuring of metabolism in response to amino acid starvation.
Functional integration of the stringent response
The array data presented here must be considered in the larger context of metabolic and structural trends evident in stationary phase cells. The medium used in our experiments contains both glucose and serine as carbon sources as well as all amino acids necessary for translation. During growth under these conditions, glucose may be used primarily for production of precursor metabolites that originate in the pentose phosphate pathway and glycolysis, i.e. nucleotides and membrane/cell wall components, while serine, in addition to being integrated into protein and providing one-carbon units for nucleotide biosynthesis, might be used primarily to drive energy generation via acetate overflow metabolism. Together, nucleic acids and membrane/cell wall components comprise about 38% of the dry weight of growing cells, while protein makes up about 50% (Neidhardt et al., 1990). Upon isoleucine starvation, the WT efficiently stops growing, shuts down ribosome synthesis, and assumes a coccoid morphology. In addition, the microarray data presented here and elsewhere (Chang et al., 2002) show a concomitant inhibition of DNA replication, and macromolecule biosynthesis, i.e., membrane/cell wall components. The starved cells switch from a primarily anabolic mode devoted to generating building blocks for biomass production, to a catabolic mode which re-assimilates unused nucleotides and fatty acids back into central metabolism (as evidenced by gene expression and Biolog assays). Accordingly, glucose consumption slows as demand for these components is reduced, while serine consumption accelerates to allow for continued energy production via fermentation to acetate. The observed changes in the expression of central metabolic genes may both reflect and effect this metabolic reorientation. Moreover, virtually all of these metabolic changes require ppGpp for their manifestation.
The data presented in this report substantiate a wide range of observations made over many years implicating ppGpp in diverse processes including peptidoglycan synthesis (Ishiguro & Ramey, 1978), cell division (Schreiber et al., 1995, Xiao et al., 1991), DNA replication (Hernandez & Bremer, 1993, Wang et al., 2007), fatty acid and phospholipid biosynthesis (Heath et al., 1994, Podkovyrov & Larson, 1996, Taguchi et al., 1980a), nucleotide biosynthesis (Turnbough, 1983), and glycogen metabolism (Taguchi et al., 1980b). In most cases, the connection of one of these processes to the stringent response was established through the activity of a given enzyme (peptidoglycan and DNA replication), synthesis of a given enzyme (nucleotide and glycogen biosynthesis), or indirectly (cell division). Our transcriptome profiles show that all of these previous observations are parts of larger trends, which are evident at the transcriptional level.
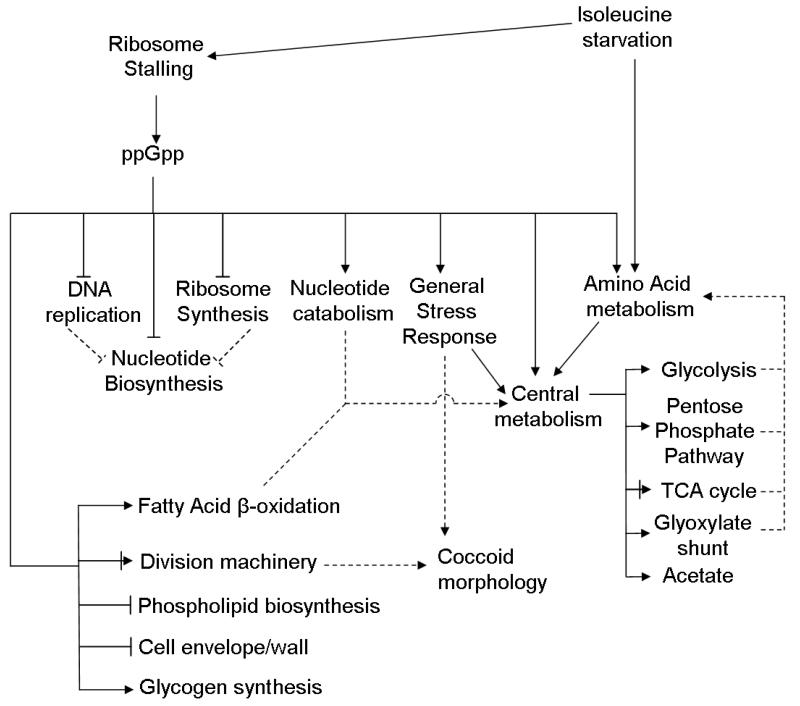
A data-driven model of the constituent processes encompassed by the ppGpp-dependent stringent response is presented in Fig. 8. We assume the changes in gene expression observed here result from two basic signal inputs: i) ribosome stalling which leads to ppGpp accumulation and ii) starvation for isoleucine, which results in allosteric and transcriptional control of branched chain amino acid biosynthetic pathways. In this model, ppGpp accumulation in response to isoleucine starvation leads to stringent down regulation of several processes including repression of DNA replication, ribosome synthesis, nucleotide biosynthesis, phospholipid biosynthesis, cell envelope synthesis, and cell division. Processes requiring ppGpp for their induction include the general stress response, central metabolism, nucleotide catabolism, fatty acid β-oxidation, and amino acid biosynthesis/catabolism genes. Down regulation of macromolecular synthesis, and induction of stationary phase morphogenes, cause stationary phase cells to take on their classical coccoid morphology. Metabolic restructuring leads to a reversal of anabolic flux (i.e., into biomass) to reassimilation of carbon previously allocated to nucleotide and fatty acid synthesis. Flux into central metabolism combined with changes in expression of central metabolic genes serves to accommodate the redistribution of metabolites into amino acid biosynthetic pathways. We also note that this extensive metabolic restructuring is accompanied by the induction of the general stress response, which prepares the cells for long-term survival in stationary phase. We expect that other regulators, especially cAMP/Crp and Lrp likely play a role in controlling the transcriptional response to isoleucine starvation. However, since the manner in which isoleucine starvation may trigger signaling through these pathways is unclear, we have not attempted to integrate them into the model presented here.
Fig. 8.
Physiological model of the ppGpp-mediated response to isoleucine starvation. Solid arrows represent positive regulation while solid lines with flat ends indicate negative regulation. Solid lines ending with both lines and arrows denote complex regulation. Dashed lines indicate functional effects (eg. Nucleotide catabolism generates intermediates which are funneled into central metabolism). Regulatory relationships are not necessarily direct.
Global transcription patterns and the regulatory mechanism of ppGpp
With the data presented in this report, we now have transcriptome profiles for cells impacted in their ability to accumulate ppGpp under two very different starvation conditions, i.e., carbon starvation (Traxler et al., 2006) and amino acid starvation (this study). A qualitative comparison reveals two trends present in both data sets: down regulation of translation apparatus genes typical of the stringent response and induction of the RpoS-dependent general stress response. Beyond these shared responses, starvation for carbon or amino acids elicited induction of different condition-specific stimulons (i.e., carbon foraging or isoleucine biosynthesis genes, respectively). Moreover, under both starvation conditions, the induction of the corresponding stimulon was ppGpp-dependent.
Based on these observations, we hypothesize that efficient global induction of all starvation-specific stimulons requires ppGpp.
ppGpp, along with DksA, is thought to exert control over global gene expression via multiple mechanisms. These include alterations in sigma factor-core RNAP interactions (Magnusson et al., 2005), direct down regulation (Paul et al., 2004, Maitra et al., 2005), passive induction by increased RNAP availability (Barker et al., 2001a), and direct activation (Paul et al., 2005). Here, we consider each of these mechanisms in light of the genome-wide expression data.
Induction of the general stress response involves ppGpp accumulation in several ways, and the data presented here (i.e., Fig. 2C) are compatible with each of them. First, ppGpp enhances the competitiveness of core RNAP for alternative sigma factors (Jishage et al., 2002), resulting in poor expression of genes controlled by alternative sigma factors in strains lacking ppGpp. Second, expression of the rpoS gene was induced 6.9-fold in the WT compared to 2.3-fold in the ppGpp0 strain (data not shown). Third, yaiB (renamed iraP) was recently found to encode a protein that stabilizes RpoS via binding to the anti-sigma factor RssB in response to carbon and phosphate starvation (Bougdour et al., 2006). Moreover, induction of iraP transcription was shown to be ppGpp-dependent (Bougdour & Gottesman, 2007). Our array data confirm that ppGpp is required for iraP induction, i.e., iraP was induced 10.7-fold in the WT compared to 2.2-fold in the ppGpp0 strain (Supplementary Table T1). Thus, poor induction of the RpoS regulon in the ppGpp0 strain likely results from lower induction of the rpoS gene, weak stabilization of RpoS by IraP, and poor competition of core RNAP for RpoS.
Perhaps the best understood mechanism of gene regulation by ppGpp/DksA applies to the direct down regulation of rRNA transcription (Paul et al., 2004). Extensive work has demonstrated that rRNA promoters form open complexes that are intrinsically unstable, which makes them particularly prone to open complex collapse, as occurs with ppGpp/DksA-bound RNAP (Haugen et al., 2006, Paul et al., 2004). Thus, when ppGpp accumulates, there is a rapid and robust cessation of stable RNA synthesis (Paul et al., 2004). An alternative model for direct down regulation was recently suggested, whereby ppGpp-bound RNAP forms dead-end complexes at stringently-controlled promoters, leading to occluded transcription (Maitra et al., 2005). Since transcription of ribosomal protein genes is regulated directly by rRNA levels, we take the comprehensive down regulation of the translation apparatus as evidence of the cessation of stable RNA synthesis. As expected, this down regulation was ppGpp-dependent (Fig. 2C). The down regulation of translation apparatus genes observed here is compatible with both proposed mechanisms of direct down regulation of rRNA synthesis; we cannot distinguish between the two models based on transcriptional output alone.
One way ppGpp might contribute to induction of the many genes observed here is by mediating the balance between RNAP engaged in transcription of the translation apparatus versus transcription of all other genes (Barker et al., 2001a). According to this passive model, the increased availability of RNAP for transcription of stringently induced promoters results from the liberation of RNAP previously sequestered in stable-RNA synthesis. We previously showed that global expression patterns observed during carbon starvation were consistent with this model (Traxler et al., 2006). However, the WT and ppGpp0 strains induced transcription of a similar number of genes in response to isoleucine starvation (Fig. 2B), despite the absence of down regulation of translation apparatus genes in the strain lacking ppGpp. Thus, we hypothesize that the slower onset of growth arrest during isoleucine starvation (about 100 min compared to <10 min for carbon starvation) relaxes the tight relationship between transcription of the translation apparatus and other genes across the genome. While the passive mechanism allows for global transcriptional flexibility during times of sudden starvation, this constraint may be loosened when starvation is encountered over longer time scales (as occurs in complex nutrient mixtures). If the passive mechanism was the driving force behind the WT transcriptional response to isoleucine starvation, the ppGpp0 strain would exhibit a similar transcriptional response if given enough time, possibly through reduced transcription initiation at rRNA gene promoters due to depletion of transcription initiating NTP pools (Paul et al., 2004). However, the transcriptional response of the ppGpp0 strain did not approximate that of the WT (Fig. 2A), suggesting an active role for ppGpp in gene induction in response to isoleucine starvation, as discussed below.
ppGpp, in concert with DksA, is known to be sufficient for direct induction of transcription of several amino acid biosynthetic operons in vitro, (Paul et al., 2005). However, we did not observe stimulation of amino acid biosynthesis genes en masse during isoleucine starvation, suggesting that the specific induction of amino acid biosynthesis genes requires ppGpp and the action of specific regulators (i.e., Lrp, LeuO, and/or IlvY, etc.). Thus, we favor a synergistic model that requires ppGpp-bound RNAP acting together with other transcription factors at a wide range of promoters to produce the stressor-specific response observed in the WT. Alternatively, the patterns of transcription observed in the WT may be the product of a regulatory cascade initiated by ppGpp accumulation and accomplished ultimately (or in part) by other transcription factors. These mechanisms are not mutually exclusive, and experiments to delineate the regulatory architecture of the stringent response are ongoing.
Concluding remarks
In this report, we examined the global response to amino acid starvation, including changes in the transcriptome, metabolic proteome, and composition of the growth medium. Our results indicated that the adjustments made by the WT in response to isoleucine starvation entail a large-scale restructuring of cellular metabolism that includes differential expression of genes involved in, and flux through, central metabolism, amino acid catabolism/anabolism, nucleotide biosynthesis/catabolism, and fatty acid metabolism. These responses were totally dependent on the alarmone ppGpp and profoundly absent in the ppGpp0 strain. We observed that cells lacking ppGpp produced significantly more RNA and biomass (but not more protein) than the WT during amino acid starvation. This observation, considered in light of the extremely aberrant patterns of transcription observed in the ppGpp0 strain, highlights the vital role of ppGpp in relaying information about the translational status of the cell not only to genes involved in translation and amino acid biosynthesis, but also to genes involved in intermediary metabolism and macromolecule synthesis. The comprehensive deficiencies of the ppGpp0 strain, as evidenced by the global measurements presented here, suggest that ppGpp is the primary signal used by E. coli cells to adjust their reproductive potential to that defined by their nutritional environment.
Experimental procedures
Bacterial strains and growth conditions
All strains used in this study were derivatives of E. coli K-12 strain MG1655. The ΔrelA, and ΔrelA ΔspoT (ppGpp0) strains were constructed for this study using a modified version of the method described by Datsenko and Wanner (Datsenko & Wanner, 2000). The ΔrelA, and ppGpp0 strains were made marker-less by removal of antibiotic cassettes using surrounding FRT sites and confirmed by sequencing and PCR. The WT and isogenic mutants were cultured in a 2-liter Biostat B fermentor (Braun Biotech) containing 1 liter of morpholinepropanesulfonic acid (MOPS) medium (Neidhardt et al., 1974) with 2.0 g/liter glucose and amino acids at the concentrations described in (Wanner et al., 1977), with the exception that isoleucine was included at 60 μM instead of the usual 400 μM. The growth medium did not contain uracil, which has been shown to stimulate growth of E. coli MG1655, which has an rph frameshift mutation (Jensen, 1993). However, inclusion of uracil had no effect on logarithmic growth, growth arrest caused by isoleucine starvation, or rescue of growth by addition of isoleucine (data not shown). The temperature was maintained at 37°C, and pH was kept constant at 7.4 by the addition of 1 M NaOH. The dissolved oxygen level was maintained above 40% of saturation by adjusting the agitation speeds in the range of 270–500 rpm with fixed 1.5 liter/min air flow. Growth was monitored as absorbance at 600 nm with a Beckman-Coulter DU 800 spectrophotometer.
Amino acid analysis
Media samples were withdrawn from the bioreactor directly into a 10 ml syringe and then filtered through 0.4 μm filters. Samples were then frozen immediately. Amino acid concentrations in the filtered culture media were determined using capillary electrophoresis-mass spectrometry (CE-MS) with a sheath-fluid electrospray interface. This method was modified from a similar method described previously (Williams et al., 2007). Briefly, CE-MS was performed using an Agilent G1600 capillary electrophoresis connected to an Agilent 1946A single quadrupole mass spectrometer with an electrospray ionization source using the Agilent capillary electrophoresis spray needle adapter (Palo Alto, CA, USA). Fused-silica capillaries were used for the separation (Polymicro Technologies, Phoenix, AZ, USA). All capillaries used in the CE-MS analysis were 50 mm I.D. and had a total length of 70 cm. Sheath liquid composed of a 1:1 (v/v) mix of 2-propanol and HPLC-grade water with 5 mM formic acid was supplied to the co-axial sheath-fluid interface at a flow rate of 4 μl/min. The nebulizer pressure was maintained at 3 psi. The separation was performed using a +25 kV potential, which produced a current of 47 μA. Samples were injected hydrostatically, at 250 mbar*s (6.1 nl). Mass spectral data was collected in the selected-ion monitoring (SIM) mode using the [M+H]+ m/z for each amino acid. The drying gas temperature was set to 70°C with a drying gas flow rate of 12.0 l/min. The potential on the MS capillary was maintained at 3800 V and the fragmentation voltage was set to 70 V. The running electrolyte was 1.0 M formic acid prepared in HPLC-grade water with no pH adjustment.
All samples and standards were diluted 1:1 in 10 mM HCl containing 200 μM ethionine (internal standard). The method of internal standards calibration was applied to each amino acid. Standards containing 10, 100, 250, and 1000 μM of each amino acid were analyzed in triplicate. All standards were commercial amino acids (Sigma Chemical, St. Louis, MO, USA). All amino acid calibration curves displayed a linearity of 0.995 or greater.
Nucleotide extraction and ppGpp quantification
Nucleotides were extracted as described, with minor modifications (Bochner & Ames, 1982). Five ml of culture was sampled directly into a 15 ml falcon tube containing 0.5 ml of 11M formic acid. The sample was vigorously mixed and chilled on ice. One ml aliquots of this mixture were incubated at 0° C on ice for 30 min. These one ml samples were centrifuged at 4° at 10000 RPM for 5 min. The supernatant was then filtered through 0.2 μm filters and stored at −20° until HPLC analysis.
ppGpp was quantified by anion exchange HPLC using a Mono Q 5/50 GL column (GE Healthcare). 250 μl of supernatant was injected under initial conditions of 95% 20mM Tris (pH 8.0) and 5% 20mM Tris + 1.5 M sodium formate (pH 8.0). This initial condition was maintained for 5 min. Absorbance at 260nm was used to detect eluted nucleotides. Over a period of 30 min, the level of sodium formate buffer was ramped up to 60%. ppGpp was identified as a peak which eluted at ~30.9 min. Samples were run in duplicate for three separate WT time course experiments. Representative results are shown in Fig 1A. ppGpp standard was purchased from TriLink Biosciences. Standard curves established that the linear range of detection of ppGpp is 50nM to 100μM.
Microarray analysis
Cells were sampled directly from the fermenter into an equal volume of ice-cold RNAlater (Ambion) and total RNA was extracted using Qiagen RNeasy Minikits with optional DNAse treatment steps. RNA was checked for integrity by gel electrophoresis and maintained in a 2:1 dilution of EtOH at −80°C until labeling. RNA was converted to cDNA by first strand synthesis using Superscript II (Invitrogen) and random hexamers, according to the manufacturer’s specifications. The cDNA was fragmented and biotinylated (Enzo Kit, Roche Diagnostics) according to the Affymetrix prokaryotic labeling protocol. The microarrays used in this study were custom built Affymetrix GeneChips containing probes for several prokaryotic genomes including E. coli K12 MG1655, E. coli O157:H7 EDL933, Bacteriodes thetaiotaomicron VPI-5482, Enterococcus faecalis V583, Salmonella typhimurium LT2, and Bacillus anthracis. Biotinylated samples were prepared according to the manufacturer’s instructions and hybridized for 16 hours at 60°C. Hybridized arrays were stained using Affymetrix protocol ProkGE_WS2v2_450. Stained microarrays were scanned and the raw data files (.cel) were further analyzed using RMA processing with quartile normalization (Irizarry et al., 2003). All samples were duplicated biologically and technically; r2 was >0.95 for all WT and ppGpp0 replicates. We considered genes to be significantly induced or repressed if the absolute value of the expression ratio was >2-fold (Wren & Conway, 2006). Hierarchical clustering algorithms were implemented in DecisionSite for Functional Genomics (Spotfire). The microarray data were deposited at Array Express (http://www.ebi.ac.uk/miamexpress/), accession # E-MEXP-1370.
Kinetic Biolog assays
Biolog assays were carried out essentially as described (Ihssen & Egli, 2005). Briefly, a volume of cells sufficient to inoculate 2 Biolog plates was harvested from the fermenter and placed in 50 ml falcon tubes containing chloramphenicol (final concentration 25 μg/ml). Samples were kept warm (37°C) through all manipulations. Cells were washed 3 times with 15 ml MOPS medium (without carbon sources) containing chloramphenicol (25 μg/ml), by centrifugation at ~3700 rpm for 8 min. After washing, cells were resuspended in the same medium to an OD 600nm of ~0.3. Biolog GN2 microplates were then loaded (150 μl in each well) and incubated for 24 hours in an Omnilog system. Dye reduction was monitored every 15 minutes. The areas under the resulting curves were then averaged as an overall indicator of substrate utilization. All Biolog measurements reported are the averages of 2 biological, and 4 technical replicates. Hierarchical clustering algorithms were implemented in DecisionSite (Spotfire).
Confocal microscopy
WT and ppGpp0 strains were grown under isoleucine starvation conditions as described above. At the indicated time points, 1 ml of culture medium containing living cells was harvested and stained by adding 20 μl of 1.0 mg/ml propidium iodide and 1.2 μl of 5mM SYTO 9. Samples were incubated for 15 minutes in darkness. Five μl of stained sample was placed in a wet mount and visualized using a Olympus FluoView 500 confocal microscope with Blue Argon (488 nm) and Green Helium Neon (543 nm) lasers for excitation of SYTO 9 and propidium iodide fluorophores, respectively. FITC and Propidium Iodide filter settings were used for fluorescence detection.
Biomass, Protein, and RNA quantification
For biomass quantification, cultures were grown as described above. Two 25 ml samples were taken at an OD of ~0.33, three 15 ml samples at the onset of stationary phase (corresponding to time point C in Fig 6 for each strain), and three 15 ml samples were taken 1.5 hours into growth arrest (corresponding to time point E in Fig 6 for each strain). Cells were chilled on ice immediately after sampling and then centrifuged for 20 min at 3700 rpm at 4° C. Supernatant was removed and the remaining biomass was resuspended in 1 ml of deionized water. The samples were transferred to pre-weighed aluminum weigh-boats. Samples were desiccated at 105° C overnight, and dry weights were obtained. The biomass values shown in Figure 6 are averages of three independent cultures for each of the WT and the ppGpp0 strains.
For protein quantification, 5 ml of living cells were harvested directly into 15 ml falcon tubes containing chloramphenicol (final concentration was 25 μg/ml) at the same time points described for biomass quantification. One ml of cells was pelleted by centrifugation for 10 min at 14000 rpm and supernatant was removed. Cells were resuspended in 1 ml of sonication buffer (10mM Tris-HCl, 0.1mM EDTA, 5% glycerol, 150mM NaCl, pH 7.65), and frozen at −20° C until further use. Thawed samples were intermittently sonicated and cooled on ice at 5 sec intervals, 3 times. Sonicated samples were pelleted by centrifugation and supernatant containing protein was assayed according to the Bradford method of protein quantification (Bradford, 1976). Standard curves for the Bradford assay were done using BSA in sonication buffer. The protein values shown in Figure 6 are averages of three independent cultures for each of the WT and the ppGpp0 strains. The assay was replicated in quadruplicate at each time point for each culture.
Total RNA quantification was carried out as described previously, with minor modifications (Brunschede et al., 1977). Briefly, 15 ml of culture was sampled into a 50 ml falcon tube containing 3 ml of 3.0 M ice-cold trichloroacetic acid. The solution was mixed by inversion and cooled briefly on ice. Three aliquots of the sample were processed separately. Six ml of sample was pipetted onto a glass fiber filter atop a vacuum flask. The filter was then rinsed 3 times with 5 ml volumes of nanopure water. The dried filter was placed in a screw-top glass vial with 2 ml of 0.2 M NaOH. The vials were incubated horizontally at RT with gentle rocking for 16-18 hours. Two ml of cold, 0.5 M perchloric acid was added to each vial and mixed by inversion. At least one ml of the resulting mixture was filtered through a 0.2 μm filter. The RNA concentration was measured spectrophotometerically on a Beckman-Coulter DU 800 using the ‘RNA’ setting. The total RNA values reported in Fig. 7 are the averages of 3 replicates from 3 independent WT cultures (9 total replicates for each timepoint) and two independent ppGpp0 strain cultures (6 total replicates for each timepoint).
Supplementary Material
Acknowledgements
The authors wish to thank Stafford Marquardt for writing and implementing the Gene Mapper program, which overlaid array data values onto the pathway diagrams in Figs. 3 and 4. Joe Grissom also provided critical bioinformatics support. Sarah Stark helped with total RNA measurement. We thank Dr. Lawrence Reitzer for critical reading of parts of the manuscript. We gratefully acknowledge Dr. Michael Cashel for theoretical advice regarding quantification of ppGpp by anion exchange and Neil Wofford for practical HPLC advice. Finally, we thank Dr. Randall Hewes for expert guidance regarding confocal microscopy. This research was supported by NIH awards RO1AI48945, P20RR016478, U24GM077905.
References
- Aberg A, Shingler V, Balsalobre C. (p)ppGpp regulates type 1 fimbriation of Escherichia coli by modulating the expression of the site-specific recombinase FimB. Molecular microbiology. 2006;60:1520–1533. doi: 10.1111/j.1365-2958.2006.05191.x. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. Journal of molecular biology. 2001a;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. Journal of molecular biology. 2001b;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- Battesti A, Bouveret E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Molecular microbiology. 2006;62:1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- Bochner BR, Ames BN. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. The Journal of biological chemistry. 1982;257:9759–9769. [PubMed] [Google Scholar]
- Bogosian G, Violand BN, Dorward-King EJ, Workman WE, Jung PE, Kane JF. Biosynthesis and incorporation into protein of norleucine by Escherichia coli. The Journal of biological chemistry. 1989;264:531–539. [PubMed] [Google Scholar]
- Bougdour A, Gottesman S. ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12896–12901. doi: 10.1073/pnas.0705561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougdour A, Wickner S, Gottesman S. Modulating RssB activity: IraP, a novel regulator of sigma(S) stability in Escherichia coli. Genes & development. 2006;20:884–897. doi: 10.1101/gad.1400306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brunschede H, Dove TL, Bremer H. Establishment of exponential growth after a nutritional shift-up in Escherichia coli B/r: accumulation of deoxyribonucleic acid, ribonucleic acid, and protein. Journal of bacteriology. 1977;129:1020–1033. doi: 10.1128/jb.129.2.1020-1033.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VJ, Vinella D. The stringent response. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: cellular and molecular biology. ASM Press; Washington, D.C.: 1996. pp. 1458–1496. [Google Scholar]
- Chang DE, Smalley DJ, Conway T. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Molecular microbiology. 2002;45:289–306. doi: 10.1046/j.1365-2958.2002.03001.x. [DOI] [PubMed] [Google Scholar]
- Chiaramello AE, Zyskind JW. Coupling of DNA replication to growth rate in Escherichia coli: a possible role for guanosine tetraphosphate. Journal of bacteriology. 1990;172:2013–2019. doi: 10.1128/jb.172.4.2013-2019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen GN, Munier R. Incorporation of structural analogues of amino acids in bacterial proteins. Biochimica et biophysica acta. 1956;21:592–593. doi: 10.1016/0006-3002(56)90207-4. [DOI] [PubMed] [Google Scholar]
- Danchin A, Dondon L, Daniel J. Metabolic alterations mediated by 2-ketobutyrate in Escherichia coli K12. Mol Gen Genet. 1984;193:473–478. doi: 10.1007/BF00382086. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. Journal of bacteriology. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichel J, Chang YY, Riesenberg D, Cronan JE., Jr. Effect of ppGpp on Escherichia coli cyclopropane fatty acid synthesis is mediated through the RpoS sigma factor (sigmaS) Journal of bacteriology. 1999;181:572–576. doi: 10.1128/jb.181.2.572-576.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry DR, Hernandez VJ, Nguyen LH, Jensen DB, Cashel M. Synthesis of the stationary-phase sigma factor sigma s is positively regulated by ppGpp. Journal of bacteriology. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyaneshwar P, Paliy O, McAuliffe J, Popham DL, Jordan MI, Kustu S. Sulfur and nitrogen limitation in Escherichia coli K-12: specific homeostatic responses. Journal of bacteriology. 2005;187:1074–1090. doi: 10.1128/JB.187.3.1074-1090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Heath RJ, Jackowski S, Rock CO. Guanosine tetraphosphate inhibition of fatty acid and phospholipid synthesis in Escherichia coli is relieved by overexpression of glycerol-3-phosphate acyltransferase (plsB) The Journal of biological chemistry. 1994;269:26584–26590. [PubMed] [Google Scholar]
- Hernandez VJ, Bremer H. Characterization of RNA and DNA synthesis in Escherichia coli strains devoid of ppGpp. The Journal of biological chemistry. 1993;268:10851–10862. [PubMed] [Google Scholar]
- Herring PA, McKnight BL, Jackson JH. Channeling behavior and activity models for Escherichia coli K-12 acetohydroxy acid synthases at physiological substrate levels. Biochemical and biophysical research communications. 1995;207:48–54. doi: 10.1006/bbrc.1995.1151. [DOI] [PubMed] [Google Scholar]
- Ihssen J, Egli T. Global physiological analysis of carbon- and energy-limited growing Escherichia coli confirms a high degree of catabolic flexibility and preparedness for mixed substrate utilization. Environmental microbiology. 2005;7:1568–1581. doi: 10.1111/j.1462-2920.2005.00846.x. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics (Oxford, England) 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Ishiguro EE, Ramey WD. Involvement of the relA gene product and feedback inhibition in the regulation of DUP-N-acetylmuramyl-peptide synthesis in Escherichia coli. Journal of bacteriology. 1978;135:766–774. doi: 10.1128/jb.135.3.766-774.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KF. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. Journal of bacteriology. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M, Kvint K, Shingler V, Nystrom T. Regulation of sigma factor competition by the alarmone ppGpp. Genes & development. 2002;16:1260–1270. doi: 10.1101/gad.227902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson J, Balsalobre C, Wang SY, Urbonaviciene J, Jin DJ, Sonden B, Uhlin BE. Nucleoid proteins stimulate stringently controlled bacterial promoters: a link between the cAMP-CRP and the (p)ppGpp regulons in Escherichia coli. Cell. 2000;102:475–485. doi: 10.1016/s0092-8674(00)00052-0. [DOI] [PubMed] [Google Scholar]
- Leavitt RI, Umbarger HE. Isoleucine and valine metabolism in Escherichia coli. XI. Valine inhibition of the growth of Escherichia coli strain K-12. Journal of bacteriology. 1962;83:624–630. doi: 10.1128/jb.83.3.624-630.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Durfee T, Cabrera JE, Zhao K, Jin DJ, Blattner FR. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. The Journal of biological chemistry. 2005;280:15921–15927. doi: 10.1074/jbc.M414050200. [DOI] [PubMed] [Google Scholar]
- Maaloe O, Kjeldgaard N. Control of Macromolecular Synthesis. W. A. Benjamin, inc; New York: 1966. [Google Scholar]
- Magnusson LU, Farewell A, Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends in microbiology. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nystrom T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. Journal of bacteriology. 2007;189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A, Shulgina I, Hernandez VJ. Conversion of Active Promoter-RNA Polymerase Complexes into Inactive Promoter Bound Complexes in E. coli by the Transcription Effector, ppGpp. Mol Cell. 2005;17:817–829. doi: 10.1016/j.molcel.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. Journal of bacteriology. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Ingraham JL, Schaechter M. Physiology of the Bacterial Cell. Sinauer Associates, Inc; Sunderland, MA: 1990. [Google Scholar]
- Oh MK, Rohlin L, Kao KC, Liao JC. Global expression profiling of acetate-grown Escherichia coli. The Journal of biological chemistry. 2002;277:13175–13183. doi: 10.1074/jbc.M110809200. [DOI] [PubMed] [Google Scholar]
- Patte J-C. Biosynthesis of Threonine and Lysine. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: cellular and molecular biology. ASM Press; Washington, D.C: 1996. [Google Scholar]
- Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- Podkovyrov SM, Larson TJ. Identification of promoter and stringent regulation of transcription of the fabH, fabD and fabG genes encoding fatty acid biosynthetic enzymes of Escherichia coli. Nucleic acids research. 1996;24:1747–1752. doi: 10.1093/nar/24.9.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G, Ron EZ, Glaser G. ppGpp-mediated regulation of DNA replication and cell division in Escherichia coli. Current microbiology. 1995;30:27–32. doi: 10.1007/BF00294520. [DOI] [PubMed] [Google Scholar]
- Stauffer GV, Brenchley JE. Evidence for the involvement of serine transhydroxymethylase in serine and glycine interconversions in Salmonella typhimurium. Genetics. 1974;77:185–198. doi: 10.1093/genetics/77.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stent GS, Brenner S. A genetic locus for the regulation of ribonucleic acid synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1961;47:2005–2014. doi: 10.1073/pnas.47.12.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks SM. Mechanism and use of the commercially available viability stain, BacLight. Cytometry A. 2004;61:189–195. doi: 10.1002/cyto.a.20069. [DOI] [PubMed] [Google Scholar]
- Taguchi M, Izui K, Katsuki H. Augmentation of cyclopropane fatty acid synthesis under stringent control in Escherichia coli. Journal of biochemistry. 1980a;88:1879–1882. [PubMed] [Google Scholar]
- Taguchi M, Izui K, Katsuki H. Augmentation of glycogen synthesis under stringent control in Escherichia coli. Journal of biochemistry. 1980b;88:379–387. doi: 10.1093/oxfordjournals.jbchem.a132983. [DOI] [PubMed] [Google Scholar]
- Tosa T, Pizer LI. Biochemical bases for the antimetabolite action of L-serine hydroxamate. Journal of bacteriology. 1971;106:972–982. doi: 10.1128/jb.106.3.972-982.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Chang DE, Conway T. Guanosine 3′,5′-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2374–2379. doi: 10.1073/pnas.0510995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbough CL., Jr. Regulation of Escherichia coli aspartate transcarbamylase synthesis by guanosine tetraphosphate and pyrimidine ribonucleoside triphosphates. Journal of bacteriology. 1983;153:998–1007. doi: 10.1128/jb.153.2.998-1007.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbarger HE. Biosynthesis of the Branched-Chain Amino Acids. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: cellular and molecular biology. ASM Press; Washington, D.C.: 1996. [Google Scholar]
- Uzan M, Danchin A. Correlation between the serine sensitivity and the derepressibility of the ilv genes in Escherichia coli relA-mutants. Mol Gen Genet. 1978;165:21–30. doi: 10.1007/BF00270372. [DOI] [PubMed] [Google Scholar]
- VanBogelen RA, Kelley PM, Neidhardt FC. Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. Journal of bacteriology. 1987;169:26–32. doi: 10.1128/jb.169.1.26-32.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D, Albrecht C, Cashel M, D’Ari R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Molecular microbiology. 2005;56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- Vinella D, D’Ari R. Overview of controls in the Escherichia coli cell cycle. Bioessays. 1995;17:527–536. doi: 10.1002/bies.950170609. [DOI] [PubMed] [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner BL, Kodaira R, Neidhart FC. Physiological regulation of a decontrolled lac operon. Journal of bacteriology. 1977;130:212–222. doi: 10.1128/jb.130.1.212-222.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Dissection of the mechanism for the stringent factor RelA. Mol Cell. 2002;10:779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- Williams BJ, Cameron CJ, Workman R, Broeckling CD, Sumner LW, Smith JT. Amino acid profiling in plant cell cultures: an inter-laboratory comparison of CE-MS and GC-MS. Electrophoresis. 2007;28:1371–1379. doi: 10.1002/elps.200600609. [DOI] [PubMed] [Google Scholar]
- Wren JD, Conway T. Meta-analysis of published transcriptional and translational fold changes reveals a preference for low-fold inductions. Omics. 2006;10:15–27. doi: 10.1089/omi.2006.10.15. [DOI] [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. The Journal of biological chemistry. 1991;266:5980–5990. [PubMed] [Google Scholar]
- Yang CR, Shapiro BE, Hung SP, Mjolsness ED, Hatfield GW. A mathematical model for the branched chain amino acid biosynthetic pathways of Escherichia coli K12. The Journal of biological chemistry. 2005;280:11224–11232. doi: 10.1074/jbc.M411471200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.