Abstract
The genus Brevicellicium encompasses wood-inhabiting corticioid fungi characterized by isodiametric subhymenial hyphae, short basidia, and smooth, often subangular spores with a distinct apiculus. Eight new LSU nrDNA sequences and 13 new ITS nrDNA of this genus, including the type species, were aligned with 47 and 42 accessions respectively of species of Trechisporales obtained from GenBank, and phylogenetic analyses were performed. The order Trechisporales was confirmed as a monophyletic group; the genera Porpomyces, Sistotremastrum, Subulicystidium and Trechispora form a highly supported clade where all Brevicellicium sequences are included. Our analyses also support that this genus belongs to Hydnodontaceae. A new species, Brevicellicium atlanticum from the Azores Archipelago, is described.
Keywords: Basidiomycota, Agaricomycetes, Corticioid fungi, ITS, LSU nrDNA, Phylogeny, Taxonomy
INTRODUCTION
Brevicellicium was described by Larsson and Hjortstam (Hjortstam & Larsson 1978) to accommodate Corticium exile. At the time, two more species were transferred to the new genus, Odontia olivascens and Athelopsis viridula. The isodiametric subhymenial hyphae, short basidia and smooth, often subangular, spores with a distinct apiculus were emphasized as important morphological characteristics of this genus of wood-inhabiting corticioid fungi.
Twelve species have been placed in this cosmopolitan genus. Brevicellicium exile, originally described from Canada (Jackson 1950) as Corticium exile, seems to be a rare species in the Northern Hemisphere (Hjortstam 2001), and is known from north Europe (Hjortstam & Larsson 1978), France (Boidin & Gilles 1990), Spain (Telleria et al. 1993, Telleria & Melo 1995), and Colombia (Hjortstam & Ryvarden 1997). Brevicellicium olivascens, described from Italy by Bresadola (1892) as Odontia olivacea, is a cosmopolitan species, widely distributed in temperate areas and less frequent in tropical and subtropical regions (Hjortstam et al. 2005); it is common in Europe including the Iberian Peninsula (Telleria & Melo 1995, Bernicchia & Gorjón 2010) and is also reported from North America (Ginns & Lefebvre 1993), South America, Burundi, and India (Hjortstam 2001, Hjortstam & Ryvarden 2007) as well as from Iran (Hallenberg 1981), and Japan (Maekawa 1993). It should also be noted that B. exile and B. olivascens were also found in the Macaronesian region: Canary Islands and Azores Archipelago (Ryvarden 1976, Hjortstam & Larsson 1978, Telleria et al. 2009a, b). Brevicellicium viridulum, transferred to the genus when it was described, was considered by Hjortstam et al. (1988) as a colour morph of B. olivascens. Brevicellicium permodicum, described from Canada by Jackson (1950) as Corticium permodicum, and also reported from New Zealand (Cunningham 1963, Hjortstam 2001), is the only species of the genus without clamps known today. Its inclusion in the genus is perhaps questionable. The other eight species have a tropical distribution: Brevicellicium mellinum, originally described from Brazil by Bresadola (1920) as Corticium mellinum, is reported from Puerto Rico and Venezuela (Hjortstam & Ryvarden 2007). Brevicellicium allantosporum, described from Tanzania (Hjortstam & Ryvarden 1980), is also known from Brazil, Colombia, Venezuela, Ecuador, and Borneo (Hjortstam 2001, Hjortstam et al. 2005, Hjortstam & Ryvarden 2008). Brevicellicium flavovirens, from Argentina and Brazil (Hjortstam 2001), is morphologically similar to B. exile but differs in basidiome colour and the shape and size of the spores. Brevicellicium molle, described from Tanzania (Hjortstam & Ryvarden 1980), is also reported from Colombia and Brazil (Hjortstam & Ryvarden 1997, Hjortstam 2001). Four species are only known from their type locality: B. asperum from Venezuela (Hjortstam et al. 2005), B. udinum from Brazil (Hjortstam 2001), B. uncinatum from Tanzania (Hjortstam & Ryvarden 1980, Hjortstam 2001), and B. vulcanense from Hawaii (Gilbertson et al. 2001). Complete or partial keys to Brevicillicium have been published by Hjortstam & Larsson (1978), Hjortstam & Ryvarden (1980), Hjortstam (2001), and Hjortstam et al. (2005).
According to Hjortstam & Larsson (1978), Brevicellicium is morphologically close to the smooth-spored species of Trechispora (e.g. Trechispora amianthina, T. cohaerens, T. confinis, T. byssinella), differing in the absence of ampullate septa on the basal hyphae. Jülich (1982), placed both genera in Hydnodontaceae, and later Larsson (2007), in his phylogenetic classification of corticioid fungi, confirmed this arrangement and included the family in Trechisporales.
Recently, the genus Brevicellopsis has been segregated from Brevicellicium (Hjorstam & Ryvarden 2008), with Brevicellicium allantosporum as type species. Both genera share similar isodiametric subhymenial hyphae, but they can be distinguished by the hymenophore appearance and shape of the spores. In Brevicellicium, the hymenophore is granular to almost smooth, and the spores are subangular or short ellipsoid, whereas Brevicellopsis has a distinctly odontioid hymenophore and allantoid spores.
The aim of this study was to identify, characterize and analyze, using morphological and molecular data, 11 collections of Brevicellicium from the Iberian Peninsula (Spain and Portugal) and the Azores Archipelago, as well as to evaluate the phylogenetic circumscription of the genus. The ITS and LSU nrDNA sequences of all collections were compared with sequences of Trechispora and Sistotremastrum generated by our research group within the framework of other studies, and with sequences deposited in GenBank, in order to establish their phylogenetic relationships. As a result, a new species is described and the phylogenetic position of Brevicellicium as member of Trechisporales is confirmed (Larsson 2007).
MATERIALS AND METHODS
Sampling, morphological studies and line drawings
Twelve specimens of Brevicellicium from the Iberian Peninsula (Spain) and the Azores Archipelago, and two from Sweden, were studied (Table 1). Vouchers are deposited in MA-Fungi, LISU, TFCMic, and GB. Measurements and drawings were made from microscopic sections mounted in 3 % aqueous solution of potassium hydroxide and examined at magnifications up to ×1250 using an Olympus BX51 microscope. The length and width of 30 spores and 10 basidia were measured from each sample. Colours of dried basidiomes are given according to the ISCC–NBS Centroid Color Charts. The drawing was made with aid of a drawing tube.
Table 1. Specimens of Brevicellicium studied with GenBank accession numbers.
| Species/Specimen | Country/Locality | Habitat |
Acc. no. |
|
|---|---|---|---|---|
| ITS | 28S | |||
| B. atlanticum sp. nov. | ||||
| LISU 178590, 9090IM | Portugal, Azores Archipelago, Terceira Island | Juniperus brevifolia subsp. azorica | HE963775 | HE963776 |
| LISU 178566, 9065IM | Portugal, Azores Archipelago, Terceira Island | Erica azorica | HE963773 | HE963774 |
| B. exile (H.S. Jacks.) K.H. Larss. & Hjortstam | ||||
| MA-Fungi 76132, 16118Tell. | Portugal, Azores Archipelago, Pico Island | Pittosporum undulatum | HE963779 | — |
| MA-Fungi 26554, 5217MD | Spain, Huesca | Buxus sempervirens | HE963777 | HE963778 |
| GB, KHL 12130 | Sweden, Västergötland | conifer | — | HE963780 |
| B. olivascens (Bres.) K.H. Larss. & Hjortstam | ||||
| MA-Fungi 75998, 17370Tell. | Portugal, Azores Archipelago, Flores Island | Pittosporum undulatum | HE963790 | — |
| TFCMic 15272 | Portugal, Azores Archipelago, Pico Island | Pittosporum undulatum | HE963791 | — |
| MA-Fungi 19016, 7743Tell. | Spain, Asturias | Quercus robur | HE963789 | — |
| MA-Fungi 13843, 3491MD | Spain, Asturias | Castanea sativa | HE963782 | HE963783 |
| MA-Fungi 5674, 239Tell. | Spain, Barcelona | Fagus sylvatica | HE963781 | — |
| MA-Fungi 21444, 3881MD | Spain, Guadalajara | Rosmarinus officinalis | HE963784 | — |
| MA-Fungi 41366, 6910MD | Spain, Madrid | Corylus avellana | HE963785 | HE963786 |
| MA-Fungi 23496, 4611MD | Spain, Toledo | Ulmus sp. | HE963787 | HE963788 |
| GB, KHL 8571 | Sweden, Bohuslän | hardwood | HE963792 | HE963793 |
DNA isolation and sequencing
Genomic DNA was extracted from 13 collections (Table 1) using the E.Z.N.A® Fungal DNA Miniprep Kit (Omega Biotek, Doraville, USA) or the DNeasy™ Plant Mini Kit (Qiagen, Valencia, CA), following the manufacturer’s instructions; lysis buffer incubation was overnight at 55 °C.
Total DNA was used for PCR amplification of the 5’-1450-base region of the large subunit (LSU nrDNA) and the internal transcribed spacer region (ITS nrDNA) of the nuclear ribosomal gene. The primers LR0R (Rehner & Samuels 1994) and LR7 (Vilgalys & Hester 1990) were used to amplify the region of the LSU nrDNA and the primers ITS1F (Gardes & Bruns 1993) and ITS4 (White et al. 1990) were used to obtain amplifications of both ITS regions, including the 5.8S of the ribosomal RNA gene cluster and flanking parts of the small subunit (SSU) and large subunit (LSU) nuclear ribosomal genes. Individual reactions to a final volume of 25 μm were carried out using illustra™ PuReTaq™ Ready-To-Go™ PCR Beads (GE Healthcare, Buckingham) with a 10 pmol μL primer concentration following the thermal cycling conditions used in Martín & Winka (2000). When these pair of primers failed, the LSU nrDNA region was amplified in two parts using LR5 and LR3R (Vilgalys & Hester 1990), in the combination LR0R/LR5 and LR3R/LR7. The ITS1 nrDNA region and the beginning of 5.8S with primers ITS1F and ITS2 (White et al. 1990), and ITS2 nrDNA region and the end of 5.8S with primer ITS3 (White et al. 1990) and ITS4.
Negative controls lacking fungal DNA were run for each experiment to check for contamination. The reactions were run with the following parameters for the LSU nrDNA: initial denaturation at 94 °C for 5 min, then 36 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s, and extension at 72 °C for 1 min and 30 s, with a final extension at 72 °C for 10 min, and 4 °C soak; for the ITS nrDNA: initial denaturation at 95 °C for 5 min, then 5 cycles of denaturation at 95 °C for 30 s, annealing at 54 °C for 30 s, and extension at 72 °C for 1 min, followed by 33 cycles of denaturation at 72 °C for 1 min, annealing at 48 °C for 30 s, and extension at 72 °C, with a final extension at 72 °C for 10 min and 4 °C soak.
The PCR products were subsequently purified using the QIAquick Gel PCR Purification (Qiagen) kit according to the manufacturer’s instructions. The purified PCR products were sequenced using the same amplification primers. When products were only faintly visible on agarose gels (less that 20 ng μL−1), cloning was conducted with a pGEM®-T Easy Vector System II cloning kit (Promega Corporation, Madison, WI). From each cloning reaction, up to six clones were selected for sequencing. To confirm that the inserted product was correct, 2 μL of the purified plasmid DNA was digested with Eco RI prior to sequencing following the instructions of the manufacturers. Both strands were sequenced separately using vector specific primers T7 and SP6 at Secugen S.L. (Madrid, Spain) or Macrogen (Seoul, Korea).
Sequencher v. 4.2 (Gene Codes Corporation, Ann Arbor, MI) was used to edit the resulting electropherograms and to assemble contiguous sequences. BLAST searches with megablast option were used to compare the sequences obtained against the sequences in the National Center of Biotechnology Information (NCBI) nucleotide databases (Altschul et al. 1997).
Sequence alignment and phylogenetic analyses
The LSU nrDNA and ITS nrDNA sequences obtained were aligned separately using Se-Al v. 2.0a11 Carbon (Rambaut 2002) for multiple sequences. The sequences were compared with homologous sequences retrieved from the EMBL/GenBank/DDBJ databases (Cochrane et al. 2011); many of the sequences were generated by our research group within the framework of other studies (Sistotremastrum: JX310442–JX310445; Trechispora and other Trechisporales: JX392812–JX392856). In the LSU nrDNA analyses, Sistotrema and Repetobasidium (cantharelloid and hymenochaetoid clade respectively, Binder et al. 2005) sequences were included as outgroups. In order to root the ITS analyses, six Sistotremastrum sequences (Sistotremastrum family in Larsson 2007) were included as outgroups because they appear as the sister group of the clade formed by Porpomyces, Subulicystidium, and Trechispora in the trechisporoid clade (Binder et al. 2005, Larsson 2007). Where ambiguities in the alignment occurred, the alignment generating the fewest potentially informative characters were chosen (Baum & Sytsma 1994). Alignment gaps were marked “–”, unresolved nucleotides and unknown sequences were indicated with “N”.
From each data set a maximum parsimony analysis (MP) was carried out; minimum length Fitch trees were constructed using heuristic searches with tree–bisection–reconnection (TBR) branch swapping, collapsing branches if maximum length was zero and with the MulTrees option on in PAUP v. 4.0b10 (Swofford 2003). Gaps were treated as missing data. Nonparametric bootstrap (bs) support (Felsenstein 1985) for each clade, based on 10 000 replicates using the fast–step option, was tested. The consistency index, CI (Kluge & Farris 1969), retention index, RI (Farris 1989), and rescaled consistency index, RC (Farris 1989) were obtained.
For each dataset a second analysis was done using a Bayesian approach (Larget & Simon 1999, Huelsenbeck et al. 2001) with MrBayes v. 3.1 (Ronquist & Huelsenbeck 2003). The analyses were performed assuming the general time reversible model (Rodríguez et al. 1990), including estimation of invariant sites and assuming a discrete gamma distribution with six categories (GTR+I+G) as selected by MrModeltest v. 2.3 (Nylander 2004). According to Rodríguez et al. (1990), only reversible models allow the calculation of the substitution rates. Two independent and simultaneous analyses starting from different random trees were run for 2 000 000 generations with four parallel chains and trees and model scores saved every 100th generation. The default priors in MrBayes were used in the analysis. Every 1 000th generation tree from the two runs was sampled to measure the similarities between them and to determine the level of convergence of the two runs. The potential scale reduction factor (PSRF) was used as a convergence diagnostic and the first 25 % of the trees were discarded as burn–in before stationary was reached. Both the 50 % majority-rule consensus tree and the posterior probability (pp) of the nodes were calculated from the remaining trees with MrBayes. Phylogenetic trees were drawn using TreeView (Page 1996).
RESULTS
In general, only amplifications in parts of the LSU nrDNA, with primers LR0R/LR5 and LR3R/LR7, and ITS nrDNA, with primers ITS1F/ITS2 and ITS3/ITS4 were successful. Weak products (faintly visible on agarose gels; less that 20 ng μL−1 after gel purification) or purified product, which sequences showed double peaks, were cloned. Thus, from LISU 178566, LISU 178590 and MA–Fungi 26554, good LSU sequences were obtained after cloning; the six cloned sequences from each fragment/collection were identical and only one was selected for analyses. The BLAST search of the LSU nrDNA and ITS nrDNA sequences obtained (both direct or after cloning), excluding uncultured/environmental samples, showed more than 100 % and 85 % similarity respectively with Trechisporales sequences published in GenBank, mainly from Larsson et al. (2004). Sequences were located in EMBL/GenBank/DDBJ and UNITE (Abarenkov et al. 2011, http://unite.ut.ee/cite.php) databases.
LSU nrDNA
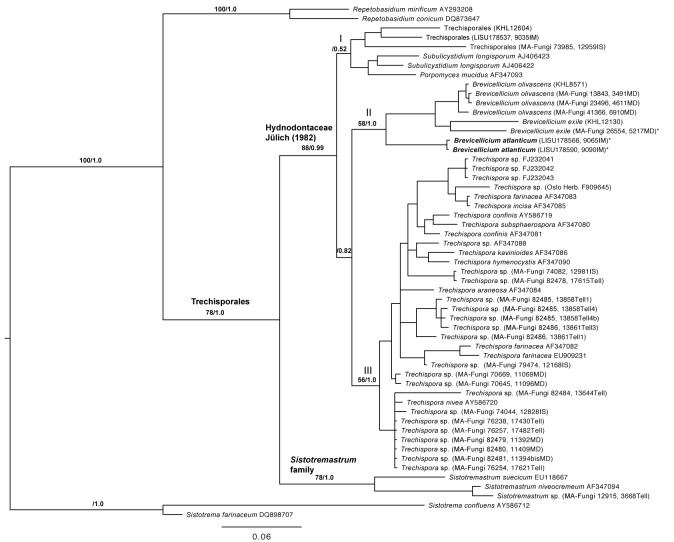
Eight LSU nrDNA sequences generated for this study were aligned with 47 sequences downloaded from GenBank to produce a matrix of 1 358 unambiguously aligned nucleotide position characters. Among them, 908 positions were constant, 167 were parsimony–uninformative and 283 were parsimony-informative. In the maximum parsimony analysis under heuristic search, 100 most parsimonious trees (MPTs) were obtained (tree length=1085, consistency index CI = 0.5512, retention index RI = 0.7105, rescaled consistency index RC = 0.3916). The trees obtained from the MP (strict consensus tree, data not shown) and the Bayesian analyses (Fig. 1) show similar topologies. In both analyses, the complete ingroup forms a highly supported monophyletic clade (bs = 78 %, pp = 1.0), and includes all Brevicellicium sequences. The Trechisporales clade is divided in two well-supported clades, one with the three Sistotremastrum accessions (bs = 78 %, pp = 1.0) and the other, with the remaining sequences (bs = 88 %, pp = 0.99). The latter are distributed over three subclades that either lack support or get support by the Bayesian analysis only. Subclade I (bs < 50 %, pp = 0.52) is formed by Porpomyces and Subulicystidium. Subclade II (bs = 58%, pp = 1.0) includes all Brevicellium collections and is the sister group of subclade III (bs = 56, pp = 1.0) formed by 34 Trechispora spp. sequences. However, this sister-group relationship is not highly supported (bs < 50 % and pp = 0.82). The Brevicellicium clade is separated in two strongly supported groups; one (bs = 100, pp = 1.0) that includes two collections from Terceira Island in Azores Archipelago (LISU 178566 and LISU 178590) and another (bs = 93, pp = 1.0) consisting of six accessions: four of B. olivascens clade (bs = 96 %, pp = 1.0) and two of B. exile (bs < 50 %, pp = 1.0).
Fig. 1.
The 50 % majority-rule consensus tree of Bayesian analysis based on nuclear D1/D2 sequences (LSU nrDNA). Bootstrap (> 50 %) and posterior probability values indicated on the branches to the main clades. The topology was rooted with Sistotrema and Repetobasidium species. The three main clades in Hydnodontaceae discussed in the text are designated I-III. The position of Brevicellicium atlanticum is indicated in bold. * Sequences obtained after cloning (from each sample, six identical sequences were obtained after cloning, but only one per sample was included in the analyses and sent to GenBank).
ITS nrDNA
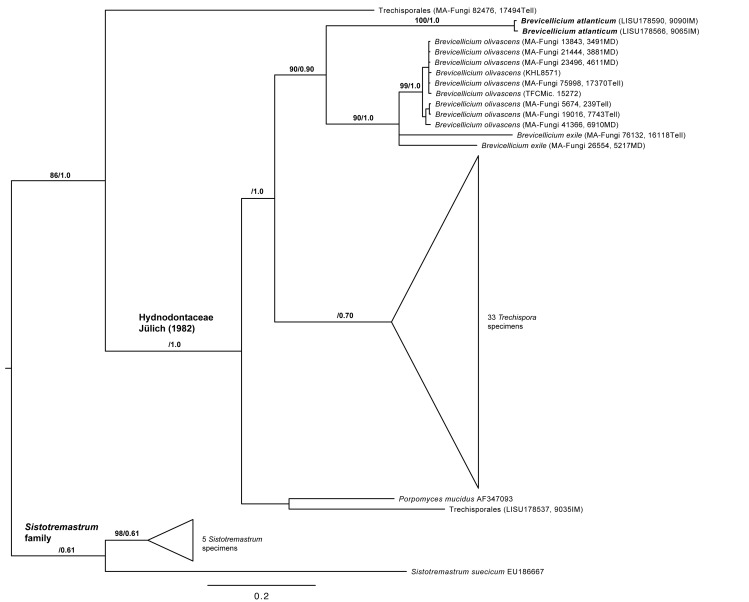
Thirteen new ITS nrDNA sequences were aligned with 42 sequences available in GenBank including six Sistotremastrum sequences serving as outgroup. The resulting matrix consisted of 871 unambiguously aligned nucleotide position characters. Among them, 314 positions were constant, 127 were parsimony–uninformative and 410 were parsimony-informative. In the maximum parsimony analysis under exhaustive search, 100 most parsimonious trees (MPTs) were obtained (tree length = 1407, CI = 0.6041, RI = 0.8113, and RC = 0.491). The trees obtained from the MP (strict consensus tree, data not shown) and Bayesian analyses show similar topologies (Fig. 2). Similar to the LSU analyses the Brevicellicium sequences form a clade (bs = 90 %, pp = 0.90), in a sister-group relationship to all Trechispora sequences (bs < 50 %, pp = 1.0). Sequences from LISU 178566 and LISU 178590 form a highly supported clade (bs = 100, pp = 1.0), sister group of B. exile and B. olivascens clade (bs = 90 %, pp = 1.0). The nine B. olivascens sequences form a highly supported clade (bs = 99 %, pp = 1.0), with low genetic variability (uncorrected “p” distances from 0.0 to 0.018), whereas the two B. exile sequences do not group together and show a high genetic variability (uncorrected “p” distance equal 0.139); apparently here are more taxonomical problems hidden that needs to be addressed.
Fig. 2.
The 50 % majority-rule consensus tree of Bayesian analysis based on ITS nrDNA sequences. Bootstrap (> 50 %) and posterior probability values indicated on the branches. The topology was rooted with Sistotremastrum species. Trechispora and Sistotremastrum clades not discussed in the text are indicated as triangles. The position of Brevicellicium atlanticum is indicated in bold.
Since LISU 178566 and LISU 178590 also have unique morphological characters we find reasons to describe them as a new species: Brevicellicium atlanticum.
TAXONOMY
Brevicellicium atlanticum Melo, Telleria, M. Dueñas & M.P. Martín, sp. nov.
MycoBank MB800016
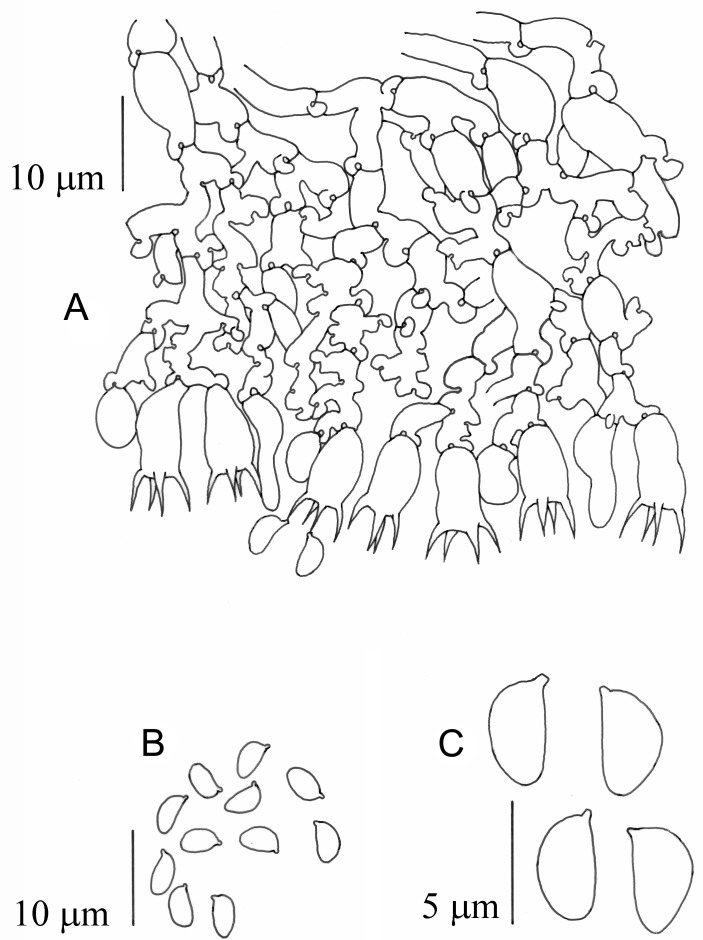
(Fig. 3)
Fig. 3.
Brevicellicium atlanticum (LISU 178566). a. Vertical section through basidiome; b–c, spores.
Etymology: The Azores Archipelago is situated in the middle of the North Atlantic and atlanticum is derived from Atlantic Ocean.
Diagnosis: Basidiome resupinate, membranaceous, smooth, whitish. Hyphal system monomitic, hyphae with clamps, subhymenial hyphae to 6.0 μm diam. Basidia clavate to short cylindrical, 8.0–10.0 × 4.5–5.5 μm, with 4 sterigmata. Basidiospores short ellipsoid with a prominent apiculus, (3.8−) 4.0–4.5 × (2.0−) 2.3–2.5 μm.
Type: Portugal: Azores: Terceira, Angra do Heroismo, Terra-Chã, Matela de Baixo, 26SMH7783, 470m asl, on live trunk of Erica azorica, 10 Mar. 2005, I. Melo & J. Cardoso 9065IM (LISU 178566 – holotype).
Description: Basidiome resupinate, effused, adnate, very thin, porose, membranaceous; hymenophore smooth, whitish; margin indeterminate. Hyphal system monomitic, hyphae with clamps, subiculum very thin, consisting of a few thin-walled, uniform, 2.5–3.5 μm diam. hyphae, subhymenial hyphae richly branched, wider, some isodiametric and up to 6.0 μm diam. Cystidia absent. Basidia short clavate to short cylindrical, basally clamped, 8.0–10.0 × 4.5–5.5 μm, with 4 sterigmata up to 4.5 μm long. Basidiospores short ellipsoid with a prominent apiculus, smooth, thin-walled, (3.8) 4.0–4.5 × (2.0) 2.3–2.5 μm, inamyloid, indextrinoid, acyanophilous.
Substratum: On live trunk of Erica azorica and on decayed branch of Juniperus brevifolia ssp. azorica, both endemic plants from the Azores Archipelago.
Additional specimen examined: Portugal: Azores: Terceira, Angra do Heroismo, Mistérios Negros, 26SMH7587, 630 m asl, on decayed branch of Juniperus brevifolia ssp. azorica, 2 Mar 2005, I. Melo & J. Cardoso 9090IM (LISU 178590).
Notes: Overall, Brevicellicium atlanticum is morphologically most similar to B. exile, but the latter has wider basidia and larger spores, 9–11 × 5–6.5 μm and 4.5–5 × 2.5–3.5 μm respectively (Jackson 1950); 9–12 × 5.5–8 μm and 4.5–6 × 2.5–3.5 μm in specimens from the Iberian Peninsula (Telleria & Melo 1995) or 10–15 × 5–6.5 μm and 5–6.5 × 3.5–4 μm from the Azores Archipelago (Telleria et al. 2009 a, b). Also, B. flavovirens and B. udinum have a similar spore morphology, but the former has a yellowish grey basidiome and wider spores (4.5–5.0 (–5.5) × 3.0–3.5 μm). In B. udinum the basidiome is thick, cracked when dry, and the spores are narrowly ellipsoid, 5.0–5.5 (–6.0) × 2.5–2.75 μm.
Boidin & Gilles (1990) reported a specimen morphologically similar to Brevicellicium exile from France, Landes, Carcen-Ponson, on Alnus glutinosa, LY 13883, but differing in the narrower basidia (8–14 × 4–5 μm) and smaller spores (3.5–4.5 × 2–2.5 μm). This material was not available to us but could well represent B. atlanticum.
DISCUSSION
Trechisporales is a rather small order described by Larsson (Hibbett et al. 2007) and placed in the subphylum Agaricomycotina, class Agaricomycetes, with three exemplar genera included in the original description: Trechispora, Sistotremastrum, and Porpomyces. In his molecular phylogenetic classification of the corticioid fungi, Larsson (2007) included sequences of Trechispora farinacea (AF347089), T. hymenocystis (AF347090), Subulicystidium (AY463468/AY586714), Porpomyces mucidus (AF347091) and Sistotremastrum niveocremeum (AF347094), and preliminarily recognized two families in the order: Hydnodontaceae (Jülich 1982) with the genera Brevicellicium, Fibriciellum, Fibrodontia, Luellia, Porpomyces, Subulicystidium, Trechispora and Tubulicium; and the Sistotremastrum family with the genus Sistotrematrum. Besides, he listed the genera Dextrinocystis, Dextrinodontia and Litchauerella as possible candidates to be included in Hydnodontaceae. The new genus Brevicellopsis, segregated from Brevicellicium (Hjortstam & Ryvarden 2008), could be another possible candidate to be included in this family.
The molecular phylogenetic analyses of the present study support Trechisporales as a monophyletic group with the species of Porpomyces, Sistotremastrum, Subulicystidium, and Trechispora forming a highly supported monophyletic clade where all Brevicellicium sequences are included. Our results also support the two families of this order: Hydnodontaceae where Brevicellicium and Trechispora are included and the Sistotremastrum family (Jülich 1982, Larsson 2007). Most species of Brevicellicium have yet to be included in molecular phylogenetic analyses. Only then can outstanding issues like the independent status of Brevicellopsis and the uncertain position for Brevicellicium permodicum be resolved.
Acknowledgments
We are grateful to Esperanza Beltrán-Tejera and J. Laura Rodríguez-Armas for kindly providing us Brevicellicium olivascens specimen from Pico Island, Azores Archipelago (TFCMic 15272), and Fátima Durán for technical assistance. Financial support was provided by DGI project CGL2009–07231.
REFERENCES
- Abarenkov K, Nilsson RH, Larsson K-H, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Ursing BM, Vrålstad T, Liimatainen K, Peintner U, Kõljalg U. (2011) The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytologist 186: 281–285 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhan J, Zhan GZ, Millar W, Lipman DJ. (1997) Gapped BLAST and PSI–BLAST: a new generation of protein database search programs. Nucleic Acid Research 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum DA, Sytsma KJ. (1994) A phylogenetic analysis of Epilobium (Onagraceae) based on nuclear ribosomal DNA sequences. Systematic Botany 19(3): 363–388 [Google Scholar]
- Bernicchia A, Gorjón SP. (2010) Corticiaceae s.l. Fungi Europaei 12 Ed. Candusso; Italia: [Google Scholar]
- Binder M, Hibbet DS, Larsson K-H, Larsson E, Langer E, Langer G. (2005) The phylogenetic distribution of resupinate forms across the major clades of mushroom-forming fungi (Homobasidiomycetes). Systematics and Biodiversity 3: 1–45 [Google Scholar]
- Boidin J, Gilles G. (1990) Corticiés s.l. intéressants ou nouveaux pour la France (Basidiomycotina). Bulletin Trimestrel de la Société Mycologique de France 106: 135–167 [Google Scholar]
- Bresadola G. (1892) Fungi Tridentini II. Ed. J. Zippei. Italia: [Google Scholar]
- Bresadola G. (1920) Selecta mycologica. Annales Mycologici 18: 26–70 [Google Scholar]
- Cochrane G, Karsch-Mizrachi I, Nakamura Y. (2010) The International Nucleotide Sequence Database Collaboration. Nucleic Acids Research 39: D15–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham GH. (1963) The Telephoraceae of Australia and New Zealand. New Zealand Department of Scientific and Industrial Research Bulletin 145: 1–359 [Google Scholar]
- Farris JS. (1989) The retention index and the rescaled consistency index. Cladistics 5: 417–419 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specifity for basidiomycetes –application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118 [DOI] [PubMed] [Google Scholar]
- Gilbertson RL, Desjardin DE, Rogers JD, Hemmes DE. (2001) Fungi from Mamane –Naio vegetation zone from Hawaii. Fungal Diversity 6: 35–69 [Google Scholar]
- Ginns J, Lefebvre MNL. (1993) Lignicolous Corticioid Fungi (Basidiomycota) of North America. Systematics, Distribution, and Ecology. Mycologia Memoir 19: 1–247 [Google Scholar]
- Hallenberg N. (1981) Synopsis of Wood-Inhabiting Aphyllophorales (Basidiomycetes) and Heterobasidiomycetes from Iran. Mycotaxon 12: 473–502 [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, Thorsten Lumbsch H, Lutzoni F, Brandon Matheny P, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai Y-C, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Kõljalg U, Kurtzman CP, Larsson K-H, Lichtwardt R, Longcore J, Miądlikowska J, Miller A, Moncalvo J-M, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schüßler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao Y-J, Zhang N. (2007) A higher-level phylogenetic classification of the Fungi. Mycological Research 111: 509–547 [DOI] [PubMed] [Google Scholar]
- Hjortstam K. (2001) Two new species of Brevicellicium and a survey of tropical and subtropical species in the genus (Basidiomycotina, Aphyllophorales). Mycotaxon 79: 181–187 [Google Scholar]
- Hjortstam K, Larsson K-H. (1978) Notes on Corticiaceae (Basidiomycetes) II. Mycotaxon 7: 117–124 [Google Scholar]
- Hjortstam K, Larsson K-H, Ryvarden L. (1988) The Corticiaceae of North Europe. Vol. 8 Fungiflora. Norway: [Google Scholar]
- Hjortstam K, Ryvarden L. (1980) Studies in Tropical Corticiaceae (Basidiomycetes) II. Mycotaxon 12: 168–184 [Google Scholar]
- Hjortstam K, Ryvarden L. (1997) Corticioid species (Basidiomycotina, Aphyllophorales) from Colombia collected by Leif Ryvarden. Mycotaxon 64: 229–241 [Google Scholar]
- Hjortstam K, Ryvarden L. (2007) Studies in corticioid fungi from Venezuela III (Basidiomycotina, Aphyllophorales). Synopsis Fungorum 23: 56–107 [Google Scholar]
- Hjortstam K, Ryvarden L. (2008) Some corticioid fungi (Basidiomycotina) from Ecuador. Synopsis Fungorum 25: 14–27 [Google Scholar]
- Hjortstam K, Ryvarden L, Iturriaga T. (2005) Studies in corticioid fungi from Venezuela II (Basidiomycotina, Aphyllophorales). Synopsis Fungorum 20: 42–78 [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294: 2310–2314 [DOI] [PubMed] [Google Scholar]
- Jackson HS. (1950) Studies of Canadian Thelephoraceae. VII. Some new species of Corticium sect. Athele. Canadian Journal of Research, Sect C, Botanical Sciences 28: 716–725 [Google Scholar]
- Jülich W. (1982) Higher Taxa of Basidiomycetes. Bibliotheca Mycologica 85: 1–485 [Google Scholar]
- Kluge AG, Farris JS. 1969. Quantitative phyletics and the evolution of anurans. Systematic Zoology 18: 1–32 [Google Scholar]
- Larget B, Simon DL. (1999) Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Molecular Biology and Evolution 16: 750–759 [Google Scholar]
- Larsson K-H. (2007) Re-thinking the classification of corticioid fungi. Mycological Research 111: 1040–1063 [DOI] [PubMed] [Google Scholar]
- Larsson K-H, Larsson E, Kõljalg U. (2004) High phylogenetic diversity among corticioid homobasidiomycetes. Mycological Research 108: 983–1002 [DOI] [PubMed] [Google Scholar]
- Maekawa N. (1993) Taxonomic study of Japanese Corticiaceae (Aphyllophorales) I. Report of the Tottori Mycological Institute 31: 1–149 [Google Scholar]
- Martín MP, Winka K. (2000) Alternative methods of extracting and amplifying DNA from lichens. Lichenologist 32: 189–196 [Google Scholar]
- Nylander JAA. (2004) MrModeltest v. 2. Program distributed by the author. Uppsala: Evolutionary Biology Center, Uppsala University; [Google Scholar]
- Page RDM. (1996) TreeView, an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2002) Se-Al: sequences alignment editor v. 2.0a11. Edinburgh: Institute of Evolutionary Biology; http://tree.bio.ed.ac.uk/software/ [Google Scholar]
- Rehner SA, Samuels GJ. (1994) Taxonomy and phylogeny of Gliocladium analyzed from nuclear large subunit ribosomal DNA sequences. Mycological Research 98: 625-634 [Google Scholar]
- Rodríguez F, Oliver JF, Martín A, Medina JR. (1990) The general stochastic model of nucleotide substitution. Journal of Theoretical Biology 142: 485–501 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Ryvarden L. (1976) Studies in the Aphyllophorales of the Canary Islands. 3. Some species from the western islands. Cuadernos de Botánica Canaria 20: 3–8 [Google Scholar]
- Swofford DL. (2003) PAUP* : phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts; USA: [Google Scholar]
- Telleria MT, Melo I. (1995) Aphyllophorales resupinatae non poroides, I. Acanthobasidium – Cystostereum. Flora Mycologica Iberica 1: 1–223 [Google Scholar]
- Telleria MT, Melo I, Dueñas M. (1993) Aphyllophorales (Basidiomycetes) of the National Park of “Ordesa y Monte Perdido” (Spain). Nova Hedwigia 57: 207–217 [Google Scholar]
- Telleria MT, Melo I, Dueñas M, Rodríguez-Armas JL, Beltrán-Tejera E, Cardoso J, Salcedo I. (2009a) Diversity and richness of corticioid fungi (Basidiomycota) on Azores Islands: a preliminary survey. Nova Hedwigia 88: 285–308 [Google Scholar]
- Telleria MT, Melo I, Dueñas M, Salcedo I, Cardoso J, Rodríguez-Armas JL, Beltrán-Tejera E. (2009b) Corticioid fungi (Basidiomycota) from Azores Islands: Flores and São Miguel. Mycotaxon 109: 141–144 [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR protocols: a guide to methods and applications: 315–322 San Diego: Academic Press; [Google Scholar]