Abstract
Background:
Invasive fungal infections cause considerable morbidity and mortality in immunocompromised hosts. Pigeon droppings could especially be a potential carrier in the spread of pathogenic yeasts and mold fungi into the environment. The objective of this study was to isolation of Cryptococcus neoformans and other opportunistic fungi from pigeon droppings.
Materials and Methods:
One hundred twenty samples of pigeon droppings were suspended 1:10 in saline solution and then cultured. Identification of C. neoformans was performed on bird seed agar, presence of a capsule on India ink preparation, urease production on urea agar medium and RapID yeast plus system. The identification of candida species was based on micro-morphological analysis on corn meal-Tween 80 agar, RapID yeast plus system and growth in CHROMagar candida. The identification of other fungi was based on macromorphologic, microscopic, biochemical and physiological characteristics.
Results:
The highest frequency of yeasts and mold fungi were observed in Candida albicans 6.6% and Penicillium spp. 25%. The frequency rate of C. neoformans isolation was 2.5%.
Conclusion:
Several types of fungi are present in pigeon droppings that can spread in environment and transmit to children and elderly as well as immunocompromised patients who are at increased risk of contracting opportunistic diseases.
Keywords: Cryptococcus neoformans, candida albicans, immunocompromised hosts, opportunistic fungi, pigeon droppings
INTRODUCTION
Immunocompromised patients are susceptible to bacterial, fungal, and viral infections that healthy immune systems usually conquer. They are also susceptible to common infections of childhood. The patient is immunocompromised either because of an acquired or inherited immune deficiency disorder. The number of immunocompromised patients is increasing due to the intensive therapy being administered those with cancer, organ transplant, and HIV infection.[1]
The incidence of invasive opportunistic mycoses has increased because of the expanding population of immunosuppressed patients, including solid-organ transplant (SOT) and hematopoietic stem cell transplant (HSCT) recipients, patients with cancer, patients with AIDS, premature neonates, elderly patients, and patients recovering from major surgery. Despite some effective treatment options, such mycoses are associated with high morbidity and mortality rates.[2,3,4]
Opportunistic mycoses show distinct regional incidence patterns throughout the world and may exhibit different epidemiologic features, depending on the geographic region; this may be particularly true for mycoses that are acquired from the environment.[1]
Avian droppings have been reported as a potential environmental source of human pathogenic yeasts. Pigeon droppings, which are abundant in public areas, could especially be a potential carrier in the spread of pathogenic yeasts i-nto the environment and subsequently humans.[5]
Since the 1960s a number of new species of yeast has been reported as pathogens for humans because of the increase in immunodeficiency syndromes and immunosuppressant chemotherapy in cancer treatment.[6]
Cryptococcus is a systemic infection caused by the yeast Cryptococcus neoformans, is the most common life-threatening systemic fungal infection occurring in HIV-infected patients.[4,5,7] Cryptococcus neoformans is an opportunistic fungal pathogen that causes meningoencephalitis as the most frequent clinical presentation in immunocompromised patients, mainly in people infected by HIV.[8]
Yeasts of the genus Candida are those most frequently isolated from blood stream infections in immunocompromised patients although they are also found in the transient normal flora of the skin and digestive tract of humans. Other important yeasts such as Rhodotorula, Trichosporon, were also frequently isolated from pigeon excreta and the cloacae of coots respectively. These fungi can cause blood infections in patients with indwelling central venous catheters and cancer.[6]
In recent years, opportunistic fungal infections have increased substantially, and the species of the genus Aspergillus, Penicillium, Mucor, Rhizopus, Paecilomyces, Fusarium, Alternaria, and Cladosporium are emerging as the cause of a variety of infections in humans.[9,10,11,12,13]
Large numbers of pigeons have inhabited different areas surrounding the hospital and can act as a source of opportunistic fungi, therefore hospitalized patients and immunocompromised persons are susceptible to a number of opportunistic infections.[8] Thus it is important for these individuals to have an understanding of the risks associated with the different species of animals and birds.
The isolation and identification of different fungi from pigeon droppings can provide useful information for ecological and epidemiological studies. Therefore, we decided to survey about the presence of C. neoformans, other yeasts and molds in pigeon droppings collected from pigeon towers.
MATERIALS AND METHODS
One hundred twenty samples, each containing about 500 grams of pigeon droppings were collected from 19 pigeon towers in urban areas of Isfahan in a period of 9 months (Sept. 2010-May 2011) and the samples were processed using a standard protocol.[14] In brief, a suspension of droppings, was spread onto the surface on Sabouraud's Dextrose Agar (SDA) medium supplemented with chloramphenicol (200 mg/l) and incubated at 32°C for 15 days.[14] The identification of isolates was established by niger seed agar (NSA) (for melanin production) medium and India ink preparation.[15] Standard strain of C. neoformans ATCC 28957 was used as a positive control in parallel with samples.
Since C. neoformans is sensitive to canavanine and cannot utilize glycine as a sole carbon and nitrogen sources, and will therefore not grow, and the media remains yellow in color, therefore, biotyping was carried out using Canavanine-Glycine-Bromothymol blue (CGB) agar growth test, as described by Kwon Chung et al.[16] Additionally, cryptococcus identification was carried out by RapID yeast plus system (RYP) which is a 4-h micro panel system that utilizes single-substrate test reactions based on performed enzymes in tested isolates to identify yeasts. Reactions are determined by color changes of chromogenic substrates within the sample wells, some of which require addition of reagents. Test reactions are grouped in triads and scored on the basis of a positive or negative reaction, resulting in a 6-digit microcode which is matched to an empirically derived database of commonly occurring microcodes for identification of the isolate.[17]
The identification of candida species was based on micro-morphological analysis on corn meal-Tween 80 agar, RapID yeast plus system and growth in CHROMagar candida.[18]
For each isolate, the identifications obtained to the species and genus level, but sometimes additional tests (low probability identifications) were required to (i) distinguish between two or more possible species, (ii) for its discrepant identifications, and (iii) for its failure to provide and identifications (no codes). When the six-digit microcode provided an identification with a low percentage of probability (<94%), the additional tests were performed by conventional methods.[19] The identification of other fungi was based on macromorphologic, microscopic, biochemical and physiological characteristics.
RESULTS
Three out of 120 samples, (2.5%), were diagnosed as Cryptococcus neoformans by using RapID yeast plus system and CGB test.
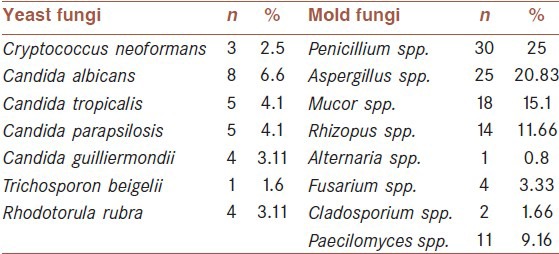
Isolated yeast species, other than Cryptococcus, were Candida albicans (n = 8), C. tropicalis (n = 5), C. parapsilosis (n = 5), C. guilliermondii (n = 4), Rhodotorula spp. (n = 4) and Trichosporon (n = 1). Mold fungi including Penicillium spp. (n = 30), Aspergillus spp. (n = 25), Mucor spp. (n = 18), Rhizopus spp. (n = 14), Alternaria spp. (n = 1), Fusarium spp. (n = 4), Cladosporium spp. (n = 2), Paecilomyces spp. (n = 11) [Table 1].
Table 1.
Frequencies of different fungi species isolated from pigeon dropping
The RapID yeast plus system is easy to use with only a few exceptions and the reactions are distinct and easy to interpret. As can be seen in Table 2, 26 of 30 strains (86.66%) were correctly identified to the species level without additional tests. Three organisms (10%) yielded the correct identification among two or more possibilities, with the correct identification upon performance of additional tests. These comprised Rhodotorula rubra with Trichosporon beigelii and Rhodotorula rubra, Candida guilliermondii with C. guilliermondii and Candida famata and finally Cryptococcus neoformans with C. neoformans and Pichia spp. Isolates [Table 3].
Table 2.
Identification of yeast and yeast like-fungi by the RapID yeast plus system
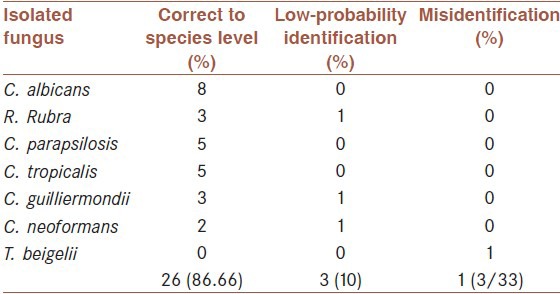
Table 3.
Details of low-probability identifications
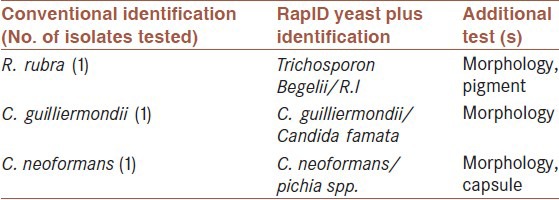
Misidentification occurred for 1 strain (3.33%), this comprised one Trichosporon beigelii identified as C. neoformans isolate.
DISCUSSION
Both acquired and congenital immunodeficiency may be associated with increased susceptibility to invasive fungal infections (IFIs), depending on the type of immune deficit. IFIs frequently occur in patients with phagocytic and cellular immune defects, but are rarely observed in those with humoral or complement defects. Aspergillus, Candida, Cryptococcus, Histoplasma and other fungal genera are variably implicated in causing invasive infections in these patients.[20]
In the past, thousands of penthouses, a home for pigeons, were made in Isfahan, and collected pigeon droppings were a valuable resource of natural fertilizer for agricultural fields. Nowadays, many of the towers are falling apart and due to the introduction of inexpensive artificial fertilizer, and extension of urbanization into countryside, only a few hundred pigeon towers remain operational. Current study was done to evaluate yeast and mold fungi frequency in some of these towers that are not completely suspended yet.
Costa et al. 2009 isolated Cryptococcus neoformans var. neoformans, C. laurentii, Candida spp, Rhodotorula mucilaginosa, and Trichosporon sp. from pigeon droppings. They confirmed that urban pigeons are a potential source of pathogenic yeasts.[21]
A.R. Khosravi 1997, 983 specimens of pigeon droppings, collected in different regions of northern Iran. Saprophytic fungi Aspergillus, Candida, Mucor and Penicillium spp. were the most frequently isolated saprophytes.[22]
In this study, C. neoformans was identified in 2 pigeon dropping samples (10/52% of total samples). Other isolated fungi were Candida albicans, C. tropicalis, C. parapsilosis, C. guilliermondii, Rhodotorula spp. and Trichosporon, Penicillium spp., Aspergillus spp., Mucor spp., Rhizopus spp., Alternaria spp., Fusarium spp., Cladosporium spp., and Paecilomyces spp.
Pigeon dropping is found in public areas, such as gardens, streets, hospitals, pigeon towers, old buildings, and etc., Our isolates were found in pigeon towers, suggesting a potential public health hazard due to human exposure to airborne cells of C. neoformans and other fungi in public areas.
The association of cryptococcosis with exposure to pigeon droppings, the best known of the putative exposure risk factors for this disease, comes almost exclusively from information about the isolation of C. neoformans from the environment and a few anecdotal reports.[23]
The incidence of cryptococcosis has shown marked increases over the last years. The number of reported cases of cryptococcosis has risen to relatively high levels, and is considered the most frequent systemic infection of opportunistic fungus in patients with Aids. These findings reinforce the hypothesis that pigeon's droppings are an important factor of yeast infection in the urban environment.[24]
Infections by opportunistic and pathogenic fungi in compromised patients, caused by candida spp., Aspergillus spp., Cryptococcus neoformans, Histoplasma capsulatum and Coccidioides immitis are increased, many fungi that occur as saprophytes in the environment and which had previously been considered to be nonpathogenic are now being encountered as causes of human infection.[25]
In a study (2001) in Isfahan and suburbs, the frequency rate of C. neoformans was detected 8.1%. The obvious decline in C. neoformans frequency in our study may be occurred because of changing environmental conditions in recent years.[26]
In our study, Penicillium spp. was the most frequently isolated saprophytes followed by Aspergillus spp., Mucor spp., Rhizopus spp., Alternaria spp., Fusarium spp., Cladosporium spp., and Paecilomyces spp. These fungi can be dangerous in immunocompromised persons.
In a study, R. rubra was frequently isolated from pigeon excreta. This yeast is dangerous in humans, since it causes blood infections in patients with indwelling central venous catheters and cancer.[6] In our study, R. rubra was isolated from four pigeon droppings samples.
These results demonstrate that pigeon droppings are an important and environmentally source of yeast and mold fungi in areas of Isfahan.
The best way to control pigeons in urban areas particularly is to reduce the food supply by not feeding them so that the large flocks disperse, and to put rubbish in secure bins so they cannot scavenge. Reducing access to nest sites will also help to limit the population.
Fecal dust drawn into hospitals through rooftop heating, ventilation and air-conditioning ducts represents a serious health hazard. Without effective bird control, a serious infectious disease problem can develop.
CONCLUSION
Further studies, will be needed to investigate the geographic distribution and population structure of fungi from pigeon droppings, as well as the possible associations of disease with exposure to environmental pigeon droppings.
Footnotes
Source of Support: We wish to thank from all personnel's Faculty of Specialized Veterinary Science, Science and Research Branch, Islamic Azad University, Tehran for supporting of this research
Conflict of Interest: None declared.
REFERENCES
- 1.Kohno S. Fungal infections in immunocompromised patients. Nihon Ishinkin Gakkai Zasshi. 2000;41:71–6. doi: 10.3314/jjmm.41.71. [DOI] [PubMed] [Google Scholar]
- 2.Nucci M, Queiroz-Telles F, Tobon AM, Restrepo A, Colombo AL. Epidemiology of opportunistic fungal infections in Latin America. Clin Infect Dis. 2010;51:561–70. doi: 10.1086/655683. [DOI] [PubMed] [Google Scholar]
- 3.Warnock DW. Trends in the epidemiology of invasive fungal infections. Nihon Ishinkin Gakkai Zasshi. 2007;48:1–12. doi: 10.3314/jjmm.48.1. [DOI] [PubMed] [Google Scholar]
- 4.Dromer F, Mathoulin-Pélissier S, Fontanet A, Ronin O, Dupont B, Lortholary O. Epidemiology of HIV-associated cryptococcosis in France (1985–2001): Comparison of the pre- and post-HAART eras. AIDS. 2004;18:555–62. doi: 10.1097/00002030-200402200-00024. [DOI] [PubMed] [Google Scholar]
- 5.Chee HY, Lee KB. Isolation of Cryptococcus neoformans var. grubii (serotype A) from pigeon droppings in Seoul, Korea. J Microbiol. 2005;43:469–72. [PubMed] [Google Scholar]
- 6.Cafarchia C, Camarda A, Romito D, Campolo M, Quaglia NC, Tullio D, et al. Occurrence of yeasts in cloacae of migratory birds. Mycopathologia. 2006;161:229–34. doi: 10.1007/s11046-005-0194-z. [DOI] [PubMed] [Google Scholar]
- 7.Chen S, Sorrell T, Nimmo G, Speed B, Currie B, Ellis D, et al. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformansin Australia and New Zealand. The Australasian Cryptococcal Study Groupa. Clin Infect Dis. 2000;31:499–508. doi: 10.1086/313992. [DOI] [PubMed] [Google Scholar]
- 8.Louisiana Office of Public Health – Infectious Disease Epidemiology Section – Annual Report. Cryptococcosis. 2010. [Last access on 2013 Jan 31]. pp. 1–8. Available from: http://new.dhh.louisiana.gov/assets/oph/Center-PHCH/Center-CH/infectious-epi/Annuals/LaIDAnnual_Cryptococcus.pdf .
- 9.Shah SS, Birnbaum BA, Jacobs JE. Disseminated aspergillosis inciting intestinal ischaemia and obstruction. Br J Radiol. 2001;74:1145–7. doi: 10.1259/bjr.74.888.741145. [DOI] [PubMed] [Google Scholar]
- 10.Dannaoui E, Garcia-Hermoso D. The Zygomycetes. In: Kavanagh K, editor. New insights in fungal pathogenicity. Dordrecht, The Netherlands: Springer Science; 2007. pp. 159–83. [Google Scholar]
- 11.Nucci M, Anaissie E. Fusarium Infections in Immunocompromised Patients. Clin Microbiol Rev. 2007;20:695–704. doi: 10.1128/CMR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courville P, Favennec L, Viacroze C, Barrel A, Young P, Abboud P, et al. Co-existent cutaneous cryptococcosis of the forearm and cutaneous alternariosis of the leg in patient with metastatic thymoma. J Cutan Pathol. 2002;29:55–8. doi: 10.1046/j.0303-6987.2001.00035.x. [DOI] [PubMed] [Google Scholar]
- 13.Aguilar C, Pujol I, Sala J, Guarro J. Antifungal susceptibilities of Paecilomycesspecies. Antimicrob Agents Chemother. 1998;42:1601–4. doi: 10.1128/aac.42.7.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarrini M, Jorfi M, Amirrajab N, Rostami M. Isolation of Cryptococcus neoformans from pigeon droppings in Ahwaz, Iran. Turk J Med Sci. 2010;40:313–6. [Google Scholar]
- 15.Horta JA, Staats CC, Casali AK, Ribeiro AM, Schrank IS, Schrank A, et al. Epidemiological aspects of clinical and environmental Cryptococcus neoformansi solates in the Brazilian state Rio Grande do Sul. Med Mycol. 2002;40:565–71. doi: 10.1080/mmy.40.6.565.571. [DOI] [PubMed] [Google Scholar]
- 16.Kwon-Chung KJ, Polacheck I, Bennett JE. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C) J Clin Microbiol. 1982;15:535–7. doi: 10.1128/jcm.15.3.535-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith MB, Daisy D, Vu H, Woods GL. Comparative performance of the RapID Yeast Plus System and the API 20C AUX Clinical Yeast System. J Clin Microbiol. 1999;37:2697–8. doi: 10.1128/jcm.37.8.2697-2698.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wadline JK, Hanko G, Stewart R, Pape J, Nachamkin I. Comparison of Three Commercial Systems for Identification of Yeasts Commonly Isolated in the Clinical Microbiology Laboratory. J Clin Microbiol. 1999;37:1967–70. doi: 10.1128/jcm.37.6.1967-1970.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espinel-Ingroff A, Stockman L, Roberts G, Pincus D, Pollack J, Marler J. Comparison of RapID Yeast Plus System with API 20C system for identification of common, new, and emerging yeast pathogens. J Clin Microbiol. 1998;36:883–6. doi: 10.1128/jcm.36.4.883-886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antachopoulos C. Invasive fungal infections in congenital immunodeficiencies. Clin Microbiol Infect. 2010;16:1335–42. doi: 10.1111/j.1469-0691.2010.03289.x. [DOI] [PubMed] [Google Scholar]
- 21.Costa AK, Sidrim JJ, Cordeiro RA, Brilhante RS, Monteiro AJ, Rocha MF. Urban pigeons (Columba livia) as a potential source of pathogenic yeasts: A focus on antifungal susceptibility of Cryptococcus strains in Northeast Brazil. Mycopathologia. 2010;169:207–13. doi: 10.1007/s11046-009-9245-1. [DOI] [PubMed] [Google Scholar]
- 22.Khosravi AR. Isolation of Cryptococcus neoformans from pigeon (Columbia livia) droppings in northern Iran. Mycopathologia. 1997;139:93–5. doi: 10.1023/a:1006863705759. [DOI] [PubMed] [Google Scholar]
- 23.Hajjeh RA, Conn LA, Stephens DS, Baughman W, Hamill R, Graviss E, et al. Cryptococcosis: Population-based multistate active surveillance and risk factors in human immunodeficiency virus–infected persons. Cryptococcal Active Surveillance Group. J Infect Dis. 1999;179:449–54. doi: 10.1086/314606. [DOI] [PubMed] [Google Scholar]
- 24.Currie BP, Casadevall A. Estimation of the prevalence of cryptococcal infection among patients infected with the human immunodeficiency virus in New York City. Clin Infect Dis. 1994;19:1029–33. doi: 10.1093/clinids/19.6.1029. [DOI] [PubMed] [Google Scholar]
- 25.Richardson MD. Opportunistic and pathogenic fungi. J Antimicrob Chemother. 1991;28:1–11. doi: 10.1093/jac/28.suppl_a.1. [DOI] [PubMed] [Google Scholar]
- 26.Isfahani BN, Shadzi SH, Pour MC, Ilchi N. Isolation and detection of Cryptococcus neoformans from pigeon droppings: Isfahan and its suburb province pigeon towers. J Res Med Sci. 2001;6:20–2. [Google Scholar]


