Abstract
Genetic alterations in kinases have been linked to multiple human pathologies. To explore the landscape of kinase genetic variation in gastric cancer (GC), we used targeted, paired-end deep sequencing to analyze 532 protein and phosphoinositide kinases in 14 GC cell lines. We identified 10,604 single-nucleotide variants (SNV) in kinase exons including greater than 300 novel nonsynonymous SNVs. Family-wise analysis of the nonsynonymous SNVs revealed a significant enrichment in mitogen-activated protein kinase (MAPK)-related genes (P < 0.01), suggesting a preferential involvement of this kinase family in GC. A potential antioncogenic role for MAP2K4, a gene exhibiting recurrent alterations in 2 lines, was functionally supported by siRNA knockdown and overexpression studies in wild-type and MAP2K4 variant lines. The deep sequencing data also revealed novel, large-scale structural rearrangement events involving kinases including gene fusions involving CDK12 and the ERBB2 receptor tyrosine kinase in MKN7 cells. Integrating SNVs and copy number alterations, we identified Hs746T as a cell line exhibiting both splice-site mutations and genomic amplification of MET, resulting in MET protein overexpression. When applied to primary GCs, we identified somatic mutations in 8 kinases, 4 of which were recurrently altered in both primary tumors and cell lines (MAP3K6, STK31, FER, and CDKL5). These results demonstrate that how targeted deep sequencing approaches can deliver unprecedented multilevel characterization of a medically and pharmacologically relevant gene family. The catalog of kinome genetic variants assembled here may broaden our knowledge on kinases and provide useful information on genetic alterations in GC.
Introduction
Protein and lipid kinases play important roles in diverse biological processes, ranging from tissue differentiation and cellular proliferation to axonal migration and organ homeostasis (1-3). As critical nodes of cellular signaling pathways, kinases frequently integrate signaling outputs of different signal transduction circuits, linking extracellular signals to nuclear gene transcription programs (1-3). In humans, genetic alterations and polymorphisms in kinases have been implicated in a wide variety of diseases, ranging from congenital defects, neurologic disease, diabetes, and cancer (4-6). On top of their medical relevance, kinases are also notable to the pharmaceutical industry as they are highly amenable to targeting by rationally designed small molecule inhibitors (7). The prominence of kinases in biology and medicine, thus, renders it essential to determine the spectrum of kinase genetic variation in healthy individuals and patients with specific diseases.
Recent advances in DNA sequencing technologies have reduced the cost of decoding human genomes by several orders of magnitude (8). At present, however, sequencing a complete human genome at sufficient depth to identify single-nucleotide variants (SNV) still represents a highly resource-intensive effort, particularly if many genomes need to be analyzed. Recently, innovative approaches have been described for integrating such massively parallel sequencing platforms with sequence capture technologies, such as molecular inversion probes (9, 10), sequence capture arrays (11-13), and solution phase hybridization (14), enabling specific genomic regions to be selected before sequencing. Such “targeted deep sequencing” approaches are particularly suitable for rapidly profiling specific genomic regions across multiple individuals and also carry the additional advantage of being able to provide insights into other levels of variation in addition to SNVs such as structural and copy number variants.
Targeted deep sequencing surveys have been reported for various disease genes (15, 16); however, to date, targeted deep sequencing of the kinome has not been reported. Here, we applied targeted deep sequencing to characterize the extent of kinome genetic variation in gastric cancer (GC), the second leading cause of global cancer-related mortality and a disease for which kinase dysregulation has been implicated (17, 18). Surveying the protein-coding regions of 537 kinases and genes across 14 commonly used GC cell lines, we detected more than 300 novel kinase SNVs and identified mitogen-activated protein kinase (MAPK) cascade genes as frequently altered kinases in GC. We also identified structural variants and splice-site alterations affecting ERBB2 and MET, 2 pharmacologically relevant receptor tyrosine kinases (RTK). Demonstrating the applicability of this approach to primary tumors, we also discovered, for the first time in GC, somatic mutations in 7 kinase genes (MAP3K6, STK31, PAK4, FER, CDKL5, INSRR, RPS6KC1). Taken collectively, our results demonstrate the power of targeted deep sequencing for generating a comprehensive catalog of molecular variants at the single-nucleotide scale, a necessary first step to exploring how such variants might affect disease susceptibility, tumorigenesis, and drug response.
Materials and Methods
GC cell lines and tissues
GC cell lines SNU5, AGS, N87, and Hs746T were obtained from the American Type Culture Collection (ATCC). Cell lines MKN1, MKN7, TMK1, IM95, AZ521, and MKN28 were obtained from the Japan Health Science Research Resource Bank (JCRB). Cell lines YCC1, YCC3, YCC11, and YCC16 were originated and obtained from Yonsei Cancer Centre (gift from Sun Yong Rha). All cell lines were tested and authenticated by the respective cell line bank (ATCC, JCRB) or originating institution (YCC) by several methods including DNA finger-printing and/or cytogenetics. Prior to the commencement of this study, we independently reauthenticated the cell lines by comparing their genome-wide copy number (array-CGH) and mutational profiles with the published literature. The cell lines were cultured as recommended. Primary gastric tissues were obtained from the SingHealth Tissue Repository, after approvals from Institutional Research Ethics Review Committees and with signed patient informed consent.
Kinome capture arrays and deep sequencing
Kinase genomic coordinates are provided (Supplementary Table 1). Customized NimbleGen 385K sequence capture arrays were fabricated using SeqCap v2 software. Sequence capture was conducted by NimbleGen and NimbleGen linkers were ligated onto captured inserts prior to Illumina adaptors. Captured DNAs (500-bp fragments) were sequenced on the Illumina GAIIx as 76-bp paired-end reads. Image analysis and base calling were performed using the Illumina pipeline (v1.4) with default parameters.
Genome mapping and coverage computation
MAQ (Mapping and Assembly with Quality) software was used to align sequence reads to NCBI Build 36.1 reference genome (hg18; ref. 19). Sequences of NimbleGen linkers were trimmed prior to alignment. Aligned sequences were imported into R/Bioconductor (20) using the Shortread package, and read depth was calculated using the IRanges package with exon coordinates exported from BioMartV (21). Coverage of an exon was defined as the total number of aligned nucleotides within the exon coordinates divided by the length of the exon in base pairs. Hilbert curves were created using HilbertVisGUI software (22).
Detection of microgenomic genetic variants
Supplementary Figure 1 shows the discovery pipeline for novel SNVs. Uniquely aligned, different start site reads with 2 mismatches or greater to the reference genome were used to identify candidate SNVs. We retained SNVs that passed additional quality filters (Supplementary Methods). Only SNVs in exons or in canonical splice sites were analyzed. We considered an SNV to be “novel” if it was not in dbSNP (v130; ref. 23) or was in COSMIC (v44; ref. 24). UCSC databases were used for gene transcript identification and annotation of amino acid changes (25). Amino acid changes corresponding to SNVs were annotated according to the largest transcript of the gene. Nonsynonymous SNVs were submitted to PolyPhen (26) and SIFT (27) for functional prediction. MAQ was used to identify candidate microindels covering 1 or a few base pairs.
Inferring kinase structural variants
Large-scale structural variants were detected using the mapview file generated by MAQ. Only read pairs in which both reads had mapping quality greater than 30 were considered. Briefly, the mapview file was searched for clusters of ≥7 spatially clustered, same strand reads for which the matched read from the other strand was separated by ≥600 bp or on a different chromosome. For such clusters, we then examined the separation among the distant-matched reads. If the matched reads were within 500 bp of each other, we considered the cluster to represent a candidate large-scale variant. Candidates were checked for their presence in the Database of Genomic Variants (28).
Inferring copy number variations
Kinases/kinase exons with abnormally high read depths were candidates for copy number gain. These regions were detected using read depths computed by the IRanges package after importing the aligned data into R/Bioconductor. A threshold of median ± 2SD was used.
Additional procedures are described in Supplementary Materials and Methods.
Results
Kinome capture and deep sequencing of GC cell lines
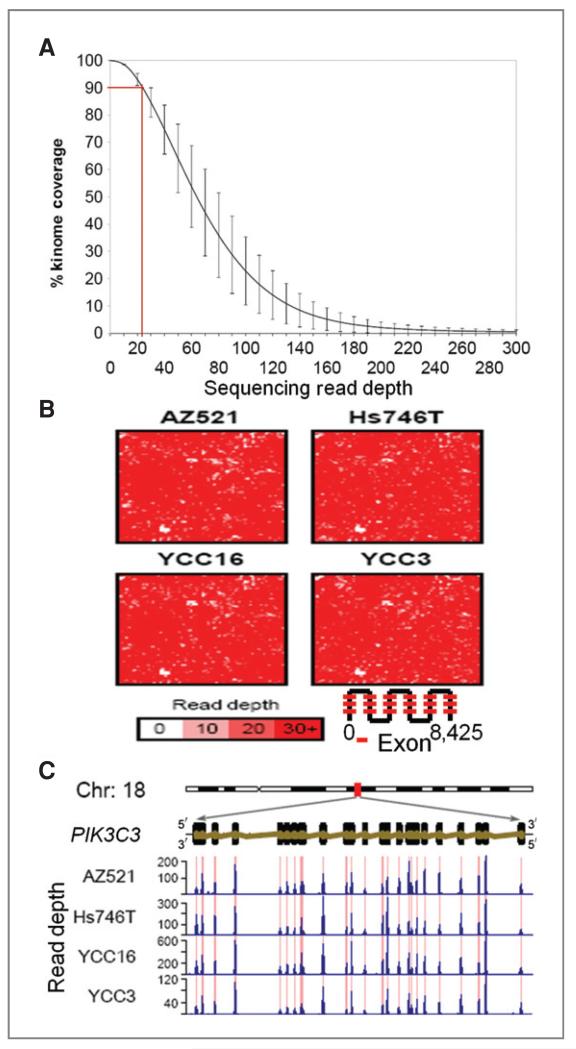
We sequenced the kinomes of 14 GC cell lines using array-based sequence capture and Illumina GAIIx sequencing. We surveyed 537 genes, including 509 protein kinases, 23 phosphoinositide kinases, and 5 cancer-related genes (TP53, KRAS, PTEN, OGG1, and MGMT). Collectively, we targeted 8,425 exons covering 1.33 Mb of nucleotide sequence (Supplementary Table 1). For each line, we generated an average of 0.75 Gb raw sequence, using 1 flowcell lane/sample (Supplementary Table 2). We achieved a read coverage of ≥20× for 90% of the targeted exons (Fig. 1A). Hilbert plots confirmed a high degree of coverage for the majority of targeted exons (Fig. 1B), with only 76 exons (0.9%) exhibiting a coverage of greater than 5× (Supplementary Table 3), our threshold for SNV calling. Compared with high-coverage exons, exons with poor coverage tended to have either extremely high (>0.64, 90 pecentile of 8,245 exons targeted) or low %GC content (<0.38, 10 percentile; P = 1.04 × 10−13; Supplementary Table 4), suggesting that the primary reason for the latter exhibiting poor coverage may be due to failed sequence capture (29). To assess capture specificity at the individual gene level, we compared exonic sequence coverage against the coverage of adjacent introns. We observed a striking accumulation of sequence reads only at the desired targeted exons and their flanking regions, but little coverage of introns within the same gene (Fig. 1C). Quantitatively, we computed a 670- to 900-fold enrichment of read coverage in targeted regions relative to nontargeted regions (Supplementary Table 5). These results confirm a sufficient degree of sequencing coverage for subsequent analysis.
Figure 1.
Genomic coverage of targeted sequence capture and deep sequencing. A, overall cumulative kinome coverage for 14 GC cell lines. Results presented represents sequences retained after filtering (see Methods). The Y-error bars indicate the standard deviation of the corresponding read depths across the 14 lines. B, Hilbert curve plots reflecting coverage of the 8,425 targeted exons in representative cell lines. The color bar indicates the depth of coverage. C, genomic organization, structure, and deep sequencing read distribution of a representative kinase, PIK3C3. Red lines indicate regions of genomic capture. Blue histograms indicate read depth per nucleotide.
SNV identification and sequencing accuracy
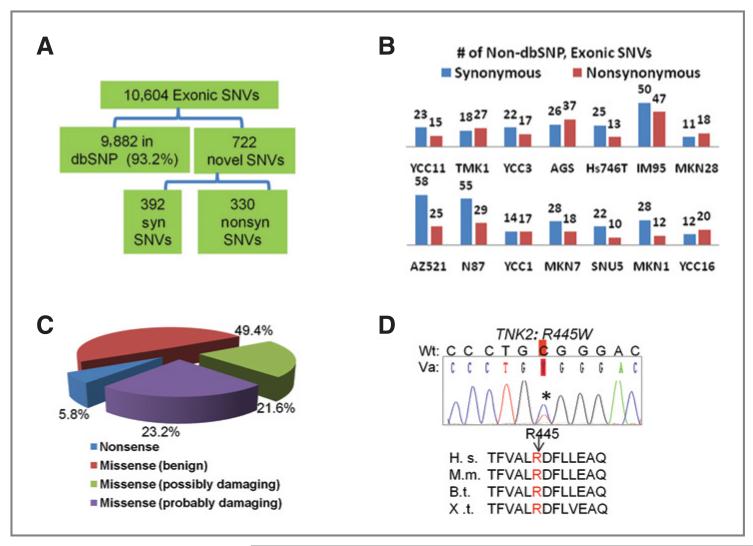
We developed a computational pipeline to discover novel SNVs (Supplementary Fig. 1). Totally, we identified 10,604 SNVs in kinase exons. Three methods were used to validate the accuracy of identified SNVs. First, we compared the SNVs with dbSNP (v130), a database of naturally occurring genetic polymorphisms (23). About 93.2% (9,882/10,604) of the SNVs were present in dbSNP (Fig. 2A). The finding that most of SNVs identified are previously observed genetic polymorphisms supports the accuracy of sequencing data.
Figure 2.
Identification of kinase microgenomic variants. A, total number of variants identified in the cell lines and their breakdown across various categories (dbSNP or novel, synonymous, syn, or nonsynonymous). B, number of novel exonic synonymous and nonsynonymous SNVs in individual cell lines. C, PolyPhen in silico assessment of functional impact of novel exonic nonsynonymous SNVs identified (see the text). D, sequencing chromatograph of a germline variant TNK2(R445W) found in 1 of 48 GC patients and in 2 cell lines (MKN28 and IM95). R445 locates in the kinase domain of TNK2 and is conserved across multiple species. H.s., Homo sapiens; M. m., Mus musculus; B.t., Bos Taurus; X.t., Xenopus tropicalis. Wt, wild-type; Va, variant.
Second, of the 722 exonic SNVs not found in dbSNP (novel SNVs), 392 correspond to synonymous SNVs, whereas the remaining 330 correspond to nonsynonymous SNVs (Fig. 2A). We selected 234 nonsynonymous SNVs for bidirectional Sanger sequencing. For SNVs with read depth ≥ 20, the false-positive rate was 6.1% (i.e., specificity = 93.9%). When the read depth was lowered to 5, the overall false-positive rate was 11.5%. Besides absolute read depth, we also found that “variant depth” (the number of reads where the variant is detected) is important for accuracy, as SNVs with high variant depth (>8) exhibited a lower false-positive rate (7%, 16/215) compared with SNVs of low variant depth 53% or 10/19). Zygosity calls based on deep sequencing were also consistent with Sanger sequencing, with 94.9% concordance.
Third, to estimate the false-negative rate (sensitivity), we compared the deep sequencing data of 2 representative cell lines (IM95 and YCC11) to genotype calls detected by Affymetrix 6.0 SNP arrays. Of 370 loci located within kinase exons that were also measurable on SNP arrays, 95% of the genotype calls were concordant with the sequencing data, indicating a false-negative rate of 5%. In addition, 9 of 9 known TP53 point mutations in our cell lines were successfully detected by deep sequencing. These results suggest that the targeted deep sequencing data are associated with a high degree of sensitivity and specificity (both >94%).
Genetic landscape of the GC kinome
We identified an average of 23.6 nonsynonymous and 28 synonymous novel exonic SNVs per cell line. The alteration rate varied across the lines, ranging from 97 in IM95 to 29 in MKN28 (Fig. 2B). Interestingly, N87, the second most altered cell line in our panel, is microsatellite instability (MSI) positive (30), whereas MSI-negative lines SNU5 and AGS exhibited relatively fewer SNVs (30, 31). The identification of more than 700 novel kinase SNVs indicates that a significant degree of genetic heterogeneity still awaits characterization within the GC kinome.
We focused on the 330 nonsynonymous kinase SNVs. After eliminating false-positive SNVs, there were 304 SNVs (Supplementary Table 6), including 289 missense and 15 nonsense substitutions, distributed across 194 genes (Supplementary Table 7). Among the 304 SNVs, only 14 (4.6%) were present in COSMIC, a public database of somatically acquired mutations in cancer (24) including 9 TP53 and 2 KRAS mutations. To identify SNVs that may impact kinase function, we used PolyPhen, a computational tool for estimating the impact of amino acid substitutions incorporating information from DNA sequence, evolutionary conservation, and structural data (26). Forty-five SNVs were classified as “uncharacterized” as PolyPhen did not give a prediction score. Of the remaining 259, 60 SNVS were classified as “probably damaging” (indicating that they are likely to affect protein structure or function) and 56 were “possibly damaging” (lower level of confidence), representing 23.2% and 21.6% of the characterized SNVs, respectively (Fig. 2C). About 5.8% of the SNVs were nonsense substitutions. Taken collectively, these results suggest that one fifth of the novel SNVs found may impact on kinase function. This figure was significantly higher compared with an analogous set of kinase germline, nonsynonymous SNVs found in dbSNP (probably damaging: P-value = 7.2 × 10−6; nonsense: P-value = 7.5 × 10−5). To benchmark the Polyphen predictions against an alternative program, we used SIFT, another in silico analysis tool (27), to analyze the same data (Supplementary Table 6). About 70% of the predictions were consistent between the 2 programs. Of the 45 SNVs uncharacterized by PolyPhen, SIFT offered predictions for 13 variants. Conversely, PolyPhen provided predictions for 9 of 41 variants for which SIFT could not classify. These results suggest that more functional calls can indeed be obtained by usage of additional prediction programs.
Eleven nonsynonymous SNVs were recurrently observed in the cell lines (Supplementary Table 6), including the TNK2 (R445W) SNV which was observed in 2 lines. A sequence analysis of TNK2(R445W) revealed that this alteration occurs at an evolutionarily conserved residue in the kinase domain (Fig. 2D). To determine if this SNVs might represent a germline variant, we screened for the TNK2(R445W) SNV in primary tissues. One of 48 normal gastric tissues exhibited the TNK2 (R445W) SNV indicating that it is likely a germline variant. The functional impact of this SNV on GC remains to be elucidated. Besides exonic SNVs, we also identified and verified 4 SNVs in canonical splice sites (Supplementary Table 6) and 15 putative microindels. Four of them were found in dbSNP, affecting ALK(SNU5), AURKAPS(MKN28), DMKN(MKN7), and ROCK1(Hs745T).
Frequent alterations of MAPK signaling genes in GC
The most frequently altered kinases were TTN (12/14 lines), followed by WNK1 (6/14), OBSCN (5/14) and SPEG (4/14; Supplementary Table 7). Notably, TTN (34,350 amino acid or aa) and OBSCN (7,968 aa) are both exceptionally large genes, which likely contributes to their being frequently mutated in human cancers (32). Of the other genes, we validated 2 WNK1 variants (I1172M, R1945C) and 1 SPEG variant (D1310N) as germline variants in primary gastric tissues (data not shown). To adjust our analysis for differences in gene size, we ranked the genes by the ratio of the number of nonsynonymous SNVs to gene size (Supplementary Table 8). TP53 and KRAS, 2 well-known oncogenes, were first and third in this ranked list whereas TTN and OBSCN fell far behind, suggesting that this approach may selectively enrich for genes with cancer-related functions.
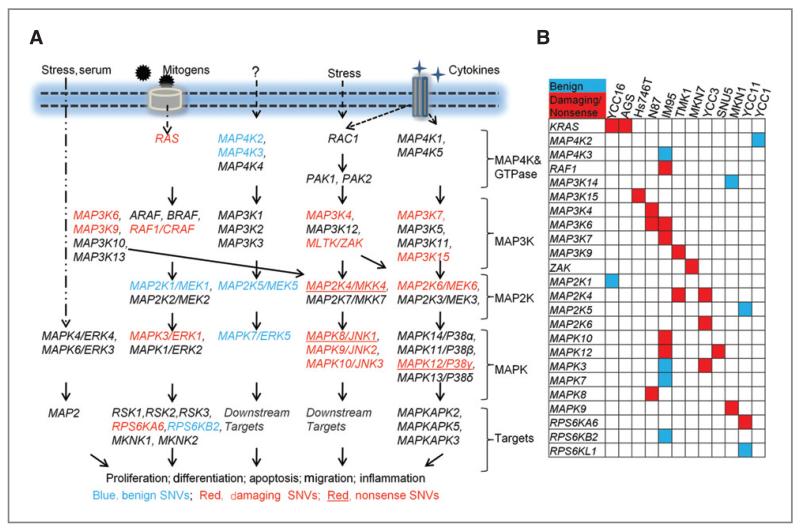
In human cells, MAPK-related pathways play important roles in the regulation of cell proliferation, stress response, apoptosis, motility, metabolism, and DNA repair. We observed several genes related to MAPK signaling at the top of the ranked list, such as KRAS, MAPK3, and MAP2K4 (Supplementary Table 8). Indeed, many components of the MAPK cascade were altered in GC lines, especially genes in the p38, ERK, and JNK pathway (Fig. 3A). Statistical analysis confirmed that when considered as a family (56 genes, cumulative protein size 38,146 aa), MAPK signaling-related genes exhibited significantly more “non-benign” alterations (1/1,908 aa) compared with other targeted genes (480 genes, 440,170aa, 1/4,310 aa; Fisher’s exact test, corrected P-value = 0.008). The significant enrichment of MAPK-signaling genes persisted even after removing the KRAS from computation (corrected P-value = 0.04). In comparison, 3 other kinase families, protein tyrosine kinases (88 genes), NF-κB pathway kinases (48 genes), and phosphoinositide kinases (PIK)3-AKT pathway kinases (31 genes) did not show similar significant enrichments of nonbenign SNVs (Supplementary Method and Table 9). Mapping the MAPK variants to the cell lines (Fig. 3B), we observed that when considered as individual genes, each MAPK-related gene exhibited alterations in only a few lines. However, when taken collectively, 11 of 14 (78%) lines exhibited “non-benign” alterations in at least 1 component of the MAPK pathway (Fig. 3B). Interestingly, the 2 lines harboring KRAS mutations (YCC16 and AGS) did not exhibit any other nonbenign MAPK gene alterations, consistent with KRAS acting as a major upstream regulator of the MAPK signaling hierarchy (Fig. 3B). These results highlight a potentially major role for perturbed MAPK signaling in GC.
Figure 3.
Frequent alterations of MAPK cascade genes in GC cell lines. A, schematic overview of novel exonic nonsynonymous SNVs in MAPK cascade genes in the cell lines. Blue, genes affected by “benign” SNVs; red, genes affected by “damaging” SNVs; red with underline, genes affected by nonsense substitutions. B, altered MAPK cascade genes in various GC cell lines. blue, Genes affected by “benign” SNVs; red, genes affected by either “damaging” missense substitutions or nonsense SNVs.
Functional consequences of MAP2K4 variants
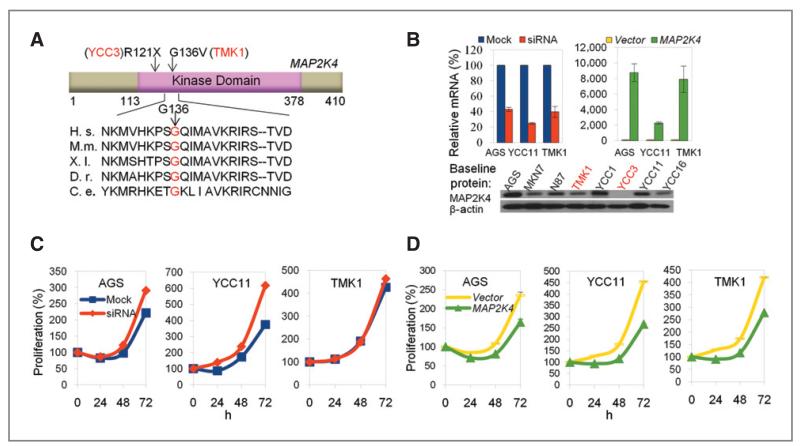
One MAPK-related gene recurrently altered was MAP2K4, a metastasis suppressor gene involved in multiple cancer types (33). We detected 2 novel MAP2K4 sequence alterations in the cell lines. The first was a homozygous nonsense C>T substitution at codon 121 in YCC3, leading to truncation of the MAP2K4 protein at codon 121 and disruption of the kinase domain (Fig. 4A). The second alteration, G136V, was a heterozygous G>T change in TMK1, converting an evolutionally conserved residue in the kinase domain (Fig. 4A). PolyPhen and SIFT classified MAP2K4(G136V) as “probably damaging” and “deleterious” respectively.
Figure 4.
Functional consequences of MAP2K4 perturbation in MAP2K4 wild-type and altered GC cell lines. A, schematic representation of the domain structure of MAP2K4 and the location of the novel SNVs found in our study (arrows). The numbers indicate the amino acid residue number. Codon G136 is strictly conserved among multiple vertebrate and invertebrate species examined. H. s., Homo sapiens; M.m., Mus musculus; X.l., Xenopus laevis; D.r., Danio rerio; D.m., D. melanogaster; C.e., C. elegans. B, validation of MAP2K4 knockdown and overexpression by quantitative PCR. Western blot shows the baseline expression level of MAP2K4 across GC cell lines. Note that YCC3 cells, which contain a MAP2K4 nonsense substitution, do not express the protein. C, siRNA-mediated silencing of MAP2K4 increases the proliferation of AGS and YCC11 cells that express wild-type MAP2K4, but not TMK1 cells that carry a MAP2K4 alteration (G136V). D, overexpression of the MAP2K4 protein suppresses cellular proliferation in AGS, YCC11, and TMK1 cells.
To explore the biological relevance of MAP2K4 in GC, we conducted knockdown and overexpression studies (Fig. 4B). Consistent with MAP2K4 playing a possible antioncogenic role, siRNA-mediated silencing of MAP2K4 significantly enhanced the growth rate of MAP2K4 wild-type cell lines (AGS and YCC11) but had minimal effects on the proliferation rate of TMK1 cells that harbor the MAP2K4(G136V) variant (Fig. 4C). This result suggests that G136V alterations may compromise MAP2K4 protein function. In the reciprocal experiment, overexpression of wild-type MAP2K4 inhibited cell proliferation in all the lines, confirming the growth inhibitory nature of MAP2K4 (Fig. 4D). Subsequent mutation screening of MAP2K4 in 48 gastric tumors and paired control tissue did not identify any somatic or germline alterations.
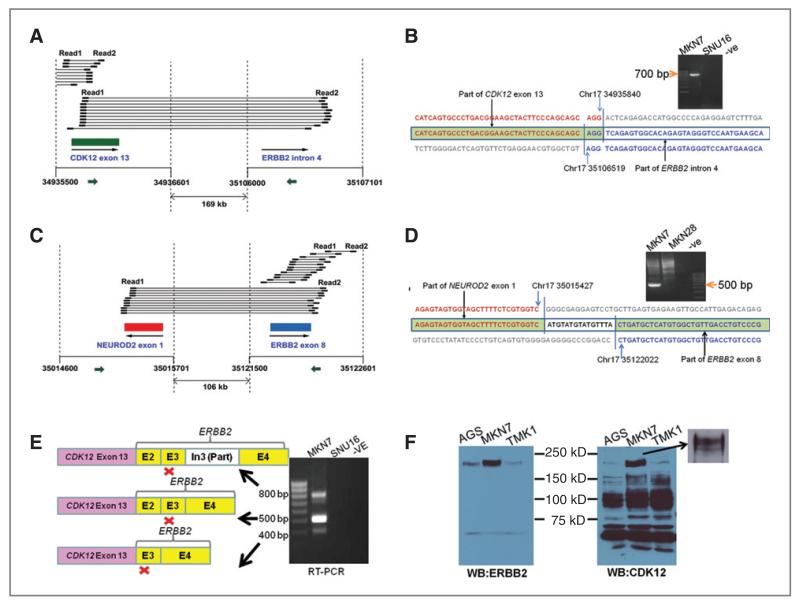
Analysis of kinase structural variants reveals ERBB2 gene fusions
Compared with conventional Sanger sequencing, 1 advantage of paired-end deep sequencing lies in its ability to identify aberrant structural variants. We examined mapped pairs of reads with inner coordinates greater than 600 bp apart (median distance = 354 bp; median absolute deviation = 40 bp). By examining such read pairs, we identified 24 potential structural variants involving kinases. Three of them (CDK11B, TYRO3, and SBK2) were found in the Database of Genomic Variants (DGV; Supplementary Table 10; ref. 28), providing confidence regarding the validity of our method. We selected 14 structural variants including 2 of the 3 in DGV for confirmatory PCR and sequencing, and validated 5 of them (35.7%). One structural variant was a 3.1 Mb deletion in chromosome X of MKN28 from positions 105,448,183 to 108,605,181, deleting GUCY2F (downstream from exon 3) and the adjacent 18 genes (Supplementary Table 10). GUCY2F catalyzes the synthesis of cGMP, a second messenger involved in multiple signaling cascades (34) and is somatically mutated in human cancer, suggesting a role for cGMP signaling in inhibiting tumorigenesis (35).
ERBB2 is a well-known proto-oncogene and the therapeutic target of trastuzumab, 1 of the first molecularly targeted therapies (36). We observed 2 genome rearrangement events involving different regions of the ERBB2 in MKN7 cells, which are known to exhibit genomic amplification of ERBB2 (37). These included a 169-kb deletion fusing CDK12 exon 13 to ERBB2 intron 4 (Fig. 5A) and a 106-kb deletion fusing NEU-ROD2 exon 1 to ERBB2 exon 8 (Fig. 5C). The ERBB2 structural variants were experimentally validated by Sanger sequencing and the breakpoints confirmed (Fig. 5B and D). To explore the potential expression of CDK12-ERBB2 fusion transcripts, we performed RT-PCR using primers targeted to CDK12 exon 13 and ERBB2 exon 5. We identified 3 CDK12-ERBB2 fusion transcripts involving CDK12 exons 13 and ERBB2 exon 2/3 and validated the identity of these transcripts by Sanger sequencing (Fig. 5E; the sequence of 3 bands is shown in Supplementary Result). Notably, whereas these results confirm the existence of CDK12-ERBB2 fusion transcripts, the expressed transcripts are not exact matches to those predicted by the structural breakpoints in Figure 5B. This may be related to 2 factors: (a) the chromosomal organization of the ERBB2 locus in MKN7 cells is likely complex due to regional genomic amplification (37) and (b) kinome resequencing does not exhaustively interrogate all possible structural arrangements in a region (e.g., rearrangements solely involving non-exonic regions will be missed). At the protein level, the CDK12-ERBB2 fusion transcripts were predicted to cause a truncated CDK12 protein but were not in-frame to ERBB2. Western blotting confirmed the presence of a 15-kD truncated CDK12 protein in MKN7 cells, whereas ERBB2 protein expression was unaltered (Fig. 5F).
Figure 5.
Identification and characterization of structural variants involving ERBB2. A, in MKN7 cells, widely separated read pairs spanning CDK12 exon 13 and ERBB2 intron 4 suggested a 169-kb deletion in chromosome 17. “Read 1” and “Read 2” indicate paired reads generated by Illumina deep sequencing. Large arrow indicates the direction of gene transcription. B, breakpoint of the deletion characterized by PCR amplification and Sanger sequencing. The primers used are in CDK12 exon 13 and ERBB2 intron 4 (small arrows in A). A 500-bp band was amplified from MKN7 genomic DNA but not control cell lines, and the sequence of this band shows the precise breakpoint. C, also in MKN7, other widely separated read pairs indicated a 105-kb deletion between NEUROD2 exon 1 and ERBB2 exon 8. D, PCR amplification and Sanger sequencing of the breakpoint. Primers used are shown as small arrows in C. E, RT-PCR using primers specific to CDK12 exon 13 and ERBB2 exon 5 amplified multiple fusion transcripts involving CDK12 and ERBB2. Red “×”s indicate the positions of premature stop codons in the fusion transcripts. F, Western Blot showing expression of CDK12 and ERBB2 proteins in MKN7 and control lines. Consistent with the predicted protein truncation resulting from the fusion transcripts in E, we noted a lower molecular weight CDK12 band in MKN7 under prolonged electrophoresis conditions (inset).
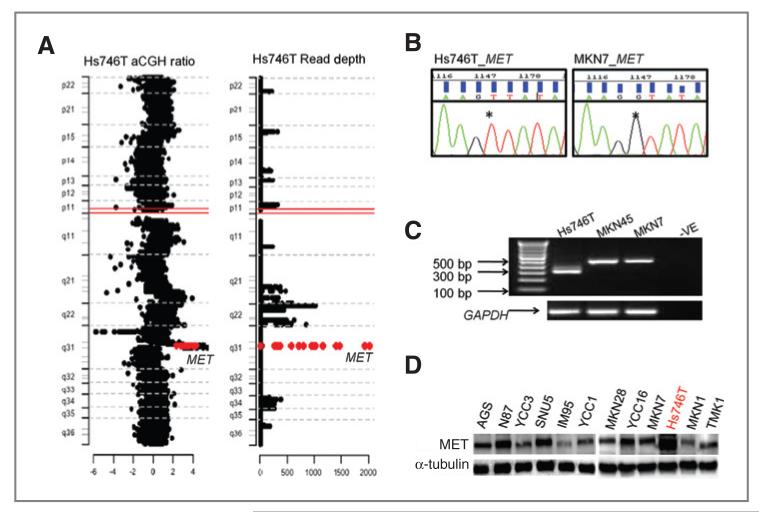
Integrative analysis of MET utilizing data from a single sequencing platform
Another intriguing aspect of targeted deep sequencing lies in the ability to integrate multiple levels of genomic information, such as SNVs, structural variants, and copy number, using data from a single sequencing platform. To investigate the association between targeted deep sequencing and copy number status, we identified kinase genes with high-read depth counts and compared these with array-CGH profiles of the same cell lines. Good overlap between the resequencing and array-CGH data were observed, as shown in Hs746T and YCC11 (Supplementary Fig. 2). Of the kinases amplified in Hs746T cells, the MET RTK is notable as MET has been shown to play an important role in cancer growth and metastasis, particularly in GC where 10% of cases have MET amplification (38). In Hs746T cells exhibiting MET genomic amplifications (Fig. 6A), we also detected a homozygous G>T substitution in MET intron13 altering the canonical splice donor site of MET exon 14 (Fig. 6B). This substitution is predicted to cause transcript skipping of exon 14, which encodes a domain important for the ubiquitination and degradation of MET protein (39). By RT-PCR, we confirmed MET transcripts in Hs746T cells are shorter due to the lack of exon 14 (Fig. 6C). The observation that MET is both altered at the splicing and copy number level suggests that MET protein should be highly overexpressed in Hs746T. Indeed, Hs746T cells exhibited a strikingly high protein level of MET (Fig. 6D).
Figure 6.
Integrative analysis of MET genetic alterations. A, plot of chromosome 7 showing a region of exceptionally high read depth corresponding to the MET locus (red labeled) from targeted deep sequencing in Hs746T (right). Confirmation of MET focal amplification in the same cells from aCGH data (left). B, sequencing chromatograph showing a homozygous G>T substitution at the canonical 5′ splice site of the intron 13 of MET in Hs746T, compared with wild-type cell lines. C, RT-PCR results showing an approximately 150-bp shorter MET transcript in Hs746T cells, compared with control cell lines. D, Western blot of MET protein in GC cell lines.
Kinome variation in primary gastric tumors
Finally, to demonstrate the utility of our approach in primary tissues, we analyzed 3 matched pairs of nonmalignant gastric tissues and gastric tumors. In benign gastric tissues, we detected an average of 19 nonsynonymous SNVs per tissue, and of the 304 nonsynonymous SNVs identified in cell lines, 2 SNVs were also observed in the normal gastric tissues [PIK3R2 (P4S) and PRKG1(H509Y)], indicating that these are likely to be germline variants. These results provide a measure of kinome variation in nonmalignant gastric mucosae. In the gastric tumors, a comparison of the 3 cancer kinomes to their nonmalignant counterparts revealed potential somatic mutations in 8 kinases: MAP3K6(S291L), ATM(R337H), STK31 (D115G), PAK4(E565K), FER(S250P), CDKL5(R722H), INSRR (R1022W), and RPS6KC1(R1025C). We subsequently validated these mutations by Sanger sequencing (Supplementary Fig. 3). Of these 8 kinases, 7 are novel in GC except for ATM gene mutations (40). Four of these kinases (MAP3K6, STK31, FER, and CDKL5) were also affected in at least 1 GC cell line.
Discussion
In this study, we performed a targeted deep sequencing analysis of the kinome in a panel of 14 GC cell lines. Our major goals of this study were to assess the utility of targeted sequencing for accurately identifying genetic variants, and to assemble a comprehensive catalog of kinome genetic variants in the 14 commonly used cell lines or experimental models of GC. We found the targeted deep sequencing methodology to be highly robust. Virtually, all the 8,425 targeted exons in our study were captured and we achieved an overall sequencing depth of ≥20× for 90% of the sequenced kinase genes. When benchmarked against Sanger sequencing and SNP genotyping arrays, we achieved an SNV detection specificity of 93.9% and sensitivity of 95%.
We identified more than 300 novel nonsynonymous SNVs in kinase exons. The novel SNVs identified likely comprise rare germline variants and somatic mutations (32). Both categories are important to document, as germline variants in certain kinases have been associated with an increased risk of cancer (41). In our study, a confirmed germline variant in TNK2 (R445W) was located at a conserved residue in the kinase domain, and found in 2 of 14 GC cell lines and 1 of 48 GC tissues, both of which are mostly Asian origin. It has been shown that TNK2 is overexpressed in breast cancers where its expression is correlated with poor prognosis (42). It would be interesting to compare the prevalence of this novel variant in the Asian healthy and GC population.
Another interesting variant we identified was a homozygous nonsense substitution (Q37X) in the STK11/LKB1 kinase in YCC16. Inactivating germline mutations in STK11/LKB1 cause Peutz–Jeghers Syndrome (PJS), an inherted disorder associated with intestinal hamartomatous polyps and frequent gastrointestinal tumors (41). To date, 4 GC-related STK11 mutations have been reported (3757–758insT, Arg297fsX38, Leu117PhefsX46, and P324L), and of these, 3 were also associated with PJS and early onset GC (43-46). Screening for STK11/LKB1 mutations in patients with early onset GC might, thus, identify additional patients with PJS.
Our study identified several interesting findings potentially related to GC tumorigenesis. We discovered that kinases related to MAPK signaling exhibited a significantly enriched tendency to harbor “non-benign” genetic alterations, with 11 of 14 lines having at least 1 affected MAPK gene. Abnormalities in MAPK signaling have been shown to impinge on many phenotypes of cancer including independence from proliferation signals, evasion of apoptosis, and unlimited replication potential (47). One of these MAP kinases was MAP2K4, which was altered in 2 lines. Expression of MAP2K4 suppressed cell growth in vitro. Inactivating mutations in MAP2K4 have been found in approximately 5% of tumors from various tissues (33) and recent evidence supports the functional role of MAP2K4 in human cancer (48). However, to date, somatic mutations in MAP2K4 have yet to be reported in GC. Our findings suggest that it might be worthwhile to characterize MAP2K4 in an expanded panel of GC tumors.
Besides SNV detection, the kinome resequencing data allowed us to identify several kinase-related structural variants, including variants affecting GUCY2F, TYRO3, SBK2, and the ERBB2 RTK. We demonstrated the expression of novel CDK12-ERBB2 fusion transcripts. However, as these transcripts are not expected to disrupt ERBB2 protein translation, their functional importance remains to be determined. Finally, the ability to infer genetic variation at multiple levels (SNVs, structural variants, and copy number) allowed us to perform integrative analysis utilizing data from a single deep sequencing platform. Using the example of the MET proto-oncogene, we identified Hs746T where MET was both amplified and associated with a splice-site point mutation. Such “dual mechanisms” of oncogene mutation and amplification have also been proposed for EGFR in lung cancer (49). Interestingly, in lung cancer, activation of MET by gene amplification and by splice-site mutations deleting the juxtamembrane domain appears to be mutually exclusive (50), suggesting that either type of the alterations may confer growth advantage to lung epithelial cells. In contrast, our results reveal that in GC, a paradigm of “dual mechanisms” (amplification and activating mutation) may impinge on MET.
In conclusion, we identified more than 300 novel micro- and macrogenomic alterations involving kinases, many of which may contribute to GC development. MAPK-signaling genes are identified as frequently altered kinases in our study, suggesting a causal role for dysregulated MAPK pathway in GC tumorigenesis. Our results may contribute to understanding the genetic architecture of this important subset of the human genome in GC and facilitate the usage of these GC models in the laboratory.
Acknowledgments
We thank Kalpana Ramnarayanan, Minghui Lee, and Jeanie Wu. We also acknowledge the generosity of the Lee Foundation who contributed to the establishment of the Lee Foundation Genome Sequencing Facility at NCCS.
Grant Support
This project was funded by grants to P. Tan from A-star, NMRC, Duke-NUS and CSIS.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Weimer JM, Anton ES. Doubling up on microtubule stabilizers: synergistic functions of doublecortin-like kinase and doublecortin in the developing cerebral cortex. Neuron. 2006;49:3–4. doi: 10.1016/j.neuron.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 3.Hunter T. A thousand and one protein kinases. Cell. 1987;50:823–9. doi: 10.1016/0092-8674(87)90509-5. [DOI] [PubMed] [Google Scholar]
- 4.Indo Y, Tsuruta M, Hayashida Y, Karim MA, Ohta K, Kawano T, et al. Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis. Nat Genet. 1996;13:485–8. doi: 10.1038/ng0896-485. [DOI] [PubMed] [Google Scholar]
- 5.West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, et al. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–7. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Odawara M, Kadowaki T, Yamamoto R, Shibasaki Y, Tobe K, Accili D, et al. Human diabetes associated with a mutation in the tyrosine kinase domain of the insulin receptor. Science. 1989;245:66–8. doi: 10.1126/science.2544998. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 8.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 9.Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–6. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 10.Krishnakumar S, Zheng J, Wilhelmy J, Faham M, Mindrinos M, Davis R. A comprehensive assay for targeted multiplex amplification of human DNA sequences. Proc Natl Acad Sci USA. 2008;105:9296–301. doi: 10.1073/pnas.0803240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–5. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 12.Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–7. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 13.Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–9. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- 14.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–9. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoischen A, Gilissen C, Arts P, Wieskamp N, van der Vliet W, Vermeer S, et al. Massively parallel sequencing of ataxia genes after array-based enrichment. Hum Mutat. 2010;31:494–9. doi: 10.1002/humu.21221. [DOI] [PubMed] [Google Scholar]
- 16.Volpi L, Roversi G, Colombo EA, Leijsten N, Concolino D, Calabria A, et al. Targeted next-generation sequencing appoints c16orf57 as clericuzio-type poikiloderma with neutropenia gene. Am J Hum Genet. 2010;86:72–6. doi: 10.1016/j.ajhg.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hohenberger P, Gretschel S. Gastric cancer. Lancet. 2003;362:305–15. doi: 10.1016/s0140-6736(03)13975-x. [DOI] [PubMed] [Google Scholar]
- 18.Yokota J, Yamamoto T, Toyoshima K, Terada M, Sugimura T, Battifora H, et al. Amplification of c-erbB-2 oncogene in human adenocarcinomas in vivo. Lancet. 1986;1:765–7. doi: 10.1016/s0140-6736(86)91782-4. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, et al. BioMart—biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anders S. Visualization of genomic data with the Hilbert curve. Bioinformatics. 2009;25:1231–5. doi: 10.1093/bioinformatics/btp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–7. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–46. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 26.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–74. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iafrate AJ, Feuk L, Rivera MN. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 29.Tewhey R, Nakano M, Wang X, Pabón-Peña C, Novak B, Giuffre A, et al. Enrichment of sequencing targets from the human genome by solution hybridization. Genome Biol. 2009;10:R116. doi: 10.1186/gb-2009-10-10-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung WK, Kim JJ, Wu L, Sepulveda JL, Sepulveda AR. Identification of a second MutL DNA mismatch repair complex (hPMS1 and hMLH1) in human epithelial cells. J Biol Chem. 2000;275:15728–32. doi: 10.1074/jbc.M908768199. [DOI] [PubMed] [Google Scholar]
- 31.Shin KH, Park JG. Microsatellite instability is associated with genetic alteration but not with low levels of expression of the human mismatch repair proteins hMSH2 and hMLH1. Eur J Cancer. 2000;36:925–31. doi: 10.1016/s0959-8049(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 32.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teng DH, Perry WL, 3rd, Hogan JK, Baumgard M, Bell R, Berry S, et al. Human mitogen-activated protein kinase kinase 4 as a candidate tumor suppressor. Cancer Res. 1997;57:4177–82. [PubMed] [Google Scholar]
- 34.Lucas KA, Pitari GM, Kazerounian S, Ruiz-Stewart I, Park J, Schulz S, et al. Guanylyl cyclases and signaling by cyclic GMP. Pharmacol Rev. 2000;52:375–414. [PubMed] [Google Scholar]
- 35.Wood LD, Calhoun ES, Silliman N, Ptak J, Szabo S, Powell SM, et al. Somatic mutations of GUCY2F, EPHA3, and NTRK3 in human cancers. Hum Mutat. 2006;27:1060–1. doi: 10.1002/humu.9452. [DOI] [PubMed] [Google Scholar]
- 36.Rebischung C, Barnoud R, Stéfani L, Faucheron JL, Mousseau M. The effectiveness of trastuzumab (Herceptin) combined with chemotherapy for gastric carcinoma with overexpression of the c-erbB-2 protein. Gastric Cancer. 2005;8:249–52. doi: 10.1007/s10120-005-0342-7. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto T, Ikawa S, Akiyama T, Semba K, Nomura N, Miyajima N, et al. Similarity of protein encoded by the human c-erb-B-2 gene to epidermal growth factor receptor. Nature. 1986;319:230–4. doi: 10.1038/319230a0. [DOI] [PubMed] [Google Scholar]
- 38.Sakakura C, Mori T, Sakabe T, Ariyama Y, Shinomiya T, Date K, et al. Gains, losses, and amplifications of genomic materials in primary gastric cancers analyzed by comparative genomic hybridization. Genes Chromosomes Cancer. 1999;24:299–305. doi: 10.1002/(sici)1098-2264(199904)24:4<299::aid-gcc2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 39.Peschard P, Park M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell. 2003;3:519–3. doi: 10.1016/s1535-6108(03)00136-3. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Jia G, Li WM, Guo RF, Cui JT, Yang L, Lu YY, et al. Alteration of the ATM gene occurs in gastric cancer cell lines and primary tumors associated with cellular response to DNA damage. Mutat Res. 2004;10(557):41–51. doi: 10.1016/j.mrgentox.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 41.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–7. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 42.Howlin J, Rosenkvist J, Andersson T. TNK2 preserves epidermal growth factor receptor expression on the cell surface and enhances migration and invasion of human breast cancer cells. Breast Cancer Res. 2008;10:R36. doi: 10.1186/bcr2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasovcák P, Puchmajerová A, Roubalík J, Krepelová A. Mutations in STK11 gene in Czech Peutz-Jeghers patients. BMC Med Genet. 2009;10:69. doi: 10.1186/1471-2350-10-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi M, Sakayori M, Takahashi S, et al. A novel germline mutation of the LKB1 gene in a patient with Peutz-Jeghers syndrome with early-onset gastric cancer. J Gastroenterol. 2004;39:1210–4. doi: 10.1007/s00535-004-1474-y. [DOI] [PubMed] [Google Scholar]
- 45.Shinmura K, Goto M, Tao H, Shimizu S, Otsuki Y, Kobayashi H, et al. A novel STK11 germline mutation in two siblings with Peutz-Jeghers syndrome complicated by primary gastric cancer. Clin Genet. 2005;67:81–6. doi: 10.1111/j.1399-0004.2005.00380.x. [DOI] [PubMed] [Google Scholar]
- 46.Park WS, Moon YW, Yang YM, Kim YS, Kim YD, Fuller BG, et al. Mutations of the STK11 gene in sporadic gastric carcinoma. Int J Oncol. 1998;13:601–4. [PubMed] [Google Scholar]
- 47.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–90. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 48.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–73. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 49.Soh J, Okumura N, Lockwood WW, Yamamoto H, Shigematsu H, Zhang W, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS ONE. 2009;4:e7464. doi: 10.1371/journal.pone.0007464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Onozato R, Kosaka T, Kuwano H, Sekido Y, Yatabe Y, Mitsudomi T. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol. 2009;4:5–11. doi: 10.1097/JTO.0b013e3181913e0e. [DOI] [PubMed] [Google Scholar]