Abstract
We present here an efficient algorithm to compute the Principal Component Analysis (PCA) of a large image set consisting of images and, for each image, the set of its uniform rotations in the plane. We do this by pointing out the block circulant structure of the covariance matrix and utilizing that structure to compute its eigenvectors. We also demonstrate the advantages of this algorithm over similar ones with numerical experiments. Although it is useful in many settings, we illustrate the specific application of the algorithm to the problem of cryo-electron microscopy.
Index Terms: EDICS Category, TEC-PRC image and video processing techniques
I. Introduction
IN image processing and computer vision applications, often one is not interested in the raw pixels of images used as input, but wishes to transform the input images into a representation that is meaningful to the application at hand. This usually comes with the added advantage of requiring less space to represent each image, effectively resulting in compression. Because less data are required to store each image, algorithms can often operate on images in this new representation more quickly.
One common transformation is to project each image onto a linear subspace of the image space. This is typically done using Principal Component Analysis (PCA), also known as the Karhunen–Loéve expansion. The PCA of a set of images produces the optimal linear approximation of these images in the sense that it minimizes the sum of squared reconstruction errors. As an added benefit, PCA often serves to reduce noise in a set of images.
PCA is typically computed as the singular value decomposition (SVD) of the data matrix X of image vectors or as the eigenvector decomposition of the covariance matrix C = XXT. One then ranks eigenvectors according to their eigenvalues and projects each image onto the linear subspace spanned by the top n eigenvectors. One can then represent each image with only n values, instead of the original number of pixel values.
There are a number of applications in which each of an image’s planar rotations are interesting as well as the original images. It is usually unreasonable due to both time and space constraints to replicate each image many times at different rotations and to compute the resulting covariance matrix for eigenvector decomposition. To cope with this, Teague, [19] and then Khotanzad and Lu [10], developed a means of creating rotation-invariant image approximations based on Zernike polynomials. While Zernike polynomials are not adaptive to the data, PCA produces an optimal data adaptive basis (in the least squares sense). This idea was first utilized in the 1990s, when, in optics, Hilai and Rubinstein [8] developed a method of computing an invariant Karhunen–Loéve expansion, which they compared with the Zernike polynomials expansion. Independently, a similar method to produce an approximate set of steerable kernels was developed by Perona [14] in the context of computer vision and machine learning. Later, Uenohara and Kanade [20] produced, and Park [12] corrected, an algorithm to efficiently compute the PCA of an image and its set of uniform rotations. Jogan et al. [9] then developed an algorithm to compute the PCA of a set of images and their uniform rotations.
The natural way to represent images when rotating them is to sample them on a polar grid consisting of Nθ radial lines at evenly spaced angles and Nr samples along each radial line. The primary advantage of [9] is that the running time of the algorithm increases nearly linearly with both the size of the image (Nr below) and the number of rotations (Nθ below). The disadvantage, however, is that the running time experiences cubic growth with respect to the number of images P in the set. The result is an algorithm with a running time of O(P2Nr Nθ log(Nr Nθ) + NθP3), which is impractical for large sets of images.
We present here an alternate algorithm that grows cubically with the size of the image, but only linearly with respect to the number of rotations and the number of images. Due to this running time, our algorithm is appropriate for computing the PCA of a large set of images with a running time of .
Although this algorithm is generally useful in many applications, we discuss its application to cryo-electron microscopy (cryo-EM) [3]. In cryo-EM, many copies of a molecule are embedded in a sheet of ice so thin that images of the molecule of interest are disjoint when viewing the sheet flat. The sheet is then imaged with an electron microscope, destroying the molecules in the process. The result is a set of projection images of the molecule taken at unknown random orientations with extremely low signal-to-noise ratio (SNR). The cryo-EM problem, then, is to reconstruct the 3-D structure of the molecule using thousands of these images as input.
Because the images are so noisy, preprocessing must be done to remove noise from the images. The first step is usually to compute the PCA of the image set [1], [21], a step which both reduces noise and compresses the image representation. This is where our algorithm comes in, because in cryo-EM every projection image is equally likely to appear in all possible in-plane rotations. After PCA, class averaging is usually performed to estimate which images are taken from similar viewing angles and to average them together in order to reduce the noise [3], [17], [22]. A popular method for obtaining class averages requires the computation of rotationally invariant distances between all P(P − 1)/2 pairs of images [13]. The principal components we compute in this paper provide a way to accelerate this large-scale computation. This particular application will be discussed in a separate publication [18].
Other applications for computing the PCA of a large set of images with their uniform rotations include those discussed in [9]. A set of full-rotation panoramic images were taken by a robot at various points around a room. Those panoramic images and their uniform rotations were then processed using PCA. A new image taken at an unknown location in the room was then template matched against the set of images already taken to determine the location in the room from which the new image was taken. Because these images are likely to be somewhat noisy, it is important to perform PCA on them to be able to effectively template match against the training set.
The remainder of this paper is organized as follows. In Section II, we derive the principles behind the algorithms. In Section III, we describe an efficient implementation of our algorithm and discuss its space and computational complexity. In Section IV, we present a method for applying this algorithm to images sampled in rectangular arrays. In Section V, we present numerical experiments comparing our algorithm to other existing algorithms. Finally, in Section VI, we present an example of its use in cryo-EM.
II. Derivation
Consider a set of P images represented in polar coordinates. Each image is conveniently stored as an Nr × Nθ matrix, where the number of columns Nθ is the number of radial lines and the number of rows Nr is the number of samples along each radial line. Note that Nθ and Nr must be the same for all images. Then, each column represents a radial line beginning in the center of the image, and each row represents a circle at a given distance from the center. The first circle is at distance 0 from the center, and so the center value is repeated Nθ times as the first row of the matrix.
We can create a rotation of an image simply by rotating the order of the columns. Moving the leftmost column to the rightmost and cyclically shifting all other columns of the matrix, for example, represents a rotation of 2π/Nθ. In this way, we can generate all Nθ rotations of each image. Thus, let matrix Ii,l represent the lth rotation of image i, 0 ≤ i ≤ P−1, 0 ≤ l ≤ Nθ−1. Note that we may assume the average pixel value in each image is 0, otherwise we subtract the mean pixel value from all pixels.
Let Ji,l be a vector created by row-major stacking of the entries of image Ii,l, that is, , 0 ≤ m ≤ Nr−1, 0 ≤ n ≤ Nθ−1. Then construct the NrNθ × PNθ data matrix X by stacking all Ji,l side by side to create
| (1) |
and set the covariance matrix C = XXT. Note that C is of size NrNθ × NrNθ. The principal components that we wish to compute are the eigenvectors of C. This can be computed directly through the eigenvector decomposition of C, or through the SVD of X. We will now detail an alternative and more efficient approach for computing these eigenvectors exploiting the block circulant structure of C.
Consider Ci, the covariance matrix given by just image i and its rotations, Ji,0 …Ji, Nθ−1 as follows:
| (2) |
Note that
| (3) |
Now, let Ci,αβ be an Nθ × Nθ block of Ci corresponding to the outer product of row α of Ii,0, …, Ii, Nθ−1in Ji,0, …, Ji, Nθ−1 with row β of Ii,0, …, Ii, Nθ−1.
We will now show that Ci is of the form
| (4) |
where each Ci,α,β is an Nθ × Nθ circulant matrix.
Because image i and its rotations comprise a full rotation of row α and a full rotation of row β, we have
| (5) |
where each row of Ii,0 is considered a circular vector, so that all indexes are taken modulo Nθ. However, due to the circular property of the vector, for any integer c, we have
| (6) |
This means that Ci,α,β is a circulant matrix. However, this analysis applies to any rows α and β. Therefore, Ci consists of Nr × Nr tiles of Nθ × Nθ circulant matrices, as in (4).
Now, the sum of circulant matrices is also a circulant matrix. Define Cα,β as
| (7) |
Thus, Cα,β is an Nθ × Nθ circulant matrix. Then C is of the form
| (8) |
A known fact is that the eigenvectors V0, V1, …, VNθ−1 of any Nθ × Nθ circulant matrix A are the columns of the discrete Fourier transform matrix [5], given by
| (9) |
where . The first coordinate of any Vk is 1, so the eigenvalues λ0, λ1, …, λNθ−1 of a circulant matrix A, for which, AVk = λkVk are given by
| (10) |
where A0 is the first row in matrix A, and Â0 is its discrete Fourier transform. These eigenvalues are, in general, complex.
It follows that, for the matrix C, there are Nθ eigenvalues associated with each of its circulant tiles. We denote these eigenvalues satisfying . Note that
| (11) |
Now construct a vector Wk,q of length NrNθ by concatenating multiples of the vector Vk Nr times, that is,
| (12) |
These Wk,q vectors will be our eigenvectors for C. The exact values of , as well as the index set for q, will be determined below.
Consider the result of the matrix-vector multiplication CWk,q. Along each tile-row of C(α = α0), each circulant submatrix Cα0,β is multiplied by an eigenvector . Thus
| (13) |
So, requiring Wk,q to be an eigenvector of C satisfying CWk,q = μk,qWk,q is equivalent to
| (14) |
However, this in turn is equivalent to the eigenvector decomposition problem for the matrix
| (15) |
This means that the vector Wk,q is an eigenvector of C with eigenvalue μk,q if the vector is an eigenvector of the matrix Λk, satisfying Λkck,q = μk,qck,q.
Next, we show that the matrix Λk is Hermitian. To that end, note that the covariance matrix C is by definition symmetric, so column γ of the Nθ × Nθ submatrix Cα,β is equal to row γ of submatrix Cβ,α. Now, set γ = 0. Then, by viewing the rows of the tile Cα,β as circular vectors, the circulant property of the tiles implies that
| (16) |
The eigenvalues of Cβ,α, then, are
| (17) |
which, by the change of variables n′ = −n, becomes
| (18) |
where refers to the complex conjugate of . Note that this property is true only because is real for all n1, n2.
Thus, Λk is a Hermitian matrix. As a result, all of Λk’s eigenvalues are real. Furthermore, these eigenvalues are the μk,q so q ∈ 0, 1, …, Nr −1 and the ck,q are complex-valued.
Thus, for some fixed k0, Λk0 has Nr linearly independent eigenvectors that in turn give Nr linearly independent eigenvectors Wk,q of C, through (12). Furthermore, the vectors V0, V1, …, VNθ−1 are linearly independent. From this, it follows that the constructed eigenvectors Wk,q are linearly independent.
Now, consider an eigenvector Wk,q, 0 ≤ k ≤ Nθ−1, 0 ≤ q ≤ Nr−1 as an eigenimage, that is, consider the eigenimage as an Nr × Nθ matrix
| (19) |
This is of the same form as the original images, in which each row is a circle and each column is a radial line, with the first row being the center of the image. The coefficients are complex, so we may write for 0 ≤ r ≤ Nr−1. Therefore, Rk,q is of the form
| (20) |
for θ = 0, 2π/Nθ, …,2π(Nθ −1)/Nθ, r = 0, 1, …, Nr−1.
The real and imaginary parts of Rk,q are eigenimages, given by
| (21) |
| (22) |
for r = 0,1, …, Nr − 1, θ = 0,2π/Nθ, …, 2π(Nθ−1)/Nθ.
Thus, each eigenimage Rk,q produces two eigenimages and . It may appear as though this gives us 2NrNθ eigenimages, which is too many. However, consider (11) for VN−1−k = V−k:
| (23) |
Therefore, , Thus
| (24) |
However, because Λk is Hermitian, μk,q is real, so . Therefore, the eigenvalues of Λk are the same as those for Λ−k, and the eigenvectors of Λk are the complex conjugates of the eigenvectors of Λ−k. Note also that k = −k if k = 0 or k = Nθ/2, which are also the values of k such that Λk has real eigenvectors. Therefore, each eigenvector from k = 0 or k = Nθ/2 contributes one real eigenimage, while eigenvectors from every other value of k each contribute two. Thus, linear combinations of the eigenvectors of the first Nθ/2 + 1 Λk matrices results in Nr × Nθ eigenimages.
Note also that, when Nθ is odd, eigenvectors for k = 0 each contribute one real eigenimage, while eigenvectors for every other k each contribute two. Thus, counting eigenimages for k = 0, 1, …, Nθ−1/2 produces Nr × Nθ eigenimages, as required.
III. Implementation and Computational Complexity
The first step in this algorithm is to compute the covariance matrix Ci of each image Ii and its rotations. However, it is not necessary to compute the actual covariance matrix. Instead, we need only compute the eigenvalues of each circulant tile Ci,αβ of the covariance matrix. From (10) and (11), it follows that the eigenvalues are equal to the discrete Fourier transform of the top row of a tile Ci,αβ.
Note that, by (5), we have
| (25) |
where m = 0, …, Nθ −1. Now, the convolution theorem for cross correlations states that, for two sequences f and g, if f ★ g indicates the cross correlation of f and g, then
| (26) |
Therefore, the eigenvalues can be computed efficiently as
| (27) |
This can be computed efficiently with the dast Fourier transform (FFT). In addition, because the covariance matrix Ci is symmetric, . We can in this way compute Nθ different Nr × Nr matrices Λi,k and sum
| (28) |
so that Λk is as defined in (15).
Note that, in some implementations, such as in Matlab, complex eigenvectors of unit norm are always returned. As a result, real eigenvectors, which occur when k = 0 or k = Nθ/2, will have twice the norm of the other eigenvectors. One must therefore normalize these eigenvectors by dividing them by .
Computation of the Fourier transforms can be performed in time O(Nθ log(Nθ)). Because we do this for every α, computation of the necessary Fourier transforms takes total time O(NrNθ log(Nθ)). In addition, it uses space O(NθNr). Computation of a matrix Λk has computational complexity , but, because we compute Nθ of them, the overall time of . In addition, the space complexity of this step is . Each of these steps must be repeated for each image i, and then the P matrices Λi,k, (i = 0,…,P −1)must be summed together for each image. Thus, the computational complexity up to this point is O(P NθNr(Nr + log(Nθ)))and the space complexity is .
Note that the computation of Λk for each image i is completely independent of each other image. Thus, these computations can be performed in parallel to further improve performance. Such a parallel implementation requires space .
At this point, we need to perform eigenvector decomposition on each Λk to determine the eigenvectors of C. The computational complexity of this step is and the space complexity is O(P NθNr). Thus, the computational complexity of the entire algorithm is
| (29) |
and the total space complexity is
| (30) |
The algorithm is summarized in Algorithm 1, and a comparison of the computational complexity of this algorithm with others is shown in Table I.1
Algorithm 1.
PCA of a Set of Images and Their Rotations
| Require: P image matrices I0,0, I1,0,…, IP−1,0of size Nr×Nθ in polar form. |
| 1: Set , α, β = 0,…, Nr−1, k = 0,…, Nθ−1 |
| 2: for i = 0,…, P − 1 do |
| 3: Compute the Fourier transforms for α = 0,…, Nr − 1, k =0,…, Nθ − 1. |
| 4: for αβ = 0,…, Nr −1 do |
| 5: Compute the eigenvalues of Ci,αβ using (27). |
| 6: end for |
| 7: Update Λk ← Λk + Λi,k for k = 0,…, Nθ −1 as in (28). |
| 8: end for |
| 9: for k = 0,…, Nθ/2 do |
| 10: Compute the eigenvector decomposition of matrix Λk to generate eigenvectors ck,q and eigenvalues μk,q for q = 0,…, Nr −1 |
| 11: for q = 0,…,Nr − 1 do |
| 12: Construct eigenvector Wk,q as in (12), using eigenvector ck,q of Λk. |
| 13: end for |
| 14: end for |
| 15: for k = 0,…, Nθ/2, q = 0,…,Nr −1 do |
| 16: Rearrange Wk,q as an image matrix as in (19) and compute its real and imaginary parts and . |
| 17: If k = 0 or k = Nθ/2, normalize eigenimages if necessary by dividing by . |
| 18: end for |
| 19: return and as in (21) and (22). |
TABLE I.
Comparison of Computational Complexities of Different PCA Algorithms
| Algorithm | Computational Complexity | |
|---|---|---|
|
| ||
| Ours |
|
|
| JZL | O(P2NrNθ log(NrNθ) + NθP3) | |
| SVD |
|
|
| Eig. Decomp. |
|
|
IV. Application to Rectangular Images
Although this algorithm is based on a polar representation of images, it can be implemented efficiently on images represented in the standard Cartesian manner efficiently and without loss of precision using the 2-D polar Fourier transform and the Radon transform.
The 2-D Radon transform of a function I(x, y) along a line through the origin is given by the projection of I(x, y) along the direction τθ [11]. The Radon transform is defined on the space of lines in ℝ2, so we may write
| (31) |
The Fourier projection-slice theorem states that the 1-D Fourier transform of the projection of an image along τθ0 is equal to the slice of the 2-D Fourier transform of the image taken along [11]. To see this, let r ∈ ℝ2 be any point on the plane, and let
| (32) |
Also, take . Note that r · w = u · w, since s · w = 0. Then
| (33) |
The Fourier projection-slice theorem provides an accurate way for computing the Radon transform by sampling the image’s 2-D Fourier transform on a polar grid and then taking inverse 1-D Fourier transforms along lines through the origin. Because the 2-D Fourier transform is discrete, performing the normal 2-D FFT and interpolating it to a polar grid leads to approximation errors. Instead, we use a nonequally spaced FFT described in [2] and [6]. This step has a computational complexity of O((mL)2log(L) + NrNθ log(NrNθ)log(1/ε)), where L is the number of pixels on the side of a square true image, Nr is the number of pixels in a radial line in polar form, Nθ is the number of radial lines, m is an oversampling factor used in the nonequally spaced FFT, and ε is the required precision.2
The Fourier transform is a unitary transformation by Parseval’s theorem. Therefore, the principal components of the normal 2-D FFT’ed images are simply the 2-D FFT of the principal component images. The 2-D polar Fourier transform, however, is not a unitary transformation, because the frequencies are nonequally spaced: they are sampled on concentric circles of Nr equally spaced radii, such that every circle has a constant number Nθ of samples instead of being proportional to its circumference, or equivalently, to its radius. In order to make it into a unitary transformation, we have to multiply the 2-D Fourier transform Î(|w|cos θ, |w| sin θ)of image I by . In other words, all images are filtered with the radially symmetric “wedge” filter or, equivalently, are convolved with the inverse 2-D Fourier transform of this filter.
After multiplying the images’ 2-D polar Fourier transforms by , we take the 1-D inverse Fourier transform of every line that passes through the origin. From the Fourier projection-slice theorem, these 1-D inverse Fourier transforms are equal to the line projections of the convolved images, that is, the collection of 1-D inverse Fourier transforms of a given “ -filtered” image, is the 2-D Radon transform of the filtered image.
These 2-D Radon transformed filtered images are real valued and are given in polar form that can be organized into matrices as described at the beginning of Section II. We can then utilize the algorithm described above to compute the principal components of such images. Moreover, since the transformation from the original images to the Radon transformed filtered images is unitary, we are guaranteed that the computed principal components are simply the Radon transform of the filtered principal components images.
After computing all of the polar Radon principal components we desire, we must convert them back to rectangular coordinates. We cannot simply invert the Radon transform because the pixels of each eigenimage are not uniformly sampled, making the problem ill-formed. However, for the inversion we can utilize the special form of the Radon eigenimages and from (21) and (22), given by
| (34) |
| (35) |
To compute the inverse Radon transform of such functions we utilize the projection-slice theorem. In principle, this is done by first computing the Fourier transform of each radial line, dividing by and then taking the 2-D inverse Fourier transform of that polar Fourier image to produce the true eigenimage.
Let us first compute the 2-D polar Fourier transform by taking 1-D discrete Fourier transform along each radial line of the function in (34). Equation (34) is only defined for r ≥ 0, but we must compute the discrete Fourier transform (DFT) along an entire line through the origin. We must therefore expand this definition to allow for any real r. Let
| (36) |
for some fixed θ0. Note that we should really denote this function , but since q is fixed throughout this section, we drop it for convenience of notation. Then
| (37) |
Define similarly for . Note that and are even functions if k is even, and odd functions if k is odd. The DFT can then be defined as
| (38) |
where r =|w|. Note that
| (39) |
because is real. Furthermore, with the change of variables n′ = −n, we find that, if k is even, then
| (40) |
and, if k is odd, then
| (41) |
Therefore, if k is even, then is real and even, and, if k is odd, then is pure imaginary and odd.
Then, the 2-D Fourier transform of the true image can be defined as (a proof of this formula can be found in Appendix A)
| (42) |
and
| (43) |
As shown above, these functions are either real or pure imaginary, depending on k.
Next we will take the 2-D inverse Fourier transform of (where r = |w| and is the filter) to obtain true eigenimages and . Define as the order k Hankel transform of
| (44) |
and define as the order k Hankel transform of :
| (45) |
where Jk is the Bessel function.
Then, the true real eigenimages can be described (in polar form) with the two equations (a proof of this formula can be found in Appendix B)
| (46) |
| (47) |
where
| (48) |
so that β(k) = 1 when k is even, and β(k) = ι when k is odd. In practice, and are computed using standard numerical procedures [4], [7], [15] and from the polar form (46)–(47), it is a straightforward manner to obtain the images’ values on the Cartesian grid.3
V. Numerical Experiments
We performed numerical experiments to test the speed and accuracy of our algorithm against similar algorithms. We performed this test on a UNIX environment with four processors, each of which was a Dual Core AMD Opteron Processor 275 running at 2.2 GHz. These experiments were performed in Matlab with randomly generated pixel data in each image. We did this because the actual output of the algorithms are not important in measuring runtime, and runtime is not affected by the content of the images. The images are assumed to be on a polar grid as described at the beginning of Section II.
We compared our algorithm against three other algorithms: 1) the algorithm described in [9], which we will refer to as JZL; 2) SVD of the data matrix; and 3) computation of the covariance matrix C and then eigenvector decomposition of C. Note that our algorithm as well as JZL compute the PCA of each image and its planar rotations, while SVD and eigenvector decomposition do not take rotations into account and are therefore not as accurate.
In Table II, one can see a comparison of the running times of various algorithms. In this experiment, images of size Nr = 100 and Nθ = 72 were used. For image counts P of at least 64 and less than 12 288, our algorithm is fastest, at which point eigenvector decomposition becomes faster. However, the goal of this paper is to work with large numbers of images, so our algorithm is not designed to compete for very small P. In addition, eigenvector decomposition does not take planar rotations into account. Thus, eigenvector decomposition only computes the principal components of P images, whereas our algorithm actually computes the principal components of P Nθ images. In addition, the calculation of the covariance matrix was made faster because Matlab makes use of parallel processors to compute matrix products.
TABLE II.
Comparison of Running Times Under Increasing Number of Images P, in Seconds Nr = 100 and Nθ = 72
| P | This Paper | JZL | SVD | Eig. Decomp. |
|---|---|---|---|---|
| 24 | 1.74 | 0.90 | 0.48 | 12.0 |
| 32 | 1.90 | 1.6 | 0.76 | 13.4 |
| 48 | 2.13 | 3.5 | 1.48 | 19.0 |
| 64 | 2.47 | 6.3 | 2.50 | 19.1 |
| 96 | 3.00 | 14.9 | 4.55 | 30.0 |
| 128 | 3.53 | 26.3 | 6.74 | 33.2 |
| 192 | 4.44 | 70.0 | 9.91 | 45.0 |
| 256 | 5.34 | 139 | 13.2 | 43.1 |
| 384 | 7.45 | 378 | 20.3 | 56.1 |
| 512 | 9.25 | 902 | 27.3 | 63.3 |
| 768 | 13.1 | 2796 | 43.7 | 73.0 |
| 1024 | 16.8 | 7926 | 63.1 | 80.2 |
| 1536 | 24.3 | N/A | 93.3 | 79.2 |
| 2048 | 31.8 | N/A | 137 | 96.8 |
| 3022 | 46.0 | N/A | 219 | 97.9 |
| 4096 | 61.9 | N/A | 317 | 122 |
| 6144 | 92.0 | N/A | 529 | 136 |
| 8192 | 122 | N/A | 703 | 151 |
| 12288 | 182 | N/A | 1116 | 160 |
| 16384 | 242 | N/A | 1739 | 197 |
In Table III, one can see a comparison of the running times of various algorithms as P = 500 and Nθ = 72 are held constant and Nr is increased. Even though the running time of our algorithm shows cubic growth here, it is still far faster than either JZL or eigenvector decomposition. SVD performs more quickly than our algorithm here; however, it is not considering image rotations. Note that the large jump in running time for eigenvector decomposition with Nr = 384 is due to the fact that, for large Nr, the NrNθ × NrNθ covariance matrix became too large to store in memory, causing the virtual memory to come into use, causing large delays.
TABLE III.
Comparison of Running TIMES Under Increasing Number of Pixels Per Radial Line Nr, in Seconds. P = 500 and Nθ = 72
| Nr | This Paper | JZL | SVD | Eig. Decomp. |
|---|---|---|---|---|
| 8 | 0.25 | 477 | 1.89 | 0.43 |
| 12 | 0.36 | 482 | 3.05 | 0.85 |
| 16 | 0.52 | 491 | 3.69 | 1.29 |
| 24 | 0.96 | 506 | 5.87 | 2.94 |
| 32 | 1.41 | 524 | 8.05 | 5.65 |
| 48 | 2.67 | 552 | 12.48 | 13.1 |
| 64 | 4.12 | 628 | 19.0 | 22.6 |
| 96 | 8.61 | 804 | 25.8 | 52.5 |
| 128 | 14.9 | 1027 | 35.1 | 87.8 |
| 192 | 34.7 | 1190 | 56.0 | 192 |
| 256 | 66.9 | 1520 | 68.9 | 417 |
| 384 | 185 | 2127 | 107 | 17916 |
| 512 | 445 | 3389 | 156 | N/A |
| 768 | 1751 | 6882 | 281 | N/A |
| 1024 | 5060 | 12149 | 386 | N/A |
In Table IV, one can see a comparison of the running times of various algorithms as P = 500 and Nr = 100 are held constant, and Nθ is increased. Again, our algorithm is significantly faster than any of the others.
TABLE IV.
Comparison of Running Times Under Increasing Number of Rotations Nθ, in Seconds. P = 500 and Nr = 100
| Nθ | This Paper | JZL | SVD | Eig. Decomp. |
|---|---|---|---|---|
| 8 | 4.84 | 299 | 3.43 | 0.91 |
| 12 | 5.21 | 325 | 4.76 | 2.01 |
| 16 | 5.64 | 351 | 5.48 | 3.11 |
| 24 | 6.27 | 500 | 8.51 | 6.71 |
| 32 | 6.77 | 459 | 11.2 | 12.9 |
| 48 | 7.78 | 585 | 18.5 | 26.0 |
| 64 | 8.70 | 708 | 23.7 | 47.3 |
| 96 | 10.7 | 988 | 40.4 | 104 |
| 128 | 12.4 | 1302 | 51.5 | 178 |
| 192 | 16.4 | 1859 | 77.4 | 479 |
| 256 | 20.1 | 2705 | 108 | 4022 |
| 384 | 27.6 | 3873 | 170 | N/A |
| 512 | 37.3 | 6976 | 257 | N/A |
| 768 | 58.9 | 11133 | 400 | N/A |
| 1024 | 69.6 | 14706 | 560 | N/A |
We now consider the numerical accuracy of our algorithm. We compare the resulting eigenvectors of SVD against those of our algorithm and eigenvector decomposition of the covariance matrix. However, these eigenvectors often occur in pairs, and any two orthonormal eigenvectors that span the correct eigensubspace are valid eigenvectors. Therefore, we cannot simply compute the distance between eigenvectors of different methods, because the paired eigenvectors produced may be different.
To measure the accuracy of the algorithms, then, we compute the Grassmannian distance between two eigensubspaces: if a subspace V is spanned by orthonormal vectors u1, …, uk then the projection matrix onto that subspace is defined by . The Grassmannian distance between two subspaces V and W is defined as the largest eigenvalue magnitude of the matrix (PV − PW).
In our first experiment, we artificially generated 50 projection images of the E. Coli 50-s ribosomal subunit, as described in the next section, and convert them into “ -filtered” radon images as described in Section IV with Nr = 100 and Nθ = 36. We then perform PCA on these Radon images and each of their 36 rotations using the four different algorithms. We then pair off principal components with identical eigenvalues, and compute the Grassmannian distance between each set of principal components and those computed by SVD.
In our second experiment, we do the same thing but using images consisting entirely of Gaussian noise with mean 0 and variance 1. The results of both experiments can be seen in Table V.
TABLE V.
Comparison of Grassmannian Distances Between SVD Eigensubspaces and Other Methods. (A)Images of Artificially Generated Projections of the E.Coli 50-s Ribosomal Subunit. (B) Images of White Noise
| (A) | (B) | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Princ. Comps | This Paper | JZL | Eig. Decomp. | Princ. Comps | This Paper | JZL | Eig. Decomp. |
| 1 | 2.25 · 10−15 | 2.09 · 10−15 | 3.11 · 10−15 | 1 & 2 | 3.73 · 10−13 | 3.71 · 10−13 | 3.41 · 10−13 |
| 2 & 3 | 5.26 · 10−15 | 8.03 · 10−15 | 9.45 · −15 | 3 & 4 | 3.75 · 10−13 | 3.71 · 10−13 | 3.44 · 10−13 |
| 4 &5 | 6.94 · 10−15 | 7.17 · 10−15 | 7.83 · 10−15 | 5 & 6 | 9.25 · 10−13 | 9.27 · 10−13 | 1.05 · 10−12 |
| 6 &7 | 9.72 · 10−15 | 1.35 · 10−14 | 7.40 · 10−15 | 7 | 1.63 · 10−12 | 1.63 · 10−12 | 2.76 · 10−12 |
| 8 &9 | 9.45 · 10−14 | 9.36 · 10−14 | 1.87 · 10−13 | 8 & 9 | 1.60 · 10−12 | 1.60 · 10−12 | 2.74 · 10−12 |
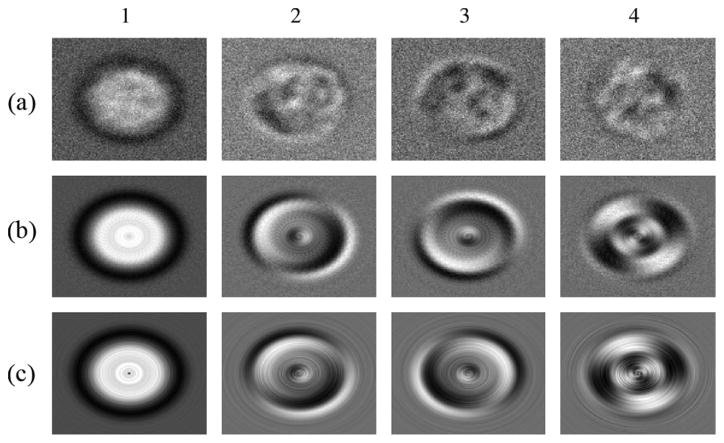
Finally, in our third experiment, we compare the true eigenimages that are produced by our algorithm using the procedure described in Section IV with those produced by performing SVD on a full set of images and rotations. To do this, we produce artificially generated 75 projection images of the E. Coli 50-s ribosomal subunit and produced 36 uniform rotations of each one.4 We then performed SVD on this set of 2700 images and compared the results with running our algorithm on the original set of 75 images. The results can be seen in Fig. 1. Eigenimages 2 and 3 are rotated differently in our algorithm than in SVD. This is because the two eigenimages have the same eigenvalue, and different rotations correspond to different eigenvectors in the 2-D subspace spanned by the eigenvectors. Eigenimage 4 is also rotated differently, which is again due to paired eigenvalues; the fifth eigenimage is not shown.
Fig. 1.
Number above each column represents which eigenimages is shown in that column. (a) Eigenimages produced by running SVD on the original set of 75 images. (b) Eigenimages produced by running SVD on the set of 2700 rotated images. (c) Eigenimages produced by our algorithm.
VI. Cryo-EM
Cryo-EM is a technique used to image and determine the three dimensional structure of molecules. Many copies of some molecule of interest are frozen in a sheet of ice that is sufficiently thin so that, when viewing the sheet from its normal direction, the molecules do not typically overlap. This sheet is then imaged using a transmission electron microscope. The result is a set of noisy projection images of the molecule of interest, taken at random orientations of the molecule. This process usually destroys the molecules of interest, preventing one from turning the sheet of ice to image a second time at a known orientation. The goal is to then to take a large set of such images, often 10 000 or more, and from that set deduce the three dimensional structure of the molecule.
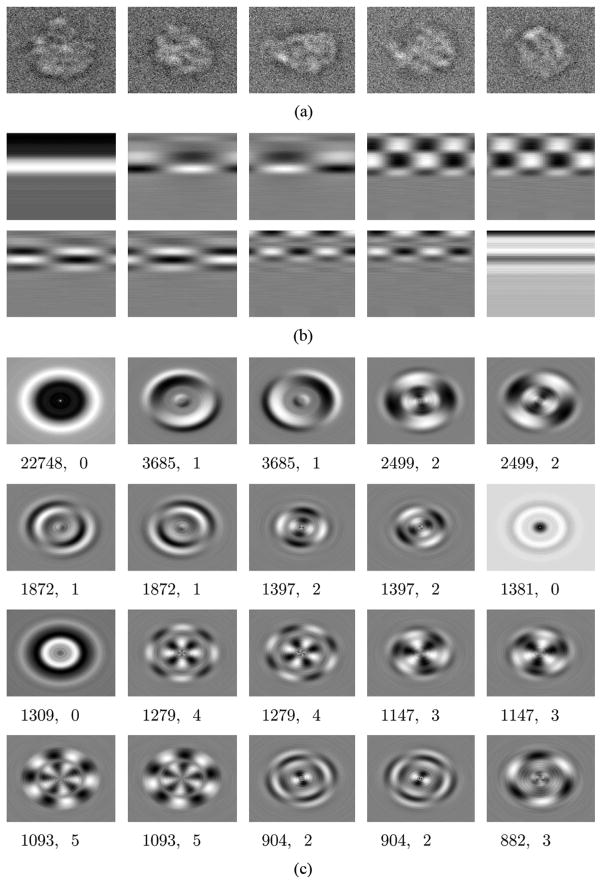
Several sample projection images are shown in Fig. 2(a). These images were artificially generated by taking projections from random orientations of the E. Coli 50-s ribosomal subunit, which has a known structure. Gaussian noise was then added to create an SNR of Var(signal)/Var(noise) = 1/5. P = 250 such images of size 129 × 129 were generated, and then transformed into radon images of size Nθ = 72 and Nr = 100. Note that, in a real application of cryo-EM, images often have much lower SNRs, and many more images are used.
Fig. 2.
(a) Artificially generated projections of the E. Coli 50-s ribosomal subunit, with Gaussian noise added to create an SNR of 1/5. (b) The first ten” -filtered” radon eigenimages. (c) The first 20 true eigenimages. Below each true eigenimage is its eigenvalue μ and frequency k, respectively.
We then applied to this set of images the algorithm described in Sections II–IV. The first 10 “ -filtered” radon eigenimages are shown in Fig. 2(b). Note that eigenimages 2 and 3 are an example of paired eigenimages. Paired eigenimages have the same eigenvalue, and occur as eigenvectors in paired Hermitian matrices Λk and Λ−k, as in (21) and (22). Note that eigenimages 1 and 6 have a frequencies of k = 0, which results in only a real eigenimage and so is not paired with another.
Fig. 2(c) shows the conversion of the Radon eigenimages in Fig. 2(b) to true eigenimages. This was done with the procedure described in Section IV. Eigenimages 2 and 3 are paired with frequency k = 1, and eigenimages 4 and 5 are paired with frequency k = 2. They are described by (46) and (47). Eigenimage 1 is described by (46) only. These images can be compared in their general shape to the eigenimages shown in [19, p. 117], where the SVD was used to compute the eigenimages for all possible rotations of KLH projection images. We remark that the true eigenimages are shown just for illustrative purposes: as will be shown in [18], the principal components of the 2-D -multiplied polar Fourier transformed images are sufficient to accelerate the computation of the rotationally invariant distances [13], but the true eigenimages are not required.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences under Award R01GM090200. The associate editor coordinating the review of this manuscript and approving it for publication was Dr. Hsueh-Ming Hang.
The authors would like to thank Y. Shkolnisky for providing us his code for computing the 2-D Fourier transform on a polar grid, as well as his code for rotating images. They would also like to thank Z. Zhao for helpful discussions and for proofreading the paper.
Biographies

Colin Ponce received the B.S.E. degree in computer science and a certificate in applied and computational mathematics from Princeton University, Princeton, NJ, in 2010. He is currently working toward the Ph.D. degree in computer science from Cornell University, Ithaca, NY.
His current research focuses on machine learning and scientific computing.
Mr. Ponce is a member of Tau Beta Pi and Sigma Xi.

Amit Singer received the B.Sc. degree in physics and mathematics and Ph.D. degree in applied mathematics from Tel Aviv University, Tel Aviv, Israel, in 1997 and 2005, respectively.
He is an Associate Professor of Mathematics and a member of the Executive Committee of the Program in Applied and Computational Mathematics (PACM) at Princeton University, Princeton, NJ. He joined Princeton University as an Assistant Professor in 2008. From 2005 to 2008, he was a Gibbs Assistant Professor in Applied Mathematics with the Department of Mathematics, Yale University, New Haven, CT. He served in the Israeli Defense Forces during 1997–2003. His current research in applied mathematics focuses on problems of massive data analysis and structural biology.
Prof. Singer was the recipient of the Alfred P. Sloan Research Fellowship (2010) and the Haim Nessyahu Prize for Best PhD in Mathematics in Israel (2007).
Appendix A. Proof of 2-D Fourier Transform Formula
Here, we prove formulas (42) and (43). To that end, consider the operation in (38):
| (49) |
Note that
| (50) |
and similarly
| (51) |
Therefore, it follows that (49) becomes
| (52) |
Therefore, define the 2-D Fourier transform of the true image as
| (53) |
A similar manipulation shows that
| (54) |
As shown above, these functions are either real or pure imaginary, depending on k.
Appendix B. Proof of the Radon Inversion Formula
Here, we show that the true eigenimages and are given by (46) and (47). To do this, we take the 2-D inverse Fourier transform of . First, let . Then
| (55) |
With the change of variables x1 = s cos α, x2 = s sin α and w1 = r cos &thetas;, w2 = r sin θ, we obtain
| (56) |
Now, with the change of variables θ′ = α + π/2− θ, we have
| (57) |
and, with the change of variables θ″ = θ − α + π/2, we have
| (58) |
where Jk is the Bessel function. Thus, becomes
| (59) |
where is the order k Hankel transform of , and is the order k Hankel transform of .
Now note that
| (60) |
As a result,
| (61) |
If k is even, then eιkπ = −1, so
| (62) |
This is a real-valued function because and are real when k is even.
If k is odd, then eιkπ = −1, so
| (63) |
While this appears imaginary, it is also real because and , and therefore and , are imaginary when k is odd. A similar manipulation shows that, when k is even, then
| (64) |
and, when k is odd, then
| (65) |
Again, both (64) and (65) are real. However, note that (62) is a complex scalar multiple of (65). Scalar multiples of real values are unimportant, so we may combine these two into the single formula
| (66) |
where
| (67) |
so that β(k) = 1 when k is even, and β(k) = ι when is odd. Similarly, (63) is a complex scalar multiple of (64), so we may combine these two into the single formula
| (68) |
Footnotes
A Matlab implementation of this algorithm can be found at http://www.cs.cornell.edu/cponce/SteerablePCA/
We would like to thank Yoel Shkolnisky for sharing with us his code for the 2-D polar Fourier transform. In his code, m = 2 and ε is single precision accuracy.
MATLAB code to convert polar Radon eigenimages into rectangular eigenimages can be found at http://www.cs.cornell.edu/~cponce/SteerablePCA/
We would like to thank Y. Shkolnisky for providing us with his code for rotating images. It produces a rotated image by upsampling using FFT, then rotating using bilinear interpolation, and finally downsampling again to the original pixels.
Contributor Information
Colin Ponce, Email: cponce@cs.cornell.edu, Department of Computer Science, Cornell University, Ithaca, NY 14850 USA.
Amit Singer, Email: amits@math.princeton.edu, Department of Mathematics and PACM, Princeton University, Princeton NJ 08544-1000 USA.
References
- 1.Borland L, van Heel M. Classification of image data in conjugate representation spaces. J Opt Soc Amer A. 1990;7(4):601–610. [Google Scholar]
- 2.Dutt A, Rokhlin V. Fast Fourier transorms for nonequispaced data. SIAM J Sci Comput. 1993;14(6):1368–1393. [Google Scholar]
- 3.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in Their Native State. 2. New York: Oxford Univ. Press; 2006. [Google Scholar]
- 4.Gardner DG, Gardner JC, Lausch G, Meinke WW. Method for the analysis of multi-component exponential decays. J Chem Phys. 1959;31:987. [Google Scholar]
- 5.Golub GH, Van Loan CF. Matrix Computations. Baltimore, MD: Johns Hopkins Univ; 1996. [Google Scholar]
- 6.Greengard L, Lee JY. Accelerating the nonuniform fast Fourier transform. SIAM Rev. 2004;46(3):443–454. [Google Scholar]
- 7.Guizar-Sicairos M, Gutierrez-Vega JC. Computation of quasi-discrete Hankel transforms of integer order for propagating optical wave fields. J Opt Soc Amer A. 2004;21:53–58. doi: 10.1364/josaa.21.000053. [DOI] [PubMed] [Google Scholar]
- 8.Hilai R, Rubinstein J. Recognition of rotated images by invariant Karhunen-Loeve expansion. J Opt Soc Amer A. 1994;11:1610–1618. [Google Scholar]
- 9.Jogan M, Zagar E, Leonardis A. Karhunen–Loéve expansion of a set of rotated templates. IEEE Trans Image Process. 2003 Jul;12(7):817–825. doi: 10.1109/TIP.2003.813141. [DOI] [PubMed] [Google Scholar]
- 10.Khotanzad A, Lu JH. Object recognition using a neural network and invariant Zernike features. Proc IEEE Conf Vis Pattern Recognit. 1989:200–205. [Google Scholar]
- 11.Natterer F. The Mathematics of Computerized Tomography. Philadelphia, PA: SIAM; 2001. [Google Scholar]
- 12.Park R. Correspondence. IEEE Trans Image Process. 2002 Mar;11(3):332–334. doi: 10.1109/83.988965. [DOI] [PubMed] [Google Scholar]
- 13.Penczek PA, Zhu J, Frank J. A common-lines based method for determining orientations for N > 3 particle projections simultaneously. Ultramicroscopy. 1996;63:205–218. doi: 10.1016/0304-3991(96)00037-x. [DOI] [PubMed] [Google Scholar]
- 14.Perona P. Deformable kernels for early vision. IEEE Trans Pattern Anal Mach Intell. 1995 May;17(5):488–499. [Google Scholar]
- 15.Siegmann AE. Quasi fast Hankel transform. Opt Lett. 1977;1:13–15. doi: 10.1364/ol.1.000013. [DOI] [PubMed] [Google Scholar]
- 16.Sigworth FJ. Classical detection theory and the Cryo-EM particle selection problem. J Structural Biol. 2004;145:111–122. doi: 10.1016/j.jsb.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 17.Singer A, Zhao Z, Shkolnisky Y, Hadani R. Viewing angle classification of cryo-electron microscopy images using eigenvectors. SIAM J Imag Sci. 2011;4(2):723–759. doi: 10.1137/090778390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singer A, Zhao Z. In Preparation. [Google Scholar]
- 19.Teague MR. Image analysis via the general theory of moments. J Opt Soc Amer. 1980;70:920–930. [Google Scholar]
- 20.Uenohara M, Kanade T. Optimal approximation of uniformly rotated images: Relationship between Karhunen–Loeve expansion and discrete cosine transform. IEEE Trans Image Process. 1998 Jan;7(1):116–119. doi: 10.1109/83.650856. [DOI] [PubMed] [Google Scholar]
- 21.van Heel M, Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6:187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- 22.van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Schatz M, Patwardhan A. Single-particle electron cryo-microscopy: Towards atomic resolution. Quarterly Rev Biophys. 2000;33(4):307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]