Abstract
Mammalian genes are regulated by the cooperative and synergistic actions of many transcription factors. In this study we recapitulate this complex regulation in human cells by targeting endogenous gene promoters, including regions of closed chromatin upstream of silenced genes, with combinations of engineered transcription activator–like effectors (TALEs). These combinations of TALE transcription factors induced substantial gene activation and allowed tuning of gene expression levels that will broadly enable synthetic biology, gene therapy and biotechnology.
Synthetic biology aims to study the control of gene expression by constructing gene regulation systems from the “bottom-up” in order to better understand natural biological systems and develop useful tools for biotechnology.1 Despite many significant accomplishments, this field has largely been limited to studying artificial promoter-transgene systems with one or two transactivators, typically in microorganisms.1-6 In contrast, the natural regulation of mammalian gene expression is extraordinarily complex and is typically achieved by the combinatorial control of each gene by many regulatory factors. This level of complexity has not yet been achieved in synthetic gene regulation systems and has not been possible for the regulation of endogenous genes. However, the recent emergence of technologies for engineering transcription activator-like effectors (TALEs) targeted to almost any DNA sequence7-14 provides a unique opportunity for recapitulating this natural complexity. In the current study, the combinatorial regulation of endogenous mammalian genes in their natural chromosomal context is achieved by engineering several TALE transcription factors (TALE-TFs) to bind nearby sites upstream of the transcriptional start site (TSS) for a target gene. The composition of these combinations of independent TALE-TFs can be manipulated to control gene activation. Synergistic regulation of gene expression by multiple transcriptional activators is known to occur via simultaneous binding and stabilization of components of the pre-initiation complex.15-17 Building on this model, we activated endogenous genes with combinations of engineered transcription factors and were able to tune gene expression levels by systematically varying these combinations.
Each TALE-TF has two distinct protein domains that carry out individual molecular functions: (i) the repeat variable diresidue region binds to DNA at user-specified sequences,7, 8 and (ii) the VP64 effector domain recruits basal transcriptional machinery9, 10 (Fig. 1a). This design permits rapid construction of synthetic transcription factors that function as autonomous units.9, 11, 12 Several TALE-TFs have recently been reported to regulate native mammalian gene expression.9, 10, 14, 18-21 However, the levels of gene activation in these studies were modest and several genes could not be induced (Supplementary Table 1). Therefore there is clear need for improvements to gene activation strategies that capitalize on the synthetic TALE-TF technology.
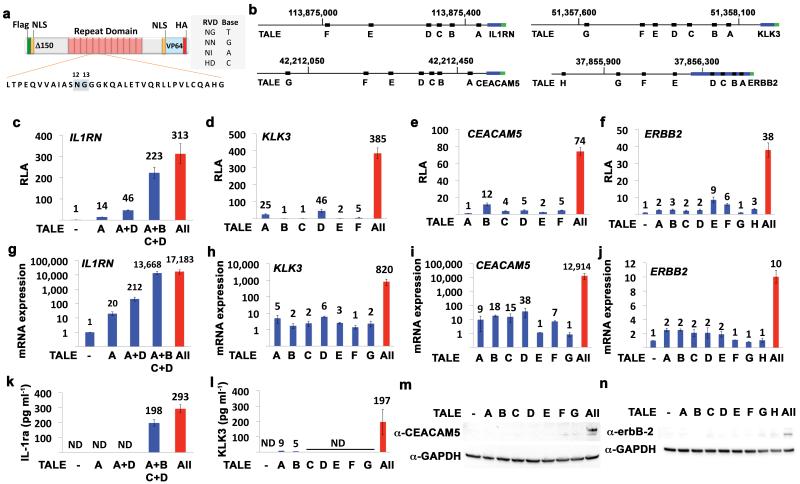
Figure 1.
Synergistic activation of gene expression by combinations of TALE-TFs. (a) Structure and sequence of TALE-TFs in this study. (b) Genomic positions of TALE-TF target sites in the CEACAM5, KLK3, IL1RN, and ERBB2 genes (hg19 coordinates) are indicated by black boxes. Transcribed and coding regions are shown in blue and green, respectively. (c-f) Relative luciferase activity (RLA) in promoter reporter assays. (g-j) Relative mRNA expression levels measured by quantitative RT-PCR and (k-n) protein expression levels assayed by ELISA or Western blot for each target gene in human cells transfected with the indicated TALE-TFs. Each gene is organized by column. n = 3 unless indicated otherwise in the online methods. Mean ± SEM and P < 0.0001 by ANOVA for all bar graphs.
We designed several TALE-TFs targeted to the promoter regions of the IL1RN, KLK3 (also known as prostate-specific antigen (PSA)), CEACAM5 (also known as CEA), and ERBB2 genes that are implicated in immunomodulation, inflammation, and cancer (Fig. 1b, Supplementary Fig. 1). TALE-TF expression plasmids were transfected into HEK293T cells and TALE-TF expression was confirmed by Western blot (Supplementary Fig. 2). TALE-TF activity was first measured in reporter assays in which luciferase is under the control of the respective gene promoter (Fig. 1c-f). Most individual TALE-TFs activated the co-transfected plasmid reporters, but only at modest levels similar to previous reports (Supplementary Table 1).9, 10, 14, 18-21 However, the delivery of combinations of TALE-TFs led to substantial synergistic effects on gene activation. Importantly, the synergistic activation of the plasmid-based reporters was recapitulated in upregulation of the native genes in their natural chromosomal context as determined by quantitative RT-PCR, including induction of mRNA levels greater than 10,000-fold (Fig. 1g-j). Detection of induced protein expression of IL-1ra, encoded by the IL1RN gene, KLK3, CEACAM5, and erbB-2 by ELISA and Western blot validated the functional outcome of the activation of these genes (Fig. 1k-n). In particular, we only reproducibly detected expression of IL-1ra, KLK3 and CEACAM5 protein in samples with combinations of TALE-TFs. We found low expression of ERBB-2 in control samples and cells transfected with single TALE-TFs, but its expression was substantially enhanced in cells transfected with all TALE-TFs (Fig. 1n).
These results are consistent with a mechanism in which the VP64 acidic activation domain of multiple transcription factors is simultaneously interacting with and stabilizing components of the pre-initiation complex.15-17 This mechanism was confirmed by demonstrating that the VP64 domain was essential to achieving the synergistic effect, indicating that the synergy is not the result of nucleosome displacement by TALEs (Supplementary Fig. 3). The synergistic gene activation was also conserved when using alternative acidic activation domains (Supplementary Fig. 4). The expression of other genes nearby IL1RN did not increase, indicating that this large synergistic activation was specific to the target gene (Supplementary Fig. 5).
The TALE-TFs used in this study were not specifically designed to target DNase-hypersensitive regions (Supplementary Fig. 6), in contrast to many other reports of synthetic transcription factors that only target open chromatin. In fact, IL1RN, KLK3, and CEACAM5 are not expressed in HEK293T cells. Interestingly, targeting chromatin inaccessible to DNase did not prevent gene activation by the engineered TALE-TFs (Fig. 1g-i). These results suggest that targeting open chromatin may not be a pre-requisite to successful TALE-TF engineering and that activation of silenced genes is possible in the absence of chromatin modifying drugs,18 particularly when using combinations of TALE-TFs. In contrast to these three genes, ERBB2 is moderately expressed in HEK293 cells and the TALE-TFs for ERBB2 regulation were targeted to open chromatin (Supplementary Fig. 6). Combinations of these TALE-TFs also led to synergistic ERBB2 activation although the effect was not as substantial relative to the other genes as a result of higher levels of basal expression (Fig. 1f,j,n).
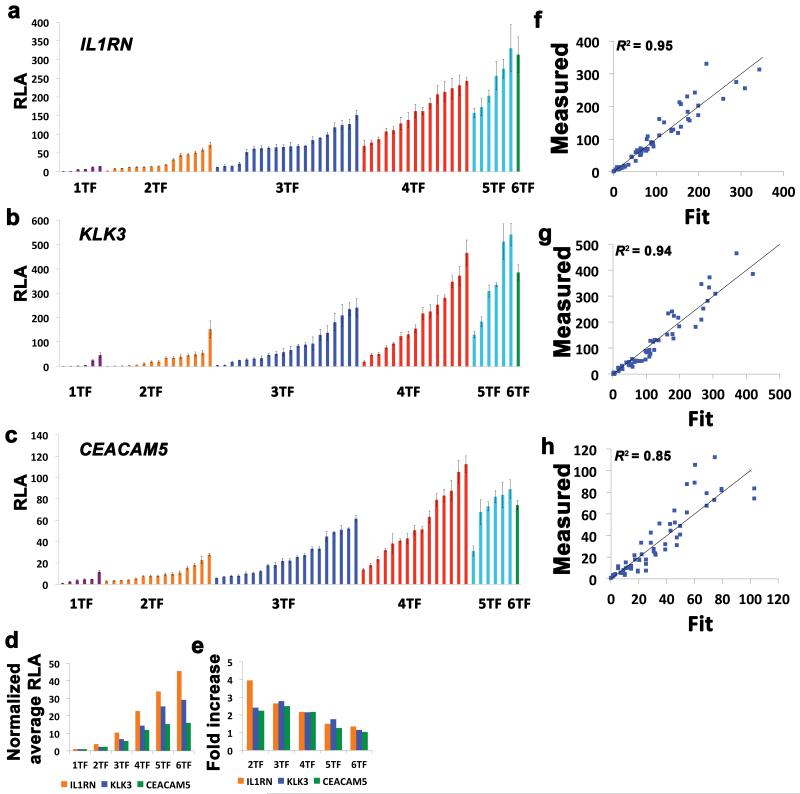
To comprehensively characterize the effects of combinatorial regulation of mammalian genes by engineered TALE-TFs, all 63 permutations of six TALE-TFs targeting three different genes were co-transfected with a corresponding luciferase reporter in HEK293T cells (Fig. 2a-c). Various combinations of TALE-TFs could be used to reproducibly achieve tunable levels of gene expression over a large dynamic range. Many TALE-TFs that did not activate the reporter when delivered alone contributed to synergistic activation of expression when combined with other TALE-TFs (Supplementary Table 2). In some cases, the addition of a TALE-TF decreased gene expression. However, for all three genes there was an increase in the average level of gene expression with increased number of TALE-TFs (Fig. 2d), and the average contribution of each additional TALE-TF decreased as the number of TALE-TFs increased (Fig. 2e).
Figure 2.
Combinatorial regulation of gene expression by TALE-TFs. (a-c) All possible 63 combinations of six TALE-TFs targeting the IL1RN, KLK3, and CEACAM5 genes were tested for activation of a luciferase reporter plasmid and ordered according to number of TALE-TFs and magnitude of relative luciferase activity (RLA). Samples receiving the same number of TALE-TFs are colored identically. Mean ± SEM (n = 3) and P < 0.0001 by ANOVA for all three data sets. (d) The average RLA for the indicated number of TALE-TFs is presented for each gene. (e) The fold increase of RLA for each number of TALE-TFs relative to the average RLA for one less TALE-TF is presented for each gene. (f-h) The measured values for all 63 combinations of TALE-TFs are plotted versus the values fit by the polynomial model along with y = x. (i) Effect coefficients and corresponding P-values resulting from multiple regression of the polynomial model are provided for each TALE-TF.
In order to assign quantitative parameters to the relative contribution of each TALE-TF to the synergistic effect across the 63 data points in these experiments, a polynomial model was applied to the data set of each gene of the form
where yj is the relative luciferase activity for the jth combination of the six TALE-TFs. The value of xi,j is 0 if the ith TALE-TF is not included in the jth combination and it is 1 if it is included. The effect coefficient wi is a fit parameter that represents the relative contribution of the ith TALE-TF to the regulation of its target promoter in the context of all permutations of the six TALE-TFs. Multiple regression was used to solve for values of wi for all TALEs for each of the three target genes. These coefficients generate an excellent fit of the experimental data (Fig. 2f-h) and are highly significant (P < 2E10−3) in accurately describing the relative contribution of each TALE (Supplementary Table 3). Importantly, the polynomial model provides a stronger description of the data than the corresponding additive and multiplicative models (Supplementary Fig. 7). This is because the additive model does not account for synergy of TALE-TF activity (Fig. 2d) and the multiplicative model does not account for the diminishing contribution of each additional TALE-TF (Fig. 2e). The superior fit of the polynomial model relative to the additive model can be mathematically explained by the second-order terms that are the product of effect coefficients for different TALE-TFs. This suggests the presence of some form of cooperativity, but cannot reveal the underlying mechanism. As discussed above, the simultaneous binding and stabilization of components of the pre-initiation complex by VP64 is likely to play a role,15-17 as well as other secondary effects of VP64-mediated gene activation on local epigenetics and chromatin structure.
Previous studies have suggested that TALE-TF activity may correspond to proximity to the transcription start site (TSS)18 or TALE RVD composition.13 In this study, there was no clear correlation of effect coefficient with TALE array length, composition, or distance to TSS that was consistent for all genes (Supplementary Fig. 1). This suggests that these TALE design parameters cannot independently be used to predict highly effective TALE-TFs. It is likely that other biological and structural components of these gene promoters, including genome folding and competition with endogenous regulatory factors, play a dominant role in determining the activity of single TALE-TFs and TALE-TF combinations.
The cooperative activation of gene expression described here presents a unique opportunity to develop tunable transcription networks that operate at different levels as a function of the number and identity of TALE-TFs. This facilitates the control of gene expression levels without the need for small molecules used in conventional chemically regulated systems. Unlike prior work in synthetic biology that has focused on the regulation of transgenes by engineered promoters customized with multiple transcription factor binding sites1-6 or gene repression or silencing,22, 23 the use of TALE-TF combinations that target endogenous promoters begins to recapitulate the complexity of natural systems in a precise and controlled manner. This approach constitutes a powerful experimental system for elucidating fundamental mechanisms of natural gene regulation that are currently poorly understood. The capacity for combinatorial regulation also provides a novel framework for engineering biocomputation systems that control endogenous genes in mammalian cells, similar to recently developed genetic logic gates that control engineered transgenes.2-6 Precise control of gene expression with multiple tunable inputs may lead to greater robustness and predictability in bioengineered systems in the context of cell-machine interfaces and gene- and cell-based therapies. For example, this could include increasing the potency of therapeutic effects of engineered transcription factors.24 In summary, this approach to gene regulation extends the capacity of synthetic biology and biological programming in mammalian systems and provides a new facile technology for regulation of native mammalian genes with widespread potential applications.
Methods
Cell culture and transfection
HEK293T cells were obtained from the American Tissue Collection Center (ATCC) through the Duke University Cancer Center Facilities and were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C with 5% CO2. HEK293T cells were transfected with Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. Transfection efficiencies were routinely higher than 95% as determined by flow cytometry following delivery of a control eGFP expression plasmid. The amount of DNA used for lipofection was 800 ng per well in 24-well plates or 200 ng per well in 96-well plates. For luciferase reporter assays in 24-well plates, 100 ng of reporter plasmid was included with 700 ng of TALE-TF expression plasmid. When comparing single TALE-TFs to the combination of all TALE-TFs (Fig. 1), the total amount of TALE-TF expression plasmid was held constant (i.e. 800 ng of single TALE-TFs vs. 800 ng of total TALE-TF expression plasmid divided equally amongst each factor). When assessing the individual contribution of each TALE-TF (Fig. 2), the amount of each TALE-TF was held constant at 116 ng, with empty expression plasmid added to a total of 700 ng. Amounts of DNA for transfections in 96-well plates were scaled accordingly.
Plasmids and TALE assembly
TAL effectors were assembled using the Golden Gate TALEN and TAL effector kit obtained from Addgene.11 A destination vector for the final assembly step was created to include a Flag epitope tag and an SV40 NLS at the N terminus, a 152 residue deletion from the N terminus of the wt TALE proteins that has previously been shown to preserve the DNA binding ability of TALEs,10 63 wt TAL amino acids following the repeat domain,9 a C-terminal SV40 NLS, a VP64 domain that contains 4 repeats of the minimal activation domain of VP16, and an HA tag at the C terminus (Fig. 1a). TAL effectors were originally designed to target within the 600 bp upstream of the transcriptional start site (Fig. 1b) based on the criteria described by Cermak et al.,11 although a subsequent study showed that these guidelines did not affect TALE functionality.12 TALEs were designed downstream of the transcriptional start site for ERBB2, but upstream of the translation start site, based on previous studies showing high levels of activity of synthetic zinc finger transcription factors targeting this region.25 The compositions of all TALE-TFs used in this study are provided in Supplementary Fig. 1.
The reporter plasmids were built by cloning PCR-amplified genomic DNA sequences upstream of the genes of interest IL1RN (chromosome 2, 113874366-113875462), ERBB2 (chromosome 17, 37855857-37856492), CEACAM5 (chromosome 19, 42211804-42212651) and KLK3 (chromosome 19, 51357466-51358177) into the vector pGL3-Basic (Promega). Coordinates are provided based on the hg19 reference genome.
Luciferase assays
Forty-eight hours after transfection, cells were collected into 96-well plates, washed with PBS once and lysed with 100 mM monobasic sodium phosphate and 0.2% Triton X-100. The lysate was incubated with Bright-Glo™ Substrate (Promega) in a 1:1 ratio and luciferase activity was measured using a Synergy 2 Multi-Mode Microplate Reader (BioTek). The results are expressed as relative luciferase activity, which is the average luciferase activity normalized to luciferase activity in samples transfected with the reporter vector and empty TALE-TF expression vector. Data is presented from at least three independent experiments performed with 2 technical replicates per experiment.
Western blot
Cells were lysed in 50 mM Tris-Cl (pH 7.4), 150 mM NaCl, 0.5% Triton X-100 and 0.1% SDS. Protein concentrations in cell lysates were measured by the BCA Protein Assay (Pierce). Lysates were mixed with loading buffer, boiled for 5 min, and equal amounts of protein were run in NuPAGE® Novex 4-12% Bis-Tris Gel polyacrylamide gels and transferred to nitrocellulose membranes. Non-specific antibody binding was blocked with 50 mM Tris/150 mM NaCl/0.1% Tween-20 (TBS-T) with 5% nonfat milk for 30 min. The membranes were incubated with primary antibodies (HRP-conjugated anti-HA (Roche, clone 3F10) in 5% milk in TBS-T diluted 1:5000 for 30 min; anti-CEACAM5 (Cell Signaling Technology, clone CB30) in 5% milk in TBS-T diluted 1:1000 overnight; anti-GAPDH (Cell Signaling Technology, clone 14C10) in 5% milk in TBS-T diluted 1:5000 for 30 min; anti-ERBB2 (Cell Signaling Technology, clone 29D8) in 5% BSA in TBS-T diluted 1:2000 for 2 h) and the membranes were washed with TBS-T for 30 min. Membranes labeled with primary antibodies were incubated with anti-rabbit HRP-conjugated antibody (Sigma-Aldrich, cat. no. A6154) diluted 1:5000 for 30 min, and washed with TBS-T for 30 minutes. Membranes were visualized using the Immun-Star WesternC™ Chemiluminescence Kit (Bio-Rad) and images were captured using a ChemiDoc™ XRS+ System and processed using ImageLab software (Bio-Rad).
ELISA
Serum-free culture media (OPTI-MEM) was collected and frozen at −80°C. Human IL-1ra and KLK3 secretion into culture media was quantified via enzyme-linked immunosorbent assay (ELISA), according to the manufacturer’s protocols (R&D Systems, Cat. No. DY280 and DKK300, respectively). For the IL-1Ra ELISA, the standard curve was prepared by diluting recombinant human IL-1ra in OPTI-MEM and the IL-1ra in culture media was measured undiluted. For the KLK3 ELISA, the standard curve was prepared by diluting recombinant KLK3 in the manufacturer’s calibrator diluent. The samples were concentrated ~8 fold via centrifugation through 3 kDa MWCO filters for 20 minutes (Amicon Ultra, Cat # UFC500396). Reported values were corrected by the concentration factor for each sample.
For both assays, optical density was measured at 450 nm with a wavelength correction at 540 nm. Each standard and sample was assayed in duplicate. The duplicate readings were averaged and normalized by subtracting the average zero standard optical density. A standard curve was generated by log transforming the data and performing a linear regression of the IL-1ra or KLK3 concentration versus the optical density. The lower limit of detection was 50 pg/ml for human IL-1ra and 32 pg/ml for human KLK3. Data reported are the mean and s.e.m. of these individual values combined from multiple experiments (n = 6 biological replicates for IL-1ra, n = 4 biological replicates for KLK3).
Quantitative RT-PCR
Total RNA was isolated using the RNeasy Plus RNA isolation kit (Qiagen). cDNA synthesis was performed using the SuperScript® VILO™ cDNA Synthesis Kit (Invitrogen). Real-time PCR using SsoFast™ EvaGreen® Supermix (Bio-Rad) was performed with the CFX96 Real-Time PCR Detection System (Bio-Rad) with 45 cycles, melting for 2 s at 95°C, and annealing and extension for 2 s at 55°C. Real-time PCR oligonucleotide primers ((ERBB2 5′-AGCCGCGAGCACCCAAGT-3′,5′-TTGGTGGGCAGGTAGGTGAGTT-3′), CEACAM5 (5′-TCCCCACAGATGGTGCAT-3′, 5′-GAACGGCGTGGATTCAATAG-3′), KLK3 (5′-CTCGTGGCAGGGCAGTCT-3′, 5′-AGCTGTGGCTGACCTGAAAT-3′), IL1RN (5′-GACCCTCTGGGAGAAAATCC-3′, 5 ′-GTCCTTGCAAGTATCCAGCA-3′) PSD4 (5′-GCAGCACCTCCTGGTCAC-3′, 5′-ATCCGACACATCCTGATTCC-3′), IL1F10 (5′-CCTCCCCATGGCAAGATACT-3′, 5′-AGCAGTTGTCTGCAACAGGA-3′) and GAPDH (5′-CAATGACCCCTTCATTGACC-3′, 5 ′-TTGATTTTGGAGGGATCTCG-3′)) were designed using Primer3Plus software and purchased from IDT. Primer specificity was confirmed by agarose gel electrophoresis and melting curve analysis. Reaction efficiencies over the appropriate dynamic range were calculated to ensure linearity of the standard curve.
Statistics
Statistical analysis was performed by single factor ANOVA with alpha equal to 0.05 in Microsoft Office Excel 2007. Effect coefficients were determined using the Regression tool in the Data Analysis Add-In to Microsoft Office Excel 2007, with the Relative Luciferase Activities (Fig. 2a-c, Supplementary Table 2) serving as the y input and an array of zeros and ones representing each TALE-TF combination as the x input.
Supplementary Material
Acknowledgments
This work was supported by an NIH Director’s New Innovator Award (DP2OD008586), NSF Faculty Early Career Development (CAREER) Award (CBET-1151035), NIH R03AR061042, The Hartwell Foundation Individual Biomedical Research Award, and a March of Dimes Basil O’Connor Starter Scholar Award to C.A.G, grants from the NIH (P50-GM081883-01) and DARPA (HR0011-09-1-0040) to A.J.H, and grants from the NIH to G.E.C. (U54HG004563) and F.G. (R01AR48852). D.G.O was supported by a predoctoral fellowship from the American Heart Association. K.A.G. was supported by an NSF Graduate Research Fellowship.
Footnotes
Author Contributions
P.P., A.M.F., G.E.C., A.J.H. and C.A.G. designed experiments. P.P., D.G.O, J.M.B., A.M.F, and K.A.G. performed the experiments. P.P., F.G., G.E.C., A.J.H. and C.A.G. analyzed the data. P.P. and C.A.G. wrote the manuscript.
References
- 1.Guido NJ, et al. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 2.Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 3.Cox RS, 3rd, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khalil AS, et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auslander S, Auslander D, Muller M, Wieland M, Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012;487:123–127. doi: 10.1038/nature11149. [DOI] [PubMed] [Google Scholar]
- 6.Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature. 2011;469:212–215. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 8.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 9.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 11.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotechnol. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 14.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carey M, Lin YS, Green MR, Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- 16.Joung JK, Koepp DM, Hochschild A. Synergistic activation of transcription by bacteriophage lambda cI protein and E. coli cAMP receptor protein. Science. 1994;265:1863–1866. doi: 10.1126/science.8091212. [DOI] [PubMed] [Google Scholar]
- 17.Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 18.Bultmann S, et al. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 2012;40:5368–5377. doi: 10.1093/nar/gks199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tremblay JP, Chapdelaine P, Coulombe Z, Rousseau J. Transcription activator-like effector proteins induce the expression of the frataxin gene. Hum Gene Ther. 2012;23:883–890. doi: 10.1089/hum.2012.034. [DOI] [PubMed] [Google Scholar]
- 20.Geissler R, et al. Transcriptional activators of human genes with programmable DNA-specificity. PLoS One. 2011;6:e19509. doi: 10.1371/journal.pone.0019509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 2012;40:7584–7595. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 23.Rinaudo K, et al. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat Biotechnol. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 24.Rebar EJ, et al. Induction of angiogenesis in a mouse model using engineered transcription factors. Nat Med. 2002;8:1427–1432. doi: 10.1038/nm1202-795. [DOI] [PubMed] [Google Scholar]
- 25.Beerli RR, Dreier B, Barbas CF. 3rd Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci U S A. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.